A Deep Learning-Based System (Microscan) for the Identification of Pollen Development Stages and Its Application to Obtaining Doubled Haploid Lines in Eggplant
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Experimental Layout and Workflow
2.3. Phase 1: Model Training
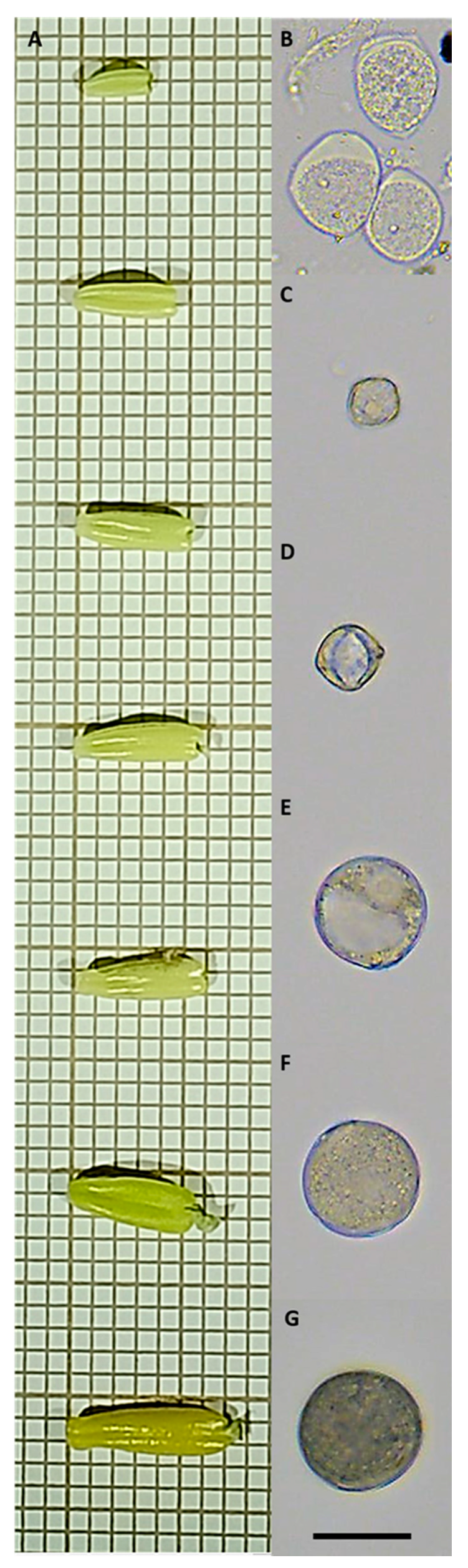
2.3.1. Microspore and Pollen Isolation
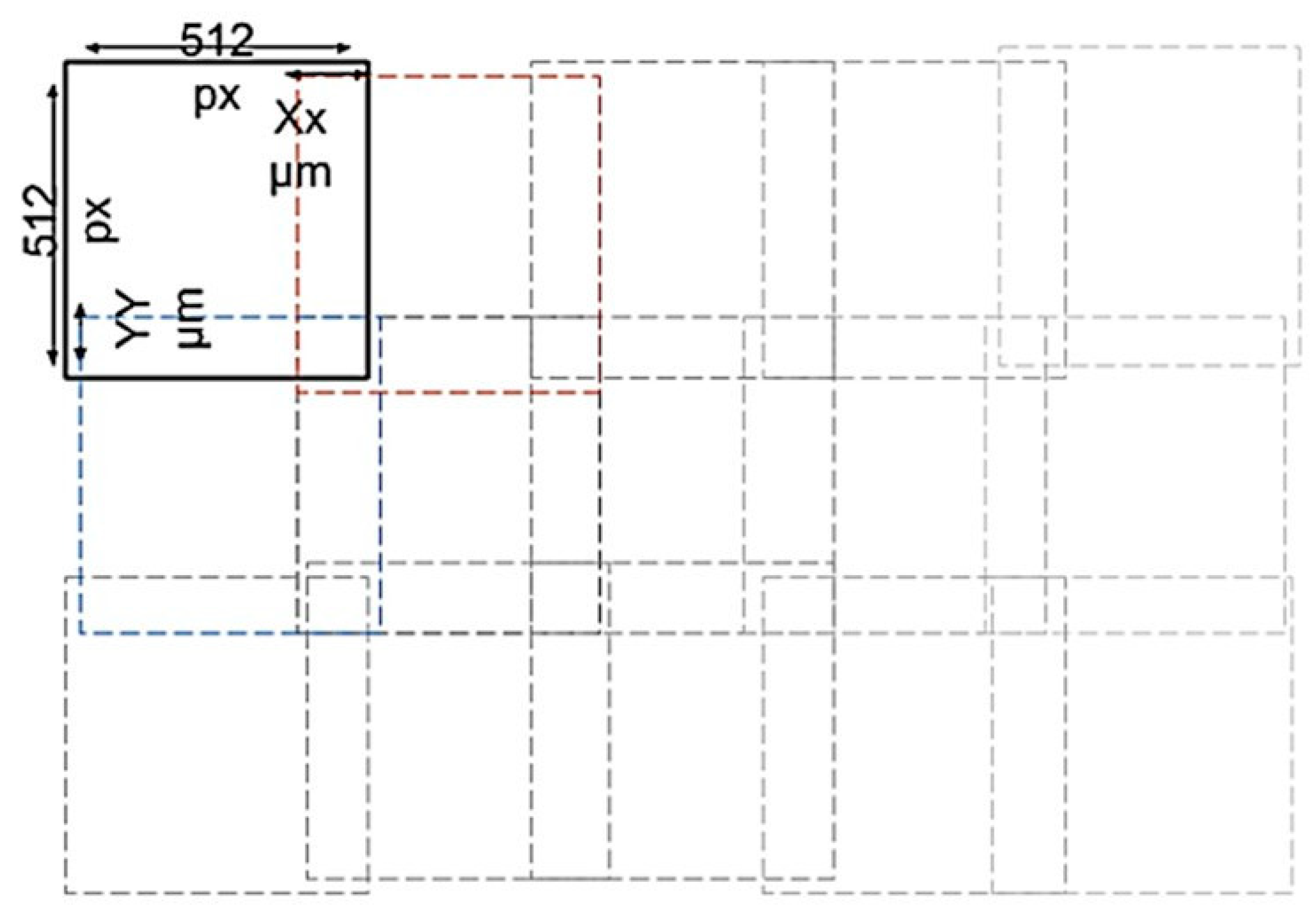
2.3.2. Digital Data Image Acquisition
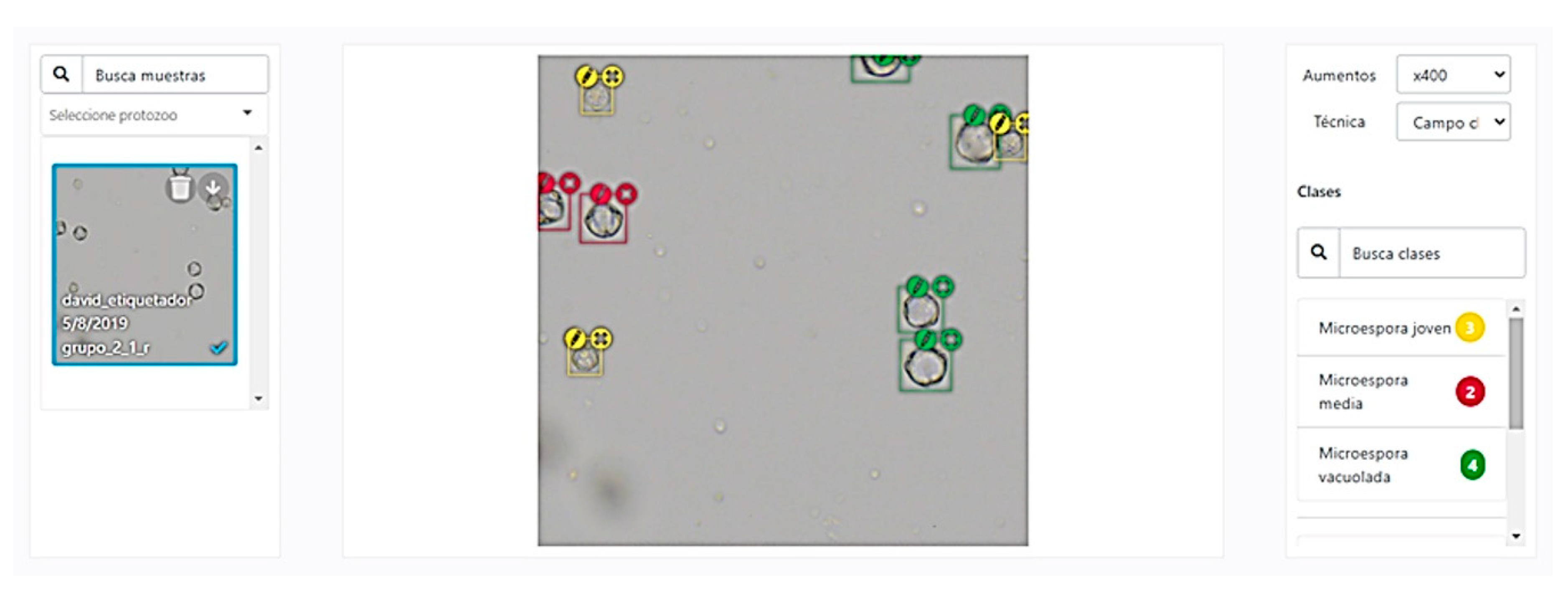
2.3.3. Image Labelling
2.3.4. Preprocessing
2.3.5. Predictive Model
2.4. Phase 2: In Vitro Androgenesis Induction Test Using the Anther Selection Software
2.4.1. E6 Protocol
2.4.2. Cb Protocol
2.4.3. Flow Cytometry
2.4.4. Single Primer Enrichment Technology (SPET) Genotyping
3. Results
3.1. Phase 1: Model Training
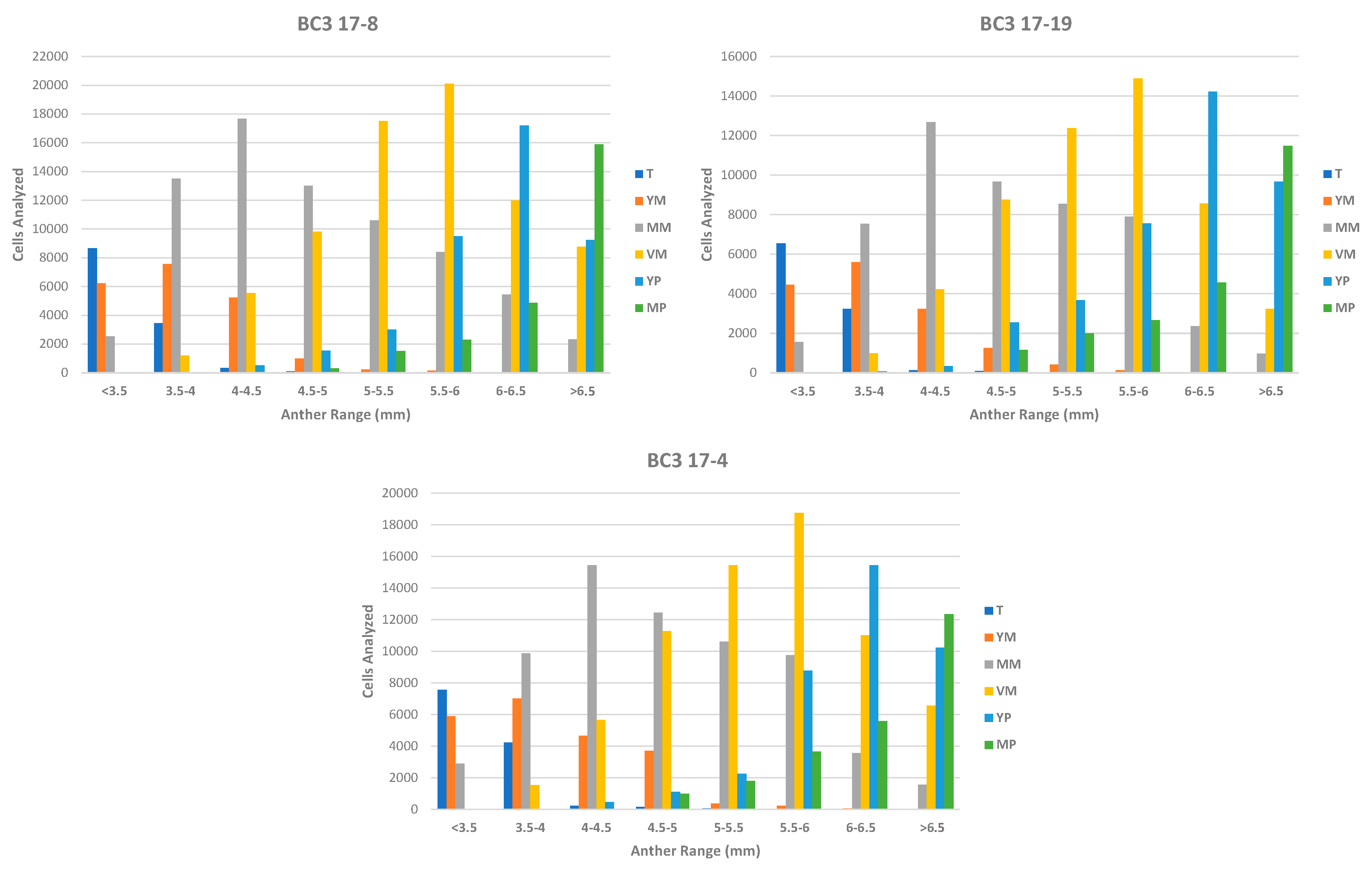
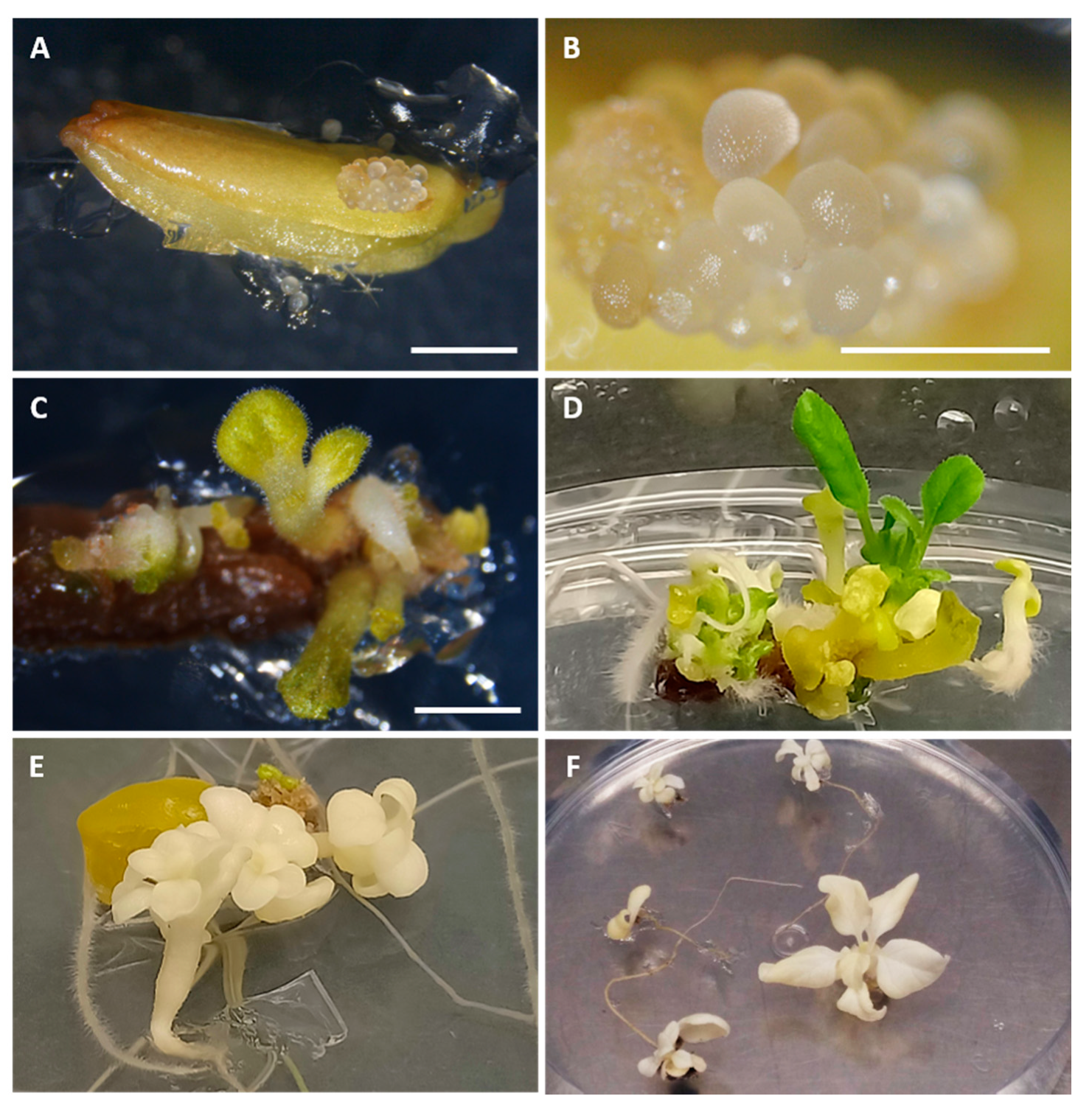
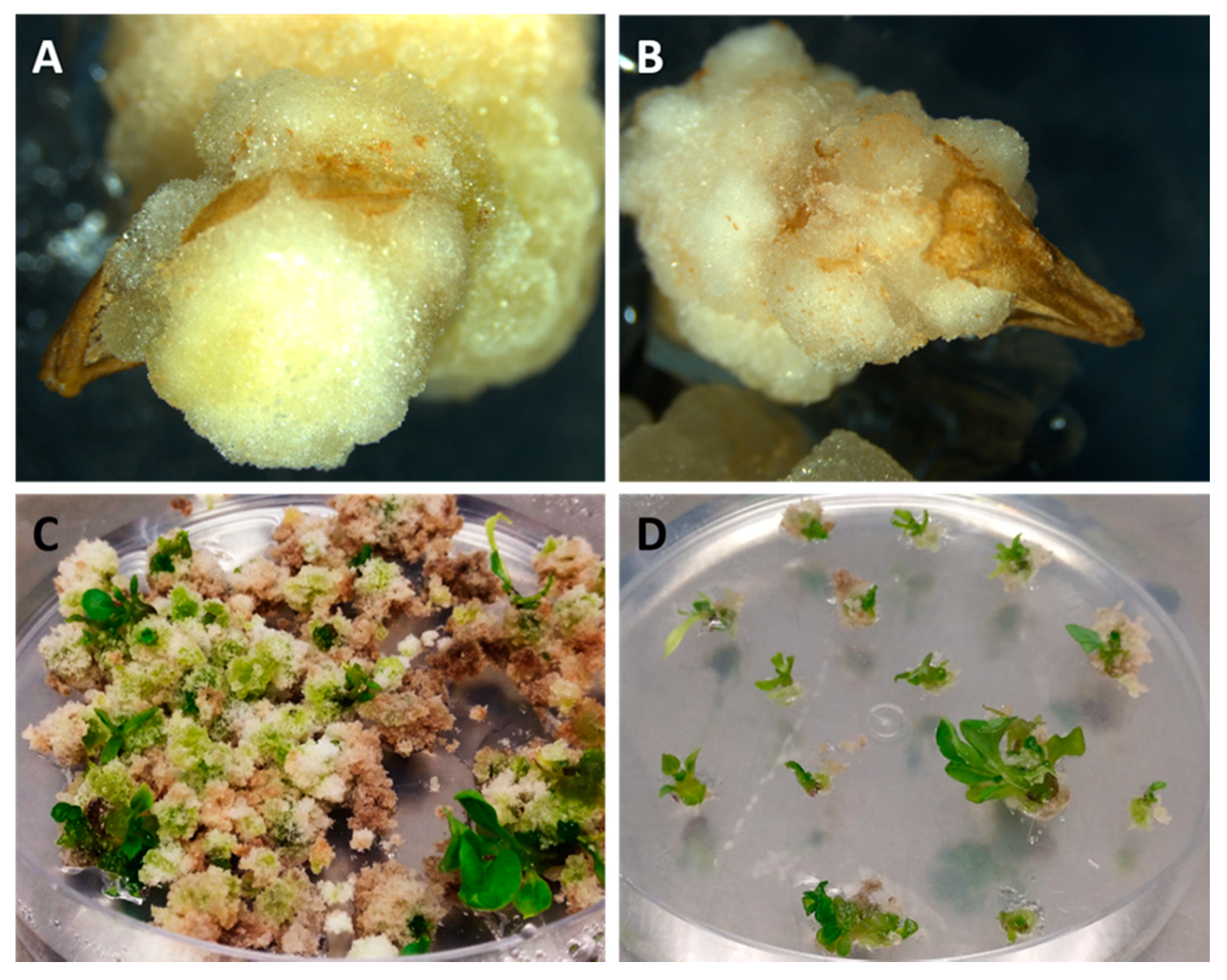
3.2. Phase 2: In Vitro Androgenesis Induction Test Using the Microscan
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Prohens, J.; Gramazio, P.; Plazas, M.; Dempewolf, H.; Kilian, B.; Díez, M.J.; Fita, A.; Herraiz, F.J.; Rodriguez-Burruezo, A.; Soler, S.; et al. Introgressiomics: A new approach for using crop wild relatives in breeding for adaptation to climate change. Euphytica 2017, 213. [Google Scholar] [CrossRef]
- Acquaah, G. Principles of Plant Genetics and Breeding, 2nd ed.; John Wiley and Sons: Chichester, UK, 2012. [Google Scholar] [CrossRef]
- Salim, M.; Gökçe, A.; Naqqash, M.N.; Bakhsh, A. Gene Pyramiding: An Emerging Control Strategy Against Insect Pests of Agronomic Crops. In Agronomics Crops; Hasanuzzaman, M., Ed.; Springer: Singapore, 2020; pp. 285–312. [Google Scholar] [CrossRef]
- Jonas, E.; De Koning, D.J. Does genomic selection have a future in plant breeding? Trends Biotechnol. 2013, 31, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, B.; Ebrahimzadeh, H. In vitro androgenesis : Spontaneous vs. artificial genome doubling and characterization of regenerants. Plant Cell Rep. 2020, 9, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.R.; Singh, K.P.; Bhatia, R.; Raju, D.V.S.; Panwar, S. Optimising protocol for successful development of haploids in marigold (Tagetes spp.) through in vitro androgenesis. Plant Cell Tiss. Org. 2019, 138, 11–28. [Google Scholar] [CrossRef]
