Nerve Segmentation with Deep Learning from Label-Free Endoscopic Images Obtained Using Coherent Anti-Stokes Raman Scattering
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
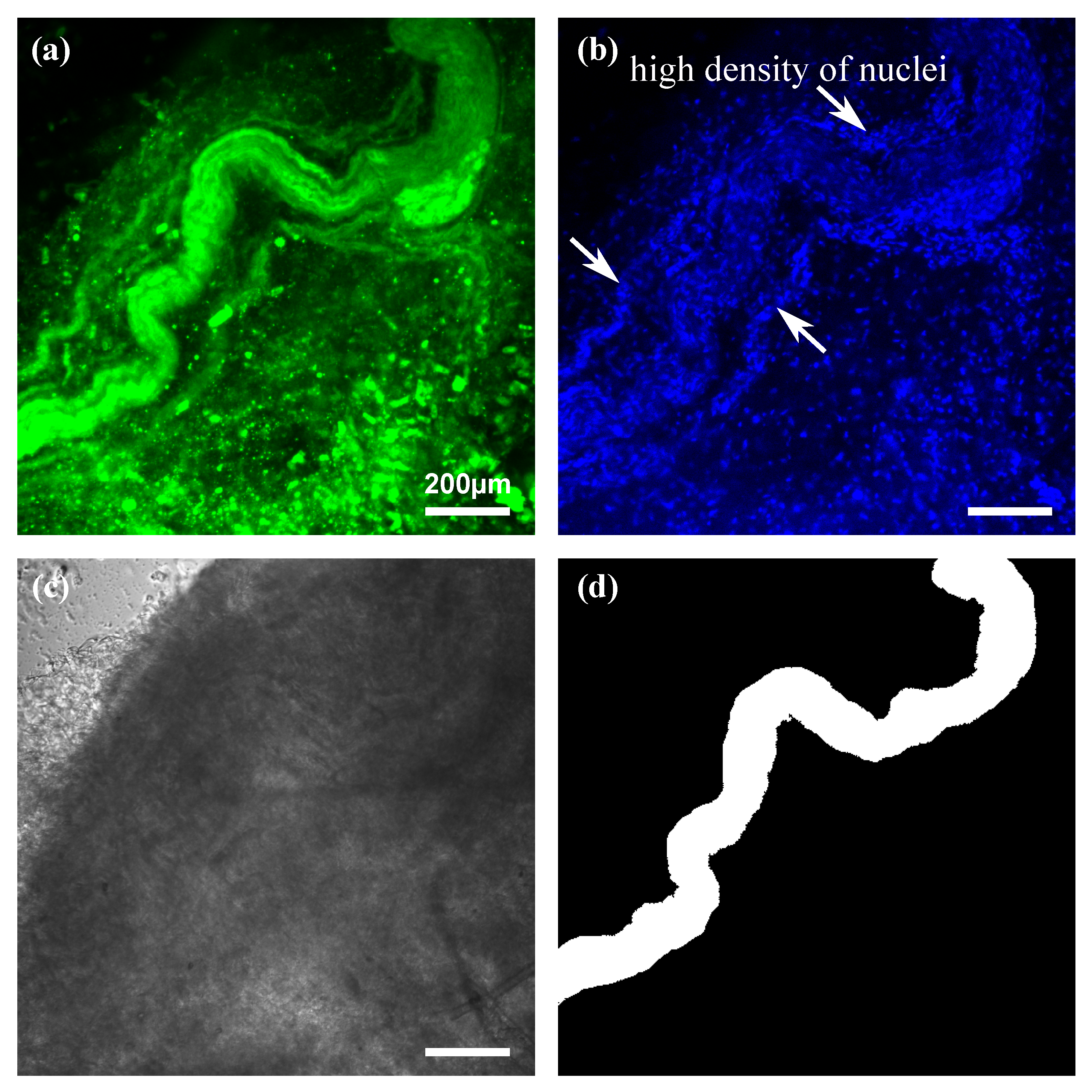
2.2. Fluorescence Images
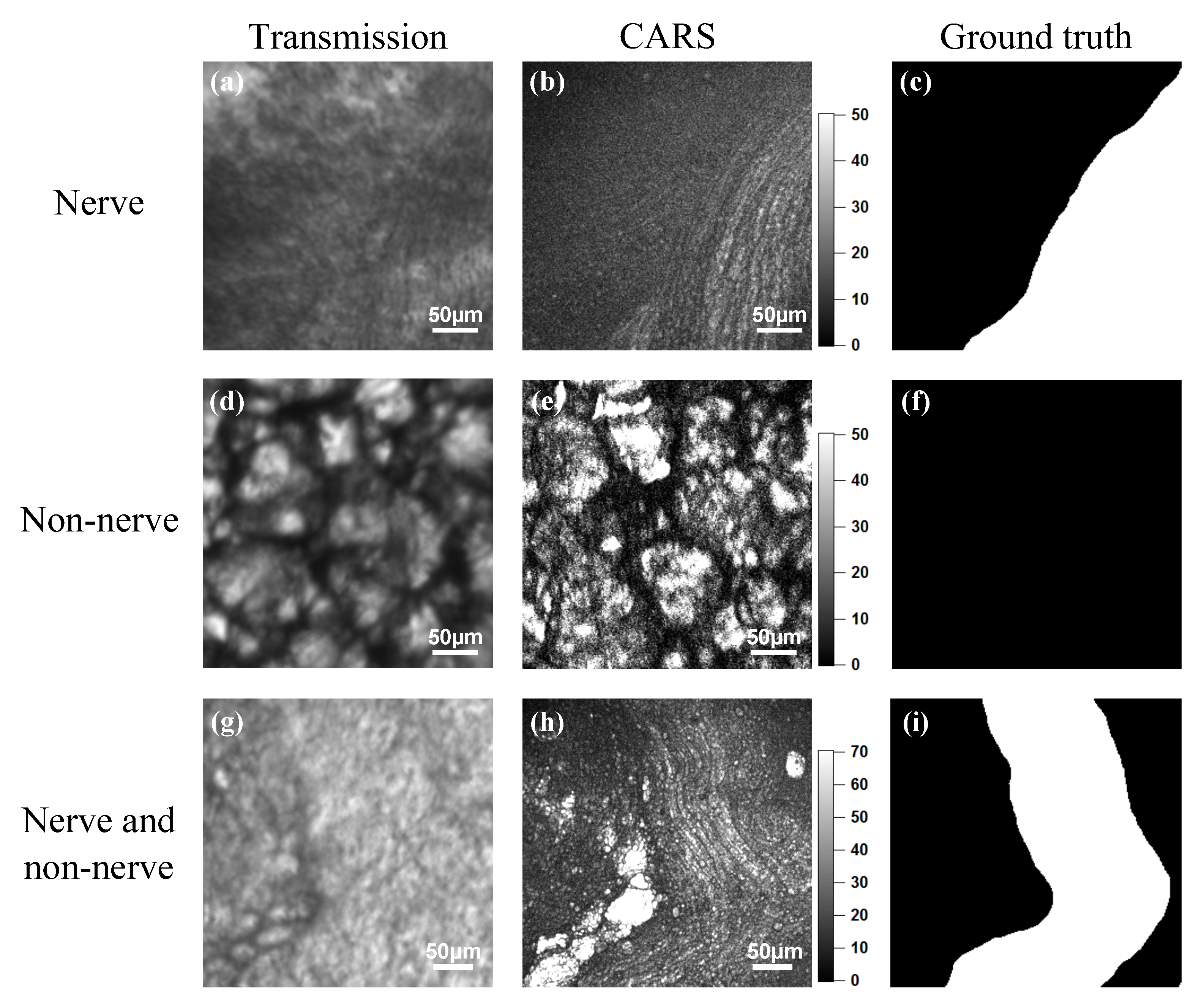
2.3. Coherent Anti-Stokes Raman Scattering Rigid Endoscopy Images
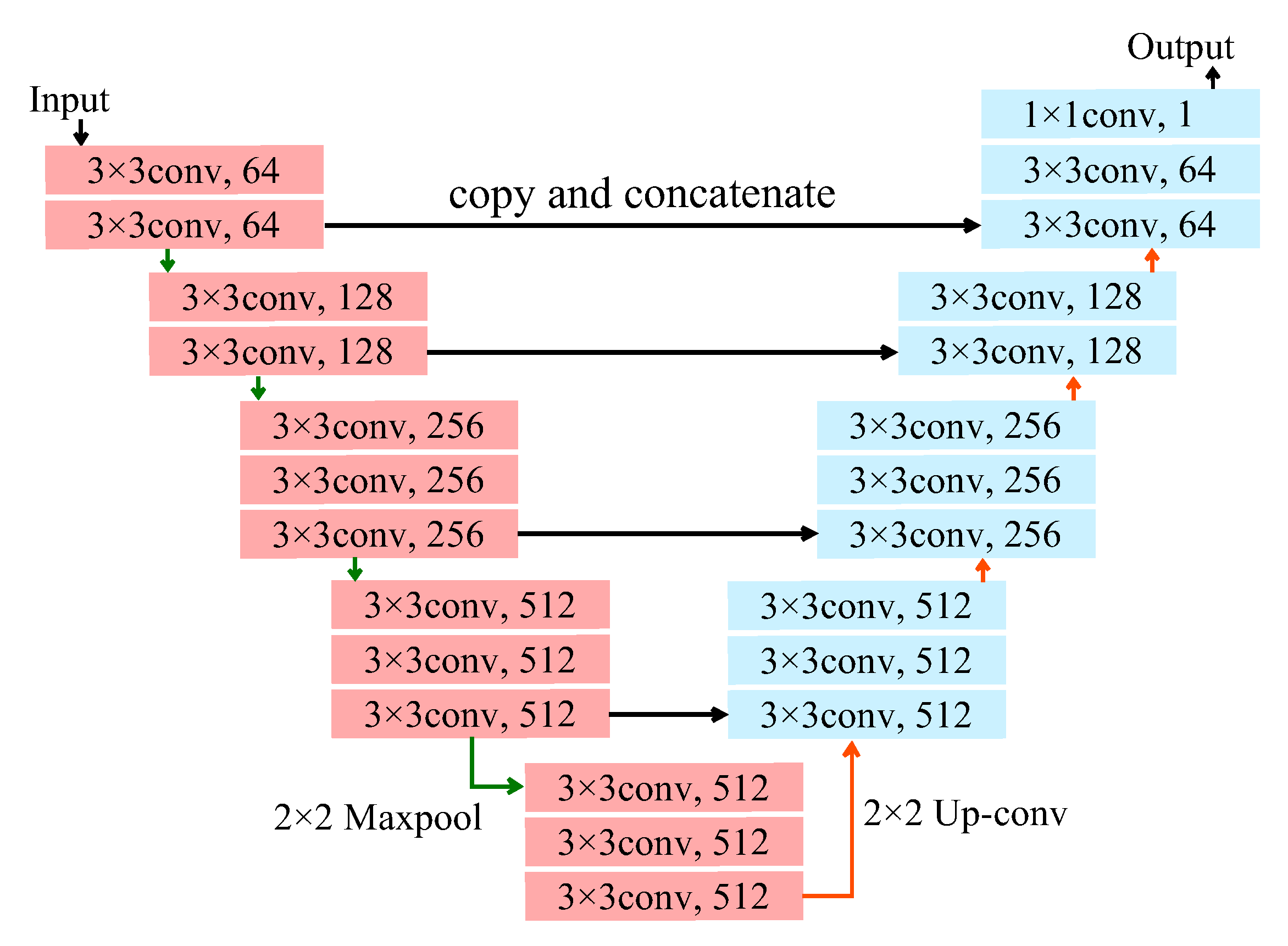
2.4. Architecture and Training of Deep Neural Network
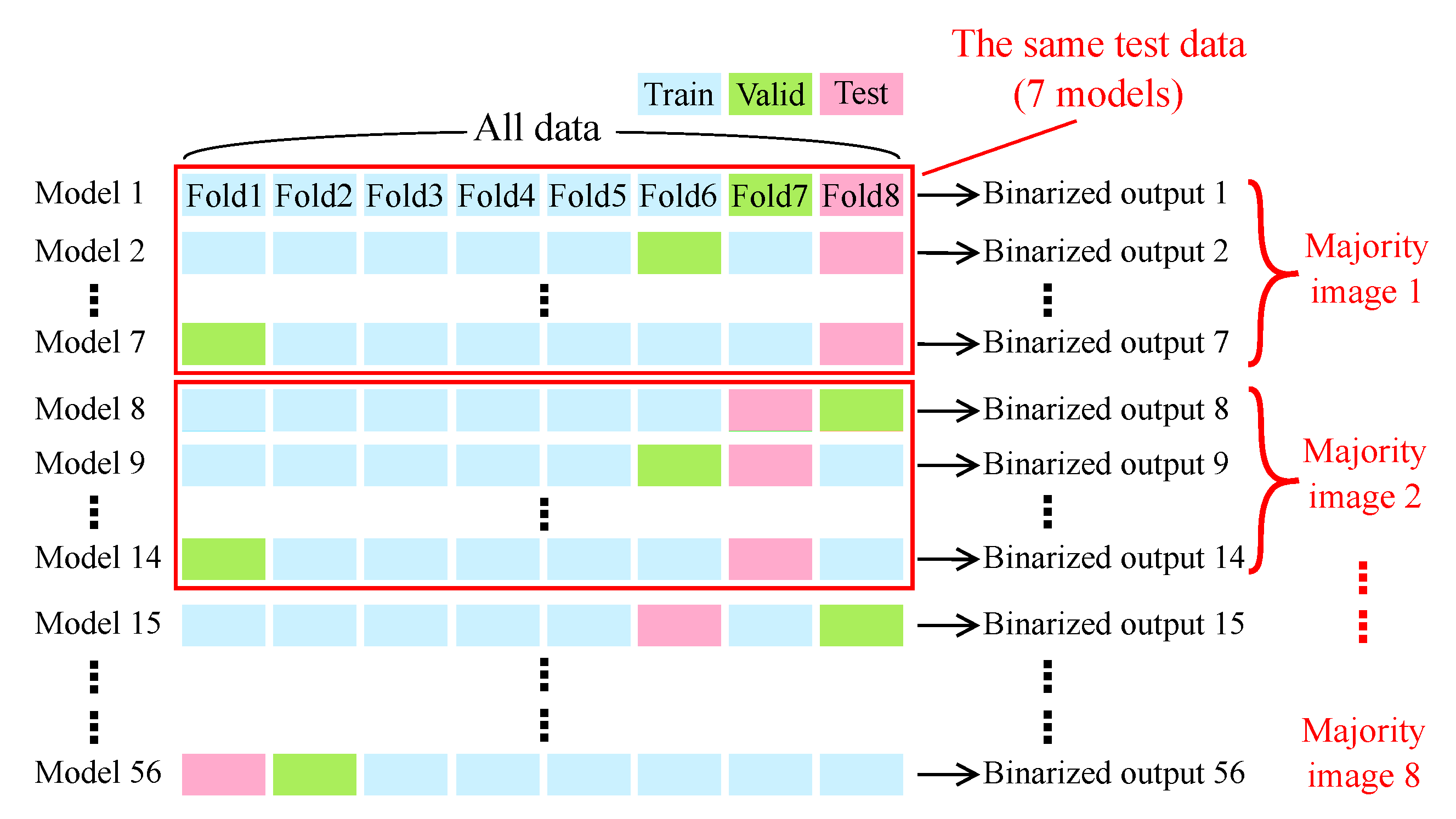
2.5. Evaluation
2.5.1. Evaluation Metrics
2.5.2. Evaluation of the Model Trained on Fluorescence Images
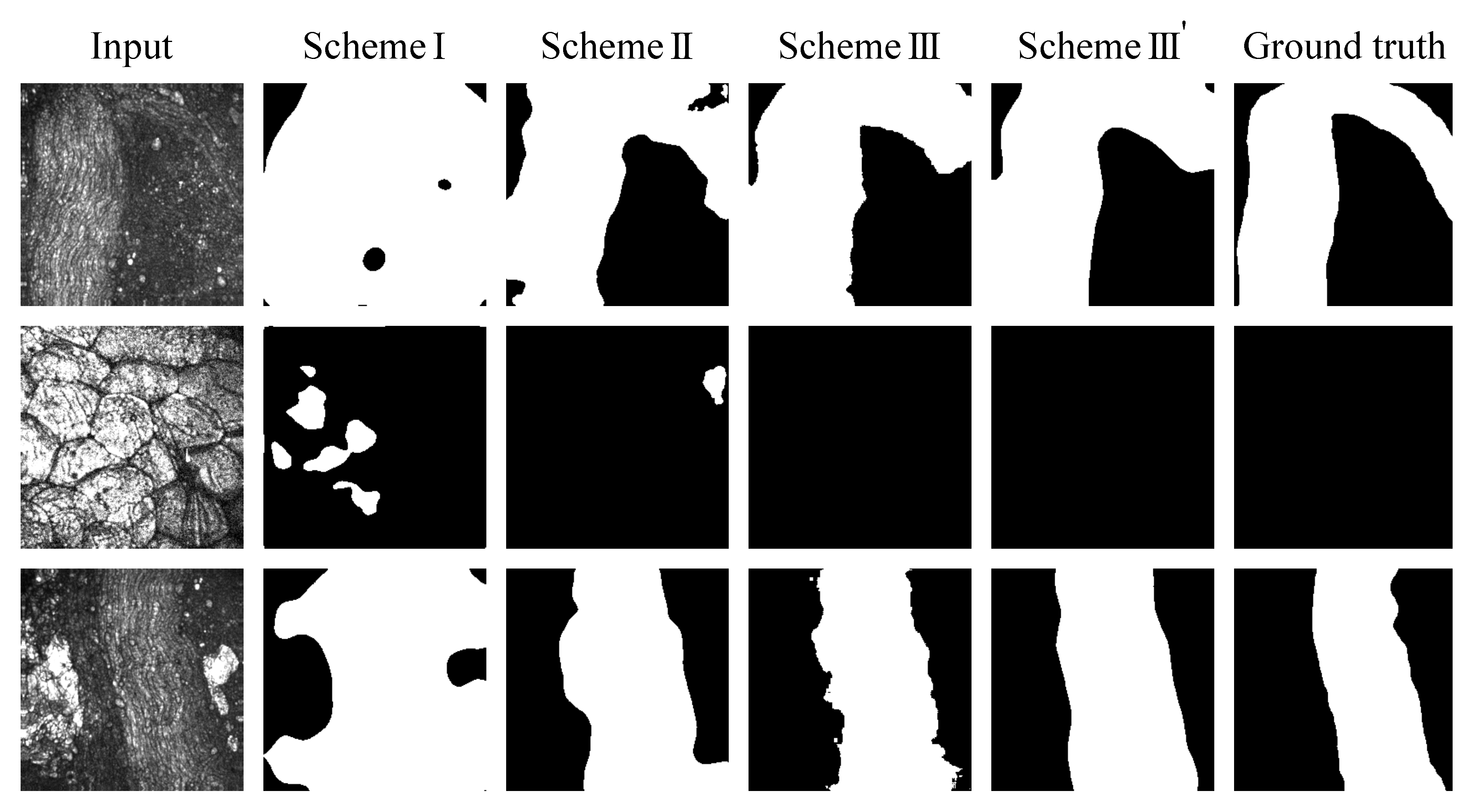
2.5.3. Evaluation of the Model Trained on the Coherent Anti-Stokes Raman Scattering Rigid Endoscopy Images (Scheme I, II, III)
2.5.4. Ensemble Learning and Median Filter for Further Performance Improvement (Scheme III’)
3. Results
3.1. Fluorescence Images
3.2. Coherent Anti-Stokes Raman Scattering Rigid Endoscopy Images
3.3. Nerve Segmentation with Deep Neural Network
