Transcriptome Analysis of Citrus Dwarfing Viroid Induced Dwarfing Phenotype of Sweet Orange on Trifoliate Orange Rootstock
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and RNA Isolation
2.2. Library Preparation and High Throughput Sequencing
2.3. Bioinformatic Analysis
2.4. Expression Analysis of Citrus mRNA Target Genes Using RT-qPCR
3. Results
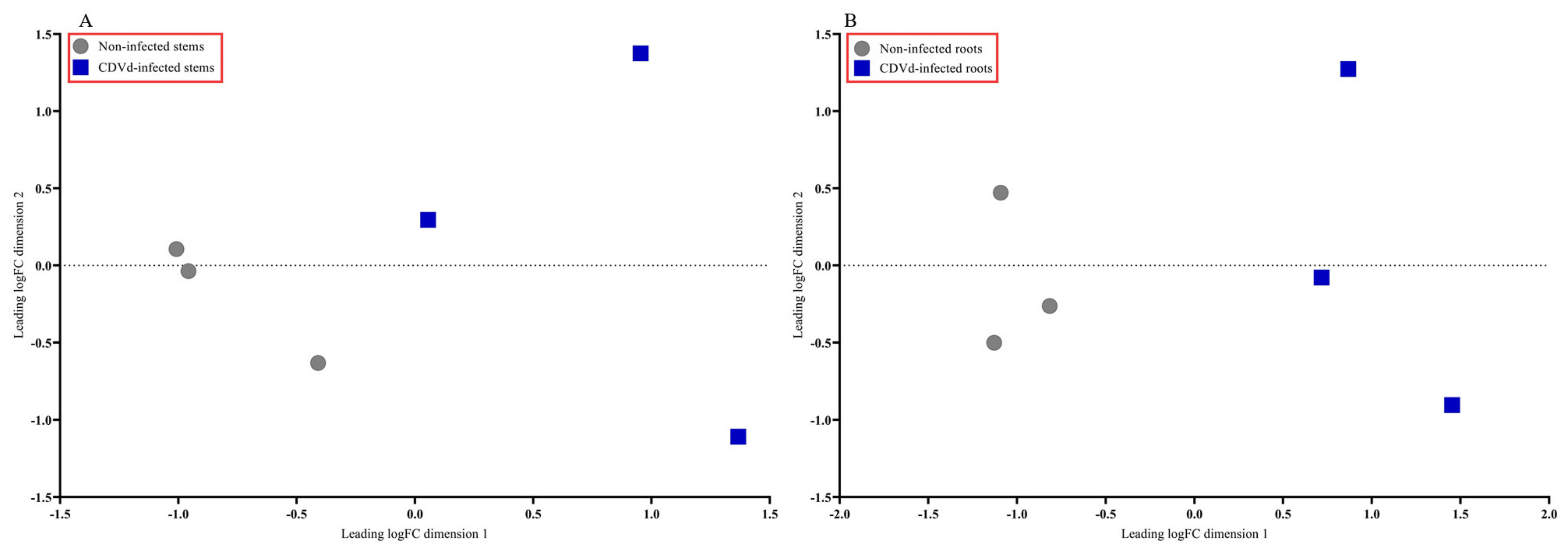
3.1. Transcriptome Assembly and Annotation
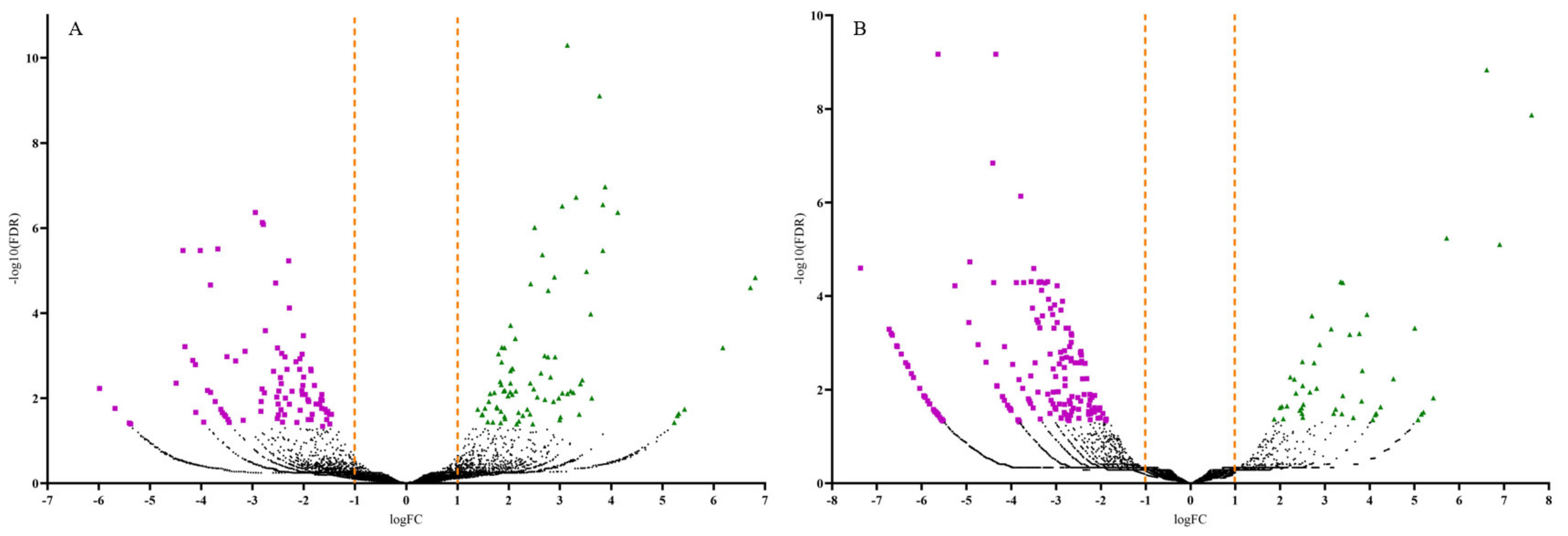
3.2. Differentially Expressed Genes (DEGs) Analysis and Identification
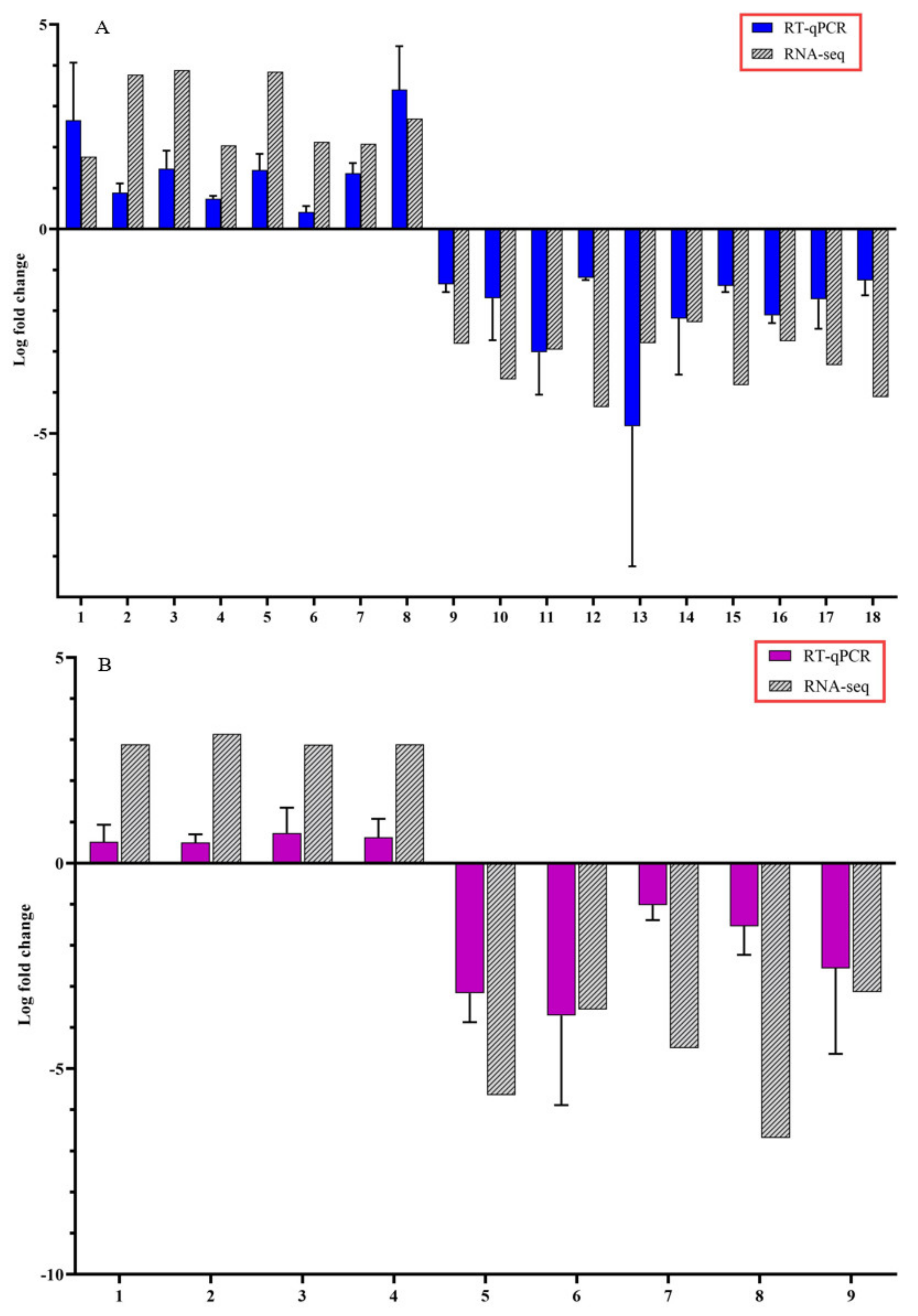
3.3. Confirmation of Candidate DEGs by RT-qPCR Analysis
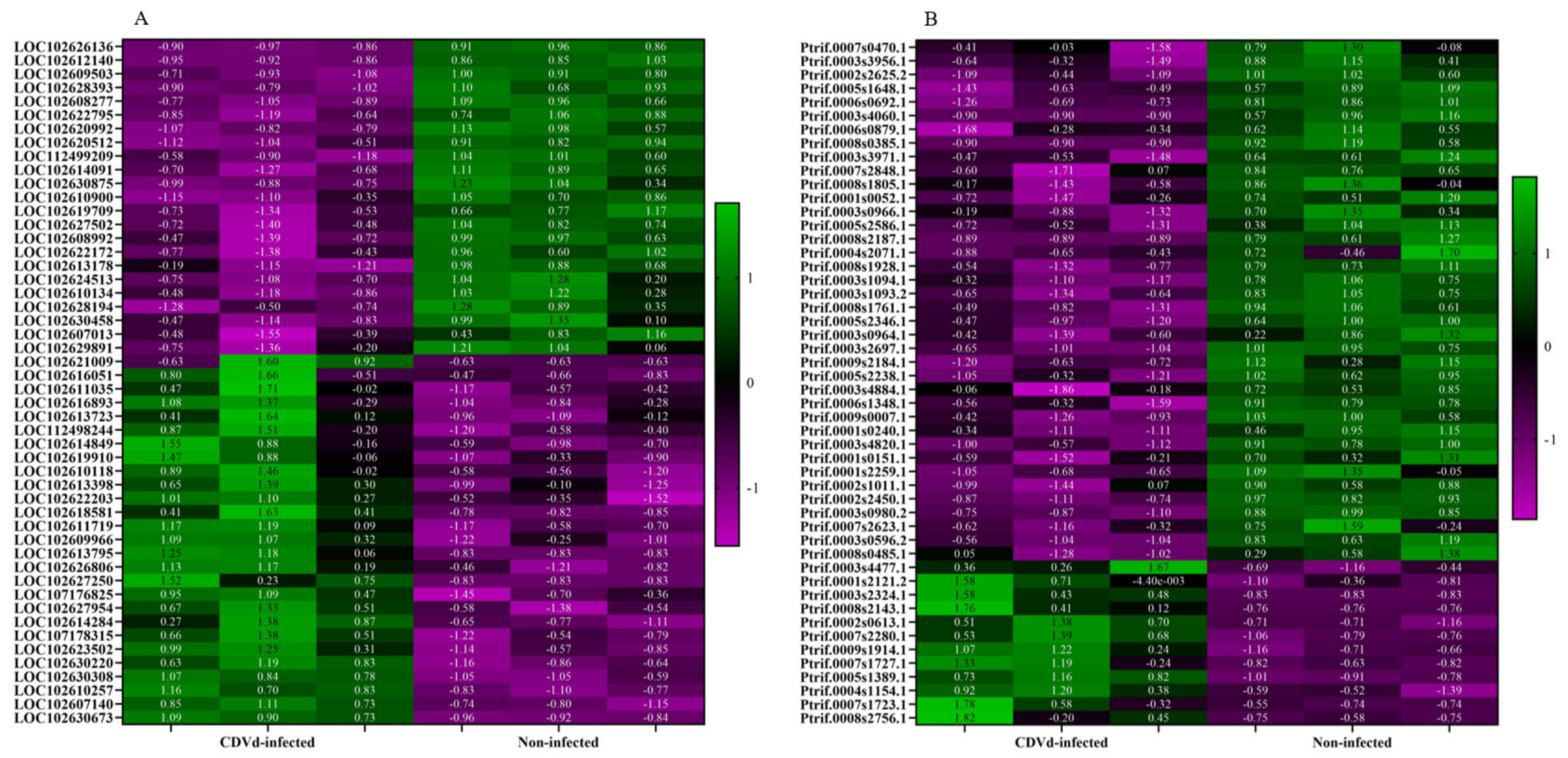
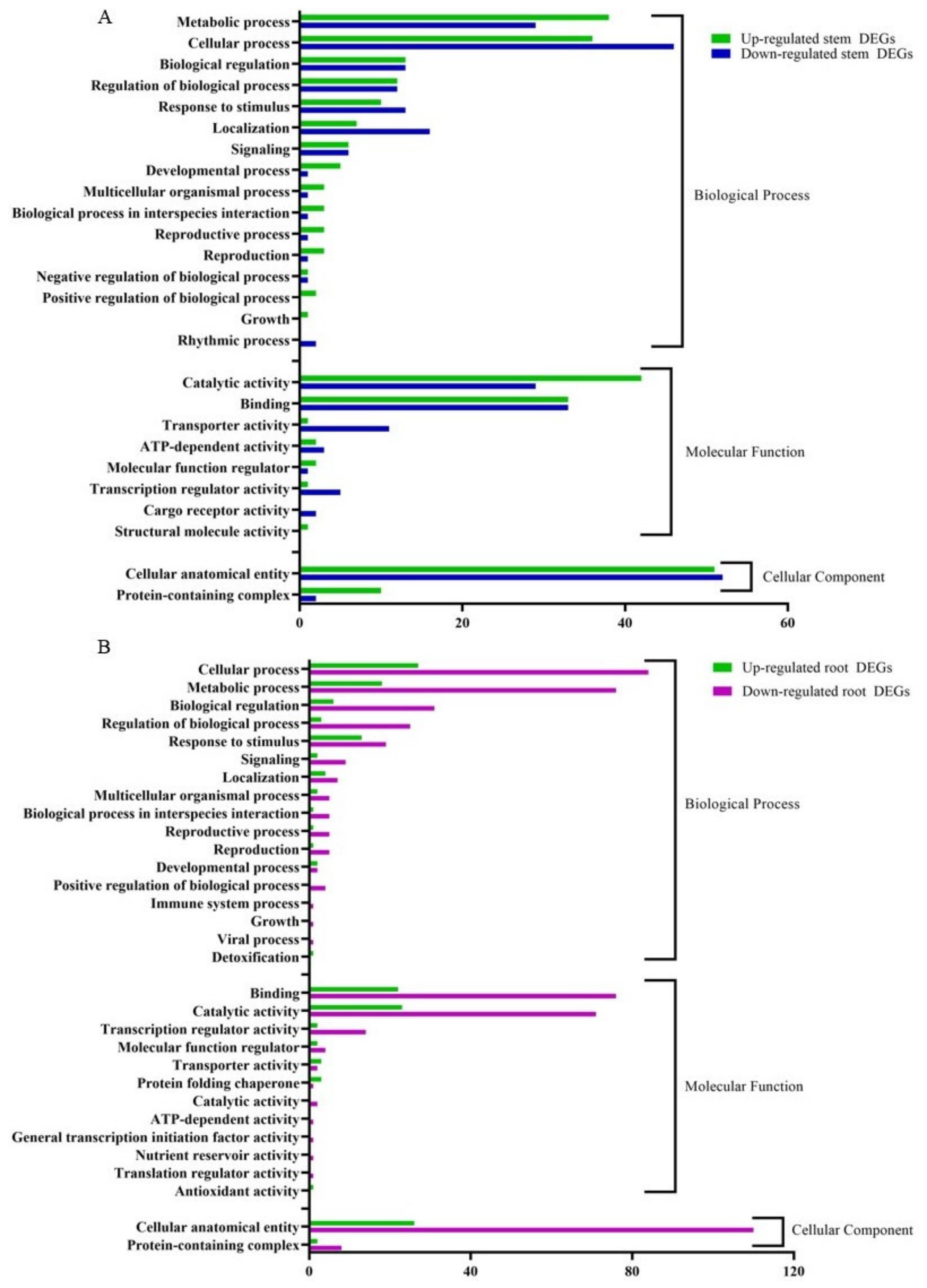
3.4. Functional Classification of DEGs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Babcock, B.A. Economic Impact of California’s Citrus Industry in 2020. J. Citrus Pathol. 2022, 9. [Google Scholar] [CrossRef]
- Van Duyn, M.A.; Pivonka, E. Overview of the Health Benefits of Fruit and Vegetable Consumption for the Dietetics Professional: Selected Literature. J. Am. Diet. Assoc. 2000, 100, 1511–1521. [Google Scholar] [CrossRef]
- Yao, L.H.; Jiang, Y.M.; Shi, J.; Tomás-Barberán, F.A.; Datta, N.; Singanusong, R.; Chen, S.S. Flavonoids in Food and Their Health Benefits. Plant Foods Hum. Nutr. 2004, 59, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Boswell, S.B. Others Tree Spacing of’Washington’navel Orange. J. Am. Soc. Hortic. Sci. 1970, 95, 523–528. [Google Scholar]
- Platt, R.G. Treatment of Frost-Inured Citrus, Avocados. Calif. Citrogr. 1973, 113–114, 140. [Google Scholar]
- Tucker, D.P.H.; Wheaton, T.A. Trends in Higher Citrus Planting Densities. Proc. Fla. State Hort. Soc. 1978, 91, 36–40. [Google Scholar]
- Schumann, A.W.; Singerman, A.; Wright, A.L.; Ferrarezi, R.S. Citrus under Protective Screen (CUPS) Production Systems. In Citrus under Protective Screen (CUPS) Production Systems; University of Florida Entomology and Nematology Department, UF/IFAS Extension: Gainesville, FL, USA, 2017. [Google Scholar]
