Known Drugs Identified by Structure-Based Virtual Screening Are Able to Bind Sigma-1 Receptor and Increase Growth of Huntington Disease Patient-Derived Cells
Abstract
1. Introduction
- (i)
- A first 2D-pharmacophore model, named Glennon–Ph, was designed in the early 90′s in the absence of structural information on the protein [27]. It was based on the structural features of a series of diphenylalkylamine σ1R ligands, and consists in one positively ionisable group (i.e., a basic amine group) and two opposite hydrophobic regions at 2.5–3.9 Å and 6–10 Å from the amine group, respectively, without any angle constraint.
- (ii)
- A second pharmacophore model was designed based on the alignment of PD144418, spipethiane, haloperidol and (+)-pentazocine [28]. This model consists in one aromatic region, one nitrogen atom that acts as hydrogen bond acceptor, and another polar feature representing one oxygen or sulphur atom.
- (iii)
- A 3D-pharmacophore model was developed based upon 23 structurally diverse molecules with Ki values for σ1R between 10 pM and 100 μM [29]. This consists in one positively ionisable group and four hydrophobic features. The model is in good agreement with the first one [27] and lacks the secondary polar binding region of the second [28].
- (iv)
- (v)
- Two new σ1R pharmacophore models were developed, by taking advantage of the information and resolution provided by the X-ray crystallographic structure of σ1R. 5HK1-PhA is based on the four most important interactions between σ1R and the PD144418 antagonist, and comprises: the amine group that interacts with Glu172 and Asp126; one hydrophobic feature for the interaction between the propyl chain of the ligand and the protein residues Ile124 and His154; and two additional hydrophobic features for the interactions of the phenyl ring and methyl group of the ligand, respectively, with Leu182, Tyr206, and Ile178. 5HK1-PhB was obtained by manually merging the two 5HK1-PhA hydrophobic features that interact with Leu182, Tyr206 and Ile178 [33].
2. Results
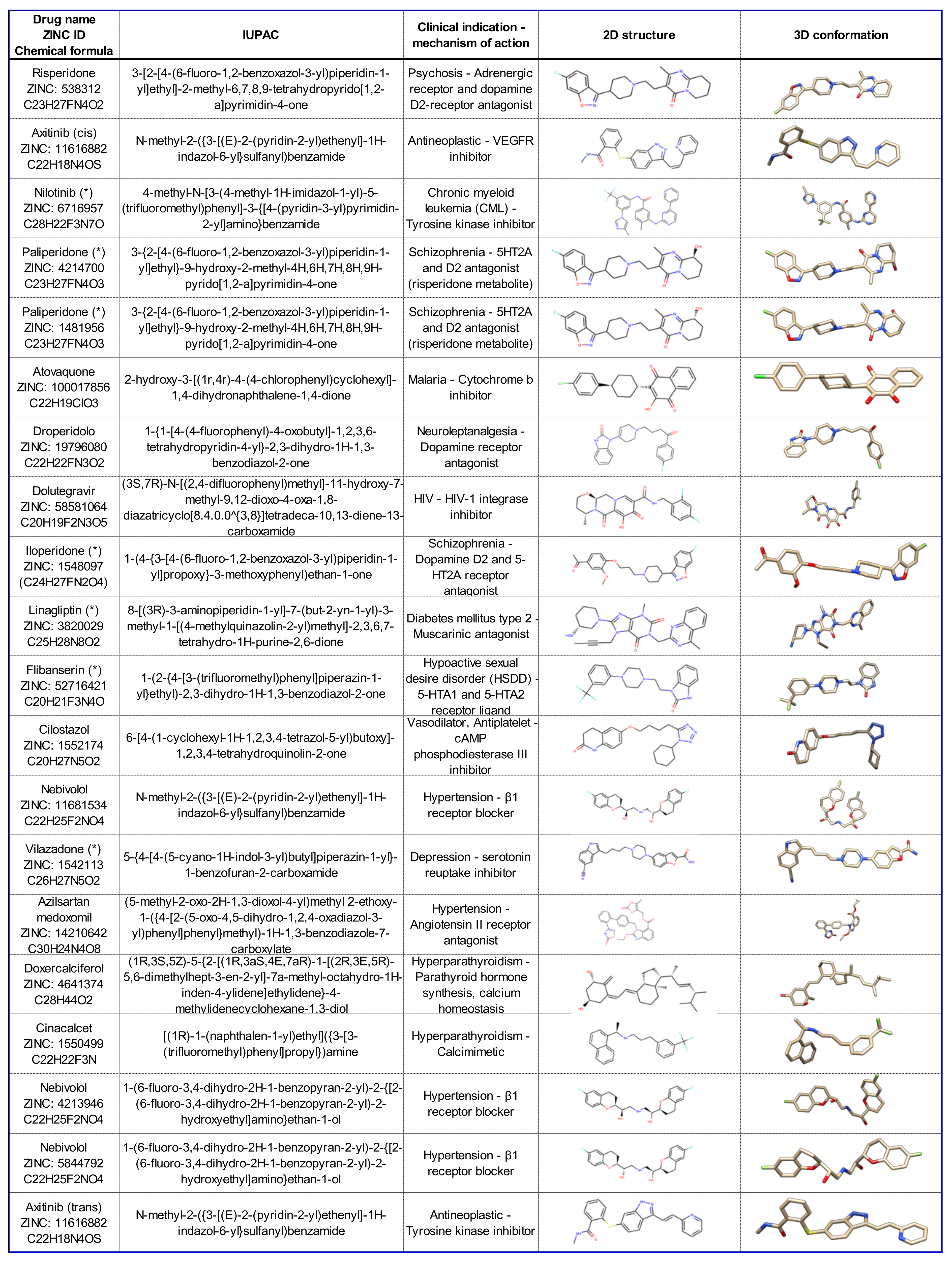
2.1. Identification of Potential σ1R Binding Drugs by Computational Methods
2.2. σ1R Expression and Purification
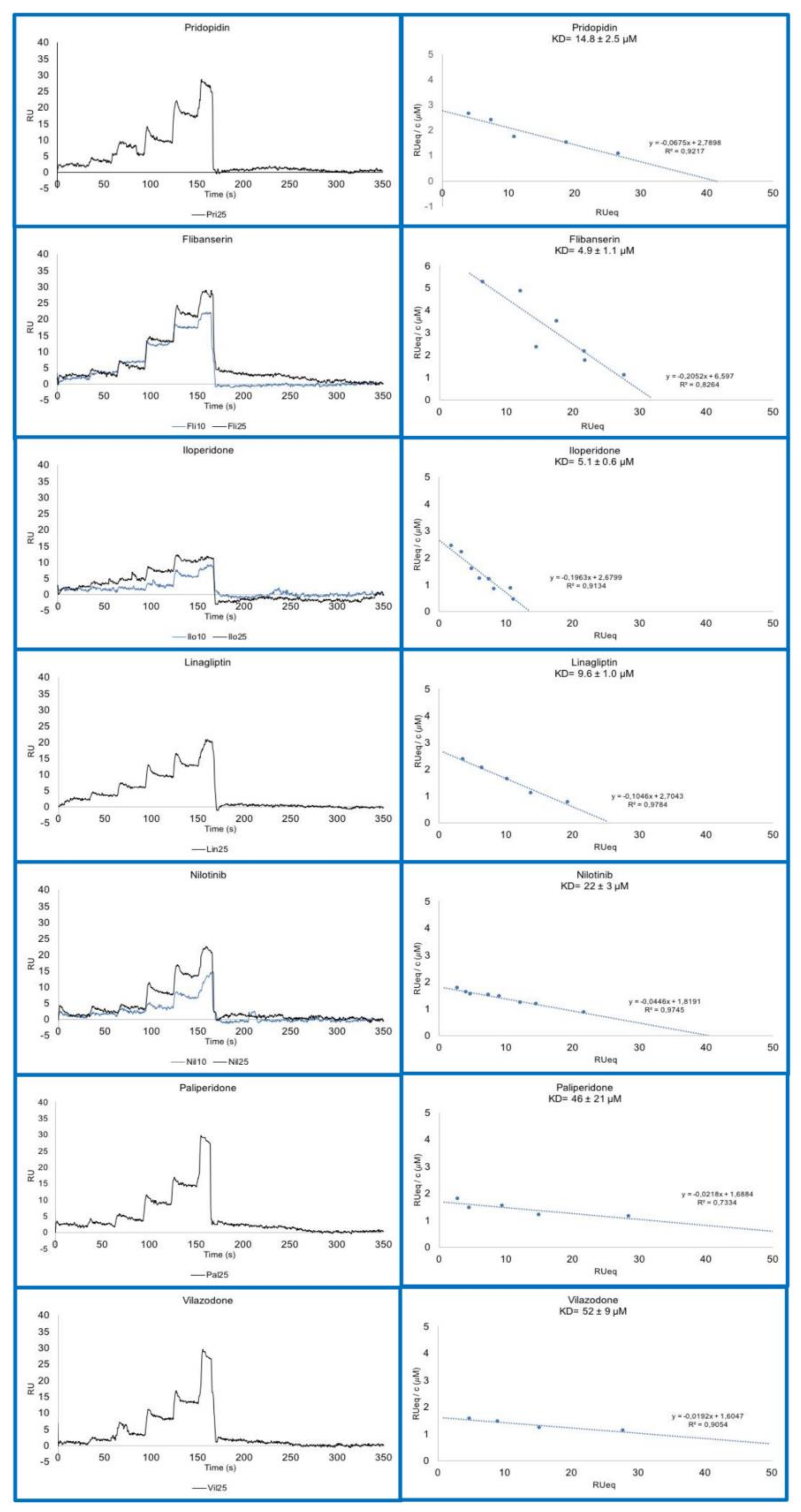
2.3. Assessment of Direct Drug Binding to σ1R In Vitro by SPR
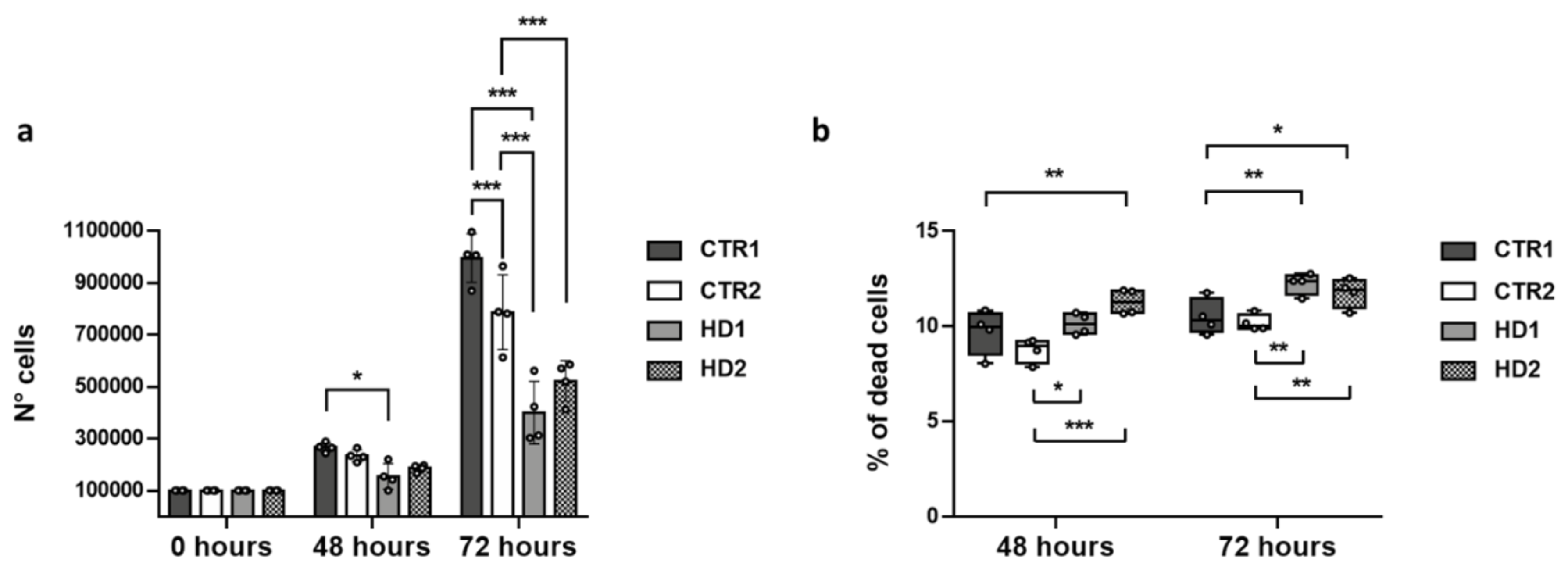
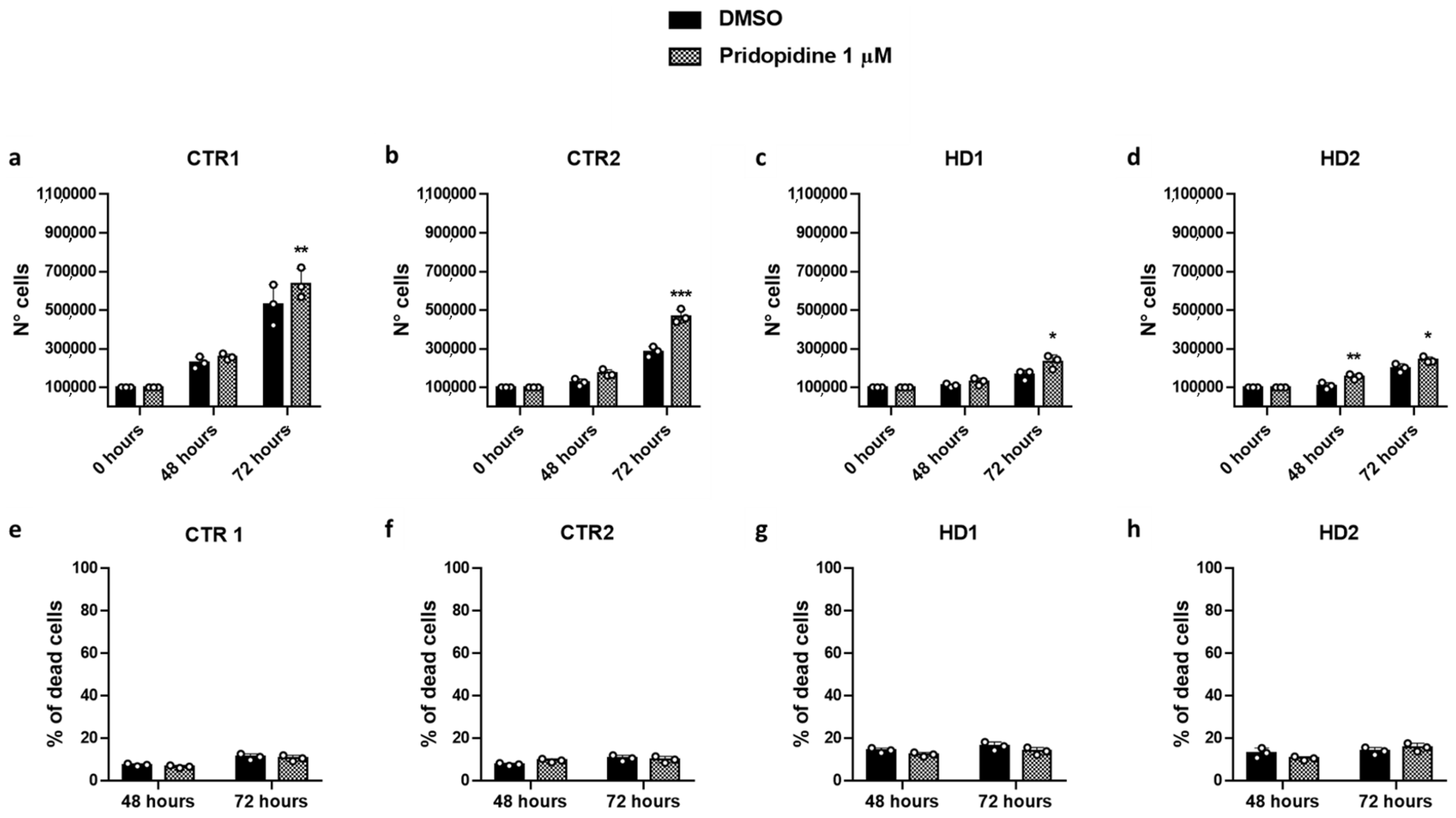
2.4. Pridopidine Effect on Healthy and HD Cell Growth
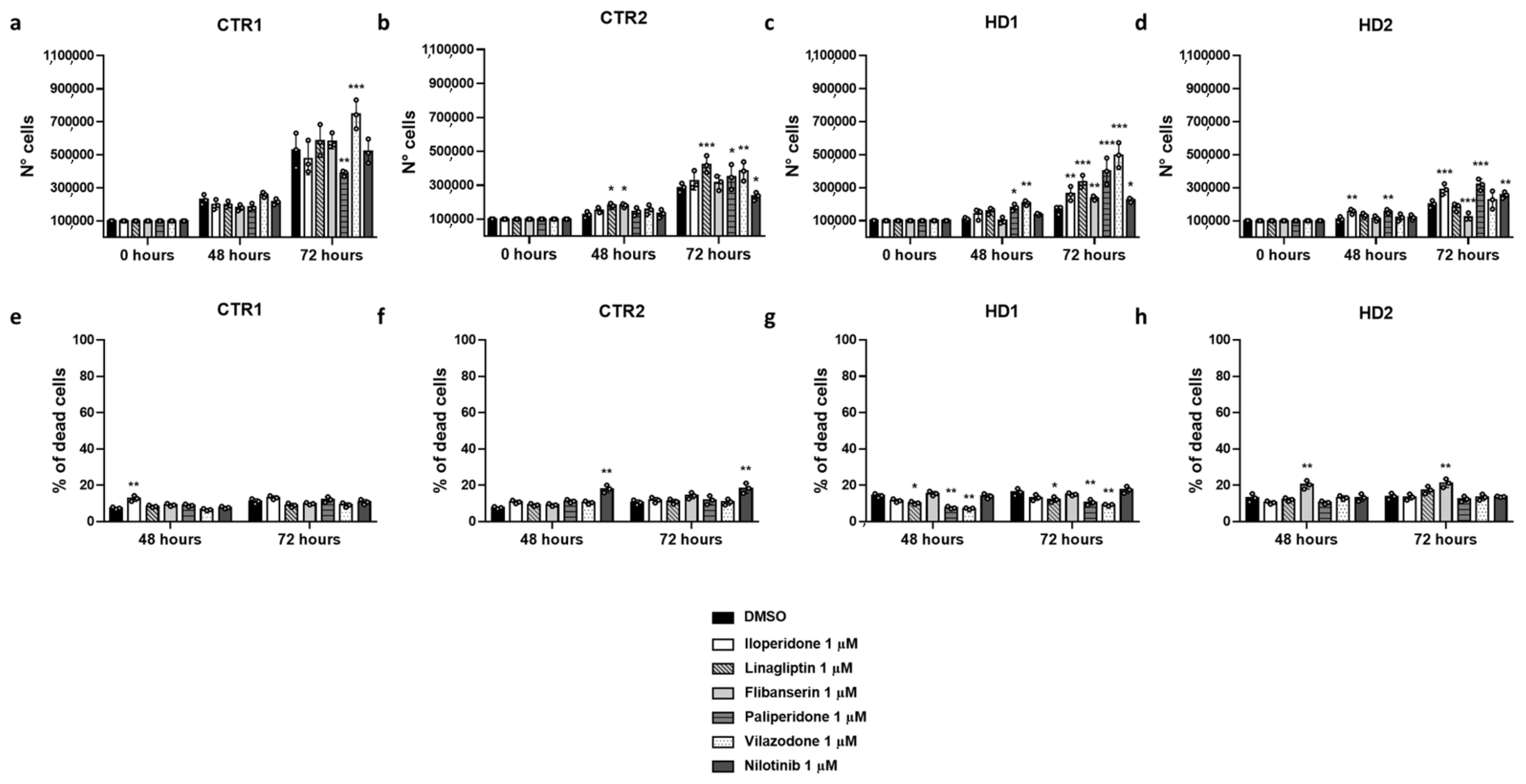
2.5. Effects of Drugs on Healthy and HD Cells Growth
2.6. Effects of Drugs on Healthy and HD Cell Death
3. Discussion
4. Materials and Methods
4.1. Identification of Potential σ1R Binding Drugs by Computational Methods
4.1.1. Receptor Preparation
4.1.2. Ligand Preparation
4.1.3. Virtual Screening
4.1.4. Molecular Docking
4.1.5. Results Analysis
4.1.6. Visual Inspection
4.2. σ1R Expression and Purification
4.3. Assessment of Direct Drug Binding to σ1R In Vitro by SPR
4.4. Ethical Approval
4.5. Skin Biopsy and Fibroblast Isolation
4.6. Growth Rate Analysis in Basal Conditions
4.7. Growth Rate Analysis after Drug Treatment
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 3D | Three-dimensional |
| AD | Alzheimer Disease |
| ADT | AutoDock Tools |
| ALS | Amyotrophic Lateral Sclerosis |
| CTR1, CTR2 | Fibroblasts from healthy subjects 1 and 2 |
| HD | Huntington disease |
| HD1, HD2 | Fibroblasts from HD patient 1 and 2 |
| KD | Dissociation constant |
| Ki | Inhibition constant |
| PD | Parkinson Disease |
| PDB | Protein Data Bank |
| RMSD | Root Mean Square Deviation |
| SPR | Surface Plasmon Resonance |
| TFC | Total Functional Capacity |
| σ1R | Sigma-1 receptor |
References
- Maurice, T.; Su, T.-P. The pharmacology of sigma-1 receptors. Pharmacol. Ther. 2009, 124, 195–206. [Google Scholar] [CrossRef]
