Challenges and Opportunities in Cytopathology Artificial Intelligence
Abstract
:1. Introduction
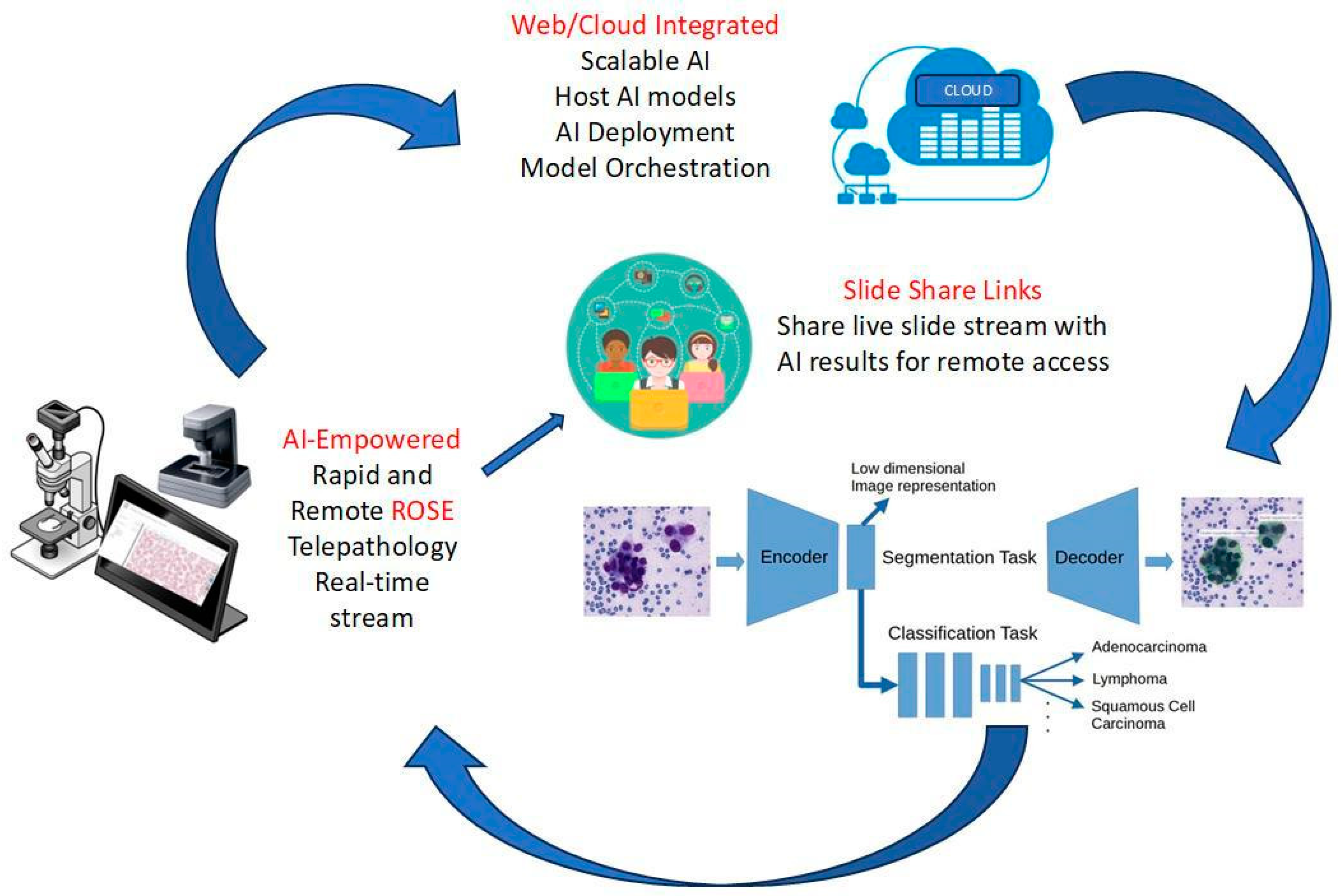
Rapid Onsite Evaluation (ROSE) of Cytology Specimens
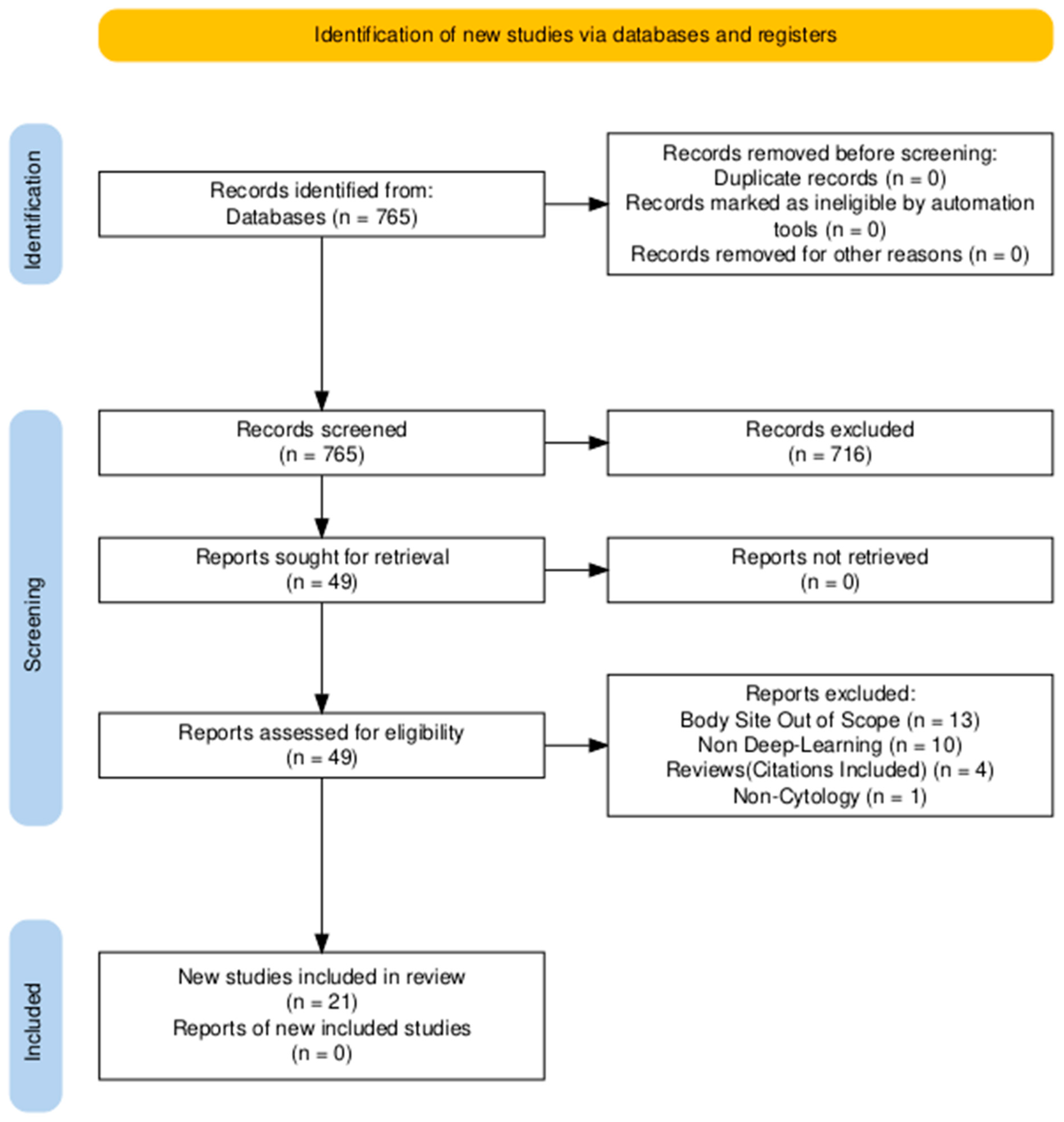
2. Methods (Search Strategy)
3. Results
3.1. Thyroid
- (1)
- (2)
- disambiguating equivocal diagnoses of atypia of undetermined significance/follicular lesions of undetermined significance (AUS-FLUS) [25].