- Lantos, C.; Bóna, L.; Nagy, É.; Békés, F.; Pauk, J. Induction of in vitro androgenesis in anther and isolated microspore culture of different spelt wheat (Triticum spelta L.) genotypes. Plant Cell Tiss. Org. 2018, 133, 385–393. [Google Scholar] [CrossRef]
- Warchoł, M.; Czyczyło-Mysza, I.; Marcińska, I.; Dziurka, K.; Noga, A.; Kapłoniak, K.; Pilipowicz, M.; Skrzypek, E. Factors inducing regeneration response in oat (Avena sativa L.) anther culture. Vitr. Cell Dev. Biol. Plant 2019, 55, 595–604. [Google Scholar] [CrossRef]
- González, J.M.; Jouve, N. Microspore development during in vitro androgenesis in triticale. Biol. Plant 2005, 49, 23–28. [Google Scholar] [CrossRef]
- Segui-Simarro, J.M.; Nuez, F. Embryogenesis induction, callogenesis, and plant regeneration by in vitro culture of tomato isolated microspores and whole anthers. J. Exp. Bot. 2007, 58, 1119–1132. [Google Scholar] [CrossRef]
- Seguí-Simarro, J.M.; Corral-Martínez, P.; Parra-Vega, V.; González-García, B. Androgenesis in recalcitrant solanaceous crops. Plant Cell Rep. 2011, 30, 765–778. [Google Scholar] [CrossRef]
- Rotino, G.L. Haploidy in eggplant. In Vitro Haploid Production in Higher Plants, Current Plant Science and Biotechnology in Agriculture; Jain, S.M., Sopory, S.K., Veilleux, R.E., Eds.; Springer: Dordrecht, The Netherlands, 1996; Volume 25, pp. 114–115. [Google Scholar] [CrossRef]
- Miyoshi, K. Callus induction and plantlet formation through culture of isolated microspores of eggplant (Solanum melongena L.). Plant Cell Rep. 1996, 15, 391–395. [Google Scholar] [CrossRef]
- Germanà, M.A. Anther culture for haploid and doubled haploid production. Plant Cell Tiss. Org. 2011, 104, 283–300. [Google Scholar] [CrossRef]
- Salas, P.; Rivas-Sendra, A.; Prohens, J.; Seguí-Simarro, J.M. Influence of the stage for anther excision and heterostyly in embryogenesis induction from eggplant anther cultures. Euphytica 2012, 184, 235–250. [Google Scholar] [CrossRef]
- Salas, P.; Prohens, J.; Seguí-Simarro, J.M. Evaluation of androgenic competence through anther culture in common eggplant and related species. Euphytica 2011, 182, 261–274. [Google Scholar] [CrossRef]
- Brinkmann, M.; Lütkemeyer, D.; Gudermann, F.; Lehmann, J. New technologies for automated cell counting based on optical image analysis “The Cellscreen”. Cytotechnology 2002, 38, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Väyrynen, J.P.; Vornanen, J.O.; Sajanti, S.; Böhm, J.P.; Tuomisto, A.; Mäkinen, M.J. An improved image analysis method for cell counting lends credibility to the prognostic significance of T cells in colorectal cancer. Virchows Arch. 2012, 460, 455–465. [Google Scholar] [CrossRef]
