Acute Intermittent Porphyria: An Overview of Therapy Developments and Future Perspectives Focusing on Stabilisation of HMBS and Proteostasis Regulators
Abstract
1. Introduction
1.1. Acute Intermittent Porphyria
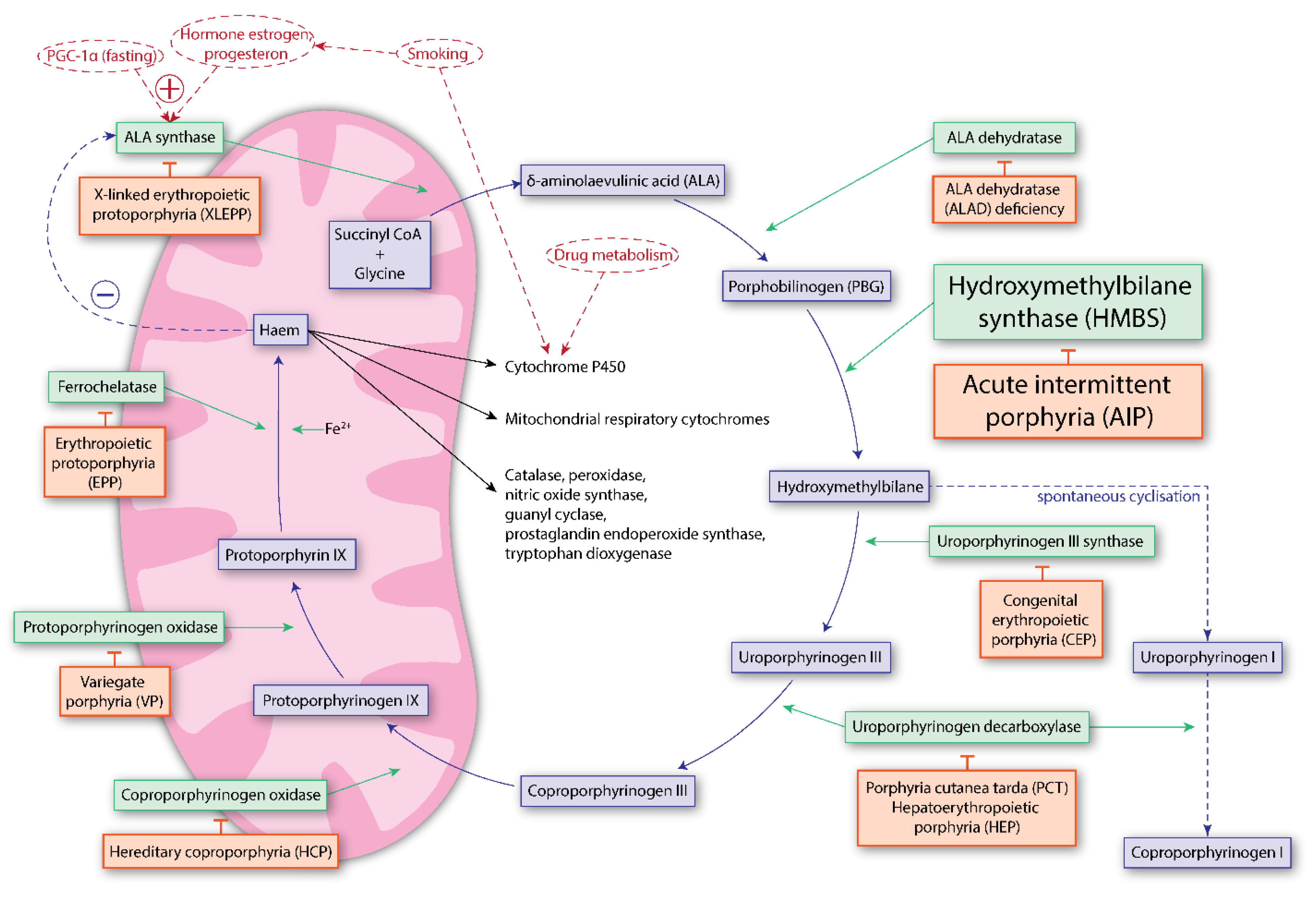
1.2. Pathophysiology of Acute Intermittent Porphyria
1.3. Hydroxymethylbilane Synthase
| PDB | Res. [Å] | Organism (UniProt) | State | Mutations | Areas NOT Defined in the Electron Density (Total Number of Residues) | Key Findings | Ref. |
|---|---|---|---|---|---|---|---|
| 1PDA | 1.76 | E. coli (P06983) | E | — | A:1–2, 49–57, 308–313 (313) | First HMBS structure description. Already suggest movement in domain 3 during the elongation process. Importance of D84 (D99 in human HMBS). Sequence comparison to higher organisms. | [65] |
| 1YPN | 2.3 | E. coli (P06983) | E | K59Q | A:1–2, 48–57, 307–313 (313) | K59Q affects the rate of intermediate formation. Crystal packing inhibits the domain movements in time-resolved study. | [67] |
| 1AH5 | 2.4 | E. coli (P06983) | E | Se-Met labelled | A: 1–2, 47–58 (313) | Seleno-methionine labelling for phasing. Structural study to confirm suitability to time-resolved experiments. | [68] |
| 2YPN | 2.3 | E. coli (P06983) | E | — | A:1–2, 43–59 (313) | Methodology based research on Laue diffraction. | [69] |
| 1GTK | 1.66 | E. coli (P06983) | E | — | A:1–2, 43–59 (313) | Highest resolution structure for E. coli | [70] |
| 3ECR | 2.18 | H. sapiens (P08397) | E | — | A: 1–17, 57–75, 259–260, 357–361 (361) B: 1–18, 57–69, 259–260, 301–312, 356–361 (361) | First to describe human HMBS structure. Confirming similarity to E. coli HMBS. Mapping disease related mutants on the human HMBS structure. Suggesting a role in domain movements for H120. | [66] |
| 3EQ1 | 2.8 | H. sapiens (P08397) | E | R167Q | A: 1–18, 56–76, 303, 307–310, 357–361 (361) B: 1–18, 56–76, 303, 307–310, 357–361 (361) | Specific activity of R167Q HMBS is lowered to 10% compared to wild-type. Accumulation of intermediates detected using native PAGE. | [71] |
| 4HTG | 1.45 | A. thaliana (Q43316) | E | — | A: 1–9, 312–320 (320) 1 | Cofactor in oxidised form. First structured active-site loop. Highest resolution HMBS structure to date. | [72] |