- (3)
3.2. Pancreatobiliary System
3.3. Lung
4. Discussion
4.1. Data Augmentation, Model Performance, and Reproducibility
4.2. Quality Control and Assurance
4.3. Tedious and Laborious Manual Annotation
4.4. Human–Machine Collaboration in Cytopathology
4.5. Integration into Cytopathology Clinical Workflow
4.6. Future Trends and Directions
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xing, L.; Giger, M.L.; Min, J.K. Artificial Intelligence in Medicine: Technical Basis and Clinical Applications; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Mezei, T.; Kolcsár, M.; Joó, A.; Gurzu, S. Image Analysis in Histopathology and Cytopathology: From Early Days to Current Perspectives. J. Imaging 2024, 10, 252. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, Y.; Tan, Y.; Wan, H.; Zheng, N.; He, Z.; Mao, L.; Ren, W.; Chen, K.; Lin, Z.; et al. Artificial intelligence enables precision diagnosis of cervical cytology grades and cervical cancer. Nat. Commun. 2024, 15, 4369. [Google Scholar] [CrossRef] [PubMed]
- Pantanowitz, J.; Manko, C.D.; Pantanowitz, L.; Rashidi, H.H. Synthetic Data and Its Utility in Pathology and Laboratory Medicine. Mod. Pathol. 2024, 104, 102095. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.C. Regulating Artificial Intelligence for a Successful Pathology Future. Arch. Pathol. Lab. Med. 2019, 143, 1175–1179. [Google Scholar] [CrossRef]
- Tolles, W.E.; Bostrom, R.C. Automatic screening of cytological smears for cancer: The instrumentation. Ann. N. Y. Acad. Sci. 1956, 63, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Karakitsos, P.; Cochand-Priollet, B.; Pouliakis, A.; Guillausseau, P.J.; Ioakim-Liossi, A. Learning vector quantizer in the investigation of thyroid lesions. Anal. Quant Cytol. Histol. 1999, 21, 201–208. [Google Scholar]
- Wang, Y.-L.; Gao, S.; Xiao, Q.; Li, C.; Grzegorzek, M.; Zhang, Y.-Y.; Li, X.-H.; Kang, Y.; Liu, F.-H.; Huang, D.-H.; et al. Role of artificial intelligence in digital pathology for gynecological cancers. Comput. Struct. Biotechnol. J. 2024, 24, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Trisolini, R.; Cancellieri, A.; Tinelli, C.; de Biase, D.; Valentini, I.; Casadei, G.; Paioli, D.; Ferrari, F.; Gordini, G.; Patelli, M.; et al. Randomized Trial of Endobronchial Ultrasound-Guided Transbronchial Needle Aspiration with and Without Rapid On-site Evaluation for Lung Cancer Genotyping. Chest 2015, 148, 1430–1437. [Google Scholar] [CrossRef]
- Jain, D.; Roy-Chowdhuri, S. Molecular Pathology of Lung Cancer Cytology Specimens: A Concise Review. Arch. Pathol. Lab. Med. 2018, 142, 1127–1133. [Google Scholar] [CrossRef]
- Fawcett, C.; Eppenberger-Castori, S.; Zechmann, S.; Hanke, J.; Herzog, M.; Prince, S.S.; Christ, E.R.; Ebrahimi, F. Effects of Rapid On-Site Evaluation on Diagnostic Accuracy of Thyroid Fine-Needle Aspiration. Acta Cytol. 2022, 66, 371–378. [Google Scholar] [CrossRef]
- Iglesias-Garcia, J.; Lariño-Noia, J.; Abdulkader, I.; Domínguez-Muñoz, J.E. Rapid on-site evaluation of endoscopic-ultrasound-guided fine-needle aspiration diagnosis of pancreatic masses. World J. Gastroenterol. 2014, 20, 9451–9457. [Google Scholar] [CrossRef]
