Abstract
Streptococcus agalactiae (Group B Streptococcus, GBS) is an opportunistic pathogen, which asymptomatically colonizes the gastrointestinal and genitourinary tract of up to one third of healthy adults. Nevertheless, GBS carriage in pregnant women may lead to several health issues in newborns causing life threatening infection, such as sepsis, pneumonia or meningitis. Recommended GBS screening in pregnant women significantly reduced morbidity and mortality in infants. Nevertheless, intrapartum antibiotic prophylaxis, recommended following the detection of carriage or in case of lack of a carriage test result for pregnant women who demonstrate certain risk factors, led to the expansion of the adverse phenomenon of bacterial resistance to antibiotics. In our paper, we reviewed some immunogenic GBS proteins, i.e., Alp family proteins, β protein, Lmb, Sip, BibA, FsbA, ScpB, enolase, elongation factor Tu, IMPDH, and GroEL, which possess features characteristic of good candidates for immunodiagnostic assays for GBS carriage detection, such as immunoreactivity and specificity. We assume that they can be used as an alternative diagnostic method to the presently recommended bacteriological cultivation and MALDI.
1. Introduction
Streptococcus agalactiae (Group B Streptococcus, GBS) is a β-hemolytic, Gram-positive bacterium, which colonizes the gastrointestinal and genitourinary tract of up to 30% of healthy adults [1]. Each year, over 21 million pregnant women worldwide are colonized with GBS, which includes approximately 18% of pregnancies [2]. This opportunistic pathogen can cause a life-threatening infection in newborns, which most often takes the form of sepsis, pneumonia and meningitis, and the first two are more common for early onset GBS disease (EOD, EOGBSD). EOD appears in the first week of life, however, the vast majority of cases concerns the first 24 h and it is a consequence of an infection acquired during natural childbirth from a GBS-colonized mother [1]. In the early 1970s, mortality in this group was very high, and even reached 55% in newborns diagnosed with GBS infection [3].
In response to this threat, in the 1990s, the American College of Obstetricians and Gynecologists (ACOG) and Centers for Disease Control and Prevention (CDC) developed guidelines to minimize this dangerous phenomenon, which led to a reduction in morbidity in newborns of over 80%, and presently the incidence of the disease caused by GBS reaches 0.23/1000 live births [4,5,6]. It included screening recommendations for women between the 36th (36-0/7) and 37th (37-0/7) week of pregnancy, by taking swabs from the vaginal introitus and the anal sphincter, followed by microbial cultivation on the appropriate growth medium [4]. This method is not flawless due to its time-consuming nature, as the waiting time for the results is up to 7 days. The results themselves can also be ambiguous due to the often-difficult differentiation of GBS from other beta streptococci using phenotypic methods. It is a severe limitation especially in the case of advanced preterm labor, before the 36th week of pregnancy, i.e., prior to the recommended carrier screening test. Therefore, as an alternative for cultivation methods, in the latest CDC guidelines, mass spectrometry MALDI is also recommended for GBS detection. The main advantage of this method is a reduction in the waiting time for results as well as the possibility to distinguish Streptococcus agalactiae from other streptococci: S. halichoeri or S. pseudoporcinus, whose pathogenic role in newborn infection is not known [4]. When a positive result from a pregnant woman is obtained and GBS is detected in the studied specimen, introducing intrapartum antibiotic prophylaxis (IAP) is recommended. This procedure significantly reduced the morbidity in the case of early onset disease, however, IAP has not reduced the incidence of late onset disease, which may appear between the 7th and 90th day of life [7]. Additionally, the side effect of the comprehensive use of antibiotics is that is leads to the expansion of the adverse phenomenon of bacterial resistance to antibiotic therapy [4]. Moreover, this solution led to an increase in the rate of infections caused by Gram-negative bacteria [8]. As an answer to the limitations mentioned above and the unfavorable phenomena, scientists conduct extensive research to find alternative methods for GBS carriage detection in pregnant women. Novel diagnostic techniques should, among others, provide rapid and unambiguous results. An immunodiagnostic assay (i.e., ELISA) based on highly immunogenic and specific bacterial proteins (antigens) for detection of anti-GBS antibodies is being considered as a good candidate for innovative GBS carriage diagnostics. In our paper, we aimed to review some immunogenic proteins representative of Streptococcus agalactiae, which demonstrate features qualifying them as potential biomarkers in an innovative immunodiagnostic assay for the detection of GBS carriage and/or infection in pregnant women. It is worth underlining that such an assay could be useful also in the diagnostic of GBS infection in adults, in which this bacterium constitutes a growing clinical problem, taking the form of urinary tracts infection (UTI), sepsis, septic arthritis, meningitis and it is also isolated from diabetic foot/ulcer [9].
2. Main Text
The first immunological research on Streptococcus agalactiae was carried out by Professor Rebecca Lancefield who, in the 1930s, classified hemolytic streptococci into sera groups, according to the differences in the polysaccharide structure present in the bacterial cell wall. Thus, Streptococcus agalactiae was qualified into Group B, and it is described as GBS (Group B Streptococcus) [10]. Next, within group B, strains, according to the differences in the capsular polysaccharide (CPS) structure, were divided into three serotypes: I, II and III [10,11]. Currently, ten GBS serotypes: Ia, Ib, II-IX are distinguished, and their distribution is related to, among others, infection type, latitude, and age [2]. Since the 1970s, the number of publications describing specific GBS immunogenic molecules, with particular emphasis on immunogenic proteins successively grows from one year to another (source: https://pubmed.ncbi.nlm.nih.gov/ after entering the phrase: “immunogenic Streptococcus agalactiae proteins”, accessed on: 16 July 2021). Below we reviewed selected immunogenic Streptococcus agalactiae proteins, which can be considered as detective antigens in immunodiagnostic assay.
2.1. Alpha-like Protein
The best-known group of GBS proteins is the Alpha-like protein family (Alp), which include the following members: αC, Alp 1 (epsilon), Alp 2, Alp 3, Alp 4, and Rib. Alp proteins are conservative, chimeric and form mosaic structures on the GBS surface [12]. These surface-anchored proteins play an important role in Streptococcus agalactiae virulence, by supporting bacterial cell adherence to infected cells of the host (Figure 1). For example, αC protein mediates GBS invasion of cervical epithelial cells by interaction with glycosaminoglycan [13].
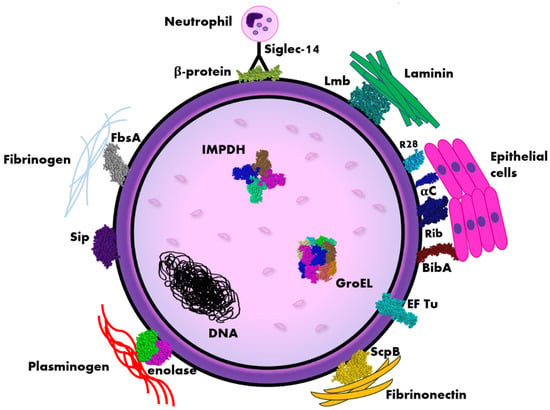
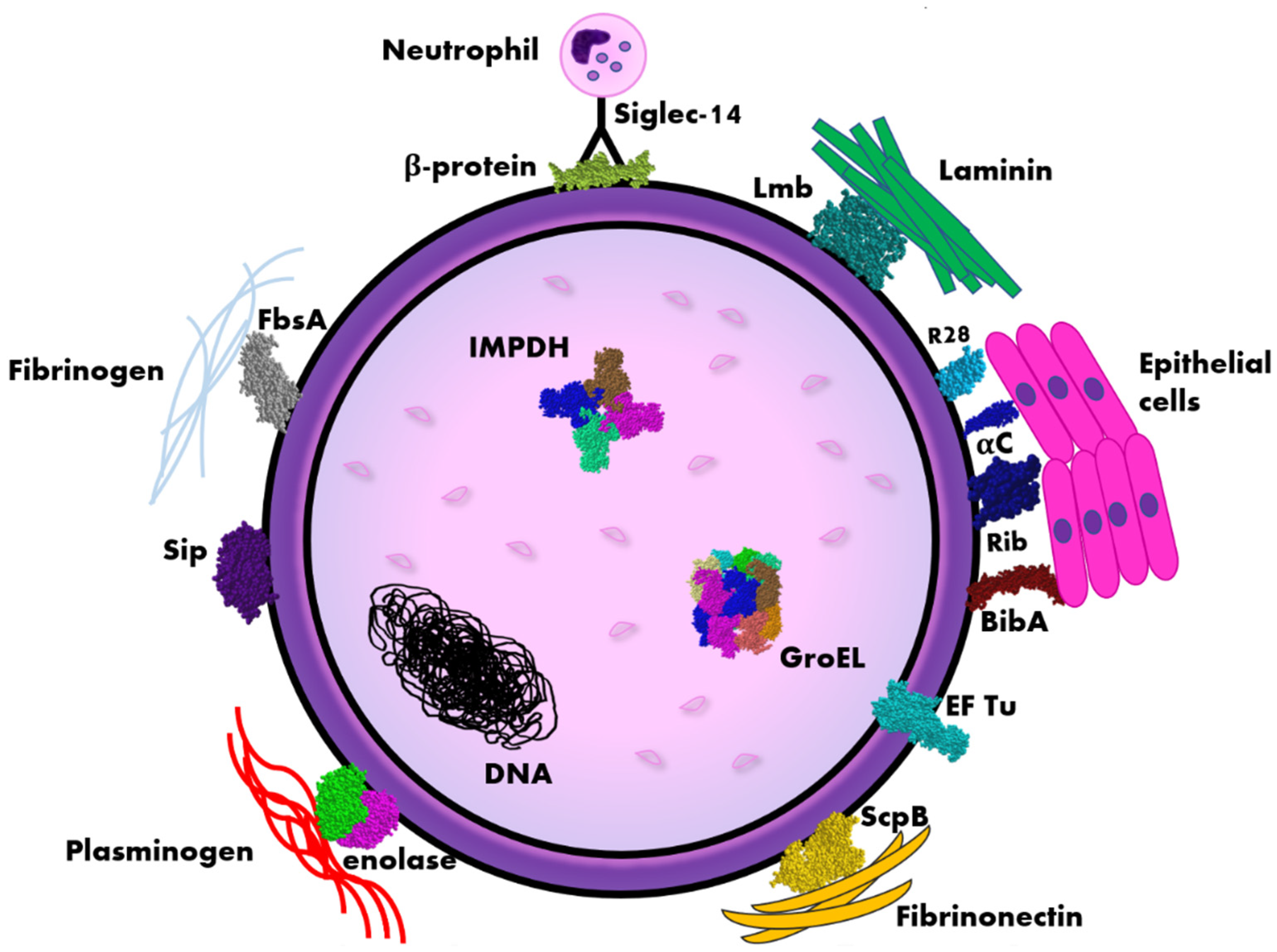
Figure 1.
Scheme of the distribution of immunogenic proteins within the Streptococcus agalactiae (GBS) cell. Legend: αC, Rib, R28 (Alp3)—surface proteins belonging to Alp-like family, BibA—Group B streptococcus immunogenic bacterial adhesin, EF Tu—elongation factor thermo unstable, FsbA—fibrinogen-binding protein, IMPDH—inosine 5′-monophosphate dehydrogenase, Lmb—laminin binding protein, ScpB—streptococcal peptidase C5a, Siglec-14—sialic acid-binding immunoglobulin-like lectin, Sip—surface immunogenic protein. The diagram is not true to scale, the individual elements have a schematic dimension.
Alp proteins consist of a major signal peptide domain, a N-terminus region, comprising 170–180 amino acids, repeat area with numerous tandem repeats (8–10) of approximately 80 amino acids each, and a C-terminus region built from 40–50 amino acids. Their molecular masses may vary among particular GBS strains, for example α mass can range from 65 kDa to 165 kDa, and it can be explained by the differences in the number of repeats [14,15]. Amino acid sequences consisting of Alp family members are very likely, which may induce cross-reactivity among the individual proteins in the family.