- Flores, R.; Hernández, C.; Martínez de Alba, A.E.; Daròs, J.-A.; Di Serio, F. Viroids and Viroid-Host Interactions. Annu. Rev. Phytopathol. 2005, 43, 117–139. [Google Scholar] [CrossRef]
- Murcia, N.; Hashemian, S.M.B.; Serra, P.; Pina, J.A.; Duran-Vila, N. Citrus Viroids: Symptom Expression and Performance of Washington Navel Sweet Orange Trees Grafted on Carrizo Citrange. Plant Dis. 2015, 99, 125–136. [Google Scholar] [CrossRef]
- Navarro, B.; Flores, R.; Di Serio, F. Advances in Viroid-Host Interactions. Annu. Rev. Virol. 2021, 8, 305–325. [Google Scholar] [CrossRef]
- Itaya, A.; Matsuda, Y.; Gonzales, R.A.; Nelson, R.S.; Ding, B. Potato Spindle Tuber Viroid Strains of Different Pathogenicity Induces and Suppresses Expression of Common and Unique Genes in Infected Tomato. Mol. Plant. Microbe Interact. 2002, 15, 990–999. [Google Scholar] [CrossRef] [Green Version]
- Rodio, M.-E.; Delgado, S.; De Stradis, A.; Gómez, M.-D.; Flores, R.; Di Serio, F. A Viroid RNA with a Specific Structural Motif Inhibits Chloroplast Development. Plant Cell 2007, 19, 3610–3626. [Google Scholar] [CrossRef] [Green Version]
- Owens, R.A.; Tech, K.B.; Shao, J.Y.; Sano, T.; Baker, C.J. Global Analysis of Tomato Gene Expression during Potato Spindle Tuber Viroid Infection Reveals a Complex Array of Changes Affecting Hormone Signaling. Mol. Plant. Microbe Interact. 2012, 25, 582–598. [Google Scholar] [CrossRef] [Green Version]
- Gillings, M.R.; Broadbent, P.; Gollnow, B.I. Viroids in Australian Citrus: Relationship to Exocortis, Cachexia and Citrus Dwarfing. Funct. Plant Biol. 1991, 18, 559–570. [Google Scholar] [CrossRef]
- Hadas, R.; Bar-Joseph, M. Variation in Tree Size and Rootstock Scaling of Grapefruit Trees Inoculated with a Complex of Citrus Viroids. In International Organization of Citrus Virologists Conference Proceedings (1957–2010); IOCV: Riverside, CA, USA, 1991; Volume 11, pp. 240–243. [Google Scholar] [CrossRef]
- Hutton, R.J.; Broadbent, P.; Bevington, K.B. Viroid Dwarfing for High Density Citrus Plantings. In Horticultural Reviews; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2000; Volume 24, pp. 277–317. ISBN 9780471333746. [Google Scholar]