- Jain, D.; Allen, T.C.; Aisner, D.L.; Beasley, M.B.; Cagle, P.T.; Capelozzi, V.L.; Hariri, L.P.; Lantuejoul, S.; Miller, R.; Mino-Kenudson, M.; et al. Rapid On-Site Evaluation of Endobronchial Ultrasound–Guided Transbronchial Needle Aspirations for the Diagnosis of Lung Cancer: A Perspective From Members of the Pulmonary Pathology Society. Arch. Pathol. Lab. Med. 2018, 142, 253–262. [Google Scholar] [CrossRef] [PubMed]
- VanderLaan, P.A.; Chen, Y.; Alex, D.; Balassanian, R.; Cuda, J.; Hoda, R.S.; Illei, P.B.; McGrath, C.M.; Randolph, M.L.; Reynolds, J.P.; et al. Results from the 2019 American Society of Cytopathology survey on rapid on-site evaluation—Part 1: Objective practice patterns. J. Am. Soc. Cytopathol. 2019, 8, 333–341. [Google Scholar] [CrossRef]
- Yan, S.; Li, Y.; Pan, L.; Jiang, H.; Gong, L.; Jin, F. The application of artificial intelligence for Rapid On-Site Evaluation during flexible bronchoscopy. Front. Oncol. 2024, 14, 1360831. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Li, X.; Shen, H.; Chen, Y.; Wang, L.; Chen, H.; Feng, J.; Liu, J. A systematic review of deep learning-based cervical cytology screening: From cell identification to whole slide image analysis. Artif. Intell. Rev. 2023, 56, 2687–2758. [Google Scholar] [CrossRef]
- Haddaway, N.R.; Page, M.J.; Pritchard, C.C.; McGuinness, L.A. An R package and Shiny app for producing PRISMA 2020-compliant flow diagrams, with interactivity for optimised digital transparency and Open Synthesis. Campbell Syst. Rev. 2022, 18, e1230. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, P.; Mukherjee, T.; Barui, S.; Das, A.; Gangopadhyay, P. Artificial Intelligence in Cytopathology: A Neural Network to Identify Papillary Carcinoma on Thyroid Fine-Needle Aspiration Cytology Smears. J. Pathol. Inform. 2018, 9, 43. [Google Scholar] [CrossRef] [PubMed]
- Range, D.D.E.; Dov, D.; Kovalsky, S.Z.; Henao, R.; Carin, L.; Cohen, J. Application of a machine learning algorithm to predict malignancy in thyroid cytopathology. Cancer Cytopathol. 2020, 128, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Dov, D.; Kovalsky, S.Z.; Feng, Q.; Assaad, S.; Cohen, J.; Bell, J.; Henao, R.; Carin, L.; Range, D.E. Use of Machine Learning–Based Software for the Screening of Thyroid Cytopathology Whole Slide Images. Arch. Pathol. Lab. Med. 2021, 146, 872–878. [Google Scholar] [CrossRef]
- Fragopoulos, C.; Pouliakis, A.; Meristoudis, C.; Mastorakis, E.; Margari, N.; Chroniaris, N.; Koufopoulos, N.; Delides, A.G.; Machairas, N.; Ntomi, V.; et al. Radial Basis Function Artificial Neural Network for the Investigation of Thyroid Cytological Lesions. J. Thyroid. Res. 2020, 2020, 5464787. [Google Scholar] [CrossRef]
- Gopinath, B.; Shanthi, N. Computer-aided diagnosis system for classifying benign and malignant thyroid nodules in multi-stained FNAB cytological images. Australas. Phys. Eng. Sci. Med. 2013, 36, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Wang, Y.; Ping, B.; Li, D.; Du, J.; Qin, Y.; Lu, H.; Wan, X.; Xiang, J. Deep convolutional neural network VGG-16 model for differential diagnosing of papillary thyroid carcinomas in cytological images: A pilot study. J. Cancer 2019, 10, 4876–4882. [Google Scholar] [CrossRef]