| 4MLQ 4MLV | 1.6 1.45 | B. megaterium (Q8GCA8) | E | — | A: 40–60, 309–310 (310) A: 40–60, 310 (310) | Mixture of oxidised and reduced form for the cofactor. | [73] |
| 5H6O | 2.7 | V. cholerae (Q9KVM1) | E | — | A:1–3, 43–61, 304–311 (311) | — | |
| 5OV4 5OV5 5OV6 | 2.69 1.81 1.87 | B. megaterium (Q8GCA8) | E E E | D82A D82E D82N | A: 40–60, 310 A: 41–60, 310 A: 40–59, 310 (310) | By mutating D82, binding/formation of the cofactor could be altered, and enzyme inactivated. D82 corresponds to D99 in humans. | [74] |
| 5M6R 5M7F 2 | 2.73 2.78 | H. sapiens (P08397) | ES2 E | — | A: 1–17, 358–361B: 1–17, 62–75, 354–361 (361) A: 18–19, 58–74, 354–361 B: 18–19, 57–74, 354–361 (344) | First ES2 crystal structure. Partially ordered structure for the active-site loop. | [75] |
| 7AAJ 7AAK | 1.8 1.7 | H. sapiens (P08397) | E ES2 | A,B: 1–17, 62–75, 354–361 (361) A: 1–17, 358–361 B: 1–17, 62–75, 354–361 (361) | Highest resolution human HMBS structure. First disease mutant structure to show accumulation of one intermediate. Partially ordered structure for active-site loop. | [76] |
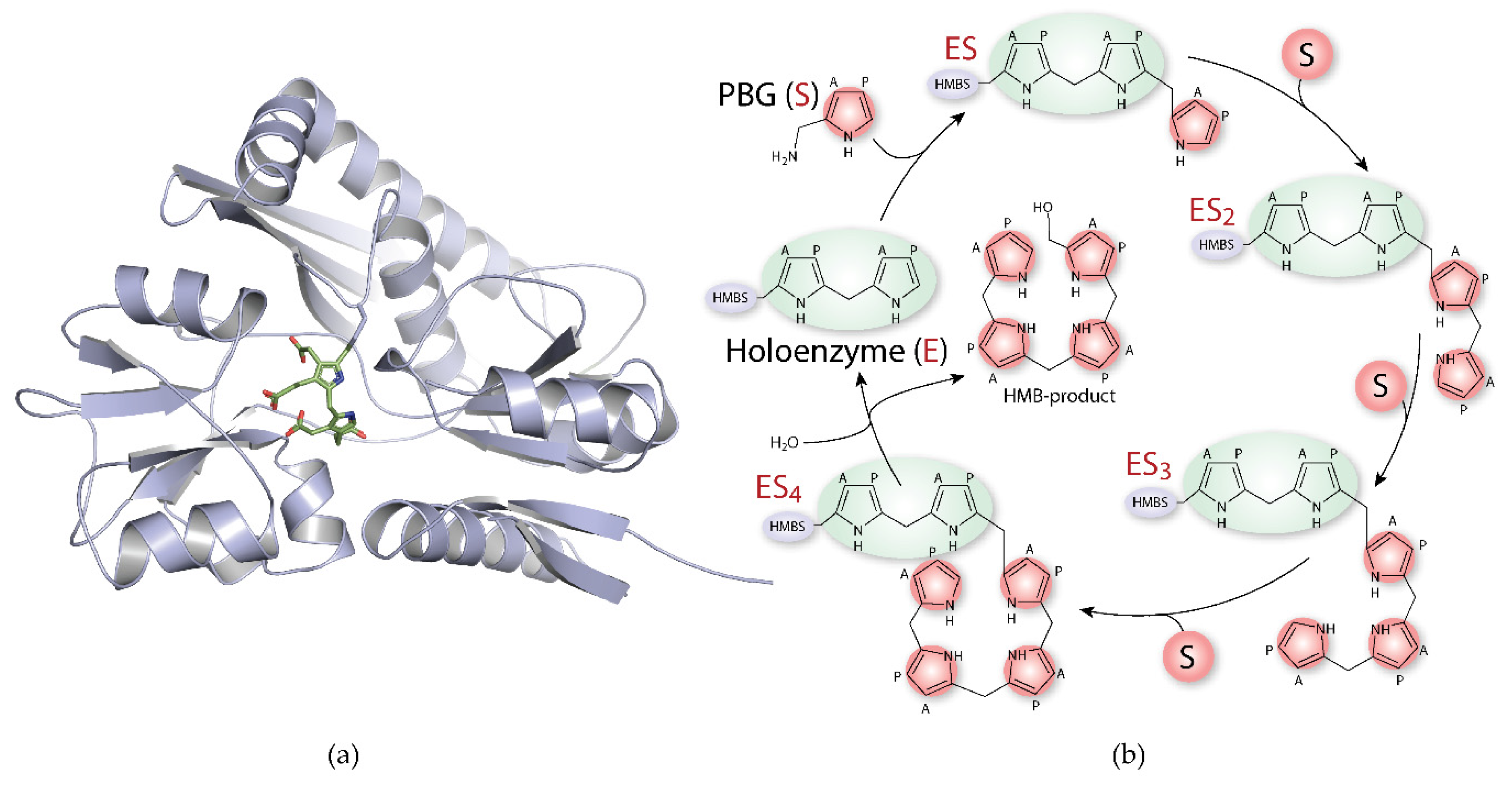
2. Current Treatment Options for Acute Intermittent Porphyria
2.1. Established Treatments for Sporadic and Recurrent Acute Attacks
2.2. Liver and Kidney Transplantation
2.3. Ribonucleic acid (RNA) Interference Therapy
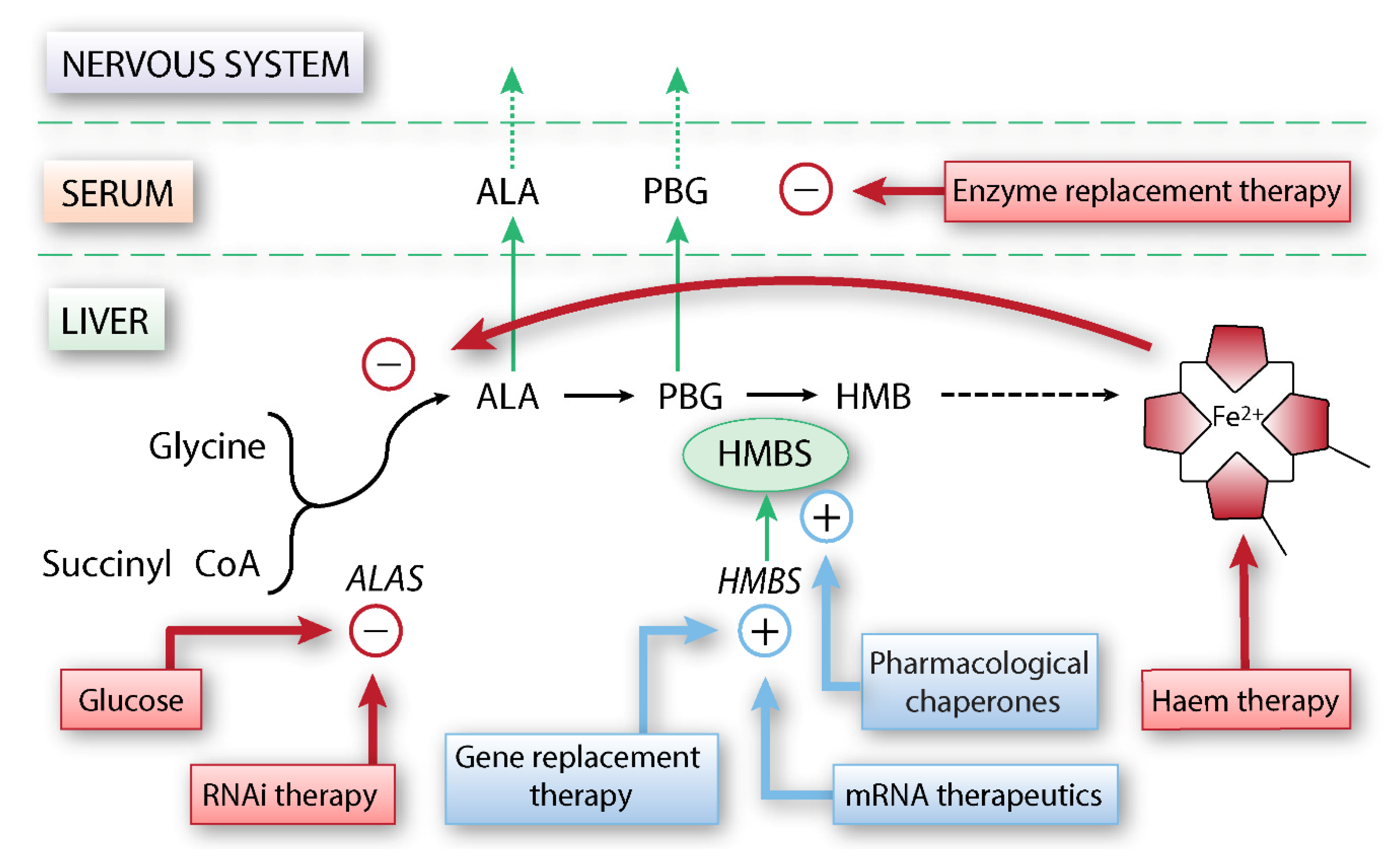
3. Other Potential Therapeutic Developments
3.1. Enzyme Replacement Therapy
3.2. Gene Replacement Therapy and mRNA Therapeutics
3.3. Hepatocyte Transplantation
4. Protein Stabilisation and Proteostasis Regulation—Emerging Treatment Opportunities
4.1. Protein Folding and Stability in HMBS
4.2. Proteostasis Regulation
4.3. Therapeutic Strategies Based on the Regulation of the Proteostasis Network
4.4. Pharmacological Chaperones
5. Structural and Mechanistic Challenges of HMBS for Therapeutic Developments
6. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AAV | adeno-associated virus |
| ALA | δ-aminolaevulinic acid |
| ALAD | δ-aminolaevulinic acid dehydratase |
| ALAS | δ-aminolaevulinic acid synthase |
| CYP | cytochrome P450-dependent oxidase |
| DPM | Dipyrromethane |