- Semancik, J.S.; Bash, J.; Gumpf, D.J. Induced Dwarfing of Citrus by Transmissible Small Nuclear RNA (TsnRNA). In International Organization of Citrus Virologists Conference Proceedings (1957–2010); IOCV: Riverside, CA, USA, 2002; Volume 15, pp. 390–394. [Google Scholar] [CrossRef]
- Semancik, J.S. Considerations for the Introduction of Viroids; Hadidi, A., Flores, R., Randles, J.W., Semancik, J.S., Eds.; CSIRO Publishing: Clayton, VIC, Australia, 2003; pp. 357–362. [Google Scholar]
- Vidalakis, G.; Pagliaccia, D.; Bash, J.A.; Afunian, M.; Semancik, J.S. Citrus Dwarfing Viroid: Effects on Tree Size and Scion Performance Specific to Poncirus Trifoliata Rootstock for High-Density Planting. Ann. Appl. Biol. 2011, 158, 204–217. [Google Scholar] [CrossRef]
- Semancik, J.S.; Rakowski, A.G.; Bash, J.A.; Gumpf, D.J. Application of Selected Viroids for Dwarfing and Enhancement of Production of ‘Valencia’ Orange. J. Hortic. Sci. 1997, 72, 563–570. [Google Scholar] [CrossRef]
- Lavagi-Craddock, I.; Campos, R.; Pagliaccia, D.; Kapaun, T.; Lovatt, C.; Vidalakis, G. Citrus Dwarfing Viroid Reduces Canopy Volume by Affecting Shoot Apical Growth of Navel Orange Trees Grown on Trifoliate Orange Rootstock. J. Citrus Pathol. 2020, 7, 1–6. [Google Scholar] [CrossRef]
- Tessitori, M.; Maria, G.; Capasso, C.; Catara, G.; Rizza, S.; De Luca, V.; Catara, A.; Capasso, A.; Carginale, V. Differential Display Analysis of Gene Expression in Etrog Citron Leaves Infected by Citrus Viroid III. Biochim. Biophys. Acta BBA Gene Struct. Expr. 2007, 1769, 228–235. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.; Qiu, Y.; Atta, S.; Zhou, C.; Cao, M. Global Transcriptomic Analysis Reveals Insights into the Response of “Etrog” Citron (Citrus medica L.) to Citrus Exocortis Viroid Infection. Viruses 2019, 11, 453. [Google Scholar] [CrossRef] [Green Version]
- Olivier, T.; Bragard, C. Innate Immunity Activation and RNAi Interplay in Citrus Exocortis Viroid—Tomato Pathosystem. Viruses 2018, 10, 587. [Google Scholar] [CrossRef] [Green Version]
- Cottilli, P.; Belda-Palazón, B.; Adkar-Purushothama, C.R.; Perreault, J.-P.; Schleiff, E.; Rodrigo, I.; Ferrando, A.; Lisón, P. Citrus Exocortis Viroid Causes Ribosomal Stress in Tomato Plants. Nucleic Acids Res. 2019, 47, 8649–8661. [Google Scholar] [CrossRef]