- Kakui, H.; Yamazaki, M.; Hamaya, N.B.; Shimizu, K.K. Pollen grain counting using a cell counter. Methods Mol. Biol. 2020, 2160, 1–11. [Google Scholar] [CrossRef]
- Sjöström, P.J.; Frydel, B.R.; Wahlberg, L.U. Artificial neural network-aided image analysis system for cell counting. Cytometry 1999, 36, 18–26. [Google Scholar] [CrossRef]
- Prematilleke, I.; Mohan, V.; Roberts, I.; Protheroe, A.; Gatter, K.; Hospital, J.R. An easy cell counting method for immunohistochemistry that does not use an image analysis program. Histopathology 2011, 59, 801–803. [Google Scholar] [CrossRef]
- Choudhry, P. High-Throughput method for automated colony and cell counting by digital image analysis based on edge detection. PLoS ONE 2016, 11, e0148469. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.C.; Kim, J.; Park, J.; Kim, E.J.; Kim, J.; Park, Y.M.; Cho, H.S.; Byun, D.; Henderson, I.R.; Copenhaver, G.P.; et al. DeepTetrad: High-throughput image analysis of meiotic tetrads by deep learning in Arabidopsis thaliana. Plant J. 2020, 101, 473–483. [Google Scholar] [CrossRef]
- Du, J.; Li, X.; Li, Q. Detection and classification of cervical exfoliated cells based on faster R-CNN*. In Proceedings of the 2019 IEEE 11th International Conference on Advanced Infocomm Technology (ICAIT), Jinan, China, 18–20 October 2019; pp. 52–57. [Google Scholar] [CrossRef]
- Chowdhury, A.B.; Roberson, J.; Hukkoo, A.; Bodapati, S.; Cappelleri, D.J. Automated complete blood cell count and malaria pathogen detection using convolution neural network. IEEE Robot Autom. Lett. 2020, 5, 1047–1054. [Google Scholar] [CrossRef]
- Hosseini, S.M.H.; Chen, H.; Jablonski, M.M. Automatic detection and counting of retina cell nuclei using deep learning. arXiv 2020, arXiv:2002.03563. [Google Scholar]
- Farooq, M.; Hafeez, A. COVID-ResNet: A Deep Learning Framework for Screening of COVID19 from Radiographs. arXiv 2020, arXiv:2003.14395. [Google Scholar]
- Elgendi, M.; Fletcher, R.; Howard, N.; Menon, C.; Ward, R. The Evaluation of Deep Neural Networks and X-Ray as a Practical Alternative for Diagnosis and Management of COVID-19. medRxiv 2020. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2999–3007. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, Y.; Oerlemans, A.; Lao, S.; Wu, S.; Lew, M.S. Deep learning for visual understanding: A review. Neurocomputing 2016, 187, 27–48. [Google Scholar] [CrossRef]