Alp proteins are also described as immunogenic proteins [12]. In the context of immunogenicity, the best-known proteins are αC and Rib proteins. The immunogenicity of αC was described as the first immunogenic GBS protein. It was described for the first time in the 1980s on mouse model, when it was shown that purified αC protein isolated from S. agalactiae cell induced immunogenic response and protected mice against infection caused by these bacteria [16]. Afterward, this observation was confirmed in other studies [17,18,19,20]. This protein, together with β protein, form the so-called C antigen, which was detected in serum against the whole bacterial cell [21,22]. It participates in GBS pathogenesis, what was examined in the mouse model. It was demonstrated that the deletion of bca gene, which encodes αC protein, led to a reduction in bacterial virulence [19].
Another well-described immunogenic Alp-like protein is Rib, which demonstrates likeliness to αC, which was demonstrated by the analysis of the N-end amino acid sequence. Surprisingly, no cross-reactivity between these proteins was noticed, even though the nucleotide sequences are identical [23,24]. The Rib protein has been studied as a component of anti-GBS vaccine as the first Group B Streptococcus protein. The results received for a prototype recombinant Alpha-like protein subunit vaccine (GBS-NN), combined with α are promising. In a randomized placebo-controlled double-blind Phase 1 trial in healthy adult women, the safety and immunogenicity of GBS-NN vaccine was proved [25]. This also indicates potential usability of these proteins as biomarkers in immunodiagnostic detection of GBS carriage and infections caused by this pathogen.
The Alp3 protein, which can be also found under the name R28 in the literature, was first described for Streptococcus pyogenes (Group A Streptococcus) and its molecular likeliness to other Alp proteins was showed [26]. Analysis of the amino acid sequences of the R28 protein showed an identity of 98% between S. pyogenes and S. agalactiae species [27]. The Alp3 protein is considered to be a chimera of three S. agalactiae proteins: α, β and Rib [28].
As it was described above, immunoreactivity of the Alp-like proteins had been proved. Additionally, their conservation allows to consider them as good candidates for markers in GBS immunodetection. Nevertheless, the distribution of individual Alp proteins within the species could be a limiting factor as long as only one Alp protein is present in a single GBS strain [29]. Therefore, constructing an immunoassay based on a single Alp protein can be insufficient and use of all Alp proteins should be considered.
2.2. β Protein
β protein (approx. 130 kDa), also called Bac, had been previously described as a component, together with αC, of antigen C, belonging to the Alp-like family. Although both proteins are encoded by genes located nearby on the chromosomes, they do not demonstrate a close relationship [15,17,30]. However, β indicates homology to the Alp3 protein, and it consists of 1159 residues [28,31]. The distinguishing feature of this protein, compared to most of the surface proteins representative of Gram-positive cocci, is lack of long repetitive tandem sequences [32]. Although the virulent nature of this protein has not been fully described yet, it is hypothesized that, due to its ability to bind to elements of the human immune system (Figure 1, Table 1), it may also be associated with virulence [33]. In addition, the β protein has an affinity for two components of the human immune system: the Fc fragment of IgA antibodies and the H factor, which regulate the alternative pathway of complement activation, so that its action is directed against the infecting pathogen, not human cells or tissues. This fact may suggest an important role of this protein in induction of immunity [15]. It was also showed that β protein binds to sialic acid-binding immunoglobulin-like lectin 5 (Siglec-5), which is an inhibitory receptor for phagocytosis, and therefore attenuates innate immune responses in the infected organism, and promotes bacterial survival [34]. Moreover, GBS β protein binds to Siglec-14 on neutrophils, and this engagement counteracts the host immune suppression induced by pathogen by activation of p38 mitogen-activated protein kinase (MAPK) and AKT signaling pathways. It is also worth to underline that Siglec-5 and Siglec-14 expression has been present in amniotic epithelium, which is the place of the initial contact of S. agalactiae with the fetus [35].

Table 1.
Summary of chosen GBS immunoreactive proteins—potential biomarkers in immunodiagnostic assays for detection of GBS carriage/infection and components of a vaccine against Streptococcus agalactiae infections.
In the experiment based on active immunization with β protein of pregnant mice, it was shown that offspring did not develop infection after contact with GBS strains containing the β protein [36]. In addition, it has been shown that IgG anti-β antibodies can cross the placenta, which may indicate the possibility of mother-to-child transmission of immunity, and thus offer protection against GBS infection [37]. Immunogenicity in the presence of IgM and IgG class antibodies was studied by natural exposure of pregnant women to β protein. Geometric mean concentration of anti-β C protein IgM and IgG antibodies was measured in an enzyme-linked immunosorbent assay (ELISA). The research was carried out on 16 pregnant women colonized with GBS, and 48 uncolonized match-age pregnant women, who constituted the control group. In the study group, 3 out of 16 women demonstrated a significant growth in IgM and IgG antibodies; therefore, it had been concluded that GBS invasion, but not colonization, induces an increase in antibody titer [38]. In summary, with no doubt, immunogenicity of β protein had been shown, however, due to insignificant growth of the antibody concentration in carriers, usability of this protein as a marker in immunoassay could be limited to detection of GBS infection but not carriage.
2.3. Lmb Protein
Laminin binding protein (Lmb, LmbP) belongs to lipoproteins by its molecular weight of 43 kDa and it is exposed on the cell surface of most Streptococcus agalactiae strains (Figure 1, Table 1). The Lmb protein, consisting of 306 amino acids, shows homology to the members of the Lra1 protein family, which are known for their role in adhesion and metal transportation in Gram-positive bacteria. Lmb is involved in colonization of the host and invasion through damaged epithelial cells [39,40]. Even though the Lmb name was limited to Streptococcus agalactiae species, an almost identical protein of Streptococcus pyogenes has been referred to as Lsp or Lbp [41,42]. For both species, the gene encoding this protein is located above the C5a-peptidase encoding gene and nucleotide sequence identity between these two species is >98% in this region. Nevertheless, contiguous sequences in the two genomes show no homology, which may indicate that the region was horizontally transferred [41,43]. The immunogenic character of Lmb had been studied for Streptococcus pyogenes (GAS), which causes various diseases ranging from pharyngitis to severe infections such as a toxic shock-like syndrome and necrotizing fasciitis, and at some points, is phylogenetically similar to Streptococcus agalactiae [44,45]. A recombinant Lmb GAS protein (rGAS-Lmb) had been studied in the presence of serum from patients with rheumatic fever and individuals with uncomplicated streptococcal infections. Antibody response for the study and control groups was examined by ELISA assay, and the differences observed in reactivity were significant, whereas no difference between infection types was noticed [46]. No data for Lmb immunoreactivity for the studied group of pregnant women colonized by Streptococcus agalactiae is available, however it can be assumed that, according to its confirmed immunoreactivity for Streptococcus pyogenes, it may also be considered as a potential candidate for GBS detection.
2.4. Sip Protein
Sip protein (surface immunogenic protein) with weight of 53 kDa was, in opposition to other previously described surface proteins, identified following immunological screening of a genomic library. Sip is present in all GBS strains, regardless of the serotype represented (Figure 1, Table 1). Analysis of the nucleotide sequences for the studied strains confirmed their 98% identity of the sip gene, which encodes the Sip protein. It indicates the conservation of this 434 amino acid protein. Moreover, Sip is also described as an immunogenic protein. Immunization in mice with the recombinant Sip protein demonstrated efficient protection against severe consequences of GBS infection caused by strains representing six serotypes (Ia, Ib, II, III, V, and VI) [47]. Immunogenicity of the Sip protein was also studied in the context of the oral vaccine administrated to tilapia, a fish species in which infection caused by Streptococcus agalactiae is common and leads to huge financial losses in fishery. The immunizing protein against Streptococcus agalactiae was expressed in Bacillus subtilis spores. It was shown that immunization indicated an effective immune response and provided protection against GBS infection [48,49]. Another promising result was obtained for the investigation of a decrease in the S. agalactiae colonization in a mouse model following oral administration of the vaccine based on Sip protein [50]. This may suggest that Sip protein induces cross-protective immunity against GBS infections, therefore, it can be considered as a potential vaccine candidate; on the other hand, its conservation qualifies the Sip protein as a potential candidate for immunodiagnostic assay. Research on an immunodiagnostic assay based on Sip protein as a detection antigen has already been carried out. Nevertheless, selectivity examined in an indirect ELISA assay for bovine mastitis detection reached 75.6% (for 45 studied serum antibodies isolated from cows, 35 were positive, whereas control examination performed by PCR gave 100% positive results) [51]. Immunoreactivity of the Sip protein was also studied as an element of fusion protein combined with two other membrane surface-associated GBS proteins, which were fibronectin (FbsA) and phosphoglycerate kinase (Pgk) in the indirect ELISA assay for detection of bovine mastitis. The obtained results indicated relatively higher sensitivity in comparison with mono-antigen fusion protein Sip [52]. Immunogenicity of the Sip protein, according to its conservation, was also investigated in the context of usage for monoclonal antibody generation to develop immunochromatographic test kit for GBS detection in pregnant women [53]. Another study was focused on examination of Sip protein as a biomarker in a rapid immunochromatographic test for detection of Group B streptococcus colonization in vaginal and/or rectal tracts in pregnant women during the 35th–37th weeks of pregnancy. The obtained results were very promising, and the developed test was characterized by high specificity, with selectivity reaching respectively 93.1% and 100% [54]. Therefore, we conclude that the Sip protein can be doubtlessly considered as a GBS detection antigen.
2.5. BibA Protein
BibA protein (Group B Streptococcus immunogenic bacterial adhesin) is an immunogenic bacterial adhesin, exhibiting molecular weight of approx. 80 kDa, which demonstrates antiphagocytic properties. BibA mediates GBS adherence to both human cervical and lung epithelial cells (Figure 1, Table 1). The protein consists of the N-end α-helix rich domain, proline-rich region, LPXTG cell-wall anchoring motif, and is composed of 594 amino acids. Due to its N-terminal helical domain, which consists of three antiparallel α-helical-bundle motifs, it is considered as unique, and thus is qualified to a new class of Gram-positive surface adhesins [55]. This protein is identified on the surface of S. agalactiae strains, but interestingly, it is also present in GBS culture supernatants [56,57]. Four allelic variants (I, II, III, IV) of this protein correlated with serotypes had been described, and what is worth underlining, variant IV, which demonstrated high likeliness to the bovine counterparts, was exclusively associated with the highly virulent ST-17 GBS strain. Therefore, BibA is considered as a multifactorial GBS virulence factor [7,58,59]. BibA expression is modulated by the CovS/CovR 2-component regulatory system, and it specifically binds to C-4 binding protein in humans, which is a regulator of the classical complement pathway. It has been demonstrated that deletion of bibA gene resulted in reduced capacity of GBS to survive in human blood as well as decreased ability to resist opsonophagocytic killing by human neutrophils. Additionally, BibA expression led to an increase in the GBS virulence in a mouse model [56,57]. While mice immunization with a recombinant BibA protein (GBS-V BibA) conferred immunity and protected them from vaginal colonization by S. agalactiae, and eventually led to decrease in mortality. Antibody response after immunization was examined in ELISA assay in which plates were coated with BibA protein. This indirect use indicates the usability of BibA protein as detection antigen in ELISA for GBS carriage and/or infection diagnosis [60]. A similar conclusion can be drawn from the research whose aim was to examine the association between antibodies against Streptococcus agalactiae surface proteins and recto-vaginal colonization during pregnancy, in which titers of IgG antibodies were measured in Luminex multiplex immunoassay [61].