| E, E(holo) | holoenzyme |
| ES1–4 | Enzyme-substrate intermediates, with 1 to 4 substrate units |
| GABA | γ-aminobutyric acid |
| GATs | GABA transporters |
| HMB | 1-hydroxymethylbilane |
| HMBS | Hydroxymethylbilane synthase |
| PBG | Porphobilinogen |
| PEPT2 | peptide transporter 2 |
| PGC-1α | peroxisome proliferator-activated receptor γ coactivator 1α |
| ROS | reactive oxygen species |
| siRNA | small interference RNA |
| Tm | half-denaturing temperature |
| UPS | ubiquitin-proteasome system |
References
- Badminton, M.; Whatley, S.; Aarsand, A.K. Porphyrins and Porphyrias. In Tietz Textbook of Clinical Chemistry and Molecular Diagnostics, 8th ed.; Rifai, N., Horvath, A.R., Wittwer, C., Eds.; Elsevier: St. Louis, MO, USA, 2018; pp. 776–880. [Google Scholar]
- Andersson, C.; Wikberg, A.; Stegmayr, B.; Lithner, F. Renal symptomatology in patients with acute intermittent porphyria. A population-based study. J. Intern. Med. 2000, 248, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Baravelli, C.M.; Sandberg, S.; Aarsand, A.K.; Nilsen, R.M.; Tollånes, M.C. Acute hepatic porphyria and cancer risk: A nationwide cohort study. J. Intern. Med. 2017, 282, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Pallet, N.; Mami, I.; Schmitt, C.; Karim, Z.; François, A.; Rabant, M.; Nochy, D.; Gouya, L.; Deybach, J.-C.; Xu-Dubois, Y.; et al. High prevalence of and potential mechanisms for chronic kidney disease in patients with acute intermittent porphyria. Kidney Int. 2015, 88, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Baumann, K.; Kauppinen, R. Penetrance and predictive value of genetic screening in acute porphyria. Mol. Genet. Metab. 2020, 130, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Hift, R.J.; Meissner, P.N. An Analysis of 112 Acute Porphyric Attacks in Cape Town, South Africa: Evidence that acute intermittent porphyria and variegate porphyria differ in susceptibility and severity. Medicine 2005, 84, 48–60. [Google Scholar] [CrossRef]
- Puy, H.; Gouya, L.; Deybach, J.-C. Porphyrias. Lancet 2010, 375, 924–937. [Google Scholar] [CrossRef]
- Fu, Y.; Jia, J.; Yue, L.; Yang, R.; Guo, Y.; Ni, X.; Shi, T. Systematically Analyzing the Pathogenic Variations for Acute Intermittent Porphyria. Front. Pharmacol. 2019, 10, 1018. [Google Scholar] [CrossRef]