- Barchi, L.; Acquadro, A.; Alonso, D.; Aprea, G.; Bassolino, L.; Demurtas, O.; Ferrante, P.; Gramazio, P.; Mini, P.; Portis, E.; et al. Single Primer Enrichment Technology (SPET) for high-throughput genotyping in tomato and eggplant germplasm. Front. Plant Sci. 2019, 10, 1005. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.D.; Ruban, A.; Rutten, T.; Zhou, Y.H.; Houben, A. Analysis of pollen grains by immunostaining and FISH in Triticeae species. Methods Mol. Biol. 2020, 2061, 347–358. [Google Scholar] [CrossRef]
- Goodman, J.W. Introduction to Fourier Optics; Roberts and Company Publishers: Englewood, IL, USA, 2005. [Google Scholar]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning; Casella, G., Fienberg, S., Olkin, I., Eds.; Springer: New York, NY, USA, 2013; Volume 112. [Google Scholar] [CrossRef]
- García-Fortea, E.; Lluch-Ruiz, A.; Pineda-Chaza, B.J.; García-Pérez, A.; Bracho-Gil, J.P.; Plazas, M.; Gramazio, P.; Vilanova, S.; Moreno, V.; Prohens, J. A highly efficient organogenesis protocol based on zeatin riboside for in vitro regeneration of eggplant. BMC Plant Biol. 2020, 20, 6. [Google Scholar] [CrossRef]
- Dumas de Vaulx, R.; Chambonnet, D.; Pochard, E. Culture in vitro d’anthères de piment (Capsicum annuum L.): Amélioration des taux d’obtention de plantes chez différents génotypes par des traitements à + 35 °C. Agron. EDP Sci. 1981, 1, 859–864. [Google Scholar] [CrossRef]
- Dpooležel, J.; Binarová, P.; Lcretti, S. Analysis of nuclear DNA content in plant cells by flow cytometry. Biol. Plant 1989, 31, 113–120. [Google Scholar] [CrossRef]
- Doyle, J. DNA Protocols for Plants. In Molecular Techniques in Taxonomy; Hewitt, G.M., Johnston, A.W.B., Young, J.P.W., Eds.; Springer: Berlin Heidelberg, Germay, 1991; Volume 57, pp. 283–293. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Akbar, S.; Martel, A.L.; Peikari, M.; Salama, S.; Nofech-Mozes, S. Determining tumor cellularity in digital slides using ResNet. In Proceedings of the SPIE Medical Imaging, Houston, TX, USA, 10–15 February 2018; p. 29. [Google Scholar] [CrossRef]
- Yan, J.; Tucci, E.; Jaffe, N. Detection of the (9;22) Chromosome translocation using deep residual neural network. J. Comput. Commun. 2019, 7, 102–111. [Google Scholar] [CrossRef]