- Savala, R.; Dey, P.; Gupta, N. Artificial neural network model to distinguish follicular adenoma from follicular carcinoma on fine needle aspiration of thyroid. Diagn. Cytopathol. 2018, 46, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Hirokawa, M.; Niioka, H.; Suzuki, A.; Abe, M.; Arai, Y.; Nagahara, H.; Miyauchi, A.; Akamizu, T. Application of deep learning as an ancillary diagnostic tool for thyroid FNA cytology. Cancer Cytopathol. 2023, 131, 217–225. [Google Scholar] [CrossRef]
- Zhu, C.; Tao, S.; Chen, H.; Li, M.; Wang, Y.; Liu, J.; Jin, M. Hybrid model enabling highly efficient follicular segmentation in thyroid cytopathological whole slide image. Intell. Med. 2021, 1, 70–79. [Google Scholar] [CrossRef]
- Ali, S.Z.; Baloch, Z.W.; Cochand-Priollet, B.; Schmitt, F.C.; Vielh, P.; VanderLaan, P.A. The 2023 Bethesda System for Reporting Thyroid Cytopathology. Thyroid 2023, 33, 1039–1044. [Google Scholar] [PubMed]
- Varlatzidou, A.; Pouliakis, A.; Stamataki, M.; Meristoudis, C.; Margari, N.; Peros, G.; Panayiotides, J.G.; Karakitsos, P. Cascaded learning vector quantizer neural networks for the discrimination of thyroid lesions. Anal. Quant. Cytol. Histol. 2011, 33, 323–334. [Google Scholar]
- Gopinath, B.; Shanthi, N. Development of an Automated Medical Diagnosis System for Classifying Thyroid Tumor Cells using Multiple Classifier Fusion. Technol. Cancer Res. Treat. 2015, 14, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Dahia, S.S.; Konduru, L.; Pandol, S.J.; Barreto, S.G. The burden of young-onset pancreatic cancer and its risk factors from 1990 to 2019: A systematic analysis of the global burden of disease study 2019. Pancreatology 2024, 24, 119–129. [Google Scholar] [CrossRef]
- Momeni-Boroujeni, A.; Yousefi, E.; Somma, J. Computer-assisted cytologic diagnosis in pancreatic FNA: An application of neural networks to image analysis. Cancer Cytopathol. 2017, 125, 926–933. [Google Scholar] [CrossRef]
- Lin, R.; Sheng, L.-P.; Han, C.-Q.; Guo, X.-W.; Wei, R.-G.; Ling, X.; Ding, Z. Application of artificial intelligence to digital-rapid on-site cytopathology evaluation during endoscopic ultrasound-guided fine needle aspiration: A proof-of-concept study. J. Gastroenterol. Hepatol. 2023, 38, 883–887. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhou, Y.; Tangb, D.; Nib, M.; Zheng, J.; Xu, G.; Peng, C.; Shen, S.; Zhan, Q.; Wang, X.; et al. A deep learning-based segmentation system for rapid onsite cytologic pathology evaluation of pancreatic masses: A retrospective, multicenter, diagnostic study. EBioMedicine 2022, 80, 104022. [Google Scholar] [CrossRef]
- Kurita, Y.; Kuwahara, T.; Hara, K.; Mizuno, N.; Okuno, N.; Matsumoto, S.; Obata, M.; Koda, H.; Tajika, M.; Shimizu, Y.; et al. Diagnostic ability of artificial intelligence using deep learning analysis of cyst fluid in differentiating malignant from benign pancreatic cystic lesions. Sci. Rep. 2019, 9, 6893. [Google Scholar] [CrossRef] [PubMed]
- Cancer Facts and Statistics. Available online: https://www.cancer.org/research/cancer-facts-statistics.html (accessed on 26 June 2024).
