Isolation of Pseudomonas Strains with Potential for Protection of Soybean Plants against Saline Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soil Sampling and Bacterial Isolation
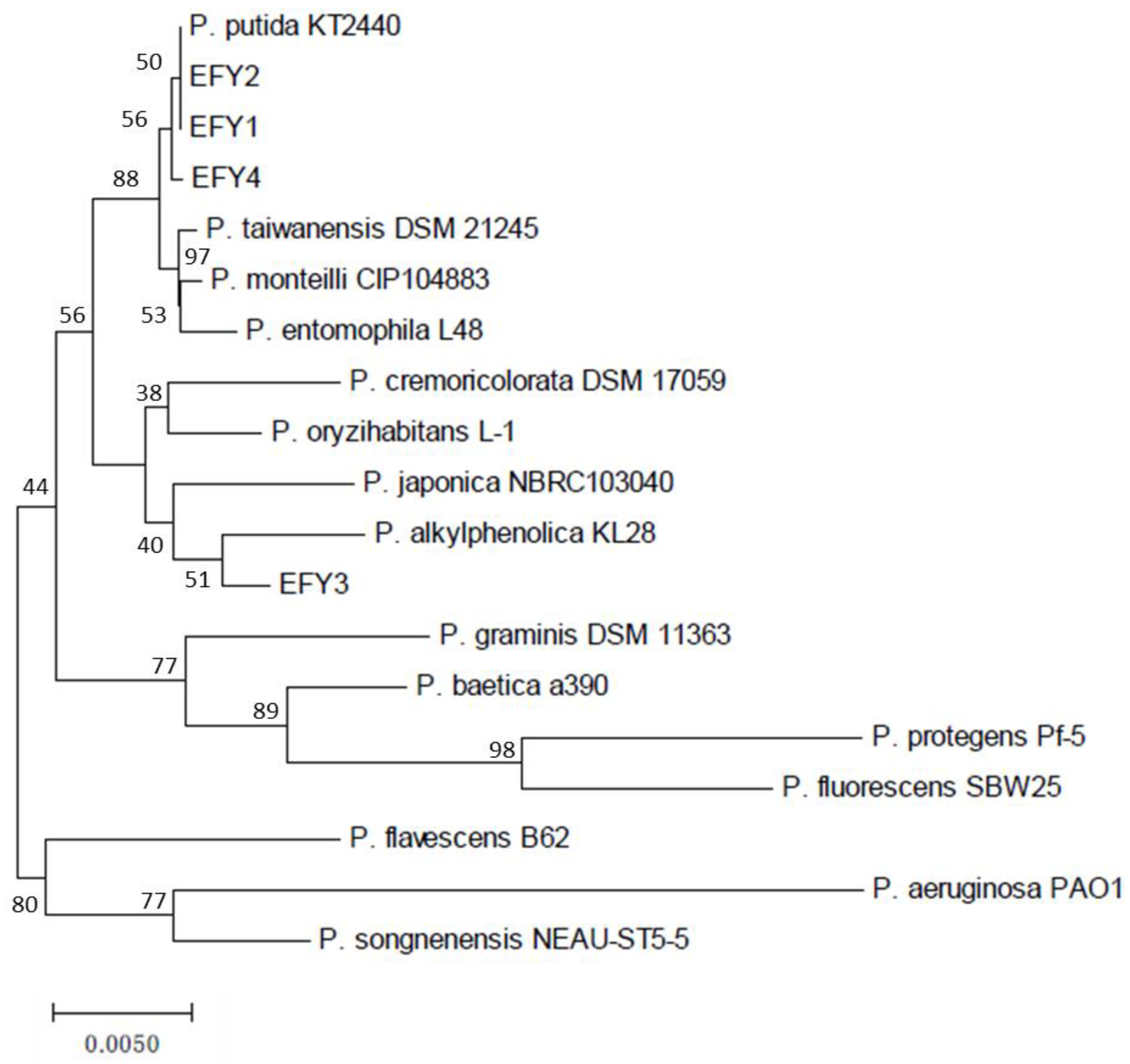
2.2. Taxonomical Identification of the Isolates
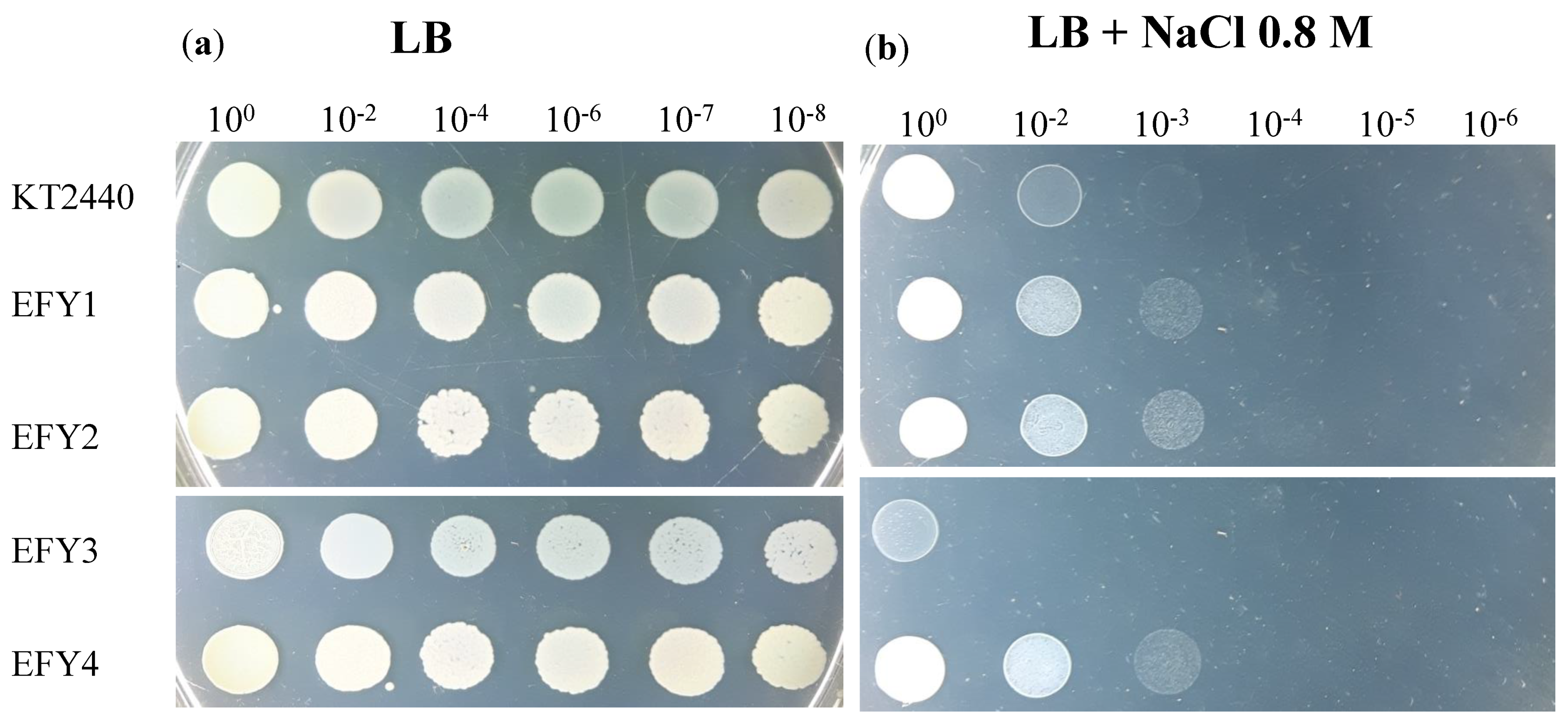
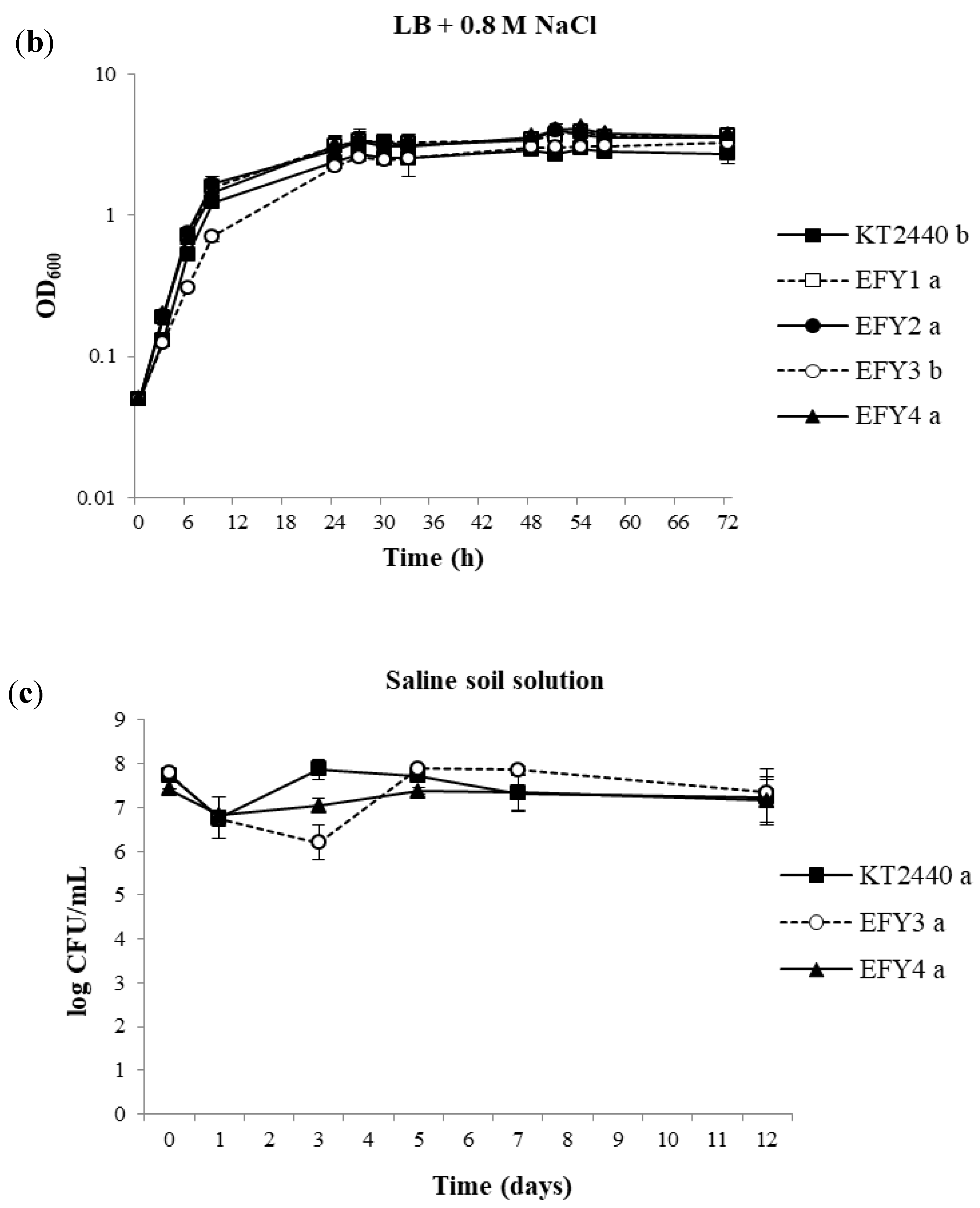
2.3. Salinity Tolerance
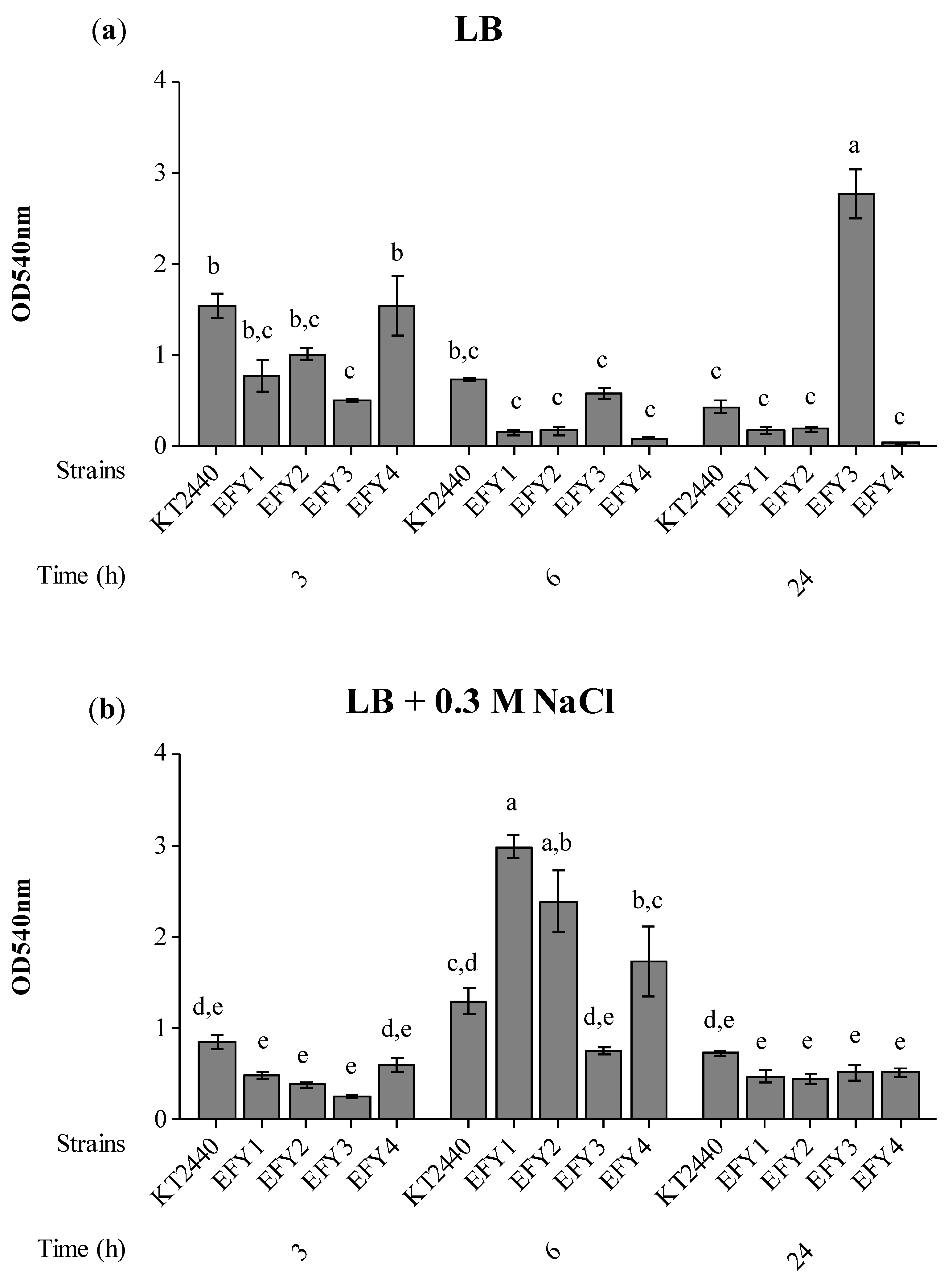
2.4. Exopolysaccharide (EPS) Production, Bacterial Surface Adhesion, Colonization and Competition in Rhizosphere
2.5. Plant Growth Promotion in Slightly Saline Soil
2.6. Statistical Analysis
3. Results and Discussion
3.1. Soil Characteristics, Bacterial Isolation, and Selection of Isolates
3.2. Identification of Pseudomonas Isolates
3.3. Salt Tolerance Assays
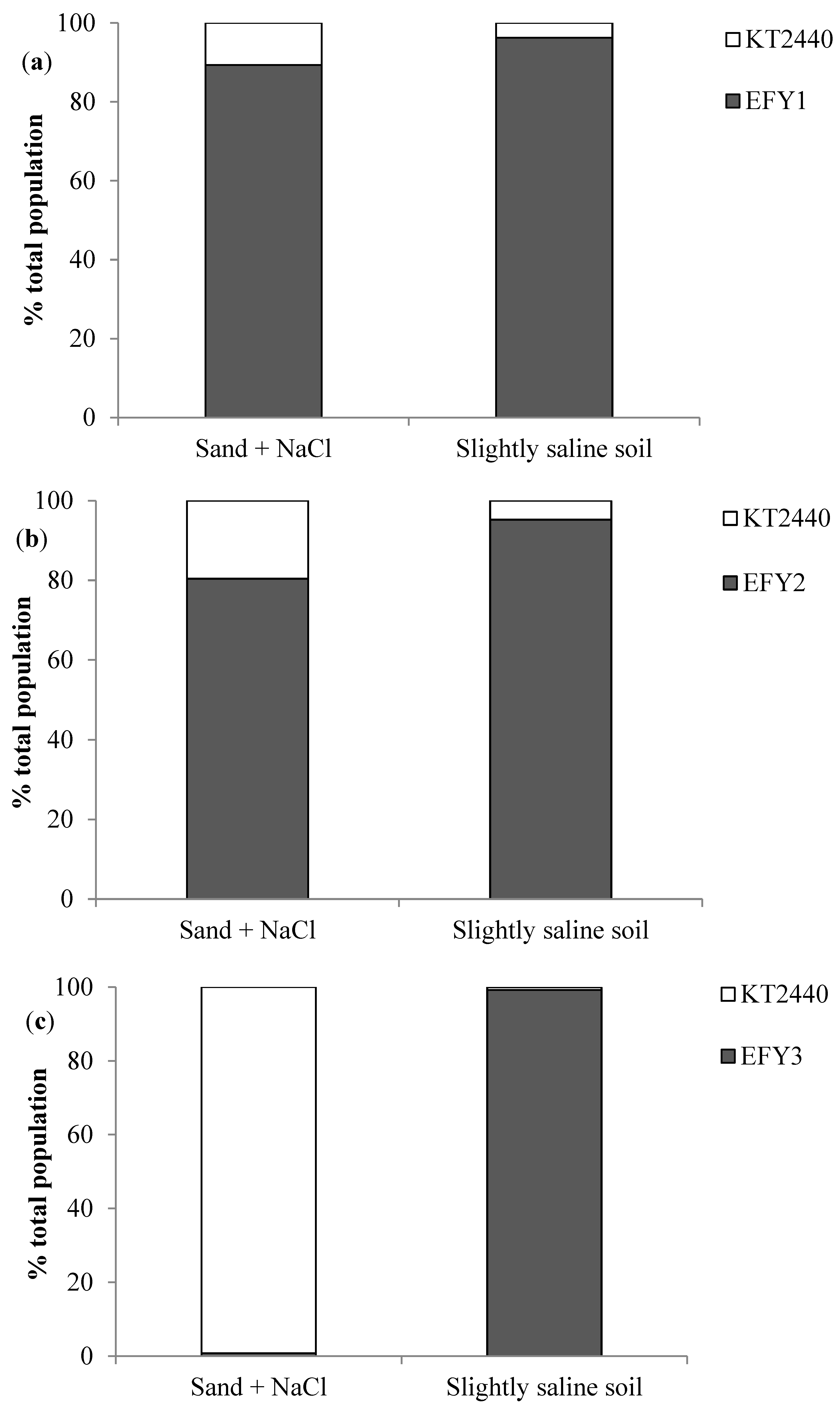
3.4. EPS Production, Surface Adhesion, Colonization, and Competition in Rhizosphere
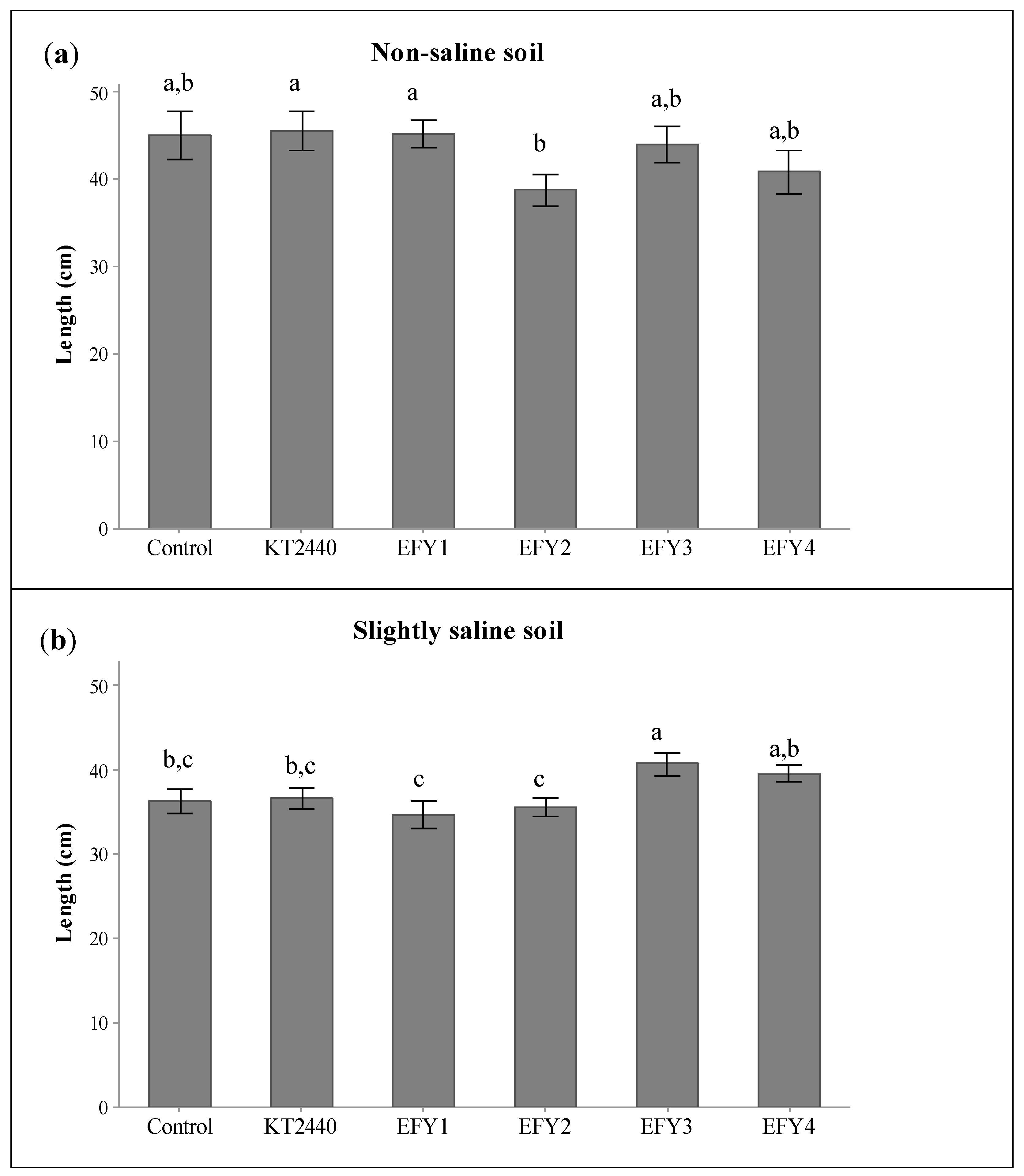
3.5. Plant Growth Promotion in Slightly Saline Soil
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Umezawa, T.; Shimizu, K.; Kato, M.; Ueda, T. Enhancement of salt tolerance in soybean with NaCl pretreatment. Physiol. Plantarum 2000, 110, 59–63. [Google Scholar] [CrossRef]
- Essa, T.A. Effect of salinity stress on growth and nutrient composition of three soybean (Glycine max L. Merrill) Cultivars. J. Agron. Crop. Sci. 2002, 188, 86–93. [Google Scholar] [CrossRef]
- Zörb, C.; Geilfus, C.M.; Dietz, K.J. Salinity and crop yield. Plant Biol. 2019, 21, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Lade, H. Plant-growth-promoting rhizobacteria to improve crop growth in saline soils: A review. Agron. Sustain. Dev. 2014, 34, 737–752. [Google Scholar] [CrossRef]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Kloepper, J.W.; Schroth, M. Plant growth-promoting rhizobacteria on radishes. Proc. 4th Int. Conf. Plant Pathog. Bact. 1978, 2, 879–882. [Google Scholar]