2.6. FsbA Protein
Fibrinogen-binding protein (FsbA), approx. 26 kDa, is one of the virulence factors of Streptococcus agalactiae, and its role is the attachment to fibrinogen, which leads to fibrinogen-dependent aggregation of platelets (Figure 1, Table 1). It was demonstrated that GBS mutants lacking fsbA gene lost the aggregation ability. Furthermore, application of monoclonal anti-FsbA antibodies impeded bacterial binding to fibrinogen as well as platelet aggregation caused by Streptococcus agalactiae [62,63,64]. FsbA is composed of 16 amino acid repetitive units. It has been demonstrated that human fibrinogen was bound by the repetitive protein region, and even a single repeat had the ability attach to fibrinogen [63]. FsbA protein consists of C-terminus cell wall anchoring motif (LPKTG), which indicates that this protein is covalently attached to the cell wall. The second GBS fibrinogen binding protein is FsbA’s analogue—FsbB, even though these proteins do not reveal significant likeliness to each other. The feature distinguishing FsbA from other fibrinogen binding proteins, representative to other bacterial species, is the LPKTG motif [65]. As long as FsbA protein structure is well described, its function is barely known, except its immunogenic role. It was shown that maternal immunization of mice with 6pGST, a protein fragment which consists of five repeats, significantly protected the offspring against lethal infection caused by Streptococcus agalactiae. It was demonstrated that the protective role of the antibodies can be obtained by administration of anti-6pGST serum from adult animals. The introduction of serum with antibodies led to protection by bacterial opsonophagocytosis or resulted in neutralization of FbsA-mediated Fng binding. Two-track action had also been noticed [66]. Other studies showed RPS (relative percentage survival) value after tilapia vaccination consisting of GBS FsbA protein reached 40.63% [67]. It allows to define FsbA as a multifunctional immunogenic protein, including immunoprotection as well as activation of the innate immune responses in the host and relevant antibody responses [68].
2.7. ScpB Protein
Streptococcal peptidase C5a (ScpB) is an enzymatic surface protein, which belongs to serine protease and is related to the subtilisin family of enzymes (Figure 1, Table 1). It was demonstrated that substitution of the following amino acids: Ser512, His193, or Asp130 with alanine confirmed their proteolytic role, which has an impact on the whole protein [69]. This protein is a large molecule, which consists of over 1100 residues, excluding the signal sequence and it weighs over 126 kDa [70,71]. A comparison of the amino acid sequence between Scp N-terminal catalytic triad and subtilisin revealed homology [69]. C-terminus of Scp includes the peptidoglycan anchor sequence, which is common to other Gram-positive bacterial surface proteins [72]. C5a peptidase is responsible for inactivation of one of the components of the human complement, i.e., decay product of C5, C5a converting enzyme [73]. C5a peptidase was first described for Streptococcus pyogenes, however, further studies have shown the presence of this enzyme also in Streptococcus agalactiae strains. Sequence analysis of both proteins revealed their 95% similarity, which is a consequence of horizontal gene transfer between both species. Therefore, in order to distinguish between them, the following nomenclature is used: C5a peptidase for the species Streptococcus pyogenes, belonging to the serological Group A, is called ScpA, while analogously for Streptococcus agalactiae, representative for Group B, this protein is described as ScpB [70,72,73,74,75,76]. Interestingly, scpB gene is common for GBS strains isolated from humans, whereas, in bovine, it is barely present [41,77]. Additionally, ScpB-related cell envelope proteases, which have a multi-domain structure, are also common to lactic acid bacteria [70,77].
C5a peptidase is described for its virulence function. It is responsible for disrupting neutrophil recruitment, resulting in a reduction in the inflammatory response elicited in the infected tissue. Interestingly, inhibition of neutrophil recruitment had not been observed in the mouse model [78,79]. This non-obvious observation should be considered during the selection of an appropriate model for research on the GBS vaccine, the component of which will be C5a peptidase, as long as Scp reveals its immunogenic character [15,80,81]. Another Scp feature, on the basis of which it is qualified as a virulence factor is the ability to bind fibrinogen, which may promote bacterial adhesion to epithelial and endothelial cells [15,82]. However, studies with mutant bacterial cells, lacking ScpB, revealed only a partial reduction in the fibrinogen binding capacity compared to the unmutated strains. It can be explained by the fact that Group B streptococci possess other proteins that are able to bind fibrinogen [83].
As it was mentioned above, both ScpA and ScpB also show strong immunogenic properties, and due to their conservation, they are considered as good vaccine candidates [84,85]. Research on mice immunized with the purified ScpB protein, which were next infected with Streptococcus agalactiae strains via the nasal route, showed a higher clearance of bacteria from the lungs [86]. Moreover, experiments on hyperimmune rabbit serum showed opsonizing activity with mouse macrophages and human whole blood. It has also been suggested that antibodies directed against ScpB antigen may protect from infection by disrupting fibrinogen binding [85]. Due to the features described above, ScpB can be considered as a biomarker in an immunodiagnostic assay, and its usability had previously been demonstrated in the enzyme-linked immunosorbent assay [87,88].
2.8. Enolase
Enolase is a dimeric protein, weighing approximately 47 kDa (approx. 430 aa), which is a glycolytic enzyme and catalyzes the penultimate stage of glycolysis, which is the dehydration reaction of 2-phosphoglycerate to phosphoenolpyruvate [89,90]. Enolase is also very conservative among the Streptococcus species. It had been demonstrated that identity of amino acid sequences exceeded 90% among the following species: Streptococcus pneumoniae, Streptococcus agalactiae, Streptococcus sobrinus, and Streptococcus mutans [91,92]. Furthermore, likeliness between human and streptococcal enolase reaches 49% [93]. There are three distinct isoforms of enolase: α, β, and γ, with the same molecular mass each. Nevertheless, the isoform typically found in bacteria is α-enolase. Enolase is a common protein for numerous bacterial species, and it plays an important role in the pathogenesis by binding plasminogen on the surface of the host cell, which mediates fibrinolysis, homeostasis, and the degradation of the extracellular matrix (Figure 1, Table 1) [94,95]. Interestingly, the investigation of amino acid sequences of the α-enolase in Streptococcus mutans, which is responsible for inducing dental caries, revealed a lack of the hexameric cell wall anchoring motif (LPXTGX) [96]. Thus, the way of transportation and cell wall attachment is not well understood yet [91]. This protein has also been described in the context of its immunogenicity for Streptococcus agalactiae [84,97,98,99,100,101]. It has been shown that streptococcal anti-enolase antibodies may react with human enolase produced after S. pyogenes infection [93]. Additionally, antibodies directed against enolase had been detected in some autoimmune diseases, such as systemic lupus erythematosus, mixed cryoglobulinemia, systemic sclerosis, and rheumatoid arthritis [102]. In our previous paper, we decided to take the next step and detected epitopes representative of enolase, which specifically recognized anti-GBS IgG antibodies, and which were studied in ELISA with the presence of umbilical cord blood serum from pregnant women qualified as GBS carriers. Presently, these epitopes are being investigated as potential components in an immunodiagnostic assay for detection of GBS carriage and/or infections in pregnant women [103]. Nevertheless, consideration of enolase as a detection antigen requires taking into account the fact of relatively close sequence similarity among streptococci as well as a possibility of cross-reactivity with human enolase.
2.9. Elongation Factor Tu
Elongation factor thermo unstable (EF Tu) is a moonlight protein, weighing approx. 44 kDa and consisting of approx. 400 residues, which is involved in several pathogenic functions, such as: adhesion, invasion, and modulation of the host immune system through stimulation of humoral immune response [104,105,106,107,108,109]. This conservative protein is the most abundant protein in bacteria, it constitutes up to 5% of the total cell content, and it is common for both prokaryotic and eukaryotic organisms [108,110]. In the bacterial cell, EF Tu constitutes part of the membrane cytoskeleton, and it is involved in the elongation phase of protein synthesis as well as in the translation process in prokaryotic cells (Figure 1, Table 1). As a GTPase, it ensures a catalysis reaction, whose aim is the correct addition of the consecutive amino acid to a growing nascent polypeptide chain [111]. Even though EF Tu is a component of the membrane cytoskeleton, it does not possess a signal secretion motif, due to its moonlight roles, it requires to localize on the cell surface [108,110,112,113]. Probably the protein is exported by several paths, such as cell lysis, extracellular vesicle secretion or via association with proteins that are secreted by the Sec machinery 54 [114,115,116]. It plays an important role in shuttling aminoacylated tRNAs to the ribosome during protein translation [104,117,118,119,120]. In Escherichia coli, EF Tu consists of three domains, whereas the first domain forms a helix structure with the Rossmann fold topology, which is characterized by tertiary protein fold composed of both α-helical and β-strands that bind nucleotides [121], the second and the third domains mostly consist of β-sheets [111,122]. Amino acid sequences of the EF Tu protein are characteristic of various bacterial species representing similarity in sequence identity; therefore, they have been used to generate a phylogenetic tree [123]. This moonlight protein interacts with several molecules, such as CD21, factor H, fibrinogen, fibronectin, laminin, nucleolin, tachykinin, plasminogen and several complement factors [107,124,125,126,127]. EF Tu is also known for its immunogenic role. Antibodies against EF Tu are detected after infections caused by Burkholderia pseudomallei, Chlamydia trachomatis, Mycoplasma capricolum, Mycoplasma hyopneumoniae, Mycoplasma ovipneumoniae, and Staphylococcus aureus [128,129,130,131,132,133]. On the other hand, EF Tu for Haemophilus influenzae is recognized by antibodies, which mediate innate immunity of the host against NTHi [134]. While, when GBS it found, the protein is common for all Streptococcus agalactiae strains, regardless of molecular differences, such as the serotypes represented, genes encoding Alp proteins or the origin [101,135,136]. Research on a subunit anti-GBS vaccine based on EF Tu revealed the capability of the indication of the immunity and protection of tilapia against infection caused by Streptococcus agalactiae, however vaccination with the subunit EF Tu vaccine generated moderate immune protection with RPS equal 70% [136]. Mouse vaccination against EF Tu of Streptococcus pneumoniae resulted in the animals’ protection against lethal challenges, and on the molecular level, increased cytokine, IgG1 and IgG2a, and CD4+ T-cell production was observed [137]. Promising results of rEF-Tu vaccination was obtained for fish as well [136]. In our previous paper, similarly to enolase described above, we aimed to detect the highly specific epitopes representative of GBS EF Tu as potential markers in an immunodiagnostic assay for detecting GBS carriage in pregnant women, and the obtained results were very promising [135]. However, it is necessary to consider some limitations, such as a tendency for cross-reactivity between different bacterial species with special emphasis on unencapsulated species, among others, various Gram-positive streptococci of the oral microbiome. Pneumococci are not detectable, what is related to the capsules of pneumococci and meningococci, which shield surface-associated EF Tu from antibody detection [134,138].
2.10. IMPDH
Inosine 5′-monophosphate dehydrogenase (IMPDH, dehydrogenase IMP), with a mass of 53 kDa, is a stable purine, which participates in catalysis of the key stage of de novo synthesis of the guanine and adenine nucleotides in all organisms, in other words, it is a crucial precursor for DNA and RNA synthesis [103,139,140]. IMPDH monomers contain 400–500 amino acids, and these differences depend on the presence of a subdomain that is not required for enzymatic activity. Each monomer is built of two domains: the α and β (α/β)8 barrel catalytic domain and the subdomain containing two CBS domains (cystathionine beta synthase, Bateman domain) [140]. The protein most often takes the form of tetramers, and it is the most stable configuration (Figure 1, Table 1) [141,142,143]. The IMPDH tetramers have square planar geometry, with the sides of the barrels at the subunit interfacing and CBS domains sticking out from the corners of the tetramer. The junction between domains is flexible, and the relative orientation can vary [144]. The IMPDH also catalyzes conversion of IMP to XMP as the first committed and rate-limiting step in guanine nucleotide biosynthesis. In turn, XMP is subsequently converted to GMP by the action of GMP synthetase (GMPS). This IMPDH/GMPS pathway is common for almost all organisms. IMPDH is also involved in glycoprotein synthesis, energy transfer, signal transduction in cells, and NAD-dependent catalysis. It is worth to mention that there are many genes encoding IMPDH [140]. In the literature, due to its crucial role in cell replication, IMPDH is frequently described as a potential target in antiviral, antibacterial and anticancer therapies. It is also considered as a part of autoimmunological disease treatment [140,145,146,147]. Unfortunately, it has been reported that some pathogens developed resistance to IMPDH inhibitors by amplifying the IMPDH gene, which significantly limits its usability as an antibiotic [140]. This protein also demonstrates immunogenic features in some bacterial species [148,149,150,151]. Immunoreactivity to Streptococcus agalactiae was also showed. Moreover, highly specific IMPDH GBS epitopes recognized by umbilical cord blood isolated from GBS-positive women were identified, and what is worth to underline, it was performed for the first time [101,103]. The obtained results allowed to consider them as potential antigens in an immunodiagnostic assay for GBS carriage detection.