- Schmidt, H.R.; Betz, R.M.; Dror, R.O.; Kruse, A.C. Structural basis for σ1 receptor ligand recognition. Nat. Struct. Mol. Biol. 2018, 25, 981–987. [Google Scholar] [CrossRef] [PubMed]
- Lupardus, P.J.; Wilke, R.A.; Aydar, E.; Palmer, C.P.; Chen, Y.; Ruoho, A.E.; Jackson, M.B. Membrane-delimited coupling between sigma re-ceptors and K+ channels in rat neurohypophysial terminals requires neither G-protein nor ATP. J. Physiol. 2000, 526, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T. Regulating ankyrin dynamics: Roles of sigma-1 receptors. Proc. Natl. Acad. Sci. USA 2001, 98, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Behrends, C.; Sowa, M.E.; Gygi, S.P.; Harper, J.W. Network organization of the human autophagy system. Nat. Cell Biol. 2010, 466, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Su, T.-P.; Hayashi, T.; Maurice, T.; Buch, S.; Ruoho, A.E. The sigma-1 receptor chaperone as an inter-organelle signaling modulator. Trends Pharmacol. Sci. 2010, 31, 557–566. [Google Scholar] [CrossRef]
- Hosp, F.; Vossfeldt, H.; Heinig, M.; Vasiljevic, D.; Arumughan, A.; Wyler, E.; Landthaler, M.; Hubner, N.; Wanker, E.E.; Lannfelt, L.; et al. Quantitative Interaction Proteomics of Neuro-degenerative Disease Proteins. Cell Rep. 2015, 11, 1134–1146. [Google Scholar] [CrossRef]
- Boldt, K.; Van Reeuwijk, J.; Lu, Q.; Koutroumpas, K.; Nguyen, T.M.T.; Texier, Y.; Van Beersum, S.E.; Horn, N.; Willer, J.R.; Mans, D.A.; et al. An organelle-specific protein landscape iden-tifies novel diseases and molecular mechanisms. Nat. Commun. 2016, 13, 7. [Google Scholar]
- Genovese, I.; Giamogante, F.; Barazzuol, L.; Battista, T.; Fiorillo, A.; Vicario, M.; D’Alessandro, G.; Cipriani, R.; Limatola, C.; Rossi, D.; et al. Sorcin is an early marker of neurodegeneration, Ca2+ dysregulation and endoplasmic reticulum stress associated to neurodegenerative diseases. Cell Death Dis. 2020, 11, 1–18. [Google Scholar] [CrossRef]
- Chu, U.B.; Ruoho, A.E. Biochemical Pharmacology of the Sigma-1 Receptor. Mol. Pharmacol. 2016, 89, 142–153. [Google Scholar] [CrossRef]
- Nguyen, L.; Lucke-Wold, B.P.; Mookerjee, S.A.; Cavendish, J.Z.; Robson, M.J.; Scandinaro, A.L.; Matsumoto, R.R. Role of sigma-1 receptors in neurodegenerative diseases. J. Pharmacol. Sci. 2015, 127, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, R.; Oliván, S.; Rando, A.; Casas, C.; Osta, R.; Navarro, X. Sigma-1R Agonist Improves Motor Function and Motoneuron Survival in ALS Mice. Neurotherapeutics 2012, 9, 814–826. [Google Scholar] [CrossRef] [PubMed]
- Maher, C.M.; Thomas, J.D.; Haas, D.A.; Longen, C.G.; Oyer, H.M.; Tong, J.Y.; Kim, F.J. Small-Molecule Sigma1 Modulator Induces Au-tophagic Degradation of PD-L1. Mol. Cancer Res. 2018, 16, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Tsai, S.-Y.; Mori, T.; Fujimoto, M.; Su, T.-P. Targeting ligand-operated chaperone sigma-1 receptors in the treatment of neuropsychiatric disorders. Expert Opin. Ther. Targets 2011, 15, 557–577. [Google Scholar] [CrossRef] [PubMed]
- Castany, S.; Gris, G.; Vela, J.M.; Verdu, E.; Boadas-Vaello, P. Critical role of sigma-1 receptors in central neuropathic pain-related behaviours after mild spinal cord injury in mice. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Bruna, J.; Videla, S.; Argyriou, A.A.; Velasco, R.; Villoria, J.; Santos, C.; Nadal, C.; Cavaletti, G.; Alberti, P.; Briani, C.; et al. Efficacy of a Novel Sigma-1 Receptor Antagonist for Oxaliplatin-Induced Neuropathy: A Randomized, Double-Blind, Placebo-Controlled Phase IIa Clinical Trial. Neurotherapeutics 2018, 15, 178–189. [Google Scholar] [CrossRef]
- Kim, F.J.; Kovalyshyn, I.; Burgman, M.; Neilan, C.; Chien, C.C.; Pasternak, G.W. σ1 Receptor modulation of G-protein-coupled re-ceptor signaling: Potentiation of opioid transduction independent from receptor binding. Mol. Pharmacol. 2010, 77, 695–703. [Google Scholar] [CrossRef]