- Bashan, Y.; Holguin, G. Proposal for the division of plant growth-promoting rhizobacteria into two classifications: Biocontrol-PGPB (Plant Growth-Promoting Bacteria) and PGPB. Soil Biol. Biochem. 1998, 30, 12251228. [Google Scholar] [CrossRef]
- Yang, J.; Kloepper, J.W.; Ryu, C.M. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 2009, 14, 1–4. [Google Scholar] [CrossRef]
- Kaymak, H.Ç.; Güvenç, I.; Yarali, F.; Dönmez, M.F. The effects of bio-priming with PGPR on germination of radish (Raphanus sativus L.) seeds under saline conditions. Turkish J. Agric. For. 2009, 33, 173–179. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Singh, D.P. Effect of salt-tolerant plant growth-promoting rhizobacteria on wheat plants and soil health in a saline environment. Plant Biol. 2015, 17, 288–293. [Google Scholar] [CrossRef]
- El-Nahrawy, S.; Yassin, M. Response of different cultivars of wheat plants (Triticum aestivum L.) to inoculation by Azotobacter sp. under salinity stress conditions. J. Adv. Microbiol. 2020, 20, 59–79. [Google Scholar] [CrossRef] [Green Version]
- Dardanelli, M.; Carletti, S.M.; Paulucci, N.S.; Medeot, D.B.; Rodriguez-Cáceres, E.A.; Vita, F.A.; Bueno, M.; Fumero, M.V.; Garcia, M.B. Benefits of plant growth-promoting rhizobacteria and rhizobia in agriculture. In Plant Growth and Health Promoting Bacteria; Microbiology Monographs; Maheshwari, D., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 18, pp. 1–20. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Singh, J.S.; Singh, D.P. Exopolysaccharide-producing plant growth-promoting rhizobacteria under salinity condition. Pedosphere 2011, 21, 214–222. [Google Scholar] [CrossRef]
- Singh, R.P.; Jha, P.; Jha, P.N. The plant-growth-promoting bacterium Klebsiella sp. SBP-8 confers induced systemic tolerance in wheat (Triticum aestivum) under salt stress. J. Plant Physiol. 2015, 184, 57–67. [Google Scholar] [CrossRef]