- Fontanellas, A.; Ávila, M.A.; Berraondo, P. Emerging therapies for acute intermittent porphyria. Expert Rev. Mol. Med. 2016, 18, e17. [Google Scholar] [CrossRef]
- Meyer, U.A.; Strand, L.J.; Doss, M.; Rees, A.C.; Marver, H.S. Intermittent Acute Porphyria—Demonstration of a Genetic Defect in Porphobilinogen Metabolism. New Engl. J. Med. 1972, 286, 1277–1282. [Google Scholar] [CrossRef]
- Badminton, M.N.; Elder, G.H. Molecular mechanisms of dominant expression in porphyria. J. Inherit. Metab. Dis. 2005, 28, 277–286. [Google Scholar] [CrossRef]
- Chen, B.; Solis-Villa, C.; Hakenberg, J.; Qiao, W.; Srinivasan, R.R.; Yasuda, M.; Balwani, M.; Doheny, D.; Peter, I.; Chen, R.; et al. Acute Intermittent Porphyria: Predicted Pathogenicity of HMBS Variants Indicates Extremely Low Penetrance of the Autosomal Dominant Disease. Hum. Mutat. 2016, 37, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Lenglet, H.; Schmitt, C.; Grange, T.; Manceau, H.; Karboul, N.; Bouchet-Crivat, F.; Robréau, A.-M.; Nicolas, G.; Lamoril, J.; Simonin, S.; et al. From a dominant to an oligogenic model of inheritance with environmental modifiers in acute intermittent porphyria. Hum. Mol. Genet. 2018, 27, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, M.; Chen, B.; Desnick, R.J. Recent advances on porphyria genetics: Inheritance, penetrance & molecular heterogeneity, including new modifying/causative genes. Mol. Genet. Metab. 2019, 128, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Elder, G.; Harper, P.; Badminton, M.; Sandberg, S.; Deybach, J.-C. The incidence of inherited porphyrias in Europe. J. Inherit. Metab. Dis. 2013, 36, 849–857. [Google Scholar] [CrossRef]
- Floderus, Y.; Sardh, E.; Möller, C.; Andersson, C.; Rejkjaer, L.; Andersson, D.E.H.; Harper, P. Variations in Porphobilinogen and 5-Aminolevulinic Acid Concentrations in Plasma and Urine from Asymptomatic Carriers of the Acute Intermittent Porphyria Gene with Increased Porphyrin Precursor Excretion. Clin. Chem. 2006, 52, 701–707. [Google Scholar] [CrossRef]
