Pneumocystis spp. in Pigs: A Longitudinal Quantitative Study and Co-Infection Assessment in Austrian Farms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Nucleic Acid Isolation
2.2. Quantitative Detection of Pulmonary Pathogens by Real Time PCR
2.3. G. parasuis Serotyping and SIV Subtyping
2.4. Assessment of Clinical Symptoms
2.5. Statistical Analysis
3. Results
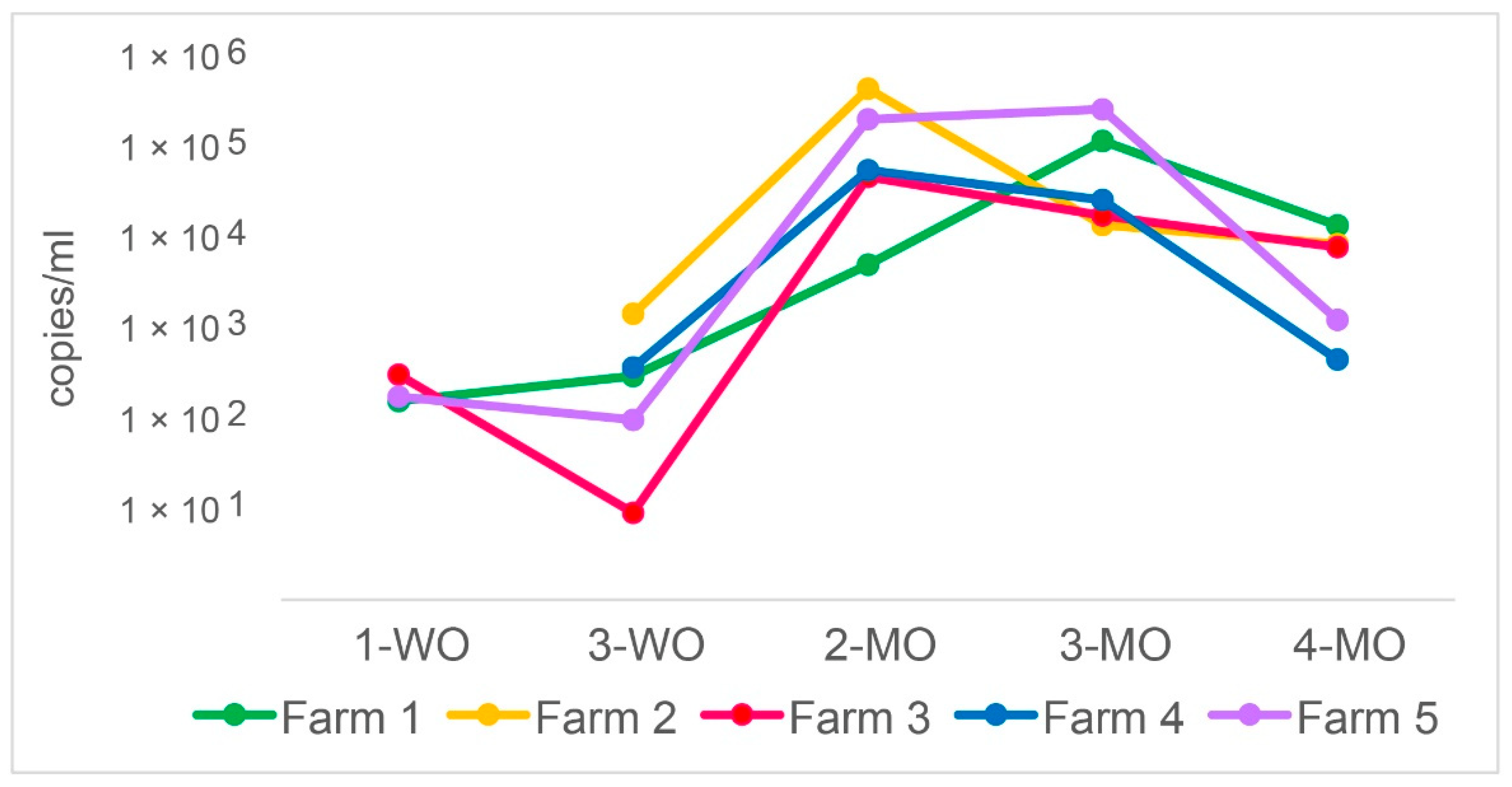
3.1. P. suis Load Varies over Time
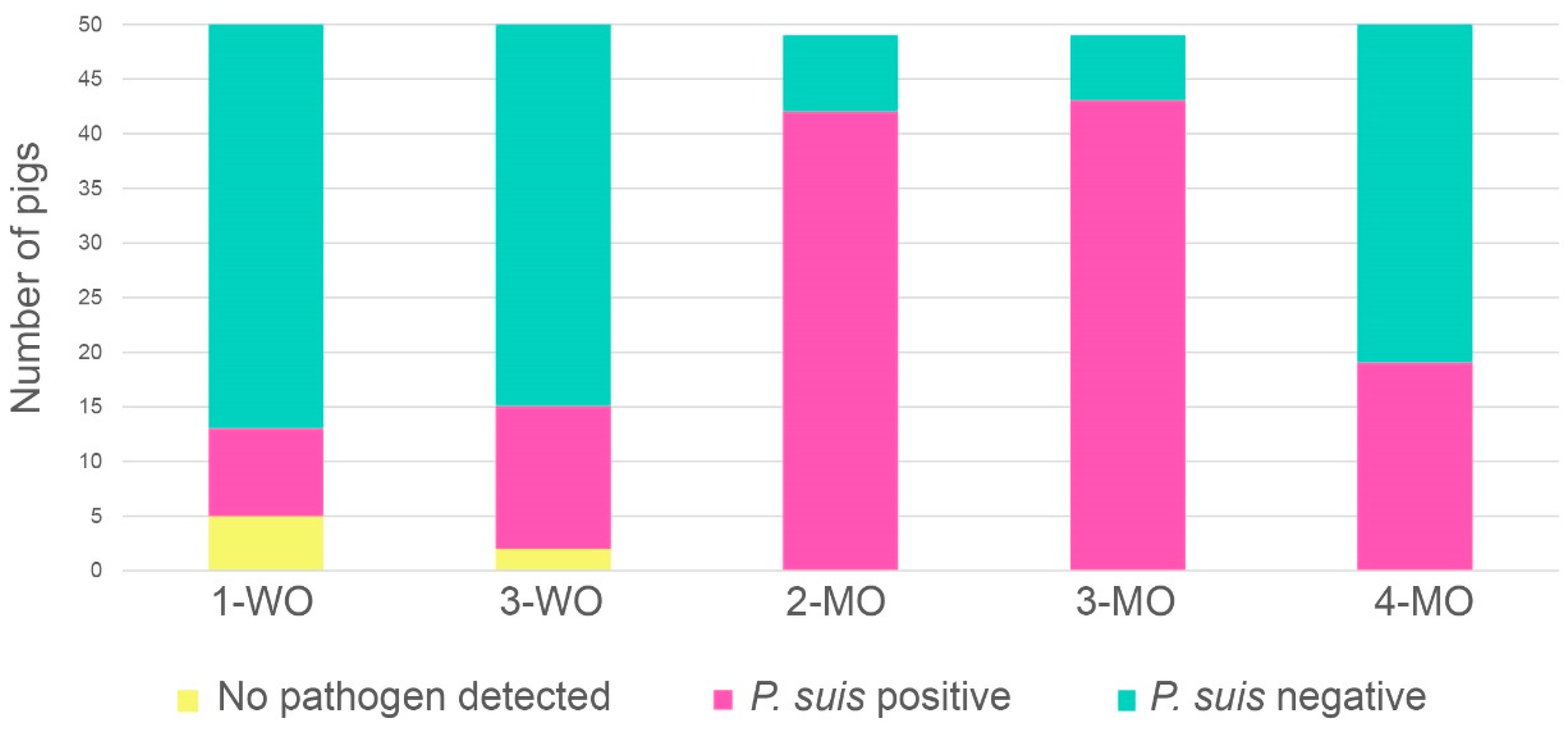
3.2. P. suis Prevalence Increases with Time
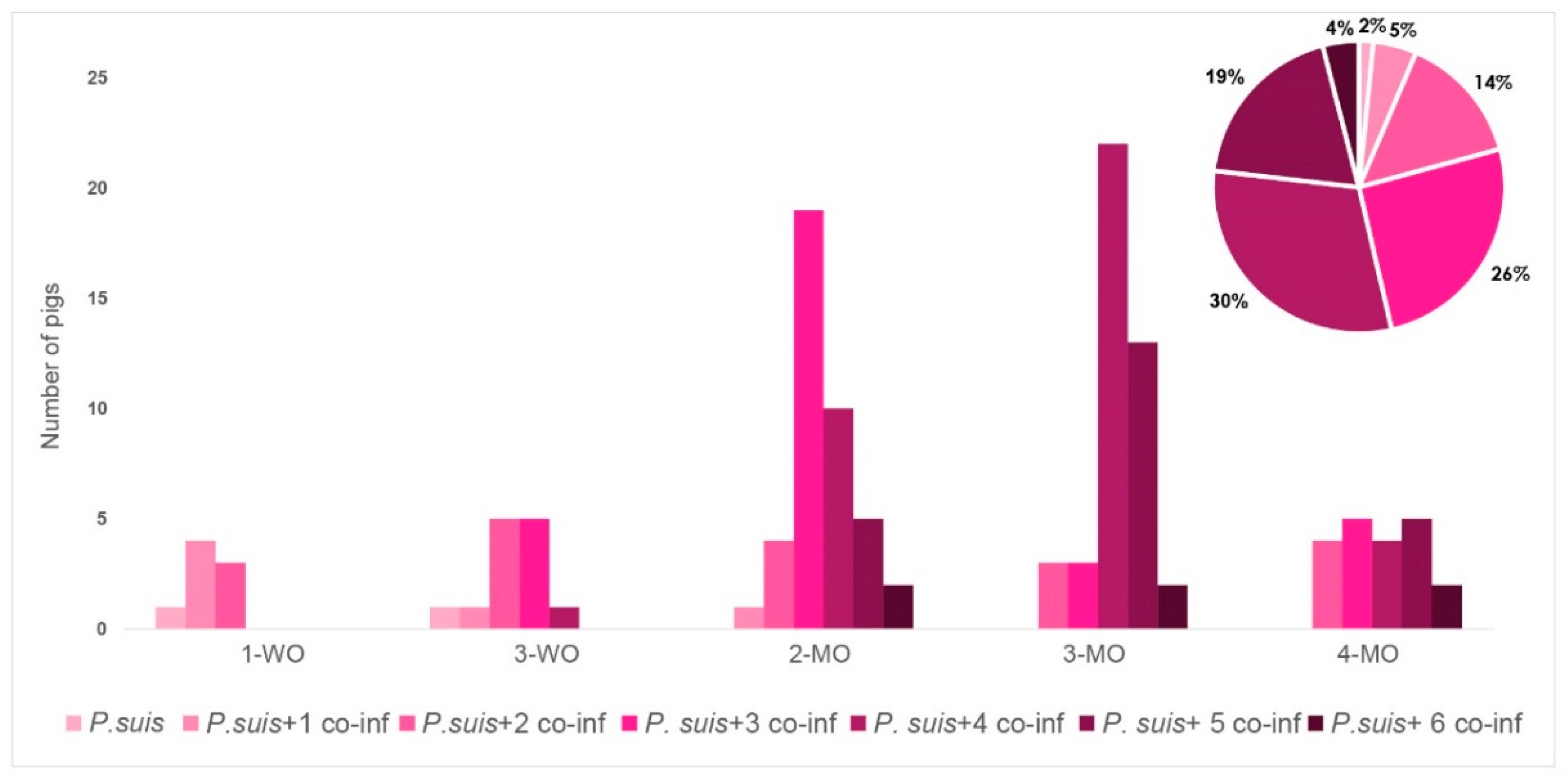
3.3. Co-Infection Patterns Are Highly Variable
3.4. P. suis Co-Infection Scenario Varies over Time
3.5. P. suis-Specific Clinical Signs Could Not Be Detected
3.6. G. parasuis Serotyping and SIV Subtyping
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chagas:, C. Nova tripanosomiae humanao. Mem. Inst. Oswaldo. Cruz. 1909, 1, 159–218. [Google Scholar]
- Delanoë, P.; Delanoë, M. Sur les rapports de kystes de Carini du poumon des rats avec le Trypanosoma lewisi. CR Hebd. Seances Acad. Sci. 1912, 155, 658–659. [Google Scholar]
- Wakefield, A.E.; Stringer, J.R.; Tamburrini, E.; Dei-Cas, E. Genetics, metabolism and host specificity of Pneumocystis carinii. Med. Mycol. 1998, 36 (Suppl. S1), 183–193. [Google Scholar] [PubMed]
- Cissé, O.H.; Ma, L.; Wei Huang, D.; Khil, P.P.; Dekker, J.P.; Kutty, G.; Bishop, L.; Liu, Y.; Deng, X.; Hauser, P.M.; et al. Comparative population genomics analysis of the mammalian fungal pathogen Pneumocystis. mBio 2018, 9, e00381-18. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, Y.; Yamada, M.; Tegoshi, T.; Yoshida, Y.; Gotoh, S.; Suzuki, J.; Matsubayashi, K. Pneumocystis infection in macaque monkeys: Macaca fuscata fuscata and Macaca fascicularis. Parasitol. Res. 1987, 73, 324–327. [Google Scholar] [CrossRef] [PubMed]
- Akbar, H.; Pinçon, C.; Aliouat-Denis, C.-M.; Derouiche, S.; Taylor, M.-L.; Pottier, M.; Carreto-Binaghi, L.-H.; González-González, A.E.; Courpon, A.; Barriel, V.; et al. Characterizing Pneumocystis in the lungs of bats: Understanding Pneumocystis evolution and the spread of Pneumocystis organisms in mammal populations. Appl. Environ. Microbiol. 2012, 78, 8122. [Google Scholar] [CrossRef] [Green Version]