- Costa-Gutierrez, S.B.; Lami, M.J.; Caram-Di Santo, M.C.; Zenoff, A.M.; Vincent, P.A.; Molina-Henares, M.A.; Espinosa-Urgel, M.; de Cristóbal, R.E. Plant growth promotion by Pseudomonas putida KT2440 under saline stress: Role of eptA. Appl. Microbiol. Biotechnol. 2020, 104, 4577–4592. [Google Scholar] [CrossRef]
- Lami, M.J.; Adler, C.; Caram-Di Santo, M.C.; Zenoff, A.M.; de Cristóbal, R.E.; Espinosa-Urgel, M.; Vincent, P.A. Pseudomonas stutzeri MJL19, a rhizosphere-colonizing bacterium that promotes plant growth under saline stress. J. Appl. Microbiol. 2020, 129, 1321–1336. [Google Scholar] [CrossRef] [PubMed]
- Al-Karhi, M.A.J.; Ismail, W.M.; Hussein, M.N. The Role of inoculation with Rhizobium leguminosarum in properties of plant (Vicia faba) under different levels of salinity. IOP Conf. Ser. Earth Environ. Sci. 2019, 01208. [Google Scholar] [CrossRef]
- Singh, R.P.; Jha, P.N. The multifarious PGPR Serratia marcescens CDP-13 augments induced systemic resistance and enhanced salinity tolerance of wheat (Triticum aestivum L.). PLoS ONE 2016, 11, e0155026. [Google Scholar] [CrossRef] [PubMed]
- Patel, T.M.; Minocheherhomji, F.P. Isolation and screening of phytohormones producing PGPR from cotton plant rhizospheric soil. J. Adv. Sci. Res. 2020, 11, 148–154. [Google Scholar]
- Granada, C.E.; Passaglia, L.M.; de Souza, E.M.; Sperotto, R.A. Is phosphate solubilization the forgotten child of plant growth-promoting rhizobacteria? Front. Microbiol. 2018, 9, 2054. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.E.; Barea, J.M.; McNeill, A.M.; Prigent-Combaret, C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 2009, 321, 305–339. [Google Scholar] [CrossRef]
- Ferreira, M.J.; Silva, H.; Cunha, A. Siderophore-producing rhizobacteria as a promising tool for empowering plants to cope with iron limitation in saline soils: A review. Pedosphere 2019, 29, 409–420. [Google Scholar] [CrossRef]
- Nadeem, S.M.; Ahmad, M.; Naveed, M.; Imran, M.; Zahir, Z.A.; Crowley, D.E. Relationship between in vitro characterization and comparative efficacy of plant growth-promoting rhizobacteria for improving cucumber salt tolerance. Arch. Microbiol. 2016, 198, 379–387. [Google Scholar] [CrossRef]