- Fraunberg, M.V.U.Z.; Pischik, E.; Udd, L.; Kauppinen, R. Clinical and Biochemical Characteristics and Genotype-Phenotype Correlation in 143 Finnish and Russian Patients with Acute Intermittent Porphyria. Medicine 2005, 84, 35–47. [Google Scholar] [CrossRef]
- Llewellyn, D.H.; Smyth, S.J.; Elder, G.H.; Hutchesson, A.C.; Rattenbury, J.M.; Smith, M.F. Homozygous acute intermittent porphyria: Compound heterozygosity for adjacent base transitions in the same codon of the porphobilinogen deaminase gene. Hum. Genet. 1992, 89, 97–98. [Google Scholar] [CrossRef]
- Solis, C.; Martínez-Bermejo, A.; Naidich, T.P.; Kaufmann, W.E.; Astrin, K.H.; Bishop, D.F.; Desnick, R.J. Acute Intermittent Porphyria—Studies of the severe homozygous dominant disease provides insights into the neurologic attacks in acute porphyrias. Arch. Neurol. 2004, 61, 1764–1770. [Google Scholar] [CrossRef]
- Balwani, M.; Singh, P.; Seth, A.; Debnath, E.M.; Naik, H.; Doheny, D.; Chen, B.; Yasuda, M.; Desnick, R.J. Acute Intermittent Porphyria in children: A case report and review of the literature. Mol. Genet. Metab. 2016, 119, 295–299. [Google Scholar] [CrossRef]
- Elder, G.H. Hepatic porphyrias in children. J. Inherit. Metab. Dis. 1997, 20, 237–246. [Google Scholar] [CrossRef]
- Hultdin, J.; Schmauch, A.; Wikberg, A.; Dahlquist, G.; Andersson, C. Acute intermittent porphyria in childhood: A population-based study. Acta Paediatr. 2003, 92, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Bonkovsky, H.L.; Dixon, N.; Rudnick, S. Pathogenesis and clinical features of the acute hepatic porphyrias (AHPs). Mol. Genet. Metab. 2019, 128, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Pischik, E.; Kauppinen, R. An update of clinical management of acute intermittent porphyria. Appl. Clin. Genet. 2015, 8, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Harper, P.; Sardh, E. Management of acute intermittent porphyria. Expert Opin. Orphan Drugs 2014, 2, 349–368. [Google Scholar] [CrossRef]
- Karim, Z.; Lyoumi, S.; Nicolas, G.; Deybach, J.-C.; Gouya, L.; Puy, H. Porphyrias: A 2015 update. Clin. Res. Hepatol. Gastroenterol. 2015, 39, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Stein, P.E.; Badminton, M.N.; Barth, J.H.; Rees, D.C.; Sarkany, R.; Stewart, M.F.; Cox, T.M. Acute intermittent porphyria: Fatal complications of treatment. Clin. Med. 2012, 12, 293–294. [Google Scholar] [CrossRef]