- Aliouat-Denis, C.M.; Chabé, M.; Demanche, C.; Aliouat, E.M.; Viscogliosi, E.; Guillot, J.; Delhaes, L.; Dei-Cas, E. Pneumocystis species, co-evolution and pathogenic power. Infect. Genet. Evol. 2008, 8, 708–726. [Google Scholar] [CrossRef]
- Sanches, E.M.C.; Borba, M.R.; Spanamberg, A.; Pescador, C.; Corbellini, L.G.; Ravazzolo, A.P.; Driemeier, D.; Ferreiro, L. Co-infection of Pneumocystis carinii f. sp. suis and porcine circovirus-2 (PCV2) in pig lungs obtained from slaughterhouses in southern and midwestern regions of Brazil. J. Eukaryot. Microbiol. 2006, 53, S92–S94. [Google Scholar] [CrossRef]
- Bondy, R. Pneumocystis carinii in human and veterinary medicine. Sborn. Ces. Akad. Zemed. Ved. 1958, 3, 199–210. [Google Scholar]
- Kucera, K.; Slesingr, L.K.A. Pneumocystosis in pigs. Folia Parasitol 1968, 15, 75–78. [Google Scholar]
- Nikolskij, S.N.; Shchetinin, A.N. Pnevmotsistoz u svineǐ. Veterinariia 1967, 44, 65. [Google Scholar]
- Binanti, D.; Mostegl, M.M.; Weissenbacher-Lang, C.; Nedorost, N.; Weissenböck, H. Detection of Pneumocystis infections by in situ hybridization in lung samples of Austrian pigs with interstitial pneumonia. J. Music Ther. 2015, 52, 196–201. [Google Scholar] [CrossRef] [Green Version]
- Esgalhado, R.; Esteves, F.; Antunes, F.; Matos, O. Study of the epidemiology of Pneumocystis carinii f. sp. suis in abattoir swine in Portugal. Med. Mycol. 2013, 51, 66–71. [Google Scholar] [CrossRef] [Green Version]
- Weissenbacher-Lang, C.; Kureljušić, B.; Nedorost, N.; Matula, B.; Schießl, W.; Stixenberger, D.; Weissenböck, H. Retrospective analysis of bacterial and viral co-infections in Pneumocystis spp. positive lung samples of Austrian pigs with pneumonia. PLoS ONE 2016, 11, e0158479. [Google Scholar] [CrossRef] [PubMed]
- Sanches, E.M.C.; Pescador, C.; Rozza, D.; Spanamberg, A.; Borba, M.R.; Ravazzolo, A.P.; Driemeier, D.; Guillot, J.; Ferreiro, L. Detection of Pneumocystis spp. in lung samples from pigs in Brazil. Med. Mycol. 2007, 45, 395–399. [Google Scholar] [CrossRef] [Green Version]
- Kondo, H.; Hikita, M.; Ito, M.; Kadota, K. Immunohistochemical study of Pneumocystis carinii infection in pigs: Evaluation of Pneumocystis pneumonia and a retrospective investigation. Vet. Rec. 2000, 147, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Settnes, O.P.; Henriksen, S.A. Pneumocystis carinii in large domestic animals in Denmark. A preliminary report. Acta Vet. Scand. 1989, 30, 437–440. [Google Scholar] [CrossRef]