2.11. GroEL
GroEL is a 57 kDa protein, which belongs to the chaperonin family. Structurally GroEL is a double ring tetradecamer, composed of seven identical 10 kDa subunits in cis and trans positions, which demonstrates ability to form barrel-like structures with hydrophilic cavities, which are isolated from each other by the equatorial domains and the C-terminal tails of each subunit (Figure 1, Table 1). Each oligomeric complex consists of fourteen identical 57kDa subunits, and each subunit can be divided into three domains: apical, intermediate, and equatorial. Apical domains are located at the outer edge of the rings and contain binding sites for GroES, with which they frequently form complex and non-native proteins. While equatorial domains adjoin each other within the individual ring, and they consist of a domain with the ATP binding site. Intermediate domain is connected with apical domain through the slender intermediate domain [152,153,154]. This protein is common for numerous bacteria, and, moreover, it is structurally and functionally closely related to the human heat shock protein (Hsp60) [155]. The key role of GroEL is folding de novo emerging proteins. Its functions, as a chaperonin protein, are recognizing, binding, and releasing other proteins. These actions are performed due to the characteristic structure of the apical domain, which consists of non-polar amino acids on the inner surface, and hydrophobic external surface, which demonstrates the ability to capture and tightly bind protein folding intermediates [156,157]. GroEL participates in folding of a wide range of proteins, with special emphasis on these, which are typically large (>20 kDa), slow folding, and prone to aggregation [158,159].
GroEL is also described as a virulence factor and participates in pathogenesis, even though the switching mechanism from folding supporting protein to virulence factor is not yet understood [160,161]. Virulence of GroEL can be carried out by promoting infection by replication and persistence followed by adhesion, invasion, evasion of host immune responses, and modification of host cell responses [162]. GroEL is described as a moonlight protein; nevertheless, its ability to change roles has not been determined yet [163]. As an example of moonlighting, GroEL from some species, such as: Lactobacillus johnsonii, Mycoplasma pneumoniae or Salmonella enterica, displays an ability to support adhesion to mucin [164]. Mucin belongs to glycoproteins, which create a gel-like layer on the mucosal surface, and thus protect the host from pathogen colonization and invasion [165]. Therefore, binding bacterial cells throughout GroEL protein to mucin promotes initiation of the colonization and invasion [163]. In turn, GroEL from Helicobacter pylori promotes binding iron, which is essential for growth of this species, and this ability could allow it to compete with other species for a colonization niche [166,167]. Another interesting and surprising function of GroEL is its toxic effect on some species of insects. For example, GroEL produced and excreted by Enterobacter aerogenes paralyzed a cockroach but not mice [168]. Some research suggest that GroEL can also be considered as a potential plant protection product, because it had been showed that GroEL from Xenorhabdus nematophila expressed in tobacco induced resistance to invading insects [169]. Even though GroEL plays its role inside a cell, it has an ability to relocate on the cell surface. As a stress response protein, it demonstrates an ability to introduce changes in the level of expression as well as in the cellular localization from the cytosol to the cell surface or the secretome. Therefore, apart from folding, GroEL is also an immunogenic protein, because it can be presented to the antibodies [163]. Mouse vaccination with rGroEL from H. pylori induced immune protection for both mother and infants [170]. Successful immunization with rGroEL had also been demonstrated for other species such as: Mycobacterium bovis [171] and Lawsonia intracellularis [172].
This molecular chaperon, as well as its epitopes, have an ability to specifically bind with anti-GBS antibodies present in both umbilical cord and vascular blood [103]. Immunogenic features of GBS GroEL were also investigated for tilapia. Immunization with recombinant GroEL induced to increase titers of anti-rGroEL antibodies and insure protection against Streptococcus agalactiae with RPS on an approximate level of 70%. While testing the titer of antibodies produced after rGroEL immunization, an ELISA in which detection antigen constituted rGroEL protein was performed [49]. It indirectly confirms the usability of this protein as a biomarker in an immunodiagnostic assay.
3. Conclusions
We conclude that the proteins described above, with special emphasis on Sip, BibA, Lmb, FsbA, ScpB IMPDH, and GroEL, due to their proven immunoreactivity and conservation, can be considered as good candidates in immunodiagnostic assays for detection of GBS carriage and infection particularly in pregnant women, but, with high probability, also in other patients in the so-called high-risk groups, such as elderly or immunosuppressed adults. Consideration of Alp proteins and the remaining three proteins, which are β-protein, enolase, and elongation factor Tu, as potential detection antigens should take into account their limitations, such as lack of universality in the case of Alp proteins, risk of cross-reactivity among other bacterial species as well as human counterparts for enolase, and EF Tu, or insignificant growth of the antibody concentration in carriers following immunization with β protein. We also believe that in the near future many more immunogenic proteins will be described and studied, and this knowledge will find its practical application, i.e., in the ELISA assay or immunochromatographic assay investigation.
Author Contributions
A.D. and M.B.-W. contributed equally. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science Centre and the National Center of Research and Development, Grant Number: TANGO2/340018/NCBR/2017.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors wish to thank Katarzyna Gąsior-Kulasiak for English language editing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rodriguez-Granger, J.; Alvargonzalez, J.C.; Berardi, A.; Berner, R.; Kunze, M.; Hufnagel, M.; Melin, P.; Decheva, A.; Orefici, G.; Poyart, C.; et al. Prevention of group B streptococcal neonatal disease revisited. The DEVANI European project. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2097–2104. [Google Scholar] [CrossRef]
- Russell, N.J.; Seale, A.C.; O’Driscoll, M.; O’Sullivan, C.; Bianchi-Jassir, F.; Gonzalez-Guarin, J.; Lawn, J.E.; Baker, C.J.; Bartlett, L.; Cutland, C.; et al. Maternal Colonization with Group B Streptococcus and Serotype Distribution Worldwide: Systematic Review and Meta-analyses. Clin. Infect. Dis. 2017, 65 (Suppl. S2), S100–S111. [Google Scholar] [CrossRef]
- Dermer, P.; Lee, C.; Eggert, J.; Few, B. A history of neonatal group B streptococcus with its related morbidity and mortality rates in the United States. J. Pediatr. Nurs. 2004, 19, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Filkins, L.; Hauser, J.; Robinson-Dunn, B.; Tibbetts, R.; Boyanton, B.; Revell, P.; on behalf of the American Society for Microbiology Clinical and Public Health Microbiology Committee, Subcommittee on Laboratory Practices. Guidelines for Detection and Identification of Group B Streptococcus; American Society for Microbiology: Washington, DC, USA, 2020; Available online: https://asm.org/Guideline/Guidelines-for-the-Detection-and-Identification-of (accessed on 16 July 2021).
- Verani, J.R.; McGee, L.; Schrag, S.J. Prevention of Perinatal Group B Streptococcal Disease. Revised Guidelines from CDC, 2010. Recomm. Rep. 2010, 19, 59. [Google Scholar]
- Schrag, S.J.; Verani, J.R. Intrapartum antibiotic prophylaxis for the prevention of perinatal group B streptococcal disease: Experience in the United States and implications for a potential group B streptococcal vaccine. Vaccine 2013, 31 (Suppl. S4), D20–D26. [Google Scholar] [CrossRef] [PubMed]
- Shabayek, S.; Spellerberg, B. Group B Streptococcal Colonization, Molecular Characteristics, and Epidemiology. Front. Microbiol. 2018, 9, 437. [Google Scholar] [CrossRef]
- Berardi, A.; Lugli, L.; Baronciani, D.; Rossi, C.; Ciccia, M.; Creti, R.; Gambini, L.; Mariani, S.; Papa, I.; Tridapalli, E.; et al. Group B Streptococcus early-onset disease in Emilia-romagna: Review after introduction of a screening-based approach. Pediatr. Infect. Dis. J. 2010, 29, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Paveenkittiporn, W.; Ungcharoen, R.; Kerdsin, A. Streptococcus agalactiae infections and clinical relevance in adults, Thailand. Diagn. Microbiol. Infect. Dis. 2020, 97, 115005. [Google Scholar] [CrossRef] [PubMed]
- Lancefield, R.C. A Serological Differentiation of Human and Other Groups of Hemolytic Streptococci. J. Exp. Med. 1933, 57, 571–595. [Google Scholar] [CrossRef]
- Lancefield, R.C. A Serological Differentiation of Specific Types of Bovine Hemolytic Streptococci (Group B). J. Exp. Med. 1934, 5, 441–458. [Google Scholar] [CrossRef]
- Maeland, J.A.; Afset, J.E.; Lyng, R.V.; Radtke, A. Survey of immunological features of the alpha-like proteins of Streptococcus agalactiae. Clin. Vaccine Immunol. 2015, 22, 153–159. [Google Scholar] [CrossRef]
- Baron, M.J.; Bolduc, G.R.; Goldberg, M.B.; Aupérin, T.C.; Madoff, L.C. Alpha C protein of group B Streptococcus binds host cell surface glycosaminoglycan and enters cells by an actin-dependent mechanism. J. Biol. Chem. 2004, 279, 24714–24723. [Google Scholar] [CrossRef]
- Navarre, W.W.; Schneewind, O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 1999, 63, 174–229. [Google Scholar] [CrossRef]