- Kang, R.; Liang, Y.; Lian, C.; Mao, Y. CNN-based automatic urinary particles recognition. arXiv 2018, arXiv:1803.02699. [Google Scholar]
- Malik, M.R.; Wang, F.; Dirpaul, J.M.; Zhou, N.; Polowick, P.L.; Ferrie, A.M.R.; Krochko, E. Transcript profiling and identification of molecular markers for early microspore embryogenesis in Brassica napus. Plant Physiol. 2007, 144, 134–154. [Google Scholar] [CrossRef]
- Heberle-Bors, E. Isolated pollen culture in tobacco: Plant reproductive development in a nutshell. Sexl. Plant Reprod. 1989, 2, 1–10. [Google Scholar] [CrossRef]
- Raghavan, V. From Microspore to Embryoid: Faces of the Angiosperm Pollen Grain. In Progress in Plant Cellular and Molecular Biology. Current Plant Science and Biotechnology in Agriculture; Nijkamp, H.J.J., Van Der Plas, L.H.W., Van Aartrijk, J., Eds.; Springer: Dordrecht, The Netherlands, 1990; Volume 9, pp. 213–221. [Google Scholar] [CrossRef]
- Emrani Dehkehan, M.; Moieni, A.; Movahedi, Z. Effects of zeatin riboside, mannitol and heat stress on eggplantn (Solanum melongena L.) anther culture. Imam Khomeini Int. Univ. Biotechnol. Soc. 2017, 6, 16–26. [Google Scholar] [CrossRef]
- Makowska, K.; Oleszczuk, S. Albinism in barley androgenesis. Plant Cell Rep. 2014, 33, 385–392. [Google Scholar] [CrossRef]
- Immonen, S.; Anttila, H. Media composition and anther plating for production of androgenetic green plants from cultivated rye (Secale cereale L.). J. Plant Physiol. 2000, 156, 204–210. [Google Scholar] [CrossRef]
- Kiviharju, E.; Puolimatka, M.; Saastamoinen, M.; Pehu, E. Extension of anther culture to several genotypes of cultivated oats. Plant Cell Rep. 2000, 19, 674–679. [Google Scholar] [CrossRef]
- Liu, W.; Zheng, M.Y.; Konzak, C.F. Improving green plant production via isolated microspore culture in bread wheat (Triticum aestivum L.). Plant Cell Rep. 2002, 20, 821–824. [Google Scholar] [CrossRef]
- Caredda, S.; Devaux, P.; Sangwan, R.S.; Proult, I.; Clément, C. Plastid ultrastructure and DNA related to albinism in androgenetic embryos of various barley (Hordeum vulgare) cultivars. Plant Cell Tiss. Org. 2004, 76, 35–43. [Google Scholar] [CrossRef]
- Kumari, M.; Clarke, H.J.; Small, I.; Siddique, K.H.M. Albinism in plants: A major bottleneck in wide hybridization, androgenesis and doubled haploid culture. CRC Crit. Rev. Plant Sci. 2009, 28, 393–409. [Google Scholar] [CrossRef]