- Ramos Vara, J.A.; Lu, J.-J.; Da Silva, A.J.; Montone, K.T.; Pieniazek, N.J.; Lee, C.-H.; Perez, L.; Steficek, B.A.; Dunstanl, R.W.; Craft, D.; et al. Histology and histopathology from cell biology to tissue engineering characterization of natural occurring Pneumocystis carinii pneumonia in pigs by histopathology, electron microscopy, in situ hybridization and PCR amplification. Histol. Histopathol. 1998, 13, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Jensen, T.K.; Boye, M.; Bille-Hansen, V. Application of fluorescent in situ hybridization for specific diagnosis of Pneumocystis carinii pneumonia in foals and pigs. Vet. Pathol. 2001, 38, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Furuta, T.; Nakajima, T.; Kurita, F.; Kaneuchi, C.; Ueda, K.; Ogata, M. Prevalence of Pneumocystis carinii in slaughtered pigs. Nippon juigaku zasshi Jpn. J. Vet. Sci. 1989, 51, 200–202. [Google Scholar] [CrossRef]
- Kureljušić, B.; Weissenbacher-Lang, C.; Nedorost, N.; Stixenberger, D.; Weissenböck, H. Association between Pneumocystis spp. and co-infections with Bordetella bronchiseptica, Mycoplasma hyopneumoniae and Pasteurella multocida in Austrian pigs with pneumonia. Vet. J. 2016, 207, 177–179. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.F.; Huang, L.; Walzer, P.D. The relationship between Pneumocystis infection in animal and human hosts, and climatological and environmental air pollution factors: A systematic review. OBM Genet. 2018, 2. [Google Scholar] [CrossRef] [PubMed]
- Brockmeier, S.L.; Halbur, P.G.; Thacker, E.L. Porcine respiratory disease complex. In Polymicrobial Diseases; Brogden, K.A., Guthmiller, J.M., Eds.; ASM Press: Washington, DC, USA, 2002. [Google Scholar]
- Holt, H.R.; Alarcon, P.; Velasova, M.; Pfeiffer, D.U.; Wieland, B. BPEX Pig Health Scheme: A useful monitoring system for respiratory disease control in pig farms? BMC Vet. Res. 2011, 7, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holtkamp, D.J.; Kliebenstein, J.B.; Neumann, E.J.; Zimmerman, J.J.; Rotto, H.F.; Yoder, T.K.; Wang, C.; Yeske, P.E.; Mowrer, C.L.; Haley, C.A. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J. Swine Heal. Prod. 2013, 21, 72–84. [Google Scholar]
- Kim, K.S.; Jung, J.Y.; Kim, J.H.; Kang, S.C.; Hwang, E.K.; Park, B.K.; Kim, D.Y.; Kim, J.H. Epidemiological characteristics of pulmonary pneumocystosis and concurrent infections in pigs in Jeju Island, Korea. J. Vet. Sci. 2011, 12, 15–19. [Google Scholar] [CrossRef]