- Kratzer, T.B.; Bandi, P.; Freedman, N.D.; Smith, R.A.; Travis, W.D.; Jemal, A.; Siegel, R.L. Lung cancer statistics, 2023. Cancer 2024, 130, 1330–1348. [Google Scholar] [CrossRef] [PubMed]
- Grigorean, V.T.; Cristian, D.A. Cancer—Yesterday, Today, Tomorrow. Medicina 2022, 59, 98. [Google Scholar] [CrossRef] [PubMed]
- Teramoto, A.; Yamada, A.; Kiriyama, Y.; Tsukamoto, T.; Yan, K.; Zhang, L.; Imaizumi, K.; Saito, K.; Fujita, H. Automated classification of benign and malignant cells from lung cytological images using deep convolutional neural network. Inform. Med. Unlocked 2019, 16, 100205. [Google Scholar] [CrossRef]
- Teramoto, A.; Tsukamoto, T.; Yamada, A.; Kiriyama, Y.; Imaizumi, K.; Saito, K.; Fujita, H. Deep learning approach to classification of lung cytological images: Two-step training using actual and synthesized images by progressive growing of generative adversarial networks. PLoS ONE 2020, 15, e0229951. [Google Scholar] [CrossRef] [PubMed]
- Asfahan, S.; Elhence, P.; Dutt, N.; Jalandra, R.N.; Chauhan, N.K. Digital-Rapid On-site Examination in Endobronchial Ultrasound-Guided Transbronchial Needle Aspiration (DEBUT): A proof of concept study for the application of artificial intelligence in the bronchoscopy suite. Eur. Respir. J. 2021, 58, 2100915. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Chang, J.; Huang, C.; Wen, Y.; Ho, C.; Cheng, Y. Effectiveness of convolutional neural networks in the interpretation of pulmonary cytologic images in endobronchial ultrasound procedures. Cancer Med. 2021, 10, 9047–9057. [Google Scholar] [CrossRef]
- Gonzalez, D.; Dietz, R.L.; Pantanowitz, L. Feasibility of a deep learning algorithm to distinguish large cell neuroendocrine from small cell lung carcinoma in cytology specimens. Cytopathology 2020, 31, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Takamatsu, M.; Ninomiya, H.; Inamura, K.; Horai, T.; Iyoda, A.; Honma, N.; Hoshi, R.; Sugiyama, Y.; Yanagitani, N.; et al. Machine learning-based gene alteration prediction model for primary lung cancer using cytologic images. Cancer Cytopathol. 2022, 130, 812–823. [Google Scholar] [CrossRef]
- Tao, Y.; Cai, Y.; Fu, H.; Song, L.; Xie, L.; Wang, K. Automated interpretation and analysis of bronchoalveolar lavage fluid. Int. J. Med. Inform. 2022, 157, 104638. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wu, Z.; Tan, C.; Liao, Y.; Wang, Z.; Chen, K.; Pan, A. Fourier Ptychographic Microscopy 10 Years on: A Review. Cells 2024, 13, 324. [Google Scholar] [CrossRef] [PubMed]
- Janowczyk, A.; Zuo, R.; Gilmore, H.; Feldman, M.; Madabhushi, A. HistoQC: An Open-Source Quality Control Tool for Digital Pathology Slides. JCO Clin. Cancer Inform. 2019, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Prodduturi, N.; Chen, D.; Gu, Q.; Flotte, T.; Feng, Q.; Hart, S. Image-to-image translation for automatic ink removal in whole slide images. J. Med. Imaging 2020, 7, 057502. [Google Scholar] [CrossRef]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Silva, V.W.K.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-H.C.; Gadepalli, K.; MacDonald, R.; Liu, Y.; Kadowaki, S.; Nagpal, K.; Kohlberger, T.; Dean, J.; Corrado, G.S.; Hipp, J.D.; et al. An augmented reality microscope with real-time artificial intelligence integration for cancer diagnosis. Nat. Med. 2019, 25, 1453–1457. [Google Scholar] [CrossRef] [PubMed]
- FDA Releases Artificial Intelligence/Machine Learning Action Plan. U.S. Food and Drug Administration. Available online: https://www.fda.gov/news-events/press-announcements/fda-releases-artificial-intelligencemachine-learning-action-plan?utm_source=chatgpt.com (accessed on 6 December 2024).