- Lindahl, G.; Stålhammar-Carlemalm, M.; Areschoug, T. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 2005, 18, 102–127. [Google Scholar] [CrossRef] [PubMed]
- Bevanger, L.; Naess, A.L. Mouse-protective antibodies against the Ibc proteins of group B streptococci. Acta. Pathol. Microbiol. Immunol. Scand. B 1985, 93, 121–124. [Google Scholar] [CrossRef]
- Lachenauer, C.S.; Creti, R.; Michel, J.L.; Madoff, L.C. Mosaicism in the alpha-like protein genes of group B streptococci. Proc. Natl. Acad. Sci. USA 2000, 97, 9630–9635. [Google Scholar] [CrossRef] [PubMed]
- Lachenauer, C.S.; Madoff, L.C. A protective surface protein from type V group B streptococci shares N-terminal sequence homology with the alpha C protein. Infect. Immun. 1996, 64, 4255–4260. [Google Scholar] [CrossRef]
- Li, J.; Kasper, D.L.; Ausubel, F.M.; Rosner, B.; Michel, J.L. Inactivation of the alpha C protein antigen gene, bca, by a novel shuttle/suicide vector results in attenuation of virulence and immunity in group B Streptococcus. Proc. Natl. Acad. Sci. USA 1997, 94, 13251–13256. [Google Scholar] [CrossRef]
- Michel, J.L.; Madoff, L.C.; Kling, D.E.; Kasper, D.L.; Ausubel, F.M. Cloned alpha and beta C-protein antigens of group B streptococci elicit protective immunity. Infect. Immun. 1991, 59, 2023–2028. [Google Scholar] [CrossRef] [PubMed]
- Hedén, L.O.; Frithz, E.; Lindahl, G. Molecular characterization of an IgA receptor from group B streptococci: Sequence of the gene, identification of a proline-rich region with unique structure and isolation of N-terminal fragments with IgA binding capacity. Eur. J. Immunol. 1991, 21, 1481–1490. [Google Scholar] [CrossRef]
- Simister, N.E. Placental transport of immunoglobulin G. Vaccine 2003, 21, 3365–3369. [Google Scholar] [CrossRef]
- Stålhammar-Carlemalm, M.; Stenberg, L.; Lindahl, G. Protein rib: A novel group B streptococcal cell surface protein that confers protective immunity and is expressed by most strains causing invasive infections. J. Exp. Med. 1993, 177, 1593–1603. [Google Scholar] [CrossRef] [PubMed]
- Heath, P.T. Status of vaccine research and development of vaccines for GBS. Vaccine 2016, 34, 2876–2879. [Google Scholar] [CrossRef] [PubMed]
- Fischer, P.; Pawlowski, A.; Cao, D.; Bell, D.; Kitson, G.; Darsley, M.; Johansson-Lindbom, B. Safety and immunogenicity of a prototype recombinant alpha-like protein subunit vaccine (GBS-NN) against Group B Streptococcus in a randomised placebo-controlled double-blind phase 1 trial in healthy adult women. Vaccine 2021, 39, 4489–4499. [Google Scholar] [CrossRef] [PubMed]
- Lancefield, R.C.; Perlmann, G.E. Preparation and properties of a protein (R antigen) occurring in streptococci of group A, type 28 and in certain streptococci of other serological groups. J. Exp. Med. 1952, 96, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Stålhammar-Carlemalm, M.; Areschoug, T.; Larsson, C.; Lindahl, G. Cross-protection between group A and group B streptococci due to cross-reacting surface proteins. J. Infect. Dis. 2000, 182, 142–149. [Google Scholar] [CrossRef]
- Stålhammar-Carlemalm, M.; Areschoug, T.; Larsson, C.; Lindahl, G. The R28 protein of Streptococcus pyogenes is related to several group B streptococcal surface proteins, confers protective immunity and promotes binding to human epithelial cells. Mol. Microbiol. 1999, 33, 208–219. [Google Scholar] [CrossRef]
- Brzychczy-Włoch, M.; Gosiewski, T.; Bodaszewska, M.; Talaga, K.; Natkaniec, J.; Adamski, J.; Heczko, P.B. Occurrence of surface protein genes from alpha-like protein (Alp) family in Streptococcus agalactiae isolates. Med. Dosw. Mikrobiol. 2011, 63, 5–14. [Google Scholar]
- Dmitriev, A.; Hu, Y.Y.; Shen, A.D.; Suvorov, A.; Yang, Y.H. Chromosomal analysis of group B streptococcal clinical strains; bac gene-positive strains are genetically homogenous. FEMS Microbiol. Lett. 2002, 208, 93–98. [Google Scholar] [CrossRef][Green Version]
- Vasilyeva, A.; Santos Sanches, I.; Florindo, C.; Dmitriev, A. Natural Mutations in Streptococcus agalactiae Resulting in Abrogation of β Antigen Production. PLoS ONE 2015, 10, e0128426. [Google Scholar] [CrossRef]
- Berner, R.; Ruess, M.; Bereswill, S.; Brandis, M. Polymorphisms in the cell wall spanning domain of the C protein beta-antigen in clinical Streptococcus agalactiae isolates are caused by genetic instability of repeating DNA sequences. Pediatr. Res. 2002, 51, 106–111. [Google Scholar] [CrossRef][Green Version]
- Nagano, N.; Nagano, Y.; Taguchi, F. High expression of a C protein beta antigen gene among invasive strains from certain clonally related groups of type Ia and Ib group B streptococci. Infect. Immun. 2002, 70, 4643–4649. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carlin, A.F.; Chang, Y.C.; Areschoug, T.; Lindahl, G.; Hurtado-Ziola, N.; King, C.C.; Varki, A.; Nizetet, V. Group B Streptococcus suppression of phagocyte functions by protein-mediated engagement of human Siglec-5. J. Exp. Med. 2009, 206, 1691–1699. [Google Scholar] [CrossRef]
- Ali, S.R.; Fong, J.J.; Carlin, A.F.; Busch, T.D.; Linden, R.; Angata, T.; Areschoug, T.; Parast, M.; Varki, N.; Murray, J.; et al. Siglec-5 and Siglec-14 are polymorphic paired receptors that modulate neutrophil and amnion signaling responses to group B Streptococcus. J. Exp. Med. 2014, 211, 1231–1242. [Google Scholar] [CrossRef] [PubMed]
- Madoff, L.C.; Michel, J.L.; Gong, E.W.; Rodewald, A.K.; Kasper, D.L. Protection of neonatal mice from group B streptococcal infection by maternal immunization with beta C protein. Infect. Immun. 1992, 60, 4989–4994. [Google Scholar] [CrossRef] [PubMed]
- Flores, A.E.; Nelson, J.A.; Wu, X.Y.; Ferrieri, P. Antibody profiles to the group B streptococcal β antigen in maternal and infant paired sera. APMIS 2003, 101, 41–49. [Google Scholar] [CrossRef]
- Pannaraj, P.S.; Kelly, J.K.; Madoff, L.C.; Rench, M.A.; Lachenauer, C.S.; Edwards, M.S.; Baker, C.J. Group B Streptococcus bacteremia elicits beta C protein-specific IgM and IgG in humans. J. Infect. Dis. 2007, 195, 353–356. [Google Scholar] [CrossRef][Green Version]
- Jenkinson, H.F. Cell surface protein receptors in oral streptococci. FEMS Microbiol. Lett. 1994, 121, 133–140. [Google Scholar] [CrossRef]
- Spellerberg, B.; Rozdzinski, E.; Martin, S.; Weber-Heynemann, J.; Schnitzler, N.; Lütticken, R.; Podbielski, A. Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infect. Immun. 1999, 67, 871–878. [Google Scholar] [CrossRef]
- Franken, C.; Haase, G.; Brandt, C.; Weber-Heynemann, J.; Martin, S.; Lämmler, C.; Podbielski, A.; Lütticken, R.; Spellerberg, B. Horizontal gene transfer and host specificity of beta-haemolytic streptococci: The role of a putative composite transposon containing scpB and lmb. Mol. Microbiol. 2001, 41, 925–935. [Google Scholar] [CrossRef]
- Terao, Y.; Kawabata, S.; Kunitomo, E.; Nakagawa, I.; Hamada, S. Novel laminin-binding protein of Streptococcus pyogenes, Lbp, is involved in adhesion to epithelial cells. Infect. Immun. 2002, 70, 993–997. [Google Scholar] [CrossRef]
- Elsner, A.; Kreikemeyer, B.; Braun-Kiewnick, A.; Spellerberg, B.; Buttaro, B.A.; Podbielski, A. Involvement of Lsp, a member of the LraI-lipoprotein family in Streptococcus pyogenes, in eukaryotic cell adhesion and internalization. Infect. Immun. 2002, 70, 4859–4869. [Google Scholar] [CrossRef] [PubMed]
- Stevens, D.L. Invasive group A streptococcal disease. Infect. Agents. Dis. 1996, 5, 157–166. [Google Scholar]
- Cartwright, K. Group A streptococcal infections in humans. Soc. Appl. Bacteriol. Symp. Ser. 1997, 26, 52S–61S. [Google Scholar] [CrossRef] [PubMed]
- Wahid, R.M.; Yoshinaga, M.; Nishi, J.; Maeno, N.; Sarantuya, J.; Ohkawa, T.; Jalil, A.M.; Kobayashi, K.; Miyata, K. Immune response to a laminin-binding protein (Lmb) in group A streptococcal infection. Pediatr. Int. 2005, 47, 196–202. [Google Scholar] [CrossRef]
- Brodeur, B.R.; Boyer, M.; Charlebois, I.; Hamel, J.; Couture, F.; Rioux, C.R.; Martin, D. Identification of Group B Streptococcal Sip Protein, Which Elicits Cross-Protective Immunity. Infect. Immun. 2000, 68, 5610–5618. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.Y.; Chen, D.D.; Cui, Z.W.; Zhang, X.Y.; Zhou, Y.Y.; Guo, X.; Li, A.H.; Zhang, Y.A. Oral vaccination of tilapia against Streptococcus agalactiae using Bacillus subtilis spores expressing Sip. Fish Shellfish Immunol. 2019, 86, 999–1008. [Google Scholar] [CrossRef]
- Li, W.; Li, Y.; Hu, Y.Z.; Mo, X.B.; Xu, G.H.; Xie, L.W.; Li, A.X. GroEL, a novel vaccine candidate of piscine Streptococcus agalactiae identified by immunoproteome. Fish Shellfish Immunol. 2019, 84, 377–383. [Google Scholar] [CrossRef]
- Diaz-Dinamarca, D.A.; Soto, D.A.; Leyton, Y.Y.; Altamirano-Lagos, M.J.; Avendaño, M.J.; Kalergis, A.M.; Vasquez, A.E. Oral vaccine based on a surface immunogenic protein mixed with alum promotes a decrease in Streptococcus agalactiae vaginal colonization in a mouse model. Mol. Immunol. 2018, 103, 63–70. [Google Scholar] [CrossRef]
- Bu, R.E.; Wang, J.L.; DebRoy, C.; Wu, J.H.; Xi, L.G.; Liu, Y.; Shen, Z.Q. Development of an indirect ELISA for bovine mastitis using Sip protein of Streptococcus agalactiae. Iran. J. Vet. Res. 2015, 16, 283–287. [Google Scholar]
- Bu, R.E.; Wang, J.L.; Wu, J.H.; Xilin, G.W.; Chen, J.L.; Wang, H. Indirect enzyme-linked immunosorbent assay method based on Streptococcus agalactiae rSip-Pgk-FbsA fusion protein for detection of bovine mastitis. Pol. J. Vet. Sci. 2017, 20, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Han, J.; Huang, Y.; Yan, Q.; Lu, G.; Yuan, Z.; Huang, G.; Zheng, J.; Liu, T. The correlation between expression of sip protein in different serotypes of group b streptococcus and diagnosis. Heliyon 2019, 5, e01899. [Google Scholar] [CrossRef] [PubMed]