- Xu, Q.; Chen, L.-L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.-B.; Hao, B.-H.; Lyon, M.P.; et al. The Draft Genome of Sweet Orange (Citrus sinensis). Nat. Genet. 2013, 45, 59–66. [Google Scholar] [CrossRef]
- Martinelli, F.; Uratsu, S.L.; Albrecht, U.; Reagan, R.L.; Phu, M.L.; Britton, M.; Buffalo, V.; Fass, J.; Leicht, E.; Zhao, W.; et al. Transcriptome Profiling of Citrus Fruit Response to Huanglongbing Disease. PLoS ONE 2012, 7, e38039. [Google Scholar] [CrossRef]
- Fu, S.; Shao, J.; Zhou, C.; Hartung, J.S. Transcriptome Analysis of Sweet Orange Trees Infected with “Candidatus Liberibacter Asiaticus” and Two Strains of Citrus Tristeza Virus. BMC Genom. 2016, 17, 349. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhou, L.; Yu, X.; Stover, E.; Luo, F.; Duan, Y. Transcriptome Profiling of Huanglongbing (HLB) Tolerant and Susceptible Citrus Plants Reveals the Role of Basal Resistance in HLB Tolerance. Front. Plant Sci. 2016, 7, 933. [Google Scholar] [CrossRef] [Green Version]
- Arce-Leal, Á.P.; Bautista, R.; Rodríguez-Negrete, E.A.; Manzanilla-Ramírez, M.Á.; Velázquez-Monreal, J.J.; Santos-Cervantes, M.E.; Méndez-Lozano, J.; Beuzón, C.R.; Bejarano, E.R.; Castillo, A.G.; et al. Gene Expression Profile of Mexican Lime (Citrus aurantifolia) Trees in Response to Huanglongbing Disease Caused by Candidatus Liberibacter Asiaticus. Microorganisms 2020, 8, 528. [Google Scholar] [CrossRef] [Green Version]
- Visser, M.; Cook, G.; Burger, J.T.; Maree, H.J. In Silico Analysis of the Grapefruit SRNAome, Transcriptome and Gene Regulation in Response to CTV-CDVd Co-Infection. Virol. J. 2017, 14, 200. [Google Scholar] [CrossRef] [Green Version]
- Dang, T.; Lavagi-Craddock, I.; Bodaghi, S.; Vidalakis, G. Next-Generation Sequencing Identification and Characterization of MicroRNAs in Dwarfed Citrus Trees Infected with Citrus Dwarfing Viroid in High-Density Plantings. Front. Microbiol. 2021, 12, 646273. [Google Scholar] [CrossRef]