- Available online: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device (accessed on 6 December 2024).

| Study | Objective | Dataset Total/Test | Method | Performance Metrics |
|---|---|---|---|---|
| Hirokawa et al., 2023 [25] | AUS classification | 393 nodule, 148,395 patch, 9782 images | EfficientNetV2-L | Sens: 94.7 Spec: 14.4 PPV: 56.3 NPV: 66.7 PR AUC: >0.95 |
| Dov et al, 2021 [20] | TBSRTC category | 254/109 | VGG11, ImageNet | AUC: 0.931 |
| Zhu et al., 2021 [26] | Semantic Segmentation vs. Patch Classification | 6900/2400 | Enhanced ASPP/ Integrated Classifier (ResNet 101 basis) | Segmentation: AUC: 99.50 Classification: Acc: 99.3 |
| Fragopoulos et al., 2020 [21] | Benign vs. Malignant | 447/223 | ANN | Nucleus Level-Acc: 86.94 Sens: 81.37 Spec: 90.01 PPV: 81.74 NPV: 83.78 Patient Level-Acc: 95.07 Sens: 95.00 Spec: 95.10 PPV 91.57 NPV 97.14 |
| Elliott Range et al., 2020 [19] | TBSRTC category | 918/109 | VGG11, ImageNet | Sens 92.0% Spec 90.5% AUC 0.932 |
| Zhu et al., 2019 [26] | AUS vs Malignant | 467 | DNN NOS | Sens: 87.91 Spec: 85.15 AUC: 0.891 Acc: 87.15 |
| Guan et al, 2019 [23] | PTC vs benign nodules | 887/128 | DCNN VGG-16 | Acc 97.66% Sens 100% Spec 94.91% PPV: 95.83% NPV: 100% |
| Savala et al., 2018 [24] | Follicular adenoma vs. carcinoma | 57/9 | ANN | Sens: 100 Spec: 100 AUC: 1.00 |
| Sanyal et al, 2018 [18] | PTC vs. Non-PTC | 418/48 | CNN | Sens: 90.48 Spec: 83.33 PPV: 63.33 NPV: 96.49 Acc: 85.06 |
| Gopinath et al., 2015 [29] | Benign vs. Malignant | 110/30 | SVM, Elman ENN | Sens: 95 Spec: 100 Acc: 96.66 |
| Gopinath et al., 2013 [22] | Benign vs. Malignant | 110/30 | ENN/SVM | Sens: 100 Spec: 80 Acc:93.33 |
| Varlatzidou et al., 2011 [28] | Benign vs. Malignant | 335/50% | LVQ43 Learning Vector Quantizer NN | Sens: 91.51 Spec: 92.43 PPV: 74.26 NPV: 97.85 |
| Study | Objective | Dataset Train/Test | Method | Performance Metrics |
|---|---|---|---|---|
| Zhang et al., 2022 [34] | Segmentation/Pancreatic Ca vs. Non-Ca | Patients: 194 Total: 5345 images Train: 2166 images, 66 patients Val: 695 images, 16 patients Test: 1162 images, 27 patients (internal); 1322 images, 85 patients (external) | UNet-based CNN | Segmentation: Acc: 0.964 F-Sc: 0.929 Prec: 0.927 Rec: 0.931 Diagnosis: Acc: 0.945 Sens: 0.987 Spec: 0.930 PPV: 0.835 NPV: 0.995 |
| Kruita et al., 2019 [35] | Benign vs. Malignant | Patients: 85 Train: 68 Test: | multi-hidden layer of neural network | AUC: 0.966 Sens:95.7% Spec:91.9% |
| Momeni-Boroujeni et al., 2017 [32] | Benign vs. Malignant | Total Dataset: 75 cases Train N: 70%Test N: 30% | MLP (feed-forward) | Sens: 80% Spec: 75% Acc: 100.0 AUC: 0.917 |
| Study | Objective | Dataset Total/Test | Method | Performance Metrics |
|---|---|---|---|---|