- Takayama, Y.; Matsui, H.; Adachi, Y.; Nihonyanagi, S.; Wada, T.; Mochizuki, J.; Unno, N.; Hanaki, H. Detection of Streptococcus agalactiae by immunochromatography with group B streptococcus-specific surface immunogenic protein in pregnant women. J. Infect. Chemother. 2017, 23, 678–682. [Google Scholar] [CrossRef] [PubMed]
- Manne, K.; Chattopadhyay, D.; Agarwal, V.; Blom, A.M.; Khare, B.; Chakravarthy, S.; Chang, C.; Ton-That, H.; Narayana, S.V.L. Novel structure of the N-terminal helical domain of BibA, a group B streptococcus immunogenic bacterial adhesin. Acta. Crystallogr. D Struct. Biol. 2020, 76, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Santi, I.; Scarselli, M.; Mariani, M.; Pezzicoli, A.; Masignani, V.; Taddei, A.; Grandi, G.; Telford, J.L.; Soriani, M. BibA: A novel immunogenic bacterial adhesin contributing to group B Streptococcus survival in human blood. Mol. Microbiol. 2007, 63, 754–767. [Google Scholar] [CrossRef] [PubMed]
- Santi, I.; Grifantini, R.; Jiang, S.M.; Brettoni, C.; Grandi, G.; Wessels, M.R.; Soriani, M. CsrRS regulates group B Streptococcus virulence gene expression in response to environmental pH: A new perspective on vaccine development. J. Bacteriol. 2009, 191, 5387–5397. [Google Scholar] [CrossRef] [PubMed]
- Santi, I.; Maione, D.; Galeotti, C.L.; Grandi, G.; Telford, J.L.; Soriani, M. BibA induces opsonizing antibodies conferring in vivo protection against group B Streptococcus. J. Infect. Dis. 2009, 200, 564–570. [Google Scholar] [CrossRef]
- Lamy, M.C.; Dramsi, S.; Billoët, A.; Réglier-Poupet, H.; Tazi, A.; Raymond, J.; Guérin, F.; Couvé, E.; Kunst, F.; Glaser, P.; et al. Rapid detection of the “highly virulent” group B Streptococcus ST-17 clone. Microbes. Infect. 2006, 8, 1714–1722. [Google Scholar] [CrossRef]
- Dos Santos, N.F.B.; da Silva, L.R.; Costa, F.J.M.D.; de Mattos, D.M.; de Carvalho, E.; de Souza Ferreira, L.C.; Rita de Cássia Café Ferreira, R. Immunization with a recombinant BibA surface protein confers immunity and protects mice against group B Streptococcus (GBS) vaginal colonization. Vaccine 2020, 38, 5286–5296. [Google Scholar] [CrossRef]
- Dzanibe, S.; Kwatra, G.; Adrian, P.V.; Kimaro-Mlacha, S.Z.; Cutland, C.L.; Madhi, S.A. Association between antibodies against group B Streptococcus surface proteins and recto-vaginal colonisation during pregnancy. Sci. Rep. 2017, 7, 16454. [Google Scholar] [CrossRef]
- Pietrocola, G.; Schubert, A.; Visai, L.; Torti, M.; Fitzgerald, J.R.; Foster, T.J.; Reinscheid, D.J.; Speziale, P. FbsA, a fibrinogen-binding protein from Streptococcus agalactiae, mediates platelet aggregation. Blood 2005, 105, 1052–1059. [Google Scholar] [CrossRef] [PubMed]
- Gutekunst, H.; Eikmanns, B.J.; Reinscheid, D.J. The Novel Fibrinogen-Binding Protein FbsB Promotes Streptococcus agalactiae Invasion into Epithelial Cells. Infect. Immun. 2004, 72, 3495–3504. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jonsson, I.M.; Pietrocola, G.; Speziale, P.; Verdrengh, M.; Tarkowski, A. Role of fibrinogen-binding adhesin expression in septic arthritis and septicemia caused by Streptococcus agalactiae. J. Infect. Dis. 2005, 192, 1456–1464. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Eisen, J.A.; Peterson, S.; Wessels, M.R.; Paulsen, I.T.; Nelson, K.E.; Margarit, I.; Read, T.D.; et al. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 2002, 99, 12391–12396. [Google Scholar] [CrossRef]
- Papasergi, S.; Cariccio, V.L.; Pietrocola, G.; Domina, M.; D’Aliberti, D.; Trunfio, M.G.; Signorino, G.; Peppoloni, S.; Biondo, C.; Mancuso, G.; et al. Immunogenic properties of Streptococcus agalactiae FbsA fragments. PLoS ONE 2013, 8, e75266. [Google Scholar] [CrossRef]
- Yi, T.; Li, Y.W.; Liu, L.; Xiao, X.X.; Li, A.X. Protection of Nile tilapia (Oreochromis niloticus L.) against Streptococcus agalactiae following immunization with recombinant FbsA and α-enolase. Aquaculture 2014, 428, 35–40. [Google Scholar] [CrossRef]
- Zhang, Z. Research Advances on Tilapia Streptococcosis. Pathogens 2021, 10, 558. [Google Scholar] [CrossRef]
- Stafslien, D.K.; Cleary, P.P. Characterization of the streptococcal C5a peptidase using a C5a-green fluorescent protein fusion protein substrate. J. Bacteriol. 2000, 182, 3254–3258. [Google Scholar] [CrossRef]
- Siezen, R.J. Multi-domain, cell-envelope proteinases of lactic acid bacteria. Lact. Acid Bact. Genet. Metab. Appl. 1999, 76, 139–155. [Google Scholar]
- Chmouryguina, I.; Suvorov, A.; Ferrieri, P.; Cleary, P.P. Conservation of the C5a peptidase genes in group A and B streptococci. Infect. Immun. 1996, 64, 2387–2390. [Google Scholar] [CrossRef]
- Cheng, Q.; Stafslien, D.; Purushothaman, S.S.; Cleary, P. The Group B Streptococcal C5a Peptidase Is Both a Specific Protease and an Invasin. Infect. Immun. 2002, 70, 2408–2413. [Google Scholar] [CrossRef]
- Wexler, D.E.; Chenoweth, D.E.; Cleary, P.P. Mechanism of action of the group A streptococcal C5a inactivator. Proc. Natl. Acad. Sci. USA 1985, 82, 8144–8148. [Google Scholar] [CrossRef]
- Bohnsack, J.F.; Mollison, K.W.; Buko, A.M.; Ashworth, J.C.; Hill, H.R. Group B streptococci inactivate complement component C5a by enzymic cleavage at the C-terminus. Biochem. J. 1991, 273, 635–640. [Google Scholar] [CrossRef][Green Version]
- Bohnsack, J.F.; Takahashi, S.; Hammitt, L.; Miller, D.V.; Aly, A.A.; Adderson, E.E. Genetic polymorphisms of group B streptococcus scpB alter functional activity of a cell-associated peptidase that inactivates C5a. Infect. Immun. 2000, 68, 5018–5025. [Google Scholar] [CrossRef]
- Wexler, D.E.; Nelson, R.D.; Cleary, P.P. Human neutrophil chemotactic response to group A streptococci: Bacteria-mediated interference with complement-derived chemotactic factors. Infect. Immun. 1983, 39, 239–246. [Google Scholar] [CrossRef]
- Fernandez-Espla, M.D.; Garault, P.; Monnet, V.; Rul, F. Streptococcus thermophilus cell wall-anchored proteinase: Release, purification, and biochemical and genetic characterization. Appl. Environ. Microbiol. 2000, 66, 4772–4778. [Google Scholar] [CrossRef] [PubMed]
- Hemming, V.G.; McCloskey, D.W.; Hill, H.R. Pneumonia in the neonate associated with group B streptococcal septicemia. Am. J. Dis. Child. 1976, 130, 1231–1233. [Google Scholar] [CrossRef] [PubMed]
- Rubens, C.E.; Raff, H.V.; Jackson, J.C.; Chi, E.Y.; Bielitzki, J.T.; Hillier, S.L. Pathophysiology and histopathology of group B streptococcal sepsis in Macaca nemestrina primates induced after intraamniotic inoculation: Evidence for bacterial cellular invasion. J. Infect. Dis. 1991, 164, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, J.F.; Chang, J.K.; Hill, H.R. Restricted ability of group B streptococcal C5a-ase to inactivate C5a prepared from different animal species. Infect Immun. 1993, 61, 1421–1426. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, J.F.; Widjaja, K.; Ghazizadeh, S.; Rubens, C.E.; Hillyard, D.R.; Parker, C.J.; Albertine, K.H.; Hill, H.R. A role for C5 and C5a-ase in the acute neutrophil response to group B streptococcal infections. J. Infect. Dis. 1997, 175, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Gibson, R.L.; Lee, M.K.; Soderland, C.; Chi, E.Y.; Rubens, C.E. Group B streptococci invade endothelial cells: Type III capsular polysaccharide attenuates invasion. Infect. Immun. 1993, 61, 478–485. [Google Scholar] [CrossRef]
- Tamura, G.S.; Rubens, C.E. Group B streptococci adhere to a variant of fibronectin attached to a solid phase. Mol. Microbiol. 1995, 15, 581–589. [Google Scholar] [CrossRef]
- Ji, Y.; Carlson, B.; Kondagunta, A.; Cleary, P.P. Intranasal immunization with C5a peptidase prevents nasopharyngeal colonization of mice by the group A Streptococcus. Infect. Immun. 1997, 65, 2080–2087. [Google Scholar] [CrossRef]
- Cheng, Q.; Carlson, B.; Pillai, S.; Eby, R.; Edwards, L.; Olmsted, S.B.; Cleary, P. Antibody against surface-bound C5a peptidase is opsonic and initiates macrophage killing of group B streptococci. Infect. Immun. 2001, 69, 2302–2308. [Google Scholar] [CrossRef]
- Cheng, Q.; Debol, S.; Lam, H.; Eby, R.; Edwards, L.; Matsuka, Y.; Olmsted, S.B.; Cleary, P.P. Immunization with C5a peptidase or peptidase-type III polysaccharide conjugate vaccines enhances clearance of group B Streptococci from lungs of infected mice. Infect. Immun. 2002, 70, 6409–6415. [Google Scholar] [CrossRef]
- Ke, X.; Li, Q.; Li, X.; Liu, Z.; Lu, M.; Yang, H. Construction and Analysis of the Immune Effects of a Streptococcus agalactiae Surface Protein ScpB Vaccine Encapsulated with Polylactic-Co-Glycolic Acid (PLGA). Open Access Libr. J. 2016, 03, 69764. [Google Scholar] [CrossRef][Green Version]
- Xue, G.; Yu, L.; Li, S.; Shen, X. Intranasal immunization with GBS surface protein Sip and ScpB induces specific mucosal and systemic immune responses in mice. FEMS Immunol. Med. Microbiol. 2010, 58, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, E.; Brewer, J.M.; Minor, W.; Carreira, L.A.; Lebioda, L. Mechanism of enolase: The crystal structure of asymmetric dimer enolase-2-phospho-D-glycerate/enolase-phosphoenolpyruvate at 2.0 A resolution. Biochemistry 1997, 36, 12526–12534. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Aronoff, D.M.; Davies, H.D.; Manning, S.D. Draft Genome Sequence of an Invasive Streptococcus agalactiae Isolate Lacking Pigmentation. Genome. Announc. 2016, 4, e00015-16. [Google Scholar] [CrossRef] [PubMed]
- Pancholi, V. Multifunctional alpha-enolase: Its role in diseases. Cell. Mol. Life. Sci. 2001, 58, 902–920. [Google Scholar] [CrossRef] [PubMed]
- Veiga-Malta, I.; Duarte, M.; Dinis, M.; Tavares, D.; Videira, A.; Ferreira, P. Enolase from Streptococcus sobrinus is an immunosuppressive protein. Cell. Microbiol. 2004, 6, 79–88. [Google Scholar] [CrossRef]
- Fontán, P.A.; Pancholi, V.; Nociari, M.M.; Fischetti, V.A. Antibodies to streptococcal surface enolase react with human alpha-enolase: Implications in poststreptococcal sequelae. J. Infect. Dis. 2000, 182, 1712–1721. [Google Scholar] [CrossRef] [PubMed]