- Peng, Z.; Bredeson, J.V.; Wu, G.A.; Shu, S.; Rawat, N.; Du, D.; Parajuli, S.; Yu, Q.; You, Q.; Rokhsar, D.S.; et al. A Chromosome-Scale Reference Genome of Trifoliate Orange (Poncirus trifoliata) Provides Insights into Disease Resistance, Cold Tolerance and Genome Evolution in Citrus. Plant J. 2020, 104, 1215–1232. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-Throughput Functional Annotation and Data Mining with the Blast2GO Suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: The Integrative Protein Signature Database. Nucleic Acids Res. 2009, 37, D211–D215. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Stover, E.; Castle, W.S.; Spyke, P. The Citrus Grove of the Future and Its Implications for Huanglongbing Management. Proc. Fla. State Hort. Soc. 2008, 121, 155–159. [Google Scholar]
- Gottwald Current Epidemiological Understanding of Citrus Huanglongbing. Annu. Rev. Phytopathol. 2010, 48, 119–139. [CrossRef] [Green Version]
- Lambin, E.F. Global Land Availability: Malthus versus Ricardo. Glob. Food Secur. 2012, 1, 83–87. [Google Scholar] [CrossRef]
- Verburg, P.H.; Mertz, O.; Erb, K.-H.; Haberl, H.; Wu, W. Land System Change and Food Security: Towards Multi-Scale Land System Solutions. Curr. Opin. Environ. Sustain. 2013, 5, 494–502. [Google Scholar] [CrossRef] [Green Version]
- da Graça, J.V.; Douhan, G.W.; Halbert, S.E.; Keremane, M.L.; Lee, R.F.; Vidalakis, G.; Zhao, H. Huanglongbing: An Overview of a Complex Pathosystem Ravaging the World’s Citrus. J. Integr. Plant Biol. 2016, 58, 373–387. [Google Scholar] [CrossRef]
- Cohen, M. Exocortis Virus as a Possible Factor in Producing Dwarf Citrus Trees. Fla. State Hort. Soc. Proc. 1968, 115–119. [Google Scholar]
- Mendel, K. Interrelations between Tree Performance and Some Virus Diseases. In International Organization of Citrus Virologists Conference Proceedings (1957–2010); IOCV: Riverside, CA, USA, 1968; Volume 4, pp. 310–313. [Google Scholar]
- Bar-Joseph, M. Citrus Viroids and Citrus Dwarfing in Israel. In Proceedings of the V International Symposium on Orchard and Plantation Systems 349; 1992; pp. 271–276. Available online: https://www.actahort.org/books/349/349_45.htm (accessed on 29 March 2022).