- Navarro, G.; Moreno, E.; Aymerich, M.; Marcellino, D.; McCormick, P.J.; Mallol, J.; Cortés, A.; Casadó, V.; Canela, E.I.; Ortiz, J.; et al. Direct involvement of σ-1 receptors in the dopamine D1 receptor-mediated effects of cocaine. Proc. Natl. Acad. Sci. USA 2010, 107, 18676–18681. [Google Scholar] [CrossRef]
- Wu, Z.; Bowen, W.D. Role of Sigma-1 Receptor C-terminal Segment in Inositol 1,4,5-Trisphosphate Receptor Activation: Constitutive enhancement of calcium signaling in MCF-7 tumor cells. J. Biol. Chem. 2008, 283, 28198–28215. [Google Scholar] [CrossRef]
- Aydar, E.; Palmer, C.P.; Klyachko, V.A.; Jackson, M.B. The Sigma Receptor as a Ligand-Regulated Auxiliary Potassium Channel Subunit. Neuron 2002, 34, 399–410. [Google Scholar] [CrossRef]
- Hong, W.C.; Yano, H.; Hiranita, T.; Chin, F.T.; McCurdy, C.R.; Su, T.-P.; Amara, S.G.; Katz, J.L. The sigma-1 receptor modulates dopamine transporter conformation and cocaine binding and may thereby potentiate cocaine self-administration in rats. J. Biol. Chem. 2017, 292, 11250–11261. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.R.; Zheng, S.; Gurpinar, E.; Koehl, A.; Manglik, A.K.A.; Kruse, A.C. Crystal structure of the human σ1 receptor. Nature 2016, 532, 527–530. [Google Scholar] [CrossRef]
- Geva, M.; Kusko, R.; Soares, H.; Fowler, K.D.; Birnberg, T.; Barash, S.; Wagner, A.M.; Fine, T.; Lysaght, A.; Weiner, B.; et al. Pridopidine activates neuroprotective pathways im-paired in Huntington Disease. Hum. Mol. Genet. 2016, 25, 3975–3987. [Google Scholar] [CrossRef]
- Ryskamp, D.A.; Wu, L.; Wu, J.; Kim, D.; Rammes, G.; Geva, M.; Hayden, M.; Bezprozvanny, I. Pridopidine stabilizes mushroom spines in mouse models of Alzheimer’s disease by acting on the sigma-1 receptor. Neurobiol. Dis. 2019, 124, 489–504. [Google Scholar] [CrossRef]
- Francardo, V.; Geva, M.; Bez, F.; Denis, Q.; Steiner, L.; Hayden, M.R.; Cenci, M.A. Pridopidine Induces Functional Neurorestoration Via the Sigma-1 Receptor in a Mouse Model of Parkinson’s Disease. Neurotherapeutics 2019, 16, 465–479. [Google Scholar] [CrossRef]
- Ionescu, A.; Gradus, T.; Altman, T.; Maimon, R.; Avraham, N.S.; Geva, M.; Hayden, M.; Perlson, E. Targeting the Sigma-1 Receptor via Pridopidine Ameliorates Central Features of ALS Pathology in a SOD1G93A Model. Cell Death Dis. 2019, 10, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Glennon, R.A.; Ablordeppey, S.Y.; Ismaiel, A.M.; El-Ashmawy, M.B.; Fischer, J.B.; Howie, K.B. Structural Features Important for σ1 Receptor Binding. J. Med. Chem. 1994, 37, 1214–1219. [Google Scholar] [CrossRef] [PubMed]
- Gund, T.M.; Floyd, J.; Jung, D. Molecular modeling of σ1 receptor ligands: A model of binding conformational and electrostatic considerations. J. Mol. Graph. Model. 2004, 22, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Laggner, C.; Schieferer, C.; Fiechtner, B.; Poles, G.; Hoffmann, R.D.; Glossmann, H.; Langer, T.; Moebius, F.F. Discovery of High-Affinity Ligands of σ1Receptor, ERG2, and Emopamil Binding Protein by Pharmacophore Modeling and Virtual Screening. J. Med. Chem. 2005, 48, 4754–4764. [Google Scholar] [CrossRef] [PubMed]
- Zampieri, D.; Mamolo, M.G.; Laurini, E.; Florio, C.; Zanette, C.; Fermeglia, M.; Posocco, P.; Paneni, M.S.; Pricl, S.; Vio, L. Synthesis, biological evaluation, and three-dimensional in silico pharmacophore model for σ1 receptor ligands based on a series of substituted benzo[d]oxa-zol-2(3H)-one derivatives. J. Med. Chem. 2009, 52, 5380–5393. [Google Scholar] [CrossRef]
- Oberdorf, C.; Schmidt, T.J.; Wünsch, B. 5D-QSAR for spirocyclic σ1 receptor ligands by Quasar receptor surface modeling. Eur. J. Med. Chem. 2010, 45, 3116–3124. [Google Scholar] [CrossRef]