- Cardinale, M.; Ratering, S.; Suarez, C.; Zapata-Montoya, A.M.; Geissler-Plaum, R.; Schnell, S. Paradox of plant growth promotion potential of rhizobacteria and their actual promotion effect on growth of barley (Hordeum vulgare L.) under salt stress. Microbiol. Res. 2015, 181, 22–32. [Google Scholar] [CrossRef]
- Zachow, C.; Müller, H.; Tilcher, R.; Donat, C.; Berg, G. Catch the best: Novel screening strategy to select stress protecting agents for crop plants. Agronomy 2013, 3, 794–815. [Google Scholar] [CrossRef]
- Amaya-Gómez, C.V.; Porcel, M.; Mesa-Garriga, L.; Gómez-Álvarez, M.I. A framework for the selection of plant growth-promoting rhizobacteria based on bacterial competence mechanisms. Appl. Environ. Microbiol. 2020, 86, e00760-20. [Google Scholar] [CrossRef]
- De Gregorio, P.R.; Michavila, G.; Muller, L.R.; De Souza-Borges, C.; Pomares, M.F.; De Sá, E.L.S.; Pereira, C.; Vincent, P.A. Beneficial rhizobacteria immobilized in nano-bers for potential application as soybean seed bioinoculants. PLoS ONE 2017, 12, e0176930. [Google Scholar] [CrossRef] [Green Version]
- Gould, W.D.; Hagedorn, C.; Bardinelli, T.R.; Zablotowicz, R.M. New selective media for enumeration and recovery of fluorescent pseudomonads from various habitats. Appl. Environ. Microbiol. 1985, 49, 28–32. [Google Scholar] [CrossRef] [Green Version]
- Scheinert, P.; Krausse, R.; Ullmann, U.; Söller, R.; Krupp, G. Molecular differentiation of bacteria by PCR amplification of the 16S–23S rRNA spacer. J. Microbiol. Methods 1996, 26, 103–117. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walter, M.V.; Barbour, K.; McDowell, M.; Seidler, R.J. A method to evaluate survival of genetically engineered bacteria in soil extracts. Curr. Microbiol. 1987, 15, 193–197. [Google Scholar] [CrossRef]