- Höfer, M.; Grafe, C.; Boudichevskaja, A.; Lopez, A.; Bueno, M.A.; Roen, D. Characterization of plant material obtained by in vitro androgenesis and in situ parthenogenesis in apple. Sci. Hortic. 2008, 117, 203–311. [Google Scholar] [CrossRef]
- Sharma, S.; Chaudhary, H.; Sethi, G. In vitro and in vivo screening for drought tolerance in winter × spring wheat doubled haploids derived through chromosome elimination. Acta Agron. Hung. 2010, 58, 301–312. [Google Scholar] [CrossRef]
- Takahira, J.; Cousin, A.; Nelson, M.N.; Cowling, W.A. Improvement in efficiency of microspore culture to produce doubled haploid canola (Brassica napus L.) by flow cytometry. Plant Cell Tiss. Org. 2011, 104, 51–59. [Google Scholar] [CrossRef]
- Garcia-Arias, F.; Sánchez-Betancourt, E.; Núñez, V. Fertility recovery of anther-derived haploid plants in cape gooseberry (Physalis peruviana L.). Agron. Colomb. 2018, 36, 201–209. [Google Scholar] [CrossRef]
- Sheng, X.; Zhao, Z.; Yu, H.; Wang, J.; Xiaohui, Z.; Gu, H. Protoplast isolation and plant regeneration of different doubled haploid lines of cauliflower (Brassica oleracea var. botrytis). Plant Cell Tiss. Org. 2011; 107, 513–520. [Google Scholar] [CrossRef]
- Keleş, D.; Özcan, C.; Pınar, H.; Ata, A.; Denli, N.; Yücel, N.K.; Taşkın, H.; Büyükalaca, S. First report of obtaining haploid plants using tissue culture techniques in spinach. HortScience 2016, 51, 742–749. [Google Scholar] [CrossRef]
- Olszewska, D.; Niklas-Nowak, A.; Nowaczyk, L. Estimation of genetic divergence within androgenic regenerants of Capsicum annuum L. ATZ1 × C. frutescens L. F1 plants using random amplified polymorphic DNA markers. BioTechnologia 2017, 98, 175–182. [Google Scholar] [CrossRef]
- Budak, H.; Shearman, R.C.; Parmaksiz, I.; Dweikat, I. Comparative analysis of seeded and vegetative biotype buffalograsses based on phylogenetic relationship using ISSRs, SSRs, RAPDs, and SRAPs. Theor. Appl. Genet 2004, 109, 280–288. [Google Scholar] [CrossRef]
- Acquadro, A.; Barchi, L.; Gramazio, P.; Portis, E.; Vilanova, S.; Comino, C.; Plazas, M.; Prohens, J.; Lanteri, S. Coding SNPs analysis highlights genetic relationships and evolution pattern in eggplant complexes. PLoS ONE 2017, 12, e0180774. [Google Scholar] [CrossRef]
- Snape, J.W. Doubled haploid breeding: Theoretical basis and practical applications. In Review of Advances in Plant Biotechnology, 1985-1988: 2nd International Symposium on Genetic Manipulation in Crops; Mujeeb-Kazi, A., y Stitch, L.A., Eds.; International Maize and Wheat Improvement Center: Mexico City, Mexico; International Rice Research Institute: Los Baños, Philippines, 1989; pp. 19–30. [Google Scholar]