- Banister, S.D.; Manoli, M.; Doddareddy, M.R.; Hibbs, D.E.; Kassiou, M. A σ1 receptor pharmacophore derived from a series of N-substituted 4-azahexacyclo [5.4.1.02,6.03,10.05,9.08,11]dodecan-3-ols (AHDs). Bioorganic Med. Chem. Lett. 2012, 22, 6053–6058. [Google Scholar] [CrossRef]
- Pascual, R.; Almansa, C.; Plata-Salamán, C.; Vela, J.M. A New Pharmacophore Model for the Design of Sigma-1 Ligands Validated on a Large Experimental Dataset. Front. Pharmacol. 2019, 10. [Google Scholar] [CrossRef]
- Gardiner, S.L.; Milanese, C.; Boogaard, M.W.; Buijsen, R.A.; Hogenboom, M.; Roos, R.A.; Mastroberardino, P.G.; van Roon-Mom, W.M.; Aziz, N.A. Bioenergetics in fibroblasts of pa-tients with Huntington disease are associated with age at onset. Neurol. Genet. 2018, 4. [Google Scholar] [CrossRef]
- Del Hoyo, P.; García-Redondo, A.; de Bustos, F.; Molina, J.A.; Sayed, Y.; Alonso-Navarro, H.; Caballero, L.; Arenas, J.; Jiménez-Jiménez, F.J. Oxidative stress in skin fibro-blasts cultures of patients with Huntington’s disease. Neurochem. Res. 2006, 31, 1103–1109. [Google Scholar] [CrossRef]
- Jędrak, P.; Mozolewski, P.; Węgrzyn, G.; Więckowski, M.R. Mitochondrial alterations accompanied by oxidative stress condi-tions in skin fibroblasts of Huntington’s disease patients. Metab. Brain Dis. 2018, 33, 2005–2017. [Google Scholar] [CrossRef]
- Marchina, E.; Misasi, S.; Bozzato, A.; Ferraboli, S.; Agosti, C.; Rozzini, L.; Borsani, G.; Barlati, S.; Padovani, A. Gene expression profile in fibroblasts of Huntington’s disease patients and controls. J. Neurol. Sci. 2014, 337, 42–46. [Google Scholar] [CrossRef]
- Squitieri, F.; Falleni, A.; Cannella, M.; Orobello, S.; Fulceri, F.; Lenzi, P.; Fornai, F. Abnormal morphology of peripheral cell tissues from patients with Huntington disease. J. Neural Transm. 2009, 117, 77–83. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Huey, R.; Morris, G.M.; Olson, A.J.; Goodsell, D.S. Software news and update a semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2007, 28, 1145–1152. [Google Scholar] [CrossRef]
- Cosconati, S.; Forli, S.; Perryman, A.L.; Harris, R.; Goodsell, D.S.; Olson, A.J. Virtual screening with AutoDock: Theory and practice. Expert Opin. Drug Discov. 2010, 5, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Sahlholm, K.; Århem, P.; Fuxe, K.; Marcellino, D. The dopamine stabilizers ACR16 and (−)-OSU6162 display nanomolar affinities at the σ-1 receptor. Mol. Psychiatry 2012, 18, 12–14. [Google Scholar] [CrossRef]
- Ashburn, T.T.; Thor, K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef]
- Squitieri, F.; Landwehrmeyer, G.B.; Reilmann, R.; Rosser, A.; De Yebenes, J.G.; Prang, A.; Ivkovic, J.; Bright, J.; Rembratt, Å. One-year safety and tolerability profile of pridopidine in patients with Huntington disease. Neurology 2013, 80, 1086–1094. [Google Scholar] [CrossRef]
- Reilmann, R.; McGarry, A.; Grachev, I.D.; Savola, J.-M.; Borowsky, B.; Eyal, E.; Gross, N.; Langbehn, D.; Schubert, R.; Wickenberg, A.T.; et al. Safety and efficacy of pridopidine in patients with Huntington’s disease (PRIDE-HD): A phase 2, randomised, placebo-controlled, multicentre, dose-ranging study. Lancet Neurol. 2019, 18, 165–176. [Google Scholar] [CrossRef]
- McGarry, A.; Leinonen, M.; Kieburtz, K.; Geva, M.; Olanow, C.W.; Hayden, M. Effects of Pridopidine on Functional Capacity in Early-Stage Participants from the PRIDE-HD Study. J. Huntington’s Dis. 2020, 9, 371–380. [Google Scholar] [CrossRef]
- Sánchez-Castañeda, C.; De Pasquale, F.; Caravasso, C.F.; Marano, M.; Maffi, S.; Migliore, S.; Sabatini, U.; Squitieri, F. Resting-state connectivity and modulated somatomotor and default-mode networks in Huntington disease. CNS Neurosci. Ther. 2017, 23, 488–497. [Google Scholar] [CrossRef]
- Ryskamp, D.A.; Wu, J.; Geva, M.; Kusko, R.; Grossman, I.; Hayden, M.R.; Bezprozvanny, I. The sigma-1 receptor mediates the beneficial effects of pridopidine in a mouse model of Huntington disease. Neurobiol. Dis. 2017, 97, 46–59. [Google Scholar] [CrossRef]
- Eddings, C.R.; Arbez, N.; Akimov, S.; Geva, M.; Hayden, M.R.; Ross, C.A. Pridopidine protects neurons from mutant-huntingtin toxicity via the sigma-1 receptor. Neurobiol. Dis. 2019, 129, 118–129. [Google Scholar] [CrossRef]
- Squitieri, F.; Di Pardo, A.; Favellato, M.; Amico, E.; Maglione, V.; Frati, L. Pridopidine, a dopamine stabilizer, improves motor performance and shows neuroprotective effects in Huntington disease R6/2 mouse model. J. Cell Mol. Med. 2015, 19, 2540–2548. [Google Scholar] [CrossRef] [PubMed]
- Van der Burg, J.M.; Björkqvist, M.; Brundin, P. Beyond the brain: Widespread pathology in Huntington’s disease. Lancet Neurol. 2009, 8, 765–774. [Google Scholar] [CrossRef]
- Fusilli, C.; Migliore, S.; Mazza, T.; Consoli, F.; De Luca, A.; Barbagallo, G.; Ciammola, A.; Gatto, E.M.; Cesarini, M.; Etcheverry, J.L.; et al. Biological and clinical manifestations of juvenile Huntington’s disease: A retrospective analysis. Lancet Neurol. 2018, 17, 986–993. [Google Scholar] [CrossRef]
- Mauri, M.; Paletta, S.; Maffini, M.; Colasanti, A.; Dragogna, F.; Di Pace, C.; Altamura, A. Clinical pharmacology of atypical antipsychotics: An update. EXCLI J. 2014, 13, 1163–1191. [Google Scholar]
- Kongsamut, S.; Roehr, J.E.; Cai, J.; Hartman, H.B.; Weissensee, P.; Kerman, L.L.; Tang, L.; Sandrasagra, A. Iloperidone binding to human and rat dopamine and 5-HT receptors. Eur. J. Pharmacol. 1996, 317, 417–423. [Google Scholar] [CrossRef]
- Grachev, I.D.; Meyer, P.M.; Becker, G.A.; Bronzel, M.; Marsteller, D.; Pastino, G.; Voges, O.; Rabinovich, L.; Knebel, H.; Zientek, F.; et al. Sigma-1 and dopamine D2/D3 receptor occu-pancy of pridopidine in healthy volunteers and patients with Huntington disease: A [18F] fluspidine and [18F] fallypride PET study. Eur. J. Nucl. Med. Mol. Imaging 2020. [Google Scholar] [CrossRef]
- Hebron, M.L.; Lonskaya, I.; Moussa, C.E.H. Nilotinib reverses loss of dopamine neurons and improvesmotorbehavior via au-tophagic degradation of α-synuclein in parkinson’s disease models. Hum. Mol. Genet. 2013, 22, 3315–3328. [Google Scholar] [CrossRef]
- Lonskaya, I.; Hebron, M.L.; Desforges, N.M.; Schachter, J.B.; Moussa, C.E.H. Nilotinib-induced autophagic changes increase endog-enous parkin level and ubiquitination, leading to amyloid clearance. J. Mol. Med. 2014, 92, 373–386. [Google Scholar] [CrossRef]
- Wang, S.-M.; Han, C.; Lee, S.-J.; Patkar, A.A.; Masand, P.S.; Pae, C.-U. A review of current evidence for vilazodone in major depressive disorder. Int. J. Psychiatry Clin. Pract. 2013, 17, 160–169. [Google Scholar] [CrossRef]
- Agarwal, R.; Baid, R. Flibanserin: A controversial drug for female hypoactive sexual desire disorder. Ind. Psychiatry J. 2018, 27, 154–157. [Google Scholar] [CrossRef]
- Greischel, A.; Binder, R.; Baierl, J. The Dipeptidyl Peptidase-4 Inhibitor Linagliptin Exhibits Time- and Dose-Dependent Localization in Kidney, Liver, and Intestine after Intravenous Dosing: Results from High Resolution Autoradiography in Rats. Drug Metab. Dispos. 2010, 38, 1443–1448. [Google Scholar] [CrossRef]
- Rios, M.B.; Ault, P. Identification of Side Effects Associated With Intolerance to BCR-ABL Inhibitors in Patients with Chronic Myeloid Leukemia. Clin. J. Oncol. Nurs. 2011, 15, 660–667. [Google Scholar] [CrossRef]
- Rm, B.; Wenning, G.K.; Kapfhammer, H.P. Huntington’s disease: Present treatments and future therapeutic modalities. Int. Clin. Psychopharmacol. 2004, 19, 51–62. [Google Scholar] [CrossRef]
- Walker, F.O. Huntington’s disease. Lancet 2007, 369, 218–228. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- PyMOL by Schrödinger. Version 2.3.2. Available online: https://www.pymol.org/ (accessed on 27 January 2021).