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Inomata, M.; Shiroshita, H.; Uchida, H.; Bandoh, T.; Akira, S.; Yamaguchi, S.; Kurokawa, Y.; Seki, Y.; Eguchi, S.; Wada, N.; et al. Current status of endoscopic surgery in Japan: The 14th National Survey of Endoscopic Surgery by the Japan Society for Endoscopic Surgery. Asian J. Endosc. Surg. 2020, 13, 7–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tewari, A.; Srivasatava, A.; Menon, M. A prospective comparison of radical retropubic and robot-assisted prostatectomy: Experience in one institution. BJU Int. 2003, 92, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Heikkinen, T.; Msika, S.; Desvignes, G.; Schwandner, O.; Schiedeck, T.H.; Shekarriz, H.; Bloechle, C.H.; Baca, I.; Weiss, O.; Morino, M.; et al. Laparoscopic surgery versus open surgery for colon cancer: Short-term outcomes of a randomised trial. Lancet Oncol. 2005, 6, 477–484. [Google Scholar]
- Trinh, Q.D.; Sammon, J.; Sun, M.; Ravi, P.; Ghani, K.R.; Bianchi, M.; Jeong, W.; Shariat, S.F.; Hansen, J.; Schmitges, J.; et al. Perioperative outcomes of robot-assisted radical prostatectomy compared with open radical prostatectomy: Results from the nationwide inpatient sample. Eur. Urol. 2012, 61, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Cotero, V.E.; Siclovan, T.; Zhang, R.; Carter, R.L.; Bajaj, A.; LaPlante, N.E.; Kim, E.; Gray, D.; Staudinger, V.P.; Yazdanfar, S.; et al. Intraoperative fluorescence imaging of peripheral and central nerves through a myelin-selective contrast agent. Mol. Imaging Biol. 2012, 14, 708–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Wu, C.; Zhu, J.; Miller, R.H.; Wang, Y. Design, synthesis, and evaluation of coumarin-based molecular probes for imaging of myelination. J. Med. Chem. 2011, 54, 2331–2340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirose, K.; Aoki, T.; Furukawa, T.; Fukushima, S.; Niioka, H.; Deguchi, S.; Hashimoto, M. Coherent anti-Stokes Raman scattering rigid endoscope toward robot-assisted surgery. Biomed. Opt. Express 2018, 9, 387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirose, K.; Fukushima, S.; Furukawa, T.; Niioka, H.; Hashimoto, M. Label-free nerve imaging with a coherent anti-Stokes Raman scattering rigid endoscope using two optical fibers for laser delivery. APL Photonics 2018, 3, 092407. [Google Scholar] [CrossRef]
- Zumbusch, A.; Holtom, G.R.; Xie, X.S. Three-dimensional vibrational imaging by coherent anti-stokes raman scattering. Phys. Rev. Lett. 1999, 82, 4142–4145. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, M.; Araki, T.; Kawata, S. Molecular vibration imaging in the fingerprint region by use of coherent anti-Stokes Raman scattering microscopy with a collinear configuration. Opt. Lett. 2000, 25, 1768. [Google Scholar] [CrossRef] [PubMed]
- Petrov, G.I.; Arora, R.; Yakovlev, V.V.; Wang, X.; Sokolov, A.V.; Scully, M.O. Comparison of coherent and spontaneous Raman microspectroscopies for noninvasive detection of single bacterial endospores. Proc. Natl. Acad. Sci. USA 2007, 104, 7776–7779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Galli, R.; Uckermann, O.; Koch, E.; Schackert, G.; Kirsch, M.; Steiner, G. Effects of tissue fixation on coherent anti-Stokes Raman scattering images of brain. J. Biomed. Opt. 2013, 19, 071402. [Google Scholar] [CrossRef] [PubMed]
- Levchenko, S.M.; Peng, X.; Liu, L.; Qu, J. The impact of cell fixation on coherent anti-stokes Raman scattering signal intensity in neuronal and glial cell lines. J. Biophotonics 2019, 12, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balakrishna, C.; Dadashzadeh, S.; Soltaninejad, S. Automatic Detection of Lumen and Media in the IVUS Images Using U-Net with VGG16 Encoder. arXiv 2018, arXiv:1806.07554. [Google Scholar]
- Falk, T.; Mai, D.; Bensch, R.; Çiçek, Ö.; Abdulkadir, A.; Marrakchi, Y.; Böhm, A.; Deubner, J.; Jäckel, Z.; Seiwald, K.; et al. U-Net: Deep learning for cell counting, detection, and morphometry. Nat. Methods 2019, 16, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Kingma, D.P.; Ba, J.L. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1026–1034. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019; pp. 8024–8035. [Google Scholar]
- Varma, S.; Simon, R. Bias in error estimation when using cross-validation for model selection. BMC Bioinform. 2006, 7, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Evaluation Metric | Learning Scheme | p Value | ||||||
|---|---|---|---|---|---|---|---|---|
| I | II | III | III’ | I vs. II | I vs. III | II vs. III | III vs. III’ | |
| sensitivity | 0.689 ± 0.391 | 0.949 ± 0.056 | 0.962 ± 0.054 | 0.977 ± 0.025 | <0.01 | <0.01 | 0.14 | 0.16 |
| specificity | 0.843 ± 0.147 | 0.930 ± 0.046 | 0.937 ± 0.054 | 0.947 ± 0.030 | <0.01 | <0.01 | 0.33 | 0.19 |
| precision | 0.469 ± 0.239 | 0.719 ± 0.123 | 0.752 ± 0.118 | 0.772 ± 0.061 | <0.01 | <0.01 | 0.06 | 0.13 |
| mean accuracy | 0.766 ± 0.156 | 0.939 ± 0.026 | 0.950 ± 0.031 | 0.962 ± 0.014 | <0.01 | <0.01 | 0.03 | 0.06 |
| value | 0.469 ± 0.243 | 0.809 ± 0.083 | 0.837 ± 0.085 | 0.860 ± 0.034 | <0.01 | <0.01 | 0.03 | 0.05 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamato, N.; Matsuya, M.; Niioka, H.; Miyake, J.; Hashimoto, M. Nerve Segmentation with Deep Learning from Label-Free Endoscopic Images Obtained Using Coherent Anti-Stokes Raman Scattering. Biomolecules 2020, 10, 1012. https://doi.org/10.3390/biom10071012
Yamato N, Matsuya M, Niioka H, Miyake J, Hashimoto M. Nerve Segmentation with Deep Learning from Label-Free Endoscopic Images Obtained Using Coherent Anti-Stokes Raman Scattering. Biomolecules. 2020; 10(7):1012. https://doi.org/10.3390/biom10071012
Chicago/Turabian StyleYamato, Naoki, Mana Matsuya, Hirohiko Niioka, Jun Miyake, and Mamoru Hashimoto. 2020. "Nerve Segmentation with Deep Learning from Label-Free Endoscopic Images Obtained Using Coherent Anti-Stokes Raman Scattering" Biomolecules 10, no. 7: 1012. https://doi.org/10.3390/biom10071012