- Huang, L.; Tang, W.; Bu, S.; Wu, W. BRM: A statistical method for QTL mapping based on bulked segregant analysis by deep sequencing. Bioinformatics 2020, 36, 2150–2156. [Google Scholar] [CrossRef] [PubMed]
- Szarejko, I.; Forster, B.P. Doubled haploidy and induced mutation. Euphytica 2007, 158, 359–370. [Google Scholar] [CrossRef]
- Ferrie, A.M.R.; Taylor, D.C.; MacKenzie, S.L.; Rakow, G.; Raney, J.P.; Keller, W.A. Microspore mutagenesis of Brassica species for fatty acid modifications: A preliminary evaluation. Plant Breed. 2008, 127, 501–506. [Google Scholar] [CrossRef]
- Goedeke, S.; Hensel, G.; Kapusi, E.; Gahrtz, M.; Kumlehn, J. Transgenic barley in fundamental research and biotechnology. Transgenic Plant J. 2007, 1, 104–117. [Google Scholar]
- Birchler, J. Heterosis: The genetic basis of hybrid vigour. Nat. Plants 2015, 1, 15020. [Google Scholar] [CrossRef] [PubMed]

| Medium | MS + Vitamins (g/L) | Prepared C (g/L) | Prepared R (g/L) | Sucrose (g/L) | Zeatin Riboside (mg/L) | Kinetin (mg/L) | 2,4-D (mg/L) | Vitamin B12 (mg/L) | Gelrite (g/L) | Bacto-Agar (g/L) |
|---|---|---|---|---|---|---|---|---|---|---|
| E0 | 2.20 | - | - | 15.00 | - | - | - | - | 7.00 | - |
| E6 | 2.20 | - | - | 15.00 | 2.00 | - | - | - | 7.00 | - |
| Cb | - | 4.55 | - | 120.00 | - | 5.00 | 5.00 | 0.20 | - | 8.00 |
| R | - | - | 4.55 | 30.00 | - | 0.10 | - | - | - | 8.00 |
| Class | Total Cellular Events | Average Precision (%) |
|---|---|---|
| Tetrad | 290 | 87.40 |
| Young Microspore | 641 | 86.28 |
| Medium Microspore | 1893 | 81.97 |
| Vacuolated Microspore | 1185 | 87.60 |
| Young Pollen | 1876 | 82.32 |
| Mature Pollen | 2186 | 92.19 |
| mAP | 86.30 |
| Protocol E6 | Protocol Cb | |||||
|---|---|---|---|---|---|---|
| Size Range/Genotype | Anthers (n) | Response (%) | Type of Response | Anthers (n) | Response (%) | Type of Response |
| 3.5–4 mm | ||||||
| BC3 17-8 | 45 | 0.00 ± 0.00 | - | 45 | 0.00 ± 0.00 | - |
| BC3 17-19 | 45 | 0.00 ± 0.00 | - | 45 | 0.00 ± 0.00 | - |
| BC3 17-4 | 45 | 0.00 ± 0.00 | - | 45 | 0.00 ± 0.00 | - |
| 5.5–6 mm | ||||||
| BC3 17-8 | 45 | 75.30 ± 0.04 | Embryo | 45 | 78.40 ± 0.04 | Callus |
| BC3 17-19 | 45 | 60.40 ± 0.05 | Embryo | 45 | 80.50 ± 0.04 | Callus |