| Ishii et al., 2022 [44] | ALK vs. EGFR vs. KRAS vs. None EP: Molecular Gene Alteration | Total: 40 cases, 114 slides, 464,378 patches, 145,468 H&E patches Train N: 20 cases Test N: 20 cases | MobileNet-V2 Transfer Learning | Sens: Patch level: 0.688 (ALK); 0.933 (EGFR); 0.942 (KRAS); 0.450 (None) Spec: Patch level: 0.778 (ALK); 0.986 (EGFR); 0.948 (KRAS); 0.961 (None) Acc: Patch level: 0.760 (ALK); 0.969 (EGFR); 0.947 (KRAS); 0.809 (None) Prec: Patch level: 0.432 (ALK); 0.968 (EGFR); 0.811 (KRAS); 0.830 (None) F-Sc: Patch level: 0.530 (ALK); 0.950 (EGFR); 0.871 (KRAS); 0.584 (None) |
| Lin et al., 2021 [42] | Semantic Segmentation/ Diagnosis: Benign vs. Malignant | Total: 499 images, 7486 patches Train N:70% Val N: 15% Test N:15% | HRNet/ResNet101 | Segmentation: AUC: 89.2 Diagnosis PL: Sens: 98.8 Spec: 98.9 PPV: 99.1 NPV: 98.3 Acc: 98.8 Diagnosis IL: Sens: 98.2 Spec: 77.8 PPV: 96.6 NPV: 87.5 Acc: 95.5 |
| Gonzalez et al., 2020 [43] | SCLC vs. LCNEC | 40 cases, 114 slides, 464,378 total patches, 59,072 Pap patches | Inception V3 | Sens: 1 Spec: 0.875 AUC: 0.875 Acc: 87.5 (SCLC); 100.0 (LCNEC) |
| Teramoto et al., 2020 [40] | Benign vs. Malignant | 60 cases, 511 images, 793 patches | Modified VGG-16 | Sens: 85.4 Spec: 85.3 Acc: 0.853 |
| Teramoto et al., 2019 [39] | Benign vs. Malignant | 47 cases/417 images/621 patches | Modified VGG-16, ImageNet Transfer Learning | Sens: 89.3 Spec: 83.3 AUC: Patch level: 0.872; Case level: 0.932 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
VandeHaar, M.A.; Al-Asi, H.; Doganay, F.; Yilmaz, I.; Alazab, H.; Xiao, Y.; Balan, J.; Dangott, B.J.; Nassar, A.; Reynolds, J.P.; et al. Challenges and Opportunities in Cytopathology Artificial Intelligence. Bioengineering 2025, 12, 176. https://doi.org/10.3390/bioengineering12020176
VandeHaar MA, Al-Asi H, Doganay F, Yilmaz I, Alazab H, Xiao Y, Balan J, Dangott BJ, Nassar A, Reynolds JP, et al. Challenges and Opportunities in Cytopathology Artificial Intelligence. Bioengineering. 2025; 12(2):176. https://doi.org/10.3390/bioengineering12020176
Chicago/Turabian StyleVandeHaar, Meredith A., Hussien Al-Asi, Fatih Doganay, Ibrahim Yilmaz, Heba Alazab, Yao Xiao, Jagadheshwar Balan, Bryan J. Dangott, Aziza Nassar, Jordan P. Reynolds, and et al. 2025. "Challenges and Opportunities in Cytopathology Artificial Intelligence" Bioengineering 12, no. 2: 176. https://doi.org/10.3390/bioengineering12020176
APA StyleVandeHaar, M. A., Al-Asi, H., Doganay, F., Yilmaz, I., Alazab, H., Xiao, Y., Balan, J., Dangott, B. J., Nassar, A., Reynolds, J. P., & Akkus, Z. (2025). Challenges and Opportunities in Cytopathology Artificial Intelligence. Bioengineering, 12(2), 176. https://doi.org/10.3390/bioengineering12020176