- Sterling, T.; Irwin, J.J. ZINC 15—Ligand Discovery for Everyone. J. Chem. Inf. Model 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Assempour, N. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.A.; James, C.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient op-timization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Ilari, A.; Fiorillo, A.; Poser, E.; Lalioti, V.S.; Sundell, G.N.; Ivarsson, Y.; Genovese, I.; Colotti, G. Structural basis of Sorcin-mediated calcium-dependent signal transduction. Sci. Rep. 2015, 5, 16828. [Google Scholar] [CrossRef] [PubMed]
- Poser, E.; Genovese, I.; Masciarelli, S.; Bellissimo, T.; Fazi, F.; Colotti, G. Surface Plasmon Resonance: A Useful Strategy for the Identification of Small Molecule Argonaute 2 Protein Binders. In Drug Target miRNA; Part of the Methods in Molecular Biology book series (MIMB); Schmidt, M., Ed.; Humana Press: New York, NY, USA, 2017; Volume 1517, pp. 223–237. ISBN1 978-1-4939-6561-8. ISBN2 978-1-4939-6563-2. [Google Scholar]
- Marder, K.; Zhao, H.; Myers, R.H.; Cudkowicz, M.; Kayson, E.; Kieburtz, K.; Orme, C.; Paulsen, J.S.; Penney, J.B.; Siemers, E.; et al. Rate of functional decline in Huntington’s disease. Neurology 2000, 54, 452. [Google Scholar] [CrossRef] [PubMed]
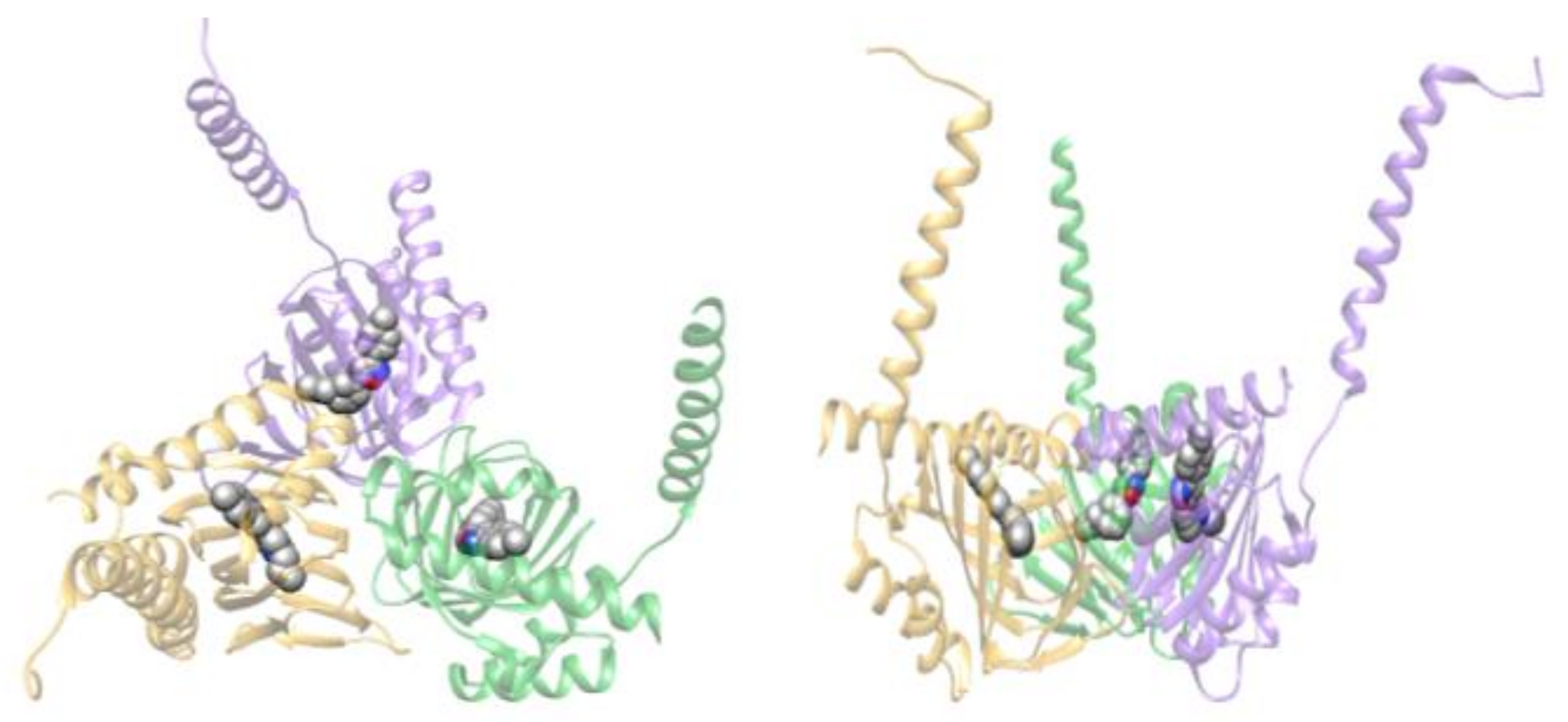

| PDB ID (Chain) | Res (Å) | Ligand Short (PDB) | Ligand Complete (Formula) | Ligand Function | Ref |
|---|---|---|---|---|---|
| 5HK1 (ABC) | 2.51 | PD144418 (61W) | 3-(4-methylphenyl)-5-(1-propyl-3,6-dihydro-2H-pyridin-5-yl)-1,2-oxazole (C18H22N2O) | Antagonist, nM binder | [22] |
| 5HK2 (ABC) | 3.20 | 4-IBP (61V) | N-(1-benzylpiperidin-4-yl)-4-iodobenzamide (C19H21IN2O) | Antagonist, nM binder | |
| 6DJZ (ABC) | 3.08 | Haloperidol (GMJ) | 4-[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]-1-(4-fluorophenyl)butan-1-one (C21H23ClFNO2) | Antagonist | [2] |
| 6DK0 (ABC) | 2.90 | NE-100 (GKY) | N-{2-[4-methoxy-3-(2-phenylethoxy)phenyl]ethyl}-N-propylpropan-1-amine (C23H33NO2) | Antagonist | |
| 6DK1 (ABC) | 3.12 | (+)-Pentazocine (GM4) | (2S,6S,11S)-6,11-dimethyl-3-(3-methylbut-2-en-1-yl)-1,2,3,4,5,6-hexahydro-2,6-methano-3-benzazocin-8-ol (C19 H27NO) | Agonist |
| Drug Name | ZINC ID Number | Vina Best E (kcal/mol) | Chimera | Vina/ATD Comparison | ATD Largest Cluster | Chimera | ATD Lowest Energy Cluster | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #hb | #cla | #con | RMSD (Å) | Best E (kcal/mol) | Mean E (kcal/mol) | #pos | #hb | #cla | #con | Best E (kcal/mol) | Mean E (kcal/mol) | #pos | |||
| Risperidone | 538312 | −12.6 | 0 | 0 | 70 | 2.9 | −11.5 | −11.1 | 37 | 0 | 1 | 99 | −11.5 | −11.1 | 37 |
| Axitinib (cis) | 11616882 | −12.6 | 3 | 0 | 59 | 0.5 | −10.9 | −10.8 | 41 | 1 | 0 | 70 | −10.9 | −10.8 | 41 |
| Nilotinib (*) | 6716957 | −12.3 | 4 | 1 | 110 | 6.4 | −7.8 | −5.5 | 16 | 2 | 11 | 148 | −9.5 | −3.8 | 6 |
| Paliperidone (*) | 4214700 | −12.2 | 0 | 0 | 72 | 3.1 | −11.5 | −10.7 | 46 | 1 | 2 | 111 | −11.5 | −10.9 | 4 |
| Paliperidone (*) | 1481956 | −12.2 | 0 | 0 | 80 | 10.7 | −11.1 | −10.6 | 14 | 1 | 1 | 93 | −11.9 | −11.2 | 4 |
| Atovaquone | 100017856 | −11.9 | 0 | 0 | 51 | 2.9 | −11.1 | −10.7 | 73 | 0 | 1 | 68 | −11.3 | −11.0 | 27 |
| Droperidolo | 19796080 | −11.9 | 0 | 0 | 58 | 3.3 | −9.8 | −9.4 | 27 | 0 | 1 | 83 | −10.2 | −9.9 | 13 |
| Dolutegravir | 58581064 | −11.9 | 3 | 0 | 58 | 9.4 | −9.4 | −9.2 | 52 | 0 | 0 | 71 | −10.3 | −9.9 | 43 |
| Iloperidone (*) | 1548097 | −11.7 | 1 | 0 | 70 | 10.7 | −10.2 | −9.7 | 39 | 0 | 2 | 84 | −10.4 | −9.6 | 7 |
| Linagliptin (*) | 3820029 | −11.7 | 0 | 1 | 97 | 0.6 | −12.4 | −10.0 | 33 | 1 | 6 | 112 | −12.4 | −10.0 | 33 |
| Flibanserin (*) | 52716421 | −11.6 | 0 | 0 | 48 | 1.0 | −9.4 | −9.2 | 71 | 0 | 0 | 59 | −10.0 | −9.4 | 11 |
| Cilostazol | 1552174 | −11.6 | 0 | 0 | 71 | 9.4 | −9.2 | −8.7 | 25 | 1 | 0 | 68 | −9.7 | −9.2 | 19 |
| Nebivolol | 11681534 | −11.6 | 2 | 0 | 62 | 10.1 | −10.0 | −8.7 | 73 | 1 | 1 | 81 | −10.0 | −8.7 | 73 |
| Vilazodone (*) | 1542113 | −11.6 | 2 | 2 | 81 | 6.1 | −9.2 | −8.2 | 38 | 2 | 7 | 126 | −9.4 | −7.6 | 7 |
| Azilsartan Medoxomil | 14210642 | −11.5 | 2 | 1 | 123 | 9.0 | −7.9 | −4.4 | 18 | 0 | 12 | 143 | −9.0 | −4.4 | 8 |
| Doxercalciferol | 4641374 | −11.5 | 0 | 2 | 96 | 3.4 | −12.2 | −10.1 | 55 | 1 | 2 | 112 | −12.2 | −10.1 | 55 |
| Cinacalcet | 1550499 | −11.5 | 0 | 0 | 52 | 8.7 | −10.4 | −9.8 | 23 | 1 | 0 | 85 | −10.4 | −9.8 | 23 |
| Nebivolol | 4213946 | −11.5 | 5 | 0 | 62 | 2.4 | −10.9 | −9.5 | 58 | 2 | 0 | 78 | −10.9 | −9.5 | 58 |
| Nebivolol | 5844792 | −11.5 | 4 | 0 | 61 | 2.5 | −11.1 | −9.3 | 62 | 4 | 0 | 84 | −11.1 | −9.3 | 62 |
| Axitinib (trans) | 11616882 | −11 | 0 | 0 | 63 | 2.3 | −7.6 | −7.6 | 75 | 0 | 0 | 61 | −7.7 | −7.6 | 15 |