| BC3 17-4 | 45 | 40.30 ± 0.05 | Embryo | 45 | 71.20 ± 0.05 | Callus |
| >6 mm | ||||||
| BC3 17-8 | 45 | 0.00 ± 0.00 | - | 45 | 3.20 ± 0.02 | Somatic callus |
| BC3 17-19 | 45 | 0.00 ± 0.00 | - | 45 | 4.00 ± 0.02 | Somatic callus |
| BC3 17-4 | 45 | 0.00 ± 0.00 | - | 45 | 4.30 ± 0.03 | Somatic callus |
| Genotype | Embryos (n) | Acclimatized Plants (n) | n | n + 2n | 2n |
|---|---|---|---|---|---|
| BC3 17-8 | 42 | 12 | 9 | 3 | 0 |
| BC3 17-19 | 26 | 12 | 8 | 1 | 3 |
| BC3 17-4 | 9 | 7 | 6 | 1 | 0 |
| Accession/Offspring | n | Missing SNPs | Heterozygosity (%) |
|---|---|---|---|
| ELE BC3 17-19 | 1 | 0.00 | 100.00 |
| BC4 (ELE BC3 17-19 × MEL3) | 6 | 4.00 (0.00–6.00) | 55.07 (39.06–65.66) |
| DH (ELE BC3 17-19 doubled haploids) | 10 | 5.00 (0.00–12.00) | 1.59 (0.19–3.95) |
| ELE BC3 17-4 | 1 | 0.00 | 100.00 |
| BC4 (ELE BC3 17-4 × MEL3) | 5 | 0.40 (0.00–2.00) | 53.87 (13.35–92.66) |
| DH (ELE BC3 17-4 doubled haploids) | 7 | 1.14 (0.00–4.00) | 0.67 (0.19–1.70) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Fortea, E.; García-Pérez, A.; Gimeno-Páez, E.; Sánchez-Gimeno, A.; Vilanova, S.; Prohens, J.; Pastor-Calle, D. A Deep Learning-Based System (Microscan) for the Identification of Pollen Development Stages and Its Application to Obtaining Doubled Haploid Lines in Eggplant. Biology 2020, 9, 272. https://doi.org/10.3390/biology9090272
García-Fortea E, García-Pérez A, Gimeno-Páez E, Sánchez-Gimeno A, Vilanova S, Prohens J, Pastor-Calle D. A Deep Learning-Based System (Microscan) for the Identification of Pollen Development Stages and Its Application to Obtaining Doubled Haploid Lines in Eggplant. Biology. 2020; 9(9):272. https://doi.org/10.3390/biology9090272
Chicago/Turabian StyleGarcía-Fortea, Edgar, Ana García-Pérez, Esther Gimeno-Páez, Alfredo Sánchez-Gimeno, Santiago Vilanova, Jaime Prohens, and David Pastor-Calle. 2020. "A Deep Learning-Based System (Microscan) for the Identification of Pollen Development Stages and Its Application to Obtaining Doubled Haploid Lines in Eggplant" Biology 9, no. 9: 272. https://doi.org/10.3390/biology9090272
APA StyleGarcía-Fortea, E., García-Pérez, A., Gimeno-Páez, E., Sánchez-Gimeno, A., Vilanova, S., Prohens, J., & Pastor-Calle, D. (2020). A Deep Learning-Based System (Microscan) for the Identification of Pollen Development Stages and Its Application to Obtaining Doubled Haploid Lines in Eggplant. Biology, 9(9), 272. https://doi.org/10.3390/biology9090272