- Broadbent, P.; Forsyth, J.B.; Hutton, R.J.; Bevington, K.B. Guidelines for the Commercial Use of Graft-Transmissible Dwarfing in Australia—Potential Benefits and Risks. Int. Citrus Congr. 1992, 7, 697–701. [Google Scholar]
- Schneider, H. The Anatomy of Citrus. In The Citrus Industry II Anatomy, Physiology, Genetics and Reproduction; Reuther, W., Batchelor, L.D., Webber, H.J., Eds.; University of California: Berkeley, CA, USA, 1968. [Google Scholar]
- Burgess, A.L.; David, R.; Searle, I.R. Conservation of TRNA and RRNA 5-Methylcytosine in the Kingdom Plantae. BMC Plant Biol. 2015, 15, 199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, G.; Bäumlein, H.; Mock, H.-P.; Himmelbach, A.; Schweizer, P. The Multigene Family Encoding Germin-like Proteins of Barley. Regulation and Function in Basal Host Resistance. Plant Physiol. 2006, 142, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Chen, X.; Zhu, F.; Li, H.; Li, L.; Yang, Q.; Chi, X.; Yu, S.; Liang, X. Characterization of Peanut Germin-like Proteins, AhGLPs in Plant Development and Defense. PLoS ONE 2013, 8, e61722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, H.; Fu, X.; Yang, C.; Tang, X.; Guo, L.; Li, C.; Xu, C.; Luo, K. Genome-Wide Investigation of Pentatricopeptide Repeat Gene Family in Poplar and Their Expression Analysis in Response to Biotic and Abiotic Stresses. Sci. Rep. 2018, 8, 2817. [Google Scholar] [CrossRef] [Green Version]
- Gidda, S.K.; Miersch, O.; Levitin, A.; Schmidt, J.; Wasternack, C.; Varin, L. Biochemical and Molecular Characterization of a Hydroxyjasmonate Sulfotransferase from Arabidopsis Thaliana. J. Biol. Chem. 2003, 278, 17895–17900. [Google Scholar] [CrossRef] [Green Version]
- Simões, I.; Faro, C. Structure and Function of Plant Aspartic Proteinases. Eur. J. Biochem. 2004, 271, 2067–2075. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.; Ni, W.; Feng, B.; Han, T.; Petrasek, M.G.; Ma, H. Members of the Arabidopsis-SKP1-like Gene Family Exhibit a Variety of Expression Patterns and May Play Diverse Roles in Arabidopsis. Plant Physiol. 2003, 133, 203–217. [Google Scholar] [CrossRef] [Green Version]
- Helariutta, Y.; Fukaki, H.; Wysocka-Diller, J.; Nakajima, K.; Jung, J.; Sena, G.; Hauser, M.T.; Benfey, P.N. The SHORT-ROOT Gene Controls Radial Patterning of the Arabidopsis Root through Radial Signaling. Cell 2000, 101, 555–567. [Google Scholar] [CrossRef] [Green Version]
- Dündar, E.; Bush, D.R. BAT1, a Bidirectional Amino Acid Transporter in Arabidopsis. Planta 2009, 229, 1047–1056. [Google Scholar] [CrossRef]
- Hanaoka, H.; Noda, T.; Shirano, Y.; Kato, T.; Hayashi, H.; Shibata, D.; Tabata, S.; Ohsumi, Y. Leaf Senescence and Starvation-Induced Chlorosis Are Accelerated by the Disruption of an Arabidopsis Autophagy Gene. Plant Physiol. 2002, 129, 1181–1193. [Google Scholar] [CrossRef] [Green Version]
- Wormit, A.; Usadel, B. The Multifaceted Role of Pectin Methylesterase Inhibitors (PMEIs). Int. J. Mol. Sci. 2018, 19, 2878. [Google Scholar] [CrossRef] [Green Version]
- Lakehal, A.; Dob, A.; Rahneshan, Z.; Novák, O.; Escamez, S.; Alallaq, S.; Strnad, M.; Tuominen, H.; Bellini, C. Ethylene Response Factor 115 Integrates Jasmonate and Cytokinin Signaling Machineries to Repress Adventitious Rooting in Arabidopsis. New Phytol. 2020, 228, 1611–1626. [Google Scholar] [CrossRef]
- Schubert, H.L.; Blumenthal, R.M.; Cheng, X. Many Paths to Methyltransfer: A Chronicle of Convergence. Trends Biochem. Sci. 2003, 28, 329–335. [Google Scholar] [CrossRef] [Green Version]
- Fedotova, A.A.; Bonchuk, A.N.; Mogila, V.A.; Georgiev, P.G. C2H2 Zinc Finger Proteins: The Largest but Poorly Explored Family of Higher Eukaryotic Transcription Factors. Acta Nat. 2017, 9, 47–58. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Liu, Z.; Wang, Z.; Ru, L.; Gonzalez, N.; Baekelandt, A.; Pauwels, L.; Goossens, A.; Xu, R.; Zhu, Z.; et al. STERILE APETALA Modulates the Stability of a Repressor Protein Complex to Control Organ Size in Arabidopsis Thaliana. PLoS Genet. 2018, 14, e1007218. [Google Scholar] [CrossRef] [Green Version]
- Dinkins, R.; Pflipsen, C.; Thompson, A.; Collins, G.B. Ectopic Expression of an Arabidopsis Single Zinc Finger Gene in Tobacco Results in Dwarf Plants. Plant Cell Physiol. 2002, 43, 743–750. [Google Scholar] [CrossRef] [Green Version]
- de Silva, K.; Laska, B.; Brown, C.; Sederoff, H.W.; Khodakovskaya, M. Arabidopsis Thaliana Calcium-Dependent Lipid-Binding Protein (AtCLB): A Novel Repressor of Abiotic Stress Response. J. Exp. Bot. 2011, 62, 2679–2689. [Google Scholar] [CrossRef] [Green Version]
- Nagano, M.; Kakuta, C.; Fukao, Y.; Fujiwara, M.; Uchimiya, H.; Kawai-Yamada, M. Arabidopsis Bax Inhibitor-1 Interacts with Enzymes Related to Very-Long-Chain Fatty Acid Synthesis. J. Plant Res. 2019, 132, 131–143. [Google Scholar] [CrossRef]
- Kruijt, M.; Brandwagt, B.F.; de Wit, P.J.G.M. Rearrangements in the Cf-9 Disease Resistance Gene Cluster of Wild Tomato Have Resulted in Three Genes that Mediate Avr9 Responsiveness. Genetics 2004, 168, 1655–1663. [Google Scholar] [CrossRef] [Green Version]