- Whatley, S.D.; Mason, N.G.; Woolf, J.R.; Newcombe, R.G.; Elder, G.H.; Badminton, M.N. Diagnostic Strategies for Autosomal Dominant Acute Porphyrias: Retrospective Analysis of 467 Unrelated Patients Referred for Mutational Analysis of the HMBS, CPOX, or PPOX Gene. Clin. Chem. 2009, 55, 1406–1414. [Google Scholar] [CrossRef]
- Marsden, J.T.; Rees, D.C. Urinary excretion of porphyrins, porphobilinogen and δ-aminolaevulinic acid following an attack of acute intermittent porphyria. J. Clin. Pathol. 2014, 67, 60–65. [Google Scholar] [CrossRef]
- Aarsand, A.K.; Petersen, P.H.; Sandberg, S. Estimation and Application of Biological Variation of Urinary δ-Aminolevulinic Acid and Porphobilinogen in Healthy Individuals and in Patients with Acute Intermittent Porphyria. Clin. Chem. 2006, 52, 650–656. [Google Scholar] [CrossRef]
- Gouya, L.; Ventura, P.; Balwani, M.; Bissell, D.M.; Rees, D.C.; Stölzel, U.; Phillips, J.D.; Kauppinen, R.; Langendonk, J.G.; Desnick, R.J.; et al. EXPLORE: A Prospective, Multinational, Natural History Study of Patients with Acute Hepatic Porphyria with Recurrent Attacks. Hepatology 2020, 71, 1546–1558. [Google Scholar] [CrossRef]
- Neeleman, R.A.; Wagenmakers, M.A.E.M.; Koole-Lesuis, R.H.; Mijnhout, G.S.; Wilson, J.H.P.; Friesema, E.C.H.; Langendonk, J.G. Medical and financial burden of acute intermittent porphyria. J. Inherit. Metab. Dis. 2018, 41, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Kauppinen, R.; Mustajoki, P. Prognosis of acute porphyria: Occurrence of acute attacks, precipitating factors, and associated diseases. Medicine 1992, 71, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Andersson, C.; Floderus, Y.; Wikberg, A.; Lithner, F. The W198X and R173W mutations in the porphobilinogen deaminase gene in acute intermittent porphyria have higher clinical penetrance than R167W. A population-based study. Scand. J. Clin. Lab. Inv. 2000, 60, 643–648. [Google Scholar]