| Ligand Name (PDB) | ZINC ID | Vina Best E (kcal/mol) | Ranking |
|---|---|---|---|
| Haloperidol (GMJ) | 537822 | −10.9 | 51 |
| 4-IBP (61V) | 1642602 | −10.7 | 71 |
| PD144418 (61W) | 5862 | −10.1 | 139 |
| NE−100 (GKY) | 598622 | −8.9 | 406 |
| Pridopidine | 22063703 | −8.7 | 458 |
| (+)-Pentazocine (GM4) | 596 | −8.6 | 516 |
| Phencyclidine | 968311 | −8.5 | 529 |
| Remoxipride | 2021799 | −7.9 | 723 |
| Captodiame | 2040210 | −7.9 | 733 |
| N,N-Dimethyltryptamine | 897457 | −7.1 | 1002 |
| FDA Name | ZINC ID | KD (μM) |
|---|---|---|
| Pridopidine | ZINC000022063703 | 14.8 ± 1.0 (*) |
| Flibanserin | ZINC000052716421 | 4.9 ± 1.1 |
| Iloperidone | ZINC000001548097 | 5.1 ± 0.6 |
| Linagliptin | ZINC000003820029 | 9.6 ± 1.0 |
| Nilotinib | ZINC000006716957 | 22.0 ± 3.0 |
| Paliperidone | ZINC000004214700 | 46.0 ± 21 |
| Vilazodone | ZINC000001542113 | 52.0 ± 9.0 |
| Cell Line | N° Cells 48 h (Mean ± SD) | N° Cells 72 h (Mean ± SD) |
|---|---|---|
| CTR1 | 2.67 × 105 ± 1.81 × 104 | 9.95 × 105 ± 9.37 × 104 |
| CTR2 | 2.34 × 105 ± 2.30 × 104 | 7.86 × 105 ± 1.43 × 105 |
| HD1 | 1.54 × 105 ± 4.95 × 104 * | 4.00 × 105 ± 1.20 × 105 *** ### |
| HD2 | 1.86 × 105 ± 1.43 × 104 | 5.22 × 105 ± 7.77 × 104 *** ### |
| Cell Line | Growth Rate 48 h (Mean ± SD) | Growth Rate 72 h (Mean ± SD) |
|---|---|---|
| CTR1 | 2.67 ± 0.18 | 3.73 ± 0.11 |
| CTR2 | 2.34 ± 0.23 | 3.34 ± 0.30 |
| HD1 | 1.54 ± 0.50 *** ## | 2.63 ± 0.34 *** ## |
| HD2 | 1.86 ± 0.14 ** # | 2.79 ± 0.21 *** ## |
| Cell Line | % Dead Cells 48 h (Mean ± SD) | % Dead Cells 72 h (Mean ± SD) |
|---|---|---|
| CTR1 | 9.71 ± 1.19 | 10.49 ± 0.93 |
| CTR2 | 8.75 ± 0.63 | 10.19 ± 0.43 |
| HD1 | 10.13 ± 0.56 # | 12.25 ± 0.55 ** ## |
| HD2 | 11.29 ± 0.66 ** ### | 11.77 ± 0.77 * ## |
| Cell Line | Drug Treatment | N° Cells 48 h (Mean ± SD) | N° Cells 72 h (Mean ± SD) |
|---|---|---|---|
| CTR1 | DMSO | 2.28 × 105 ± 3.01 × 104 | 5.28 × 105 ± 1.05 × 105 ## |
| Pridopidine | 2.58 × 105 ± 1.56 × 104 | 6.37 × 105 ± 7.71 × 104 ** | |
| Flibanserin | 1.80 × 105 ± 1.80 × 104 # | 5.82 × 105 ± 4.50 × 104 | |
| Iloperidone | 1.98 × 105 ± 3.02 × 104 | 4.78 × 105 ± 9.89 × 104 ## | |
| Linagliptin | 1.98 × 105 ± 1.93 × 104 | 5.85 × 105 ± 9.45 × 104 | |
| Nilotinib | 2.16 × 105 ± 1.80 × 104 | 5.20 × 105 ± 7.30 × 104 ## | |
| Paliperidone | 1.86 × 105 ± 1.82 × 104 | 3.89 × 105 ± 1.70 × 104 ** ### | |
| Vilazodone | 2.58 × 105 ± 1.60 × 104 | 7.45 × 105 ± 8.74 × 104 *** ## | |
| CTR2 | DMSO | 1.26 × 105 ± 1.80 × 104 | 2.86 × 105 ± 2.59 × 104 ### |
| Pridopidine | 1.74 × 105 ± 1.82 × 104 | 4.69 × 105 ± 3.44 × 104 *** | |
| Flibanserin | 1.80 × 105 ± 1.11 × 104 * | 3.14 × 105 ± 4.26 × 104 ### | |
| Iloperidone | 1.50 × 105 ± 1.57 × 104 | 3.25 × 105 ± 5.37 × 104 ### | |
| Linagliptin | 1.80 × 105 ± 1.21 × 104 * | 4.22 × 105 ± 5.00 × 104 *** | |
| Nilotinib | 1.32 × 105 ± 2.40 × 104 | 2.35 × 105 ± 2.35 × 104 * ### | |
| Paliperidone | 1.44 × 105 ± 2.30 × 104 | 3.48 × 105 ± 7.39 × 104 * ### | |
| Vilazodone | 1.56 × 105 ± 2.80 × 104 | 3.82 × 105 ± 5.64 × 104 ** ## | |
| HD1 | DMSO | 1.10 × 105 ± 1.00 × 104 | 1.65 × 105 ± 2.49 × 104 # |
| Pridopidine | 1.30 × 105 ± 1.86 × 104 | 2.34 × 105 ± 3.52 × 104 * | |
| Flibanserin | 1.00 × 105 ± 1.92 × 104 | 2.36 × 105 ± 1.28 x 104 ** | |
| Iloperidone | 1.40 × 105 ± 3.21 × 104 | 2.63 × 105 ± 4.29 × 104 ** | |
| Linagliptin | 1.60 × 105 ± 1.92 × 104 | 3.35 × 105 ± 3.71 × 104 *** ## | |
| Nilotinib | 1.38 × 105 ± 8.49 × 103 | 2.24 × 105 ± 1.76 × 104 * | |
| Paliperidone | 1.80 × 105 ± 2.83 × 104 * | 4.00 × 105 ± 1.13 × 105 *** ### | |
| Vilazodone | 2.06 × 105 ± 1.98 × 104 ** # | 4.97 × 105 ± 1.06 × 105 *** ### | |
| HD2 | DMSO | 1.08 × 105 ± 1.70 × 104 ## | 2.01 × 105 ± 2.21 × 104 # |
| Pridopidine | 1.56 × 105 ± 1.37 × 104 ** | 2.43 × 105 ± 1.53 × 104 * | |
| Flibanserin | 1.14 × 105 ± 1.60 × 104 # | 1.23 × 105 ± 2.16 × 104 *** ### | |
| Iloperidone | 1.56 × 105 ± 1.54 × 104 ** | 2.90 × 105 ± 3.30 × 104 *** ## | |
| Linagliptin | 1.32 × 105 ± 1.30 × 104 | 1.83 × 105 ± 2.46 ×104 ## | |
| Nilotinib | 1.20 × 105 ± 1.55 × 104 # | 2.56 × 105 ± 1.92 × 104 ** | |
| Paliperidone | 1.56 × 105 ± 1.20 × 104 ** | 3.19 × 105 ± 3.46 × 104 *** ### | |
| Vilazodone | 1.20 × 105 ± 2.20 × 104 # | 2.26 × 105 ± 5.46 × 104 |
| Cell Line | Drug Treatment | Growth Rate 48 h (Mean ± SD) | Growth Rate 72 h (Mean ± SD) |
|---|---|---|---|
| CTR1 | DMSO | 2.28 ± 0.30 | 2.30 ± 0.17 |
| Pridopidine | 2.58 ± 0.16 | 2.46 ± 0.15 | |
| Flibanserin | 1.80 ± 0.18 ** ### | 3.24 ± 0.14 *** ### | |
| Iloperidone | 1.98 ± 0.30 ## | 2.40 ± 0.14 | |
| Linagliptin | 1.98 ± 0.19 ## | 2.94 ± 0.21 ** ## | |
| Nilotinib | 2.16 ± 0.18 ## | 2.40 ± 0.14 | |
| Paliperidone | 1.86 ± 0.18 ** ### | 2.10 ± 0.16 # | |