- Friedman, L.; Kolter, R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 2004, 186, 4457–4465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Toole, G.A.; Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis. Mol. Microbiol. 1998, 28, 449–461. [Google Scholar] [CrossRef] [PubMed]
- de Lorenzo, V.; Herrero, M.; Jakubzik, U.; Timmis, K.N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 1990, 172, 6568–6572. [Google Scholar] [CrossRef] [Green Version]
- Espinosa-Urgel, M.; Salido, A.; Ramos, J.L. Genetic analysis offunc- tions involved in adhesion of Pseudomonas putida to seeds. J. Bacteriol. 2000, 182, 2363–2369. [Google Scholar] [CrossRef] [Green Version]
- Ajithkumar, P.V.; Gangadhara, K.P.; Manilal, P.; Kunhi, A.A.M. Soil inoculation with Pseudomonas aeruginosa 3MT eliminates the inhibitory effect of 3-chloro- and 4-chlorobenzoate on tomato seed germination. Soil Biol. Biochem. 1998, 30, 1053–1059. [Google Scholar] [CrossRef]
- Safdar, H.; Amin, A.; Shafiq, Y.; Ali, A.; Yasin, R. A review: Impact of salinity on plant growth. Nat. Sci. 2019, 17, 34–40. [Google Scholar] [CrossRef]
- Raimondo, E.E.; Aparicio, J.D.; Briceño, G.E.; Fuentes, M.S.; Benimeli, C.S. Lindane bioremediation in soils of different textural classes by an Actinobacteria consortium. J. Soil Sci. Plant Nutr. 2019, 19, 29–41. [Google Scholar] [CrossRef]
- Marrero, M.A.; Agaras, B.; Wall, L.G.; Valverde, C. Enriquecimiento diferencial de Pseudomonas spp. en el rizoplano de distintas especies cultivadas. Rev. Argent. Microbiol. 2015, 47, 132–137. [Google Scholar] [CrossRef] [Green Version]
- Costa-Gutierrez, S.B. Microflora of the Soybean Rhizosphere in Conditions of High Salinity: ISOLATION and Study of Pseudomonas putida Strains Benefits Against Saline Stress. Ph.D. Thesis, National University of Tucumán, Tucumán, Argentina, 2019. [Google Scholar]
- Costa-Gutierrez, S.B.; Raimondo, E.E.; Lami, M.J.; Vincent, P.A.; Espinosa-Urgel, M.; de Cristóbal, R.E. Inoculation of Pseudomonas mutant strains can improve growth of soybean and corn plants in soils under salt stress. Rhizosphere 2020, 16, 100255. [Google Scholar] [CrossRef]