- Patil, R.V.; Pawar, K.D. Comparative de Novo Flower Transcriptome Analysis of Polygamodioecious Tree Garcinia Indica. 3 Biotech 2019, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Abdullah-Zawawi, M.-R.; Ahmad-Nizammuddin, N.-F.; Govender, N.; Harun, S.; Mohd-Assaad, N.; Mohamed-Hussein, Z.-A. Comparative Genome-Wide Analysis of WRKY, MADS-Box and MYB Transcription Factor Families in Arabidopsis and Rice. Sci. Rep. 2021, 11, 19678. [Google Scholar] [CrossRef] [PubMed]
- Ellinger, D.; Stingl, N.; Kubigsteltig, I.I.; Bals, T.; Juenger, M.; Pollmann, S.; Berger, S.; Schuenemann, D.; Mueller, M.J. Dongle and Defective in Anther Dehiscence1 Lipases Are Not Essential for Wound- and Pathogen-Induced Jasmonate Biosynthesis: Redundant Lipases Contribute to Jasmonate Formation. Plant Physiol. 2010, 153, 114–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boij, P.; Patel, R.; Garcia, C.; Jarvis, P.; Aronsson, H. In Vivo Studies on the Roles of Tic55-Related Proteins in Chloroplast Protein Import in Arabidopsis Thaliana. Mol. Plant 2009, 2, 1397–1409. [Google Scholar] [CrossRef] [PubMed]
- Okuda, K.; Hammani, K.; Tanz, S.K.; Peng, L.; Fukao, Y.; Myouga, F.; Motohashi, R.; Shinozaki, K.; Small, I.; Shikanai, T. The Pentatricopeptide Repeat Protein OTP82 Is Required for RNA Editing of Plastid NdhB and NdhG Transcripts. Plant J. 2010, 61, 339–349. [Google Scholar] [CrossRef]
- Kirik, V.; Kölle, K.; Wohlfarth, T.; Miséra, S.; Bäumlein, H. Ectopic Expression of a Novel MYB Gene Modifies the Architecture of the Arabidopsis Inflorescence. Plant J. 1998, 13, 729–742. [Google Scholar] [CrossRef]
- Levy, M.; Wang, Q.; Kaspi, R.; Parrella, M.P.; Abel, S. Arabidopsis IQD1, a Novel Calmodulin-Binding Nuclear Protein, Stimulates Glucosinolate Accumulation and Plant Defense. Plant J. 2005, 43, 79–96. [Google Scholar] [CrossRef]
- Bürstenbinder, K.; Savchenko, T.; Müller, J.; Adamson, A.W.; Stamm, G.; Kwong, R.; Zipp, B.J.; Dinesh, D.C.; Abel, S. Arabidopsis Calmodulin-Binding Protein IQ67-Domain 1 Localizes to Microtubules and Interacts with Kinesin Light Chain-Related Protein-1. J. Biol. Chem. 2013, 288, 1871–1882. [Google Scholar] [CrossRef] [Green Version]
- Basu, D.; Tian, L.; Wang, W.; Bobbs, S.; Herock, H.; Travers, A.; Showalter, A.M. A Small Multigene Hydroxyproline-O-Galactosyltransferase Family Functions in Arabinogalactan-Protein Glycosylation, Growth and Development in Arabidopsis. BMC Plant Biol. 2015, 15, 295. [Google Scholar] [CrossRef] [Green Version]
- Murgia, I.; Tarantino, D.; Soave, C.; Morandini, P. Arabidopsis CYP82C4 Expression Is Dependent on Fe Availability and Circadian Rhythm, and Correlates with Genes Involved in the Early Fe Deficiency Response. J. Plant Physiol. 2011, 168, 894–902. [Google Scholar] [CrossRef]
- Léon-Kloosterziel, K.M.; Verhagen, B.W.M.; Keurentjes, J.J.B.; VanPelt, J.A.; Rep, M.; VanLoon, L.C.; Pieterse, C.M.J. Colonization of the Arabidopsis Rhizosphere by Fluorescent Pseudomonas Spp. Activates a Root-Specific, Ethylene-Responsive PR-5 Gene in the Vascular Bundle. Plant Mol. Biol. 2005, 57, 731–748. [Google Scholar] [CrossRef] [Green Version]
- Che, P.; Lall, S.; Nettleton, D.; Howell, S.H. Gene Expression Programs during Shoot, Root, and Callus Development in Arabidopsis Tissue Culture. Plant Physiol. 2006, 141, 620–637. [Google Scholar] [CrossRef] [Green Version]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [Green Version]
- Diaz, M.; Sanchez-Barrena, M.J.; Gonzalez-Rubio, J.M.; Rodriguez, L.; Fernandez, D.; Antoni, R.; Yunta, C.; Belda-Palazon, B.; Gonzalez-Guzman, M.; Peirats-Llobet, M.; et al. Calcium-Dependent Oligomerization of CAR Proteins at Cell Membrane Modulates ABA Signaling. Proc. Natl. Acad. Sci. USA 2016, 113, E396–E405. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; Bhatt, D. The Circadian Clock and Defence Signalling in Plants. Mol. Plant Pathol. 2015, 16, 210–218. [Google Scholar] [CrossRef]
- Roden, L.C.; Ingle, R.A. Lights, Rhythms, Infection: The Role of Light and the Circadian Clock in Determining the Outcome of Plant-Pathogen Interactions. Plant Cell 2009, 21, 2546–2552. [Google Scholar] [CrossRef] [Green Version]
- Tauzin, A.S.; Giardina, T. Sucrose and Invertases, a Part of the Plant Defense Response to the Biotic Stresses. Front. Plant Sci. 2014, 5, 293. [Google Scholar] [CrossRef]
- Gutierrez, C.K.; Matsui, G.Y.; Lincoln, D.E.; Lovell, C.R. Production of the Phytohormone Indole-3-Acetic Acid by Estuarine Species of the Genus Vibrio. Appl. Environ. Microbiol. 2009, 75, 2253–2258. [Google Scholar] [CrossRef] [Green Version]
- Krueger, R.R.; Vidalakis, G. Study and Detection of Citrus Viroids in Woody Hosts. Methods Mol. Biol. 2022, 2316, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Vidalakis, G.; Gumpf, D.J.; Bash, J.A.; Semancik, J.S. Finger Imprint of Poncirus Trifoliata: A Specific Interaction of a Viroid, a Host, and Irrigation. Plant Dis. 2004, 88, 709–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vernière, C.; Perrier, X.; Dubois, C.; Dubois, A.; Botella, L.; Chabrier, C.; Bové, J.M.; Vila, N.D. Interactions between Citrus Viroids Affect Symptom Expression and Field Performance of Clementine Trees Grafted on Trifoliate Orange. Phytopathology 2006, 96, 356–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murcia, N.; Bernad, L.; Serra, P.; Hashemian, S.M.B.; Duran-Vila, N. Molecular and Biological Characterization of Natural Variants of Citrus Dwarfing Viroid. Arch. Virol. 2009, 154, 1329–1334. [Google Scholar] [CrossRef]