- Sanches, E.M.; Ferreiro, L.; Borba, M.; Spanamberg, A.; Ravazzolo, A.; Santurio, J.; Driemeir, D.; Barcellos, D.E.S.; Berthelemy, M.; Guillot, J. Phylogenetic analysis of Pneumocystis from pig lungs obtained from slaughterhouses in southern and midwestern regions of Brazil. Arq. Bras. Med. Veterinária Zootec. 2011, 63, 1154–1159. [Google Scholar] [CrossRef]
- Le, T.M.; Le, Q.V.C.; Truong, D.M.; Lee, H.J.; Choi, M.K.; Cho, H.; Chung, H.J.; Kim, J.H.; Do, J.T.; Song, H.; et al. β2-microglobulin gene duplication in cetartiodactyla remains intact only in pigs and possibly confers selective advantage to the species. PLoS ONE 2017, 12, e182322. [Google Scholar] [CrossRef] [Green Version]
- Weissenbacher-Lang, C.; Nedorost, N.; Knecht, C.; Hennig-Pauka, I.; Huber, M.; Voglmayr, T.; Weissenböck, H. Comparison of Pneumocystis nucleic acid and antibody profiles and their associations with other respiratory pathogens in two Austrian pig herds. PLoS ONE 2017, 12, e0185387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wernike, K.; Hoffmann, B.; Dauber, M.; Lange, E.; Schirrmeier, H.; Beer, M. Detection and typing of highly pathogenic porcine reproductive and respiratory syndrome virus by multiplex real-time rt-PCR. PLoS ONE 2012, 7, e38251. [Google Scholar] [CrossRef] [Green Version]
- Bonin, E.; Quéguiner, S.; Woudstra, C.; Gorin, S.; Barbier, N.; Harder, T.C.; Fach, P.; Hervé, S.; Simon, G. Molecular subtyping of European swine influenza viruses and scaling to high-throughput analysis. Virol. J. 2018, 15, 7. [Google Scholar] [CrossRef]
- Turni, C.; Pyke, M.; Blackall, P.J. Validation of a real-time PCR for Haemophilus parasuis. J. Appl. Microbiol. 2010, 108, 1323–1331. [Google Scholar] [CrossRef]
- Fourour, S.; Fablet, C.; Tocqueville, V.; Dorenlor, V.; Eono, F.; Eveno, E.; Kempf, I.; Marois-Créhan, C. A new multiplex real-time TaqMan® PCR for quantification of Mycoplasma hyopneumoniae, M. hyorhinis and M. flocculare: Exploratory epidemiological investigations to research mycoplasmal association in enzootic pneumonia-like lesions in slaughtered pigs. J. Appl. Microbiol. 2018, 125, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Tocqueville, V.; Kempf, I.; Paboeuf, F.; Marois-Créhan, C. Quantification of Pasteurella multocida in experimentally infected pigs using a real-time PCR assay. Res. Vet. Sci. 2017, 112, 177–184. [Google Scholar] [CrossRef]