- Compant, S.; Clément, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef] [Green Version]
- Ansari, F.A.; Ahmad, I. Biofilm development, plant growth promoting traits and rhizosphere colonization by Pseudomonas entomophila FAP1: A promising PGPR. Adv. Microbiol. 2018, 8, 235–251. [Google Scholar] [CrossRef] [Green Version]
- Molina, L.Á.; Ramos, C.; Duque, E.; Ronchel, M.C.; García, J.M.; Wyke, L.; Ramos, J.L. Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol. Biochem. 2000, 32, 315–321. [Google Scholar] [CrossRef]
- Bashan, Y. Inoculants of plant growth-promoting bacteria for use in agriculture. Biotechnol. Adv. 1998, 16, 729–770. [Google Scholar] [CrossRef]
- Adhikari, A.; Khan, M.A.; Lee, K.E.; Kang, S.M.; Dhungana, S.K.; Bhusal, N.; Lee, I.J. The halotolerant rhizobacterium—Pseudomonas koreensis MU2 enhances inorganic silicon and phosphorus use efficiency and augments salt stress tolerance in soybean (Glycine max L.). Microorganisms 2020, 8, 1256. [Google Scholar] [CrossRef] [PubMed]
- Kasotia, A.; Varma, A.; Tuteja, N.; Choudhary, D.K. Amelioration of soybean plant from saline-induced condition by exopolysaccharide producing Pseudomonas-mediated expression of high affinity K+-transporter (HKT1) gene. Curr. Sci. 2016, 1961–1967. [Google Scholar] [CrossRef]
- Lami, M.J. Rhizospheric Bacteria for Plant Growth Promotion and Salt Stress Tolerance in Soybean Crops. Ph.D. Thesis, National University of Tucumán, Tucumán, Argentina, 2018. [Google Scholar]


| Properties | Slightly Saline Soil | Strongly Saline Soil | Non-Saline Soil |
|---|---|---|---|
| Silt a (%) | 66.8 | 61.2 | 63.7 |
| Clay a (%) | 5.7 | 14.5 | 5.0 |
| Sand a (%) | 27.5 | 24.3 | 31.3 |
| pH b | 8.4 | 8.8 | 8.1 |
| EC c (dS/m) | 1.5 | 29.8 | 0.5 |
| Sum of cations d (cmol/Kg) | 26.0 | 66.8 | 17.7 |
| Na+ (ppm) | 498.9 | 6496.9 | 124.1 |
| K+ (ppm) | 1049.1 | 1450.8 | 624.0 |