- Pancholi, V.; Fischetti, V.A. alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 1998, 273, 14503–14515. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Catt, D.M.; Gregory, R.L. Streptococcus mutans surface alpha-enolase binds salivary mucin MG2 and human plasminogen. Infect. Immun. 2004, 72, 6748–6752. [Google Scholar] [CrossRef] [PubMed]
- Fischetti, V.A.; Pancholi, V.; Schneewind, O. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol. Microbiol. 1990, 4, 1603–1605. [Google Scholar] [CrossRef]
- McNeely, T.B.; Cope, L.; Smith, S.; Joshi, A.; Pak, I.; Friedman, A. Enolase Peptide Conjugate Vaccines against Staphylococcus. Aureus. Patent No. US9447403B2, 11 November 2011. [Google Scholar]
- Rahi, A.; Matta, S.K.; Dhiman, A.; Garhyan, J.; Gopalani, M.; Chandra, S.; Bhatnagar, R. Enolase of Mycobacterium tuberculosis is a surface exposed plasminogen binding protein. Biochim. Biophys. Acta. Gen. Subj. 2017, 1861, 3355–3364. [Google Scholar] [CrossRef] [PubMed]
- Hughes, M.J.G.; Moore, J.C.; Lane, J.D.; Wilson, R.; Pribul, P.K.; Younes, Z.N.; Dobson, R.J.; Everest, P.; Reason, A.J.; Redfern, J.M.; et al. Identification of major outer surface proteins of Streptococcus agalactiae. Infect. Immun. 2002, 70, 1254–1259. [Google Scholar] [CrossRef]
- Fluegge, K.; Schweier, O.; Schiltz, E.; Batsford, S.; Berner, R. Identification and immunoreactivity of proteins released from Streptococcus agalactiae. Eur. J. Clin. Microbiol. Infect. Dis. 2004, 23, 818–824. [Google Scholar] [CrossRef]
- Brzychczy-Wloch, M.; Gorska, S.; Brzozowska, E.; Gamian, A.; Heczko, P.B.; Bulanda, M. Identification of high immunoreactive proteins from Streptococcus agalactiae isolates recognized by human serum antibodies. FEMS Microbiol. Lett. 2013, 349, 61–70. [Google Scholar] [CrossRef][Green Version]
- Pratesi, F.; Moscato, S.; Sabbatini, A.; Chimenti, D.; Bombardieri, S.; Migliorini, M. Autoantibodies specific for alpha-enolase in systemic autoimmune disorders. J. Rheumatol. 2000, 27, 109–115. [Google Scholar]
- Dobrut, A.; Brzozowska, E.; Górska, S.; Pyclik, M.; Gamian, A.; Bulanda, M.; Brzychczy-Włoch, M. Epitopes of Immunoreactive Proteins of Streptococcus agalactiae: Enolase, Inosine 5′-Monophosphate Dehydrogenase and Molecular Chaperone GroEL. Front. Cell. Infect. Microbiol. 2018, 8, 349. [Google Scholar] [CrossRef] [PubMed]
- Harvey, K.L.; Jarocki, V.M.; Charles, I.G.; Djordjevic, S.P. The Diverse Functional Roles of Elongation Factor Tu (EF-Tu) in Microbial Pathogenesis. Front. Microbiol. 2019, 10, 2351. [Google Scholar] [CrossRef]
- Kunert, A.; Losse, J.; Gruszin, C.; Hühn, M.; Kaendler, K.; Mikkat, S.; Volke, D.; Hoffmann, R.; Jokiranta, S.T.; Harald Seeberger, H.; et al. Immune evasion of the human pathogen Pseudomonas aeruginosa: Elongation factor Tu is a factor H and plasminogen binding protein. J. Immunol. 2007, 179, 2979–2988. [Google Scholar] [CrossRef] [PubMed]
- Araújo, F.R.; Costa, C.M.; Ramos, C.A.; Farias, T.A.; Souza, I.I.; Melo, E.S.; Elisei, C.; Rosinha, G.M.S.; Soares, C.O.; Fragoso, S.P.; et al. IgG and IgG2 antibodies from cattle naturally infected with Anaplasma marginale recognize the recombinant vaccine candidate antigens VirB9, VirB10, and elongation factor-Tu. Mem. Inst. Oswaldo. Cruz. 2008, 103, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Mohan, S.; Hertweck, C.; Dudda, A.; Hammerschmidt, S.; Skerka, C.; Hallström, T.; Zipfel, P.F. Tuf of Streptococcus pneumoniae is a surface displayed human complement regulator binding protein. Mol. Immunol. 2014, 62, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Widjaja, M.; Harvey, K.L.; Hagemann, L.; Berry, I.J.; Jarocki, V.M.; Raymond, B.B.A.; Tacchi, J.L.; Gründel, A.; Steele, J.R.; Padula, M.P.; et al. Elongation factor Tu is a multifunctional and processed moonlighting protein. Sci. Rep. 2017, 7, 11227. [Google Scholar] [CrossRef]
- Flores, A.R.; Galloway-Peña, J.; Sahasrabhojane, P.; Saldaña, M.; Yao, H.; Su, X.; Ajami, N.J.; Holder, M.E.; Petrosino, J.F.; Thompson, E.; et al. Sequence type 1 group B Streptococcus, an emerging cause of invasive disease in adults, evolves by small genetic changes. Proc. Natl. Acad. Sci. USA 2015, 112, 6431–6436. [Google Scholar] [CrossRef] [PubMed]
- Granato, D.; Bergonzelli, G.E.; Pridmore, R.D.; Marvin, L.; Rouvet, M.; Corthésy-Theulaz, I.E. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect. Immun. 2004, 72, 2160–2169. [Google Scholar] [CrossRef]
- Sprinzl, M. Elongation factor Tu: A regulatory GTPase with an integrated effector. Trends Biochem. Sci. 1994, 19, 245–250. [Google Scholar] [CrossRef]
- Hempel, K.; Pané-Farré, J.; Otto, A.; Sievers, S.; Hecker, M.; Becher, D. Quantitative cell surface proteome profiling for SigB-dependent protein expression in the human pathogen Staphylococcus aureus via biotinylation approach. J. Proteome Res. 2010, 9, 1579–1590. [Google Scholar] [CrossRef]
- Monteiro, R.; Hébraud, M.; Chafsey, I.; Chambon, C.; Viala, D.; Torres, C.; Poeta, P.; Igrejas, G. Surfaceome and exoproteome of a clinical sequence type 398 methicillin resistant Staphylococcus aureus strain. Biochem. Biophys. Rep. 2015, 3, 7–13. [Google Scholar] [CrossRef]
- Dallo, S.F.; Zhang, B.; Denno, J.; Hong, S.; Tsai, A.; Haskins, W.; Ye, J.Y.; Weitaoet, T. Association of Acinetobacter baumannii EF-Tu with cell surface, outer membrane vesicles, and fibronectin. Sci. World J. 2012, 2012, 128705. [Google Scholar] [CrossRef]
- Turnbull, L.; Toyofuku, M.; Hynen, A.L.; Kurosawa, M.; Pessi, G.; Petty, N.K.; Osvath, S.R.; Cárcamo-Oyarce, G.; Gloag, E.S.; Shimoni, R.; et al. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 2016, 7, 1122. [Google Scholar] [CrossRef] [PubMed]
- Ebner, P.; Prax, M.; Nega, M.; Koch, I.; Dube, L.; Yu, W.; Rinker, J.; Popella, P.; Flötenmeyer, M.; Götz, F. Excretion of cytoplasmic proteins (ECP) in Staphylococcus aureus. Mol. Microbiol. 2015, 97, 775–789. [Google Scholar] [CrossRef] [PubMed]
- Cacan, E.; Kratzer, J.T.; Cole, M.F.; Gaucher, E.A. Interchanging functionality among homologous elongation factors using signatures of heterotachy. J. Mol. Evol. 2013, 76, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Surve, M.V.; Anil, A.; Kamath, K.G.; Bhutda, S.; Sthanam, L.K.; Pradhan, A.; Srivastava, R.; Basu, B.; Dutta, S.; Sen, S.; et al. Membrane Vesicles of Group B Streptococcus Disrupt Feto-Maternal Barrier Leading to Preterm Birth. PLoS Pathog. 2016, 12, e1005816. [Google Scholar] [CrossRef]
- Barbier, M.; Owings, J.P.; Martínez-Ramos, I.; Damron, F.H.; Gomila, R.; Blázquez, J.; Goldberg, J.B.; Albertí, S. Lysine trimethylation of EF-Tu mimics platelet-activating factor to initiate Pseudomonas aeruginosa pneumonia. mBio 2013, 4, e00207–e00213. [Google Scholar] [CrossRef]
- Mayer, F. Cytoskeletal elements in bacteria Mycoplasma pneumoniae, Thermoanaerobacterium sp., and Escherichia coli as revealed by electron microscopy. J. Mol. Microbiol. Biotechnol. 2006, 11, 228–243. [Google Scholar] [CrossRef]
- Hanukoglu, I. Proteopedia: Rossmann fold: A beta-alpha-beta fold at dinucleotide binding sites. Biochem. Mol. Biol. Educ. 2015, 43, 206–209. [Google Scholar] [CrossRef]
- Clark, B.F.; Kjeldgaard, M.; la Cour, T.F.; Thirup, S.; Nyborg, J. Structural determination of the functional sites of E. coli elongation factor Tu. Biochim. Biophys. Acta. 1990, 1050, 203–208. [Google Scholar] [CrossRef]
- Baldauf, S.L.; Palmer, J.D.; Doolittle, W.F. The root of the universal tree and the origin of eukaryotes based on elongation factor phylogeny. Proc. Natl. Acad. Sci. USA 1996, 93, 7749–7754. [Google Scholar] [CrossRef]
- Barel, M.; Charbit, A. Detection of the interaction between host and bacterial proteins: Eukaryotic nucleolin interacts with Francisella elongation factor Tu. Methods. Mol. Biol. 2014, 1197, 123–139. [Google Scholar] [CrossRef]
- Wolff, D.G.; Castiblanco-Valencia, M.M.; Abe, C.M.; Monaris, D.; Morais, Z.M.; Souza, G.O.; Vasconcellos, S.A.; Isaac, L.; Abreu, P.A.E.; Barbosa, A.S. Interaction of Leptospira elongation factor Tu with plasminogen and complement factor H: A metabolic leptospiral protein with moonlighting activities. PLoS ONE 2013, 8, e81818. [Google Scholar] [CrossRef]
- Li, Q.; Liu, H.; Du, D.; Yu, Y.; Ma, C.; Jiao, F.; Yao, Y.; Lu, C.; Zhang, W. Identification of Novel Laminin- and Fibronectin-binding Proteins by Far-Western Blot: Capturing the Adhesins of Streptococcus suis Type 2. Front. Cell. Infect. Microbiol. 2015, 5, 82. [Google Scholar] [CrossRef]
- N’Diaye, A.; Mijouin, L.; Hillion, M.; Diaz, S.; Konto-Ghiorghi, Y.; Percoco, G.; Chevalier, S.; Lefeuvre, L.; Harmer, N.J.; Lesouhaitier, O.; et al. Effect of Substance P in Staphylococcus aureus and Staphylococcus epidermidis Virulence: Implication for Skin Homeostasis. Front. Microbiol. 2016, 7, 506. [Google Scholar] [CrossRef] [PubMed]
- Kloppot, P.; Selle, M.; Kohler, C.; Stentzel, S.; Fuchs, S.; Liebscher, V.; Müller, E.; Kale, D.; Ohlsen, K.; Bröker, B.M.; et al. Microarray-based identification of human antibodies against Staphylococcus aureus antigens. Proteomics. Clin. Appl. 2015, 9, 1003–1111. [Google Scholar] [CrossRef]
- Churchward, C.P.; Rosales, R.S.; Gielbert, A.; Domínguez, M.; Nicholas, R.A.J.; Ayling, R.D. Immunoproteomic characterisation of Mycoplasma mycoides subspecies capri by mass spectrometry analysis of two-dimensional electrophoresis spots and western blot. J. Pharm. Pharmacol. 2015, 67, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; He, J.; Navarro-Alvarez, N.; Xu, J.; Li, X.; Li, P.; Wu, W. Elongation Factor Tu and Heat Shock Protein 70 Are Membrane-Associated Proteins from Mycoplasma ovipneumoniae Capable of Inducing Strong Immune Response in Mice. PLoS ONE 2016, 11, e0161170. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Campillo, M.; Bini, L.; Comanducci, M.; Raggiaschi, R.; Marzocchi, B.; Pallini, V.; Ratti, G. Identification of immunoreactive proteins of Chlamydia trachomatis by Western blot analysis of a two-dimensional electrophoresis map with patient sera. Electrophoresis 1999, 20, 2269–2279. [Google Scholar] [CrossRef]