- Kielstein, P.; Rapp-Gabrielson, V. Designation of 15 serovars of Haemophilus parasuis on the basis of immunodiffusion using heat-stable antigen extracts. J. Clin. Microbiol. 1992, 30, 862–865. [Google Scholar] [CrossRef] [Green Version]
- Schuwerk, L.; Hoeltig, D.; Waldmann, K.-H.; Strutzberg-Minder, K.; Valentin-Weigand, P.; Rohde, J. Serotyping and pathotyping of Glaesserella parasuis isolated 2012–2019 in Germany comparing different PCR-based methods. Vet. Res. 2020, 51, 1–14. [Google Scholar] [CrossRef]
- Chiapponi, C.; Moreno, A.; Barbieri, I.; Merenda, M.; Foni, E. Multiplex RT-PCR assay for differentiating European swine influenza virus subtypes H1N1, H1N2 and H3N2. J. Virol. Methods 2012, 184, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Henritzi, D.; Zhao, N.; Starick, E.; Simon, G.; Krog, J.S.; Larsen, L.E.; Reid, S.M.; Brown, I.H.; Chiapponi, C.; Foni, E.; et al. Rapid detection and subtyping of European swine influenza viruses in porcine clinical samples by haemagglutinin- and neuraminidase-specific tetra- and triplex real-time RT-PCRs. Influenza Other Respi. Viruses 2016, 10, 504–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, M. The use of ranks to avoid the assumption of normality implicit in the analysis of variance. J. Am. Stat. Assoc. 1937, 32, 675–701. [Google Scholar] [CrossRef]
- Garvy, B.A.; Harmsen, A.G. Susceptibility to Pneumocystis carinii infection: Host responses of neonatal mice from immune or naive mothers and of immune or naive adults. Infect. Immun. 1996, 64, 3987–3992. [Google Scholar] [CrossRef] [Green Version]
- Icenhour, C.R.; Rebholz, S.L.; Collins, M.S.; Cushion, M.T. Early acquisition of Pneumocystis carinii in neonatal rats as evidenced by PCR and oral swabs. Eukaryot. Cell 2002, 1, 414–419. [Google Scholar] [CrossRef] [Green Version]
- Tamburrini, E.; Ortona, E.; Visconti, E.; Mencarini, P.; Margutti, P.; Zolfo, M.; Barca, S.; Peters, S.E.; Wakefield, A.E.; Siracusano, A. Pneumocystis carinii infection in young non-immunosuppressed rabbits. Kinetics of infection and of the primary specific immune response. Med. Microbiol. Immunol. 1999, 188, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Vargas, S.L.; Ponce, C.A.; Hughes, W.T.; Wakefield, A.E.; Weitz, J.C.; Donoso, S.; Ulloa, A.V.; Madrid, P.; Gould, S.; Latorre, J.J.; et al. Association of primary Pneumocystis carinii infection and sudden infant death syndrome. Clin. Infect. Dis. 1999, 29, 1489–1493. [Google Scholar] [CrossRef] [PubMed]