| Treatment | Avg. Total Reads | Avg. Uniquely Mapped Reads | Avg. Percent Mapped Reads | Avg. Mapped Length |
|---|---|---|---|---|
| Non-infected stem | 57,720,333 ± 7,556,534 | 40,055,889 ± 10,591,488 | 71.63% | 199.04 ± 0.25 |
| CDVd-infected stem | 36,254,830 ± 15,692,505 | 31,769,508 ± 13,584,944 | 87.87% | 199.51 ± 0.32 |
| Non-infected root | 58,030,198 ± 9,821,953 | 48,928,990 ± 10,744,466 | 83.95% | 199.8 ± 0.2 |
| CDVd-infected roots | 25,671,752 ± 1,148,290 | 22,553,155 ± 1,891,667 | 87.78% | 199.98 ± 0.09 |
| Tissue Type | Upregulated | Downregulated | Total |
|---|---|---|---|
| Stem | 83 | 92 | 175 |
| Root | 48 | 186 | 234 |
| Accession | Product | Fold Change (FC) | LogFC | Log Count Per Million (CPM) | p-Value | FDR | |
|---|---|---|---|---|---|---|---|
| Upregulated stem | LOC102607140 | probable 28S rRNA (cytosine-C(5))-methyltransferase | 8.83 | 3.14 | 11.04 | 2.70 × 10−15 | 5.05 × 10−11 |
| LOC102611719 | germin-like protein subfamily T member | 13.66 | 3.77 | 7.41 | 8.39 × 10−14 | 7.84 × 10−10 | |
| LOC102610118 | pentatricopeptide repeat-containing protein At2g17525 | 14.72 | 3.88 | 6.52 | 1.73 × 10−11 | 1.08 × 10−7 | |
| LOC102616051 | cytosolic sulfotransferase 15-like | 17.47 | 4.13 | 6.04 | 1.62 × 10−10 | 4.29 × 10−7 | |
| LOC102614284 | aspartic proteinase Asp1-like | 5.67 | 2.50 | 9.39 | 5.71 × 10−10 | 9.70 × 10−7 | |
| Downregulated stem | LOC102627502 | pentatricopeptide repeat-containing protein At4g21300 | −7.70 | −2.95 | 7.86 | 1.84 × 10−10 | 4.29 × 10−7 |
| LOC102620512 | SKP1-like protein 21 | −7.01 | −2.81 | 8.07 | 3.56 × 10−10 | 7.40 × 10−7 | |
| LOC102610900 | protein SHORT-ROOT-like | −6.90 | −2.79 | 8.06 | 4.41 × 10−10 | 8.24 × 10−7 | |
| LOC102620992 | amino-acid permease BAT1-like | −12.78 | −3.68 | 6.34 | 1.99 × 10−9 | 3.11 × 10−6 | |
| LOC102628393 | serine/threonine-protein kinase ATG1a | −16.21 | −4.02 | 6.03 | 2.69 × 10−9 | 3.38 × 10−6 |
| Gene ID | Alt Gene ID | Product | Fold Change (FC) | LogFC | Log Count Per Million (CPM) | p-Value | FDR | |
|---|---|---|---|---|---|---|---|---|
| Upregulated roots | Ptrif.0009s1914.1 | AT5G62350/LOC_Os06g49760 | Plant invertase/pectin methylesterase inhibitor superfamily protein | 15.33 | 3.94 | 7.00 | 3.62 × 10−7 | 2.48 × 10−4 |
| Ptrif.0004s1154.1 | AT5G07310/Cre14.g620500/LOC_Os04g32620 | Integrase-type DNA-binding superfamily protein | 6.54 | 2.71 | 12.63 | 4.04 × 10−7 | 2.64 × 10−4 | |
| Ptrif.0005s1389.1 | Cre05.g240400/LOC_Os12g25450 | S-adenosyl-L-methionine-dependent O-methyltransferase/ethylene-responsive transcription factor ERF114 | 8.82 | 3.14 | 7.80 | 1.02 × 10−6 | 5.05 × 10−4 | |
| Ptrif.0005s2465.1 | AT5G52010/LOC_Os06g07020 | C2H2-like zinc finger protein | 7.38 | 2.88 | 8.03 | 2.69 × 10−6 | 1.09 × 10−3 | |
| Ptrif.0006s1595.1 | AT5G03800 | Pentatricopeptide repeat (PPR) superfamily protein | 7.43 | 2.89 | 8.06 | 2.73 × 10−6 | 1.09 × 10−3 | |
| Downregulated roots | Ptrif.0001s0240.1 | AT4G32295 | Kinase-inducible domain interacting 9, Kix9 | −49.72 | −5.64 | 9.20 | 6.04 × 10−14 | 6.75 × 10−10 |
| Ptrif.0003s0980.2 | LOC_Os04g48160 | IQ calmodulin-binding motif family protein, putative, expressed | −13.83 | −3.79 | 10.36 | 1.96 × 10−10 | 7.31 × 10−7 | |
| Ptrif.0006s0692.1 | AT2G37740/LOC_Os05g20930 | zinc-finger protein 10 | −30.39 | −4.93 | 7.76 | 7.47 × 10−9 | 1.85 × 10−05 | |
| Ptrif.0003s4060.1 | AT4G01200 | Calcium-dependent lipid-binding (CaLB domain) family protein | −164.81 | −7.36 | 7.11 | 1.13 × 10−8 | 2.53 × 10−5 | |
| Ptrif.0005s2586.1 | AT1G75000/LOC_Os12g43890 | GNS1/SUR4 membrane protein family | −11.31 | −3.50 | 9.35 | 1.27 × 10−8 | 2.58 × 10−5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lavagi-Craddock, I.; Dang, T.; Comstock, S.; Osman, F.; Bodaghi, S.; Vidalakis, G. Transcriptome Analysis of Citrus Dwarfing Viroid Induced Dwarfing Phenotype of Sweet Orange on Trifoliate Orange Rootstock. Microorganisms 2022, 10, 1144. https://doi.org/10.3390/microorganisms10061144
Lavagi-Craddock I, Dang T, Comstock S, Osman F, Bodaghi S, Vidalakis G. Transcriptome Analysis of Citrus Dwarfing Viroid Induced Dwarfing Phenotype of Sweet Orange on Trifoliate Orange Rootstock. Microorganisms. 2022; 10(6):1144. https://doi.org/10.3390/microorganisms10061144
Chicago/Turabian StyleLavagi-Craddock, Irene, Tyler Dang, Stacey Comstock, Fatima Osman, Sohrab Bodaghi, and Georgios Vidalakis. 2022. "Transcriptome Analysis of Citrus Dwarfing Viroid Induced Dwarfing Phenotype of Sweet Orange on Trifoliate Orange Rootstock" Microorganisms 10, no. 6: 1144. https://doi.org/10.3390/microorganisms10061144
APA StyleLavagi-Craddock, I., Dang, T., Comstock, S., Osman, F., Bodaghi, S., & Vidalakis, G. (2022). Transcriptome Analysis of Citrus Dwarfing Viroid Induced Dwarfing Phenotype of Sweet Orange on Trifoliate Orange Rootstock. Microorganisms, 10(6), 1144. https://doi.org/10.3390/microorganisms10061144