| Vilazodone | 2.58 ± 0.16 | 2.88 ± 0.16 ** ## | |
| CTR2 | DMSO | 1.26 ± 0.18 ## | 2.28 ± 0.12 ## |
| Pridopidine | 1.74 ± 0.18 ** | 2.70 ± 0.12 ** | |
| Flibanserin | 1.80 ± 0.11 ** | 1.74 ± 0.13 ** ### | |
| Iloperidone | 1.50 ± 0.16 | 2.16 ± 0.15 ## | |
| Linagliptin | 1.80 ± 0.12 ** | 2.34 ± 0.12 # | |
| Nilotinib | 1.32 ± 0.24 ## | 1.80 ± 0.15 ** ### | |
| Paliperidone | 1.44 ± 0.23 # | 2.4 ± 0.13 # | |
| Vilazodone | 1.56 ± 0.28 * | 2.46 ± 0.08 | |
| HD1 | DMSO | 1.10 ± 0.10 | 1.50 ± 0.13 |
| Pridopidine | 1.30 ± 0.19 | 1.80 ± 0.15 | |
| Flibanserin | 1.00 ± 0.19 | 2.40 ± 0.35 *** ## | |
| Iloperidone | 1.40 ± 0.32 | 1.91 ± 0.23 * | |
| Linagliptin | 1.60 ± 0.19 ** | 2.10 ± 0.20 ** | |
| Nilotinib | 1.38 ± 0.09 | 1.62 ± 0.03 | |
| Paliperidone | 1.80 ± 0.28 ** # | 2.20 ± 0.28 ** | |
| Vilazodone | 2.06 ± 0.20 *** ## | 2.40 ± 0.28 *** ## | |
| HD2 | DMSO | 1.08 ± 0.17 ### | 1.87 ± 0.09 ## |
| Pridopidine | 1.56 ± 0.14 *** | 1.56 ± 0.09 ** | |
| Flibanserin | 1.14 ± 0.16 ## | 1.08 ± 0.09 *** ### | |
| Iloperidone | 1.56 ± 0.15 *** | 1.86 ± 0.06 ## | |
| Linagliptin | 1.32 ± 0.13 * # | 1.38 ± 0.08 *** | |
| Nilotinib | 1.20 ± 0.16 ## | 2.14 ± 0.12 * ### | |
| Paliperidone | 1.56 ± 0.12 *** | 2.04 ± 0.07 ### | |
| Vilazodone | 1.20 ± 0.22 ## | 1.87 ± 0.12 ## |
| Cell Line | Drug Treatment | % Dead Cells 48 h (Mean ± SD) | % Dead Cells 72 h (Mean ± SD) |
|---|---|---|---|
| CTR1 | DMSO | 7.43 ± 0.89 | 11.23 ± 2.02 |
| Pridopidine | 6.62 ± 0.80 | 10.58 ± 1.91 | |
| Flibanserin | 9.24 ± 1.08 | 9.95 ± 0.88 | |
| Iloperidone | 12.81 ± 2.26 ** ## | 13.24 ± 1.48 | |
| Linagliptin | 8.46 ± 1.00 | 9.01 ± 1.66 | |
| Nilotinib | 7.82 ± 0.93 | 10.81 ± 1.95 | |
| Paliperidone | 8.97 ± 1.05 | 12.17 ± 2.16 | |
| Vilazodone | 6.63 ± 0.80 | 9.18 ± 1.69 | |
| CTR2 | DMSO | 7.69 ± 0.91 | 10.58 ± 1.91 |
| Pridopidine | 9.52 ± 1.11 | 9.97 ± 2.12 | |
| Flibanserin | 9.25 ± 1.08 | 14.32 ± 2.48 | |
| Iloperidone | 10.88 ± 1.25 | 11.87 ± 2.12 | |
| Linagliptin | 9.24 ± 1.08 | 11.06 ± 1.99 | |
| Nilotinib | 18.04 ± 3.00 ** ## | 18.40 ± 3.92 ** ## | |
| Paliperidone | 11.28 ± 1.29 | 11.86 ± 3.43 | |
| Vilazodone | 10.51 ± 1.21 | 10.84 ± 2.28 | |
| HD1 | DMSO | 14.27 ± 1.58 | 16.23 ± 2.75 |
| Pridopidine | 12.35 ± 1.39 | 13.91 ± 2.42 | |
| Flibanserin | 15.48 ± 1.68 | 15.04 ± 1.06 | |
| Iloperidone | 11.57 ± 1.32 | 13.24 ± 2.33 | |
| Linagliptin | 10.27 ± 1.19 * | 12.17 ± 2.16 * | |
| Nilotinib | 14.00 ± 1.89 | 17.54 ± 2.71 | |
| Paliperidone | 7.78 ± 0.99 ** # | 10.64 ± 2.20 ** | |
| Vilazodone | 7.10 ± 0.54 ** ## | 9.18 ± 0.36 ** # | |
| HD2 | DMSO | 13.06 ± 3.23 | 13.91 ± 2.42 |
| Pridopidine | 10.51 ± 1.21 | 15.71 ± 2.68 | |
| Flibanserin | 20.3 ± 3.28 ** ## | 21.19 ± 3.38 ** # | |
| Iloperidone | 10.52 ± 1.20 | 13.52 ± 2.37 | |
| Linagliptin | 12.18 ± 1.38 | 17.40 ± 2.91 | |
| Nilotinib | 13.24 ± 1.48 | 13.77 ± 0.41 | |
| Paliperidone | 10.51 ± 1.21 | 12.48 ± 2.21 | |
| Vilazodone | 13.24 ± 1.48 | 13.46 ± 2.36 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Battista, T.; Pascarella, G.; Staid, D.S.; Colotti, G.; Rosati, J.; Fiorillo, A.; Casamassa, A.; Vescovi, A.L.; Giabbai, B.; Semrau, M.S.; et al. Known Drugs Identified by Structure-Based Virtual Screening Are Able to Bind Sigma-1 Receptor and Increase Growth of Huntington Disease Patient-Derived Cells. Int. J. Mol. Sci. 2021, 22, 1293. https://doi.org/10.3390/ijms22031293
Battista T, Pascarella G, Staid DS, Colotti G, Rosati J, Fiorillo A, Casamassa A, Vescovi AL, Giabbai B, Semrau MS, et al. Known Drugs Identified by Structure-Based Virtual Screening Are Able to Bind Sigma-1 Receptor and Increase Growth of Huntington Disease Patient-Derived Cells. International Journal of Molecular Sciences. 2021; 22(3):1293. https://doi.org/10.3390/ijms22031293
Chicago/Turabian StyleBattista, Theo, Gianmarco Pascarella, David Sasah Staid, Gianni Colotti, Jessica Rosati, Annarita Fiorillo, Alessia Casamassa, Angelo Luigi Vescovi, Barbara Giabbai, Marta Stefania Semrau, and et al. 2021. "Known Drugs Identified by Structure-Based Virtual Screening Are Able to Bind Sigma-1 Receptor and Increase Growth of Huntington Disease Patient-Derived Cells" International Journal of Molecular Sciences 22, no. 3: 1293. https://doi.org/10.3390/ijms22031293
APA StyleBattista, T., Pascarella, G., Staid, D. S., Colotti, G., Rosati, J., Fiorillo, A., Casamassa, A., Vescovi, A. L., Giabbai, B., Semrau, M. S., Fanelli, S., Storici, P., Squitieri, F., Morea, V., & Ilari, A. (2021). Known Drugs Identified by Structure-Based Virtual Screening Are Able to Bind Sigma-1 Receptor and Increase Growth of Huntington Disease Patient-Derived Cells. International Journal of Molecular Sciences, 22(3), 1293. https://doi.org/10.3390/ijms22031293