- Sipos, W.; Duvigneau, C.; Pietschmann, P.; Höller, K.; Hartl, R.; Wahl, K.; Steinborn, R.; Gemeiner, M.; Willheim, M.; Schmoll, F. Parameters of humoral and cellular immunity following vaccination of pigs with a European modified-live strain of porcine reproductive and respiratory syndrome virus (PRRSV). Viral Immunol. 2003, 16, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Cushion, M.T.; Tisdale-Macioce, N.; Sayson, S.G.; Porollo, A. The Persistent Challenge of Pneumocystis Growth Outside the Mammalian Lung: Past and Future Approaches. Front. Microbiol. 2021, 12, 1120. [Google Scholar] [CrossRef] [PubMed]
- Cushion, M.T.; Smulian, A.G.; Slaven, B.E.; Sesterhenn, T.; Arnold, J.; Staben, C.; Porollo, A.; Adamczak, R.; Meller, J. Transcriptome of Pneumocystis carinii during fulminate infection: Carbohydrate metabolism and the concept ofa compatible parasite. PLoS ONE 2007, 2, e423. [Google Scholar] [CrossRef] [Green Version]
- Cissé, O.H.; Pagni, M.; Hauser, P.M. Comparative genomics suggests that the human pathogenic fungus Pneumocystis jirovecii acquired obligate biotrophy through gene loss. Genome Biol. Evol. 2014, 6, 1938–1948. [Google Scholar] [CrossRef] [Green Version]
- Porollo, A.; Sesterhenn, T.M.; Collins, M.S.; Welge, J.A.; Cushion, M.T. Comparative genomics of Pneumocystis species suggests the absence of genes for myo-inositol synthesis and reliance on inositol transport and metabolism. mBio 2014, 5, e01834-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauser, P.M. Genomic insights into the fungal pathogens of the genus Pneumocystis: Obligate biotrophs of humans and other mammals. PLoS Pathog 2014, 10, e1004425. [Google Scholar] [CrossRef] [Green Version]
- Opriessnig, T.; Giménez-Lirola, L.G.; Halbur, P.G. Polymicrobial respiratory disease in pigs. Anim. Heal. Res. Rev. 2011, 12, 133–148. [Google Scholar] [CrossRef]
- Sipos, W.; Dobrokes, B.; Meppiel, L.; Sailer, J.; Friedmann, U.; Cvjetković, V. Association of lung score findings from slaughter pigs with their vaccination status against Mycoplasma hyopneumoniae and PCV2. Berl. Munch. Tierarztl. Wochenschr. 2020, 133, 1–5. [Google Scholar] [CrossRef]
- Evans, H.M.; Bryant, G.L.; Garvy, B.A. The life cycle stages of Pneumocystis murina have opposing effects on the immune response to this opportunistic fungal pathogen. Infect. Immun. 2016, 84, 3195–3205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Opriessnig, T.; Fenaux, M.; Thomas, P.; Hoogland, M.; Rothschild, M.; Meng, X.; Halbur, P. Evidence of breed-dependent differences in susceptibility to porcine circovirus type-2-associated disease and lesions. Vet. Pathol. 2006, 43, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Halbur, P.G.; Rothschild, M.F.; Thacker, B.J.; Meng, X.J.; Paul, P.S.; Bruna, J.D. Differences in susceptibility of Duroc, Hampshire, and Meishan pigs to infection with a high virulence strain (VR2385) of porcine reproductive and respiratory syndrome virus (PRRSV). J. Anim. Breed. Genet. 1998, 115, 181–189. [Google Scholar] [CrossRef]
- Sipos, W.; Schmoll, F.; Stumpf, I. Minipigs and potbellied pigs as pets in the veterinary practice--a retrospective study. J. Vet. Med. A Physiol. Pathol. Clin. Med. 2007, 54, 504–511. [Google Scholar] [CrossRef]
- Salak-Johnson, J.L.; Webb, S.R. Short- and long-term effects of weaning age on pig innate immune status. Open J. Anim. Sci. 2018, 8, 137–150. [Google Scholar] [CrossRef] [Green Version]
- Segalés, J.; Domingo, M.; Sega, J. Postweaning mulstisystemic wasting syndrome (PMWS) in pigs. A review. Vet. Q. 2002, 24, 109–124. [Google Scholar] [CrossRef]
- Sipos, W. Shifts in porcine PBMC populations from adolescence to adulthood. Vet. Immunol. Immunopathol. 2019, 211, 35–37. [Google Scholar] [CrossRef]

| Species | Gene | Forward primer 5′-3′ | Reverse Primer 5′-3′ | Probe 5′-3′ | Annealing Temperature | Reference |
|---|---|---|---|---|---|---|
| P. suis | sodA1 | GGCGAGTTAGCTGCAATTCAA | AGCACATTATAGGACCATTGTTGT | FAM-ACAAGCCAACACCACCCACTTCC-TAMRA | 55 °C | This study |
| sodA2 | GGCAGTGTAAATCAATTTAT | AAACGAGTCAAATAAATACA | FAM-AGGAAGTGGATGGTGTTGGC-TAMRA | 55 °C | ||
| PCV2 | capsid protein | GGTACTCCTCAACTGCTGTCC | GGGAAAGGGTGACGAACTGG | FAM-ACAGAACAATCCACGGAGGAAGGG-TAMRA | 60 °C | Weissenbacher-Lang 2017 [29] |
| PRRSV-EU | N (nucleocapsid protein) | GCACCACCTCACCCRRAC | CAGTTCCTGCRCCYTGAT | FAM-CCTCTGYYTGCAATCGATCCAGAC-BHQ1 | 55 °C | Wernike et al., 2012 [30]* This study |
| PRRSV-US | N (nucleocapsid protein) | ATRATGRGCTGGCATTC | ACACGGTCGCCCTAATTG | JOE-TGTGGTGAATGGCACTGATTGACA-BHQ1 * | 55 °C | |
| PRRSV-HP | pp1a | CCGCGTAGAACTGTGACAAC | TCCAGGATGCCCATGTTCTG | CY5-ACGCACCAGGATGAGCCTCTGGAT-TAMRA | 55 °C | |
| SIV | matrix protein M | AGATGAGTCYTCTAACCGAGGTCG | TGCAAARACAYYTTCMAGTCTCTG | FAM-TCAGGCCCCCTCAAAGCCGA-TAMRA | 60 °C | Bonin et al., 2018 [31] |
| A. pleuropneumoniae | apxIVA | GTGGTTTGGAAAGTATTATC | TTTAAACCTTGTTTCGTCTA | FAM-CGGTATCGGTGGAACGGTAA-TAMRA | 55 °C | This study |
| G. parasuis | infB | CGACTTACTTGAAGCCATTCTTCTT | CCGCTTGCCATACCCTCTT | FAM-GCACTTAATTCTAATACTTCCGAT-TAMRA | 55 °C | Turni et al., 2010 [32] |
| M. flocculare | fruA | TTAGCAGTTCCAATTTTATCAG | AAACCATAGGTATCTTTAAGTTG | FAM-CAATTCCGCCAACTACAAATCCAG-BHQ1 | 55 °C | Fourour et al., 2018 [33]* This study |
| M. hyopneumoniae | P102 | TAAGGGTCAAAGTCAAAGTC | AAATTAAAAGCTGTTCAAATGC | CY5-AACCAGTTTCCACTTCATCGCC-BHQ2 | 55 °C | |
| M. hyorhinis | P37 | TTCTATTTTCATCTATATTTTCGC | TCATTGACCTTGACTAACTG | CY3-CAGGAGTAGTCAAGCAAGAGGATG-BHQ2 * | 55 °C | |
| P. multocida | sodA | GGAAGCCTTCCAAGCAGAATTTG | CCGCAATAGCTTTACCCATTACAG | CY5a-CGGCAGCAACCCGTTTCGGTTCAG-BHQ2b | 60 °C | Tocqueville et al., 2017 [34] |
| S. suis | rsmG | TGAAGCATTTCTACGATTCT | GCGGAAAGATAATCTTCATA | FAM-TTGGAGCTGGAGCTGGATTC-TAMRA | 52 °C | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blasi, B.; Sipos, W.; Knecht, C.; Dürlinger, S.; Ma, L.; Cissé, O.H.; Nedorost, N.; Matt, J.; Weissenböck, H.; Weissenbacher-Lang, C. Pneumocystis spp. in Pigs: A Longitudinal Quantitative Study and Co-Infection Assessment in Austrian Farms. J. Fungi 2022, 8, 43. https://doi.org/10.3390/jof8010043
Blasi B, Sipos W, Knecht C, Dürlinger S, Ma L, Cissé OH, Nedorost N, Matt J, Weissenböck H, Weissenbacher-Lang C. Pneumocystis spp. in Pigs: A Longitudinal Quantitative Study and Co-Infection Assessment in Austrian Farms. Journal of Fungi. 2022; 8(1):43. https://doi.org/10.3390/jof8010043
Chicago/Turabian StyleBlasi, Barbara, Wolfgang Sipos, Christian Knecht, Sophie Dürlinger, Liang Ma, Ousmane H. Cissé, Nora Nedorost, Julia Matt, Herbert Weissenböck, and Christiane Weissenbacher-Lang. 2022. "Pneumocystis spp. in Pigs: A Longitudinal Quantitative Study and Co-Infection Assessment in Austrian Farms" Journal of Fungi 8, no. 1: 43. https://doi.org/10.3390/jof8010043