- Nieves, W.; Heang, J.; Asakrah, S.; Höner zu Bentrup, K.; Roy, C.J.; Morici, L.A. Immunospecific responses to bacterial elongation factor Tu during Burkholderia infection and immunization. PLoS ONE 2010, 5, e14361. [Google Scholar] [CrossRef] [PubMed]
- Pinto, P.M.; Chemale, G.; de Castro, L.A.; Metz Costa, A.P.; Kich, J.D.; Vainstein, M.H.; Zaha, A.; Ferreira, H.B. Proteomic survey of the pathogenic Mycoplasma hyopneumoniae strain 7448 and identification of novel post-translationally modified and antigenic proteins. Vet. Microbiol. 2007, 121, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Thofte, O.; Su, Y.C.; Brant, M.; Littorin, N.; Duell, B.L.; Alvarado, V.; Jalalvand, F.; Riesbeck, K. EF-Tu From Non-typeable Haemophilus influenzae Is an Immunogenic Surface-Exposed Protein Targeted by Bactericidal Antibodies. Front. Immunol. 2018, 9, 2910. [Google Scholar] [CrossRef] [PubMed]
- Pyclik, M.; Górska, S.; Brzozowska, E.; Dobrut, A.; Ciekot, J.; Gamian, A.; Brzychczy-Włoch, M. Epitope Mapping of Streptococcus agalactiae Elongation Factor Tu Protein Recognized by Human Sera. Front. Microbiol. 2018, 9, 125. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, J.; Wang, K.; Liu, T.; Zhu, L.; He, S.; Geng, Y.; Chen, D.; Huang, X.; Ou-yang, P. Evaluation of immunogenicity and protective efficacy of the elongation factor Tu against Streptococcus agalactiae in tilapia. Aquaculture 2018, 492, 184–189. [Google Scholar] [CrossRef]
- Nagai, K.; Domon, H.; Maekawa, T.; Hiyoshi, T.; Tamura, H.; Yonezawa, D.; Habuka, R.; Saitoh, A.; Terao, Y. Immunization with pneumococcal elongation factor Tu enhances serotype-independent protection against Streptococcus pneumoniae infection. Vaccine 2019, 37, 160–168. [Google Scholar] [CrossRef]
- Kolberg, J.; Hammerschmidt, S.; Frank, R.; Jonák, J.; Sanderová, H.; Aase, A. The surface-associated elongation factor Tu is concealed for antibody binding on viable pneumococci and meningococci. FEMS Immunol. Med. Microbiol. 2008, 53, 222–230. [Google Scholar] [CrossRef]
- Cohen, M.B.; Sadee, W. Contributions of the depletions of guanine and adenine nucleotides to the toxicity of purine starvation in the mouse T lymphoma cell line. Cancer Res. 1983, 43, 1587–1591. [Google Scholar]
- Hedstrom, L. IMP dehydrogenase: Structure, mechanism, and inhibition. Chem. Rev. 2009, 109, 2903–2928. [Google Scholar] [CrossRef]
- Ji, Y.; Gu, J.; Makhov, A.M.; Griffith, J.D.; Mitchell, B.S. Regulation of the interaction of inosine monophosphate dehydrogenase with mycophenolic Acid by GTP. J. Biol. Chem. 2006, 281, 206–212. [Google Scholar] [CrossRef]
- Gunter, J.H.; Thomas, E.C.; Lengefeld, N.; Kruger, S.J.; Worton, L.; Gardiner, E.M.; Jones, A.; Barnett, N.L.; Whitehead, J.P. Characterisation of inosine monophosphate dehydrogenase expression during retinal development: Differences between variants and isoforms. Int. J. Biochem. Cell Biol. 2008, 40, 1716–1728. [Google Scholar] [CrossRef] [PubMed]
- Whitby, F.G.; Luecke, H.; Kuhn, P.; Somoza, J.R.; Huete-Perez, J.A.; Phillips, J.D.; Hill, C.P.; Fletterick, R.J.; Wang, C.C. Crystal structure of Tritrichomonas foetus inosine-5′-monophosphate dehydrogenase and the enzyme-product complex. Biochemistry 1997, 36, 10666–10674. [Google Scholar] [CrossRef]
- Colby, T.D.; Vanderveen, K.; Strickler, M.D.; Markham, G.D.; Goldstein, B.M. Crystal structure of human type II inosine monophosphate dehydrogenase: Implications for ligand binding and drug design. Proc. Natl. Acad. Sci. USA 1999, 96, 3531–3536. [Google Scholar] [CrossRef]
- Ratcliffe, A.J. Inosine 5′-monophosphate dehydrogenase inhibitors for the treatment of autoimmune diseases. Curr. Opin. Drug Discov. Dev. 2006, 9, 595–605. [Google Scholar]
- Juda, P.; Smigová, J.; Kováčik, L.; Bártová, E.; Raška, I. Ultrastructure of cytoplasmic and nuclear inosine-5′-monophosphate dehydrogenase 2 "rods and rings" inclusions. J. Histochem. Cytochem. 2014, 62, 739–750. [Google Scholar] [CrossRef]
- Shah, C.P.; Kharkar, P.S. Inosine 5′-monophosphate dehydrogenase inhibitors as antimicrobial agents: Recent progress and future perspectives. Future Med. Chem. 2015, 7, 1415–1429. [Google Scholar] [CrossRef] [PubMed]
- Pitarch, A.; Díez-Orejas, R.; Molero, G.; Pardo, M.; Sánchez, M.; Gil, C.; Nombela, C. Analysis of the serologic response to systemic Candida albicans infection in a murine model. Proteomics 2001, 1, 550–559. [Google Scholar] [CrossRef]
- Köhler, G.A.; White, T.C.; Agabian, N. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 1997, 179, 2331–2338. [Google Scholar] [CrossRef]
- Seelig, H.P.; Appelhans, H.; Bauer, O.; Blüthner, M.; Hartung, K.; Schranz, P.; Schultze, D.; Seelig, C.A.; Volkmann, M. Autoantibodies against inosine-5′- monophosphate dehydrogenase 2-characteristics and prevalence in patients with HCV-infection. Clin. Lab. 2011, 57, 753–765. [Google Scholar] [PubMed]
- Duan, Z.T.; He, K.W.; Zhang, X.H.; Ni, X.X.; Lu, C.P. Cloning and characterization of the gene encoding IMPDH of Streptococcus suis serotype 2. Wei Sheng Wu Xue Bao 2006, 46, 730–733. [Google Scholar] [PubMed]
- Zeilstra-Ryalls, J.; Fayet, O.; Georgopoulos, C. The universally conserved GroE (Hsp60) chaperonins. Annu. Rev. Microbiol. 1991, 45, 301–325. [Google Scholar] [CrossRef] [PubMed]
- Mayhew, M.; da Silva, A.C.; Martin, J.; Erdjument-Bromage, H.; Tempst, P.; Hartl, F.U. Protein folding in the central cavity of the GroEL-GroES chaperonin complex. Nature 1996, 379, 420–426. [Google Scholar] [CrossRef]
- Braig, K.; Otwinowski, Z.; Hegde, R.; Boisvert, D.C.; Joachimiak, A.; Horwich, A.L.; Sigler, P.B. The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature 1994, 371, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Horwich, A.L.; Fenton, W.A.; Chapman, E.; Farr, G.W. Two families of chaperonin: Physiology and mechanism. Annu. Rev. Cell Dev. Biol. 2007, 23, 115–145. [Google Scholar] [CrossRef] [PubMed]
- Fenton, W.A.; Kashi, Y.; Furtak, K.; Horwich, A.L. Residues in chaperonin GroEL required for polypeptide binding and release. Nature 1994, 371, 614–619. [Google Scholar] [CrossRef] [PubMed]
- Farr, G.W.; Furtak, K.; Rowland, M.B.; Ranson, N.A.; Saibil, H.R.; Kirchhausen, T.; Horwich, A.L. Multivalent binding of nonnative substrate proteins by the chaperonin GroEL. Cell 2000, 100, 561–573. [Google Scholar] [CrossRef]
- Kerner, M.J.; Naylor, D.J.; Ishihama, Y.; Maier, T.; Chang, H.C.; Stines, A.P.; Georgopoulos, C.; Frishman, D.; Hayer-Hartl, M.; Mannet, M.; et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 2005, 122, 209–220. [Google Scholar] [CrossRef]
- Zand, L.; Rye, H.S. GroEL-Mediated Protein Folding: Making the Impossible, Possible. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 211–239. [Google Scholar] [CrossRef]
- Hayer-Hartl, M.; Bracher, A.; Hartl, F.U. The GroEL-GroES Chaperonin Machine: A Nano-Cage for Protein Folding. Trends Biochem. Sci. 2016, 41, 62–76. [Google Scholar] [CrossRef]
- Chen, C.; Zabad, S.; Liu, H.; Wang, W.; Jeffery, C. MoonProt 2.0: An expansion and update of the moonlighting proteins database. Nucleic. Acids. Res. 2018, 46, D640–D644. [Google Scholar] [CrossRef] [PubMed]
- Webb, S.A.R.; Kahler, C.M. Bench-to-bedside review: Bacterial virulence and subversion of host defences. Crit. Care 2008, 12, 234. [Google Scholar] [CrossRef]
- Fourie, K.R.; Wilson, H.L. Understanding GroEL and DnaK Stress Response Proteins as Antigens for Bacterial Diseases. Vaccines 2020, 8, 773. [Google Scholar] [CrossRef]
- Hagemann, L.; Gründel, A.; Jacobs, E.; Dumke, R. The surface-displayed chaperones GroEL and DnaK of Mycoplasma pneumoniae interact with human plasminogen and components of the extracellular matrix. Pathog. Dis. 2017, 75, ftx017. [Google Scholar] [CrossRef]
- Kim, J.J.; Khan, W.I. Goblet cells and mucins: Role in innate defense in enteric infections. Pathogens 2013, 2, 55–70. [Google Scholar] [CrossRef]
- González-López, M.A.; Velázquez-Guadarrama, N.; Romero-Espejel, M.E.; de Jesús Olivares-Trejo, J. Helicobacter pylori secretes the chaperonin GroEL (HSP60), which binds iron. FEBS Lett. 2013, 587, 1823–1828. [Google Scholar] [CrossRef]
- Senkovich, O.; Ceaser, S.; McGee, D.J.; Testerman, T.L. Unique host iron utilization mechanisms of Helicobacter pylori revealed with iron-deficient chemically defined media. Infect. Immun. 2010, 78, 1841–1849. [Google Scholar] [CrossRef]
- Yoshida, N.; Oeda, K.; Watanabe, E.; Mikami, T.; Fukita, Y.; Nishimura, K.; Komai, K.; Matsudaet, K. Protein function. Chaperonin turned insect toxin. Nature 2001, 411, 44. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Kant, S.; Zaman, S.; Mahapatro, G.K.; Banerjee, N.; Sarin, N.B. A novel insecticidal GroEL protein from Xenorhabdus nematophila confers insect resistance in tobacco. Transgenic Res. 2014, 23, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Karachaliou, M.; Chatzi, L.; Michel, A.; Kyriklaki, A.; Kampouri, M.; Koutra, K.; Roumeliotaki, T.; Chalkiadaki, G.; Stiakaki, E.; Pawlita, M.; et al. Helicobacter pylori Seropositivity and Childhood Neurodevelopment, the Rhea Birth Cohort in Crete, Greece. Paediatr. Perinat. Epidemiol. 2017, 31, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Zügel, U.; Kaufmann, S.H. Activation of CD8 T cells with specificity for mycobacterial heat shock protein 60 in Mycobacterium bovis bacillus Calmette-Guérin-vaccinated mice. Infect. Immun. 1997, 65, 3947–3950. [Google Scholar] [CrossRef]
- Obradovic, J.; Pasternak, A.; Ng, S.H.; Allan, B.; Brownlie, R.; Wilson, H.L. Immunoproteomic analysis of Lawsonia intracellularis identifies candidate neutralizing antibody targets for use in subunit vaccine development. Vet. Microbiol. 2019, 235, 270–279. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).