- Schneider-Yin, X.; Ulbrichová, D.; Mamet, R.; Martasek, P.; Marohnic, C.C.; Goren, A.; Minder, E.I.; Schoenfeld, N. Characterization of two missense variants in the hydroxymethylbilane synthase gene in the Israeli population, which differ in their associations with acute intermittent porphyria. Mol. Genet. Metab. 2008, 94, 343–346. [Google Scholar] [CrossRef]
- To-Figueras, J.; Badenas, C.; Carrera, C.; Muñoz, C.; Mila, M.; Lecha, M.; Herrero, C. Genetic and biochemical characterization of 16 acute intermittent porphyria cases with a high prevalence of the R173W mutation. J. Inherit. Metab. Dis. 2006, 29, 580–585. [Google Scholar] [CrossRef]
- Bustad, H.J.; Vorland, M.; Ronneseth, E.; Sandberg, S.; Martinez, A.; Toska, K. Conformational stability and activity analysis of two hydroxymethylbilane synthase mutants, K132N and V215E, with different phenotypic association with acute intermittent porphyria. Biosci. Rep. 2013, 33, e00056. [Google Scholar] [CrossRef]
- Lang, E.; Schäfer, M.; Schwender, H.; Neumann, N.J.; Frank, J. Occurrence of Malignant Tumours in the Acute Hepatic Porphyrias. JIMD Rep. 2015, 22, 17–22. [Google Scholar] [CrossRef]
- Lithner, F.; Wetterberg, L. Hepatocellular Carcinoma in Patients with Acute Intermittent Porphyria. Acta Med. Scand. 2009, 215, 271–274. [Google Scholar] [CrossRef]
- Andant, C.; Puy, H.; Bogard, C.; Faivre, J.; Soulé, J.-C.; Nordmann, Y.; Deybach, J.-C. Hepatocellular carcinoma in patients with acute hepatic porphyria: Frequency of occurrence and related factors. J. Hepatol. 2000, 32, 933–939. [Google Scholar] [CrossRef]
- Innala, E.; Andersson, C. Screening for hepatocellular carcinoma in acute intermittent porphyria: A 15-year follow-up in northern Sweden. J. Intern. Med. 2011, 269, 538–545. [Google Scholar] [CrossRef]
- Strand, L.J.; Felsher, B.F.; Redeker, A.G.; Marver, H.S. Heme Biosynthesis in Intermittent Acute Porphyria: Decreased Hepatic Conversion of Porphobilinogen to Porphyrins and Increased Delta Aminolevulinic Acid Synthetase Activity. Proc. Natl. Acad. Sci. USA 1970, 67, 1315–1320. [Google Scholar] [CrossRef]
- Bissell, D.M.; Lai, J.C.; Meister, R.K.; Blanc, P.D. Role of Delta-aminolevulinic Acid in the Symptoms of Acute Porphyria. Am. J. Med. 2015, 128, 313–317. [Google Scholar] [CrossRef]