| Ca++ (ppm) | 3834.0 | 1342.0 | 2750.0 |
| Mg++ (ppm) | 230.4 | 50.4 | 219.6 |
| Medium | Soybean Rhizosphere | Slightly Saline Soil Sample | Strongly Saline Soil Sample |
|---|---|---|---|
| LB 1× | 1.6 × 107 ± 5.0 × 106 a | 3.0 × 106 ± 3.0 × 105 b | 3.6 × 105 ± 8.0 × 104 b |
| LB 1× + NaCl | 6.0 × 105 ± 4.0 × 104 a | 6.6 × 105 ± 1.2 × 105 a | 1.0 × 105 ± 1.5 × 104 b |
| LB 0.1× | 1.8 ×107 ± 2.4 × 106 a | 4.2 × 106 ± 3.0 × 105 b | 5.7 × 105 ± 3.0 × 104 c |
| LB 0.1× + NaCl | 3.0 × 105 ± 8.0 × 104 a | 4.5 × 105 ± 7.0 × 104 a | 1.4 × 105 ±1.2 × 104 b |
| M9-glucose | 9.9 × 106 ± 7.0 × 105 a | 1.5 × 106 ± 3.1 × 105 b | 3.0 × 105 ± 4.0 × 104 b |
| M9-glucose + NaCl | 5.4 × 105 ± 6.0 × 104 a | 4.2 × 105 ± 5.0 × 104 b | 4.5 × 104 ± 1.5 × 104 c |
| M9-citrate | 9.0 × 106 ± 1.0 × 106 a | 1.4 × 106 ± 2.0 × 105 b | 1.8 × 105 ± 1.3 × 104 b |
| M9-citrate + NaCl | 1.5 × 106 ± 2.0 × 105 a | 4.2 × 105 ± 7.0 × 104 b | 6.0 × 104 ± 4.0 × 103 c |
| M9-benzoate | 6.5 × 105 ± 5.0 × 104 a | 3.9 × 105 ± 2.0 × 104 b | 4.5 × 105 ± 5.0 × 104 b |
| M9-benzoate + NaCl | 6.0 × 106 ± 8.0 × 105 a | 4.8 × 105 ± 3.0 × 104 b | 2.0 × 103 ± 5.0 × 101 c |
| Modified S1 | 1.2 × 106 ± 8.0 × 105 a | 6.0 × 103 ± 3.0 × 102 b | 4.2 × 102 ± 1.1 × 101 c |
| Strains | Soybean Seeds (UFC/mL) | Soybean Roots in Sand + NaCl (CFU/g Root) | Soybean Roots in Slightly Saline Soil (CFU/g Root) |
|---|---|---|---|
| KT2440 | 1.6 106 ± 6.8 105 b | 3.6 106 ± 4.1 106 b | 2.4 106 ± 1.5 106 a |
| EFY1 | 3.6 106 ± 2.6 106 b | 2.9 108 ± 3.8 108 a | 5.0 106 ± 7.6 105 a |
| EFY2 | 6.4 106 ± 4.0 105 b | 8.9 108 ± 9.3 108 a | 3.8 106 ± 2.0 105 a |
| EFY3 | 8.1 106 ± 2.8 106 a, b | 6.1 108 ± 1.0 108 a | 2.8 106 ± 4.3 106 a |
| EFY4 | 1.6 107 ± 6.7 106 a | 7.9 108 ± 1.2 108 a | 7.1 106 ± 4.8 106 a |
| Strains | Germination Rate (%) | Vigor Index | V2 (% of Plants in Phenological State V2) | |||
|---|---|---|---|---|---|---|
| Non-Saline | Saline | Non-Saline | Saline | Non-Saline | Saline | |
| Control | 80.0 ± 5.6 c | 60.0 ± 5.6 c | 360.4 ± 82.9 c | 217.8 ± 38.2 c | 36.6 ± 4.7 b | 19.5 ± 3.0 c |
| KT2440 | 96.0 ± 0.0 b | 94.0 ± 2.8 b | 438.0 ± 106.0 a,b | 344.9 ± 58.0 b | 47.9 ± 2.9a | 18.7 ± 2.9c |
| EFY1 | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 451,8 ± 77.9 a | 347.3 ± 72.5 b | 38.0 ± 2.8 b | 30.0 ± 2.8 b |
| EFY2 | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 388,2 ± 88.4 b,c | 356.5 ± 49.0 b | 14.0 ± 2.8 c | 10.0 ± 2.8 d |
| EFY3 | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 440,5 ± 100.4 a,b | 407.8 ± 68,0 a | 38.0 ± 2.8 b | 64.5 ± 2.9 a |
| EFY4 | 100.0 ± 0.0 a | 100.0 ± 0.0 a | 408.9 ± 123.0 a,b,c | 396.2 ± 46.5 a | 34.0 ± 2.8 b | 31.2 ± 2.9 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa-Gutierrez, S.B.; Caram-Di Santo, M.C.d.V.; Zenoff, A.M.; Espinosa-Urgel, M.; de Cristóbal, R.E.; Vincent, P.A. Isolation of Pseudomonas Strains with Potential for Protection of Soybean Plants against Saline Stress. Agronomy 2021, 11, 2236. https://doi.org/10.3390/agronomy11112236
Costa-Gutierrez SB, Caram-Di Santo MCdV, Zenoff AM, Espinosa-Urgel M, de Cristóbal RE, Vincent PA. Isolation of Pseudomonas Strains with Potential for Protection of Soybean Plants against Saline Stress. Agronomy. 2021; 11(11):2236. https://doi.org/10.3390/agronomy11112236
Chicago/Turabian StyleCosta-Gutierrez, Stefanie Bernardette, María Carolina del Valle Caram-Di Santo, Ana María Zenoff, Manuel Espinosa-Urgel, Ricardo Ezequiel de Cristóbal, and Paula Andrea Vincent. 2021. "Isolation of Pseudomonas Strains with Potential for Protection of Soybean Plants against Saline Stress" Agronomy 11, no. 11: 2236. https://doi.org/10.3390/agronomy11112236
APA StyleCosta-Gutierrez, S. B., Caram-Di Santo, M. C. d. V., Zenoff, A. M., Espinosa-Urgel, M., de Cristóbal, R. E., & Vincent, P. A. (2021). Isolation of Pseudomonas Strains with Potential for Protection of Soybean Plants against Saline Stress. Agronomy, 11(11), 2236. https://doi.org/10.3390/agronomy11112236