- Lin, C.S.-Y.; Lee, M.-J.; Park, S.B.; Kiernan, M.C. Purple pigments: The pathophysiology of acute porphyric neuropathy. Clin. Neurophysiol. 2011, 122, 2336–2344. [Google Scholar] [CrossRef] [PubMed]
- Pischik, E.; Kauppinen, R. Neurological manifestations of acute intermittent porphyria. Cell. Mol. Biol. 2009, 55, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Meyer, U.A.; Schuurmans, M.M.; Lindberg, R.L.P. Acute Porphyrias: Pathogenesis of Neurological Manifestations. Semin. Liver Dis. 1998, 18, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, H.P.; Abdalla, D.S.P.; Faljoni-Alário, A.; Bechara, E.J.H. Generation of active oxygen species during coupled autoxidation of oxyhemoglobin and δ-aminolevulinic acid. Biochim. Biophys. Acta BBA Gen. Subj. 1986, 881, 100–106. [Google Scholar] [CrossRef]
- Bechara, E.J.H.; Medeiros, M.H.G.; Monteiro, H.P.; Hermes-Lima, M.; Pereira, B.; Demasi, M.; Costa, C.A.; Abdalla, D.S.; Onuki, J.; Wendel, C.M.A.; et al. A free radical hypothesis of lead poisoning and inborn porphyrias associated with 5-aminolevulinic acid overload. Quimica Nova 1993, 16, 385–392. [Google Scholar]
- Monteiro, H.P.; Abdalla, D.S.P.; Augusto, O.; Bechara, E.J.H. Free radical generation during δ-Aminolevulinic acid autoxidation: Induction by hemoglobin and connections with porphyrinpathies. Arch. Biochem. Biophys. 1989, 271, 206–216. [Google Scholar] [CrossRef]
- Laafi, J.; Homedan, C.; Jacques, C.; Gueguen, N.; Schmitt, C.; Puy, H.; Reynier, P.; Martinez, M.C.; Malthièry, Y. Pro-oxidant effect of ALA is implicated in mitochondrial dysfunction of HepG2 cells. Biochimie 2014, 106, 157–166. [Google Scholar] [CrossRef]
- Onuki, J.; Teixeira, P.C.; Medeiros, M.H.G.; Dörnemann, D.; Douki, T.; Cadet, J.; Di Mascio, P. Is 5-aminolevulinic acid involved in the hepatocellular carcinogenesis of acute intermittent porphyria? Cell. Mol. Biol. 2002, 48, 17–26. [Google Scholar]
- Baglo, Y.; Gabrielsen, M.; Sylte, I.; Gederaas, O.A. Homology Modeling of Human γ-Butyric Acid Transporters and the Binding of Pro-Drugs 5-Aminolevulinic Acid and Methyl Aminolevulinic Acid Used in Photodynamic Therapy. PLoS ONE 2013, 8, e65200. [Google Scholar] [CrossRef] [PubMed]
- Novak, B.; Schulten, R.; Lübbert, H. δ-Aminolevulinic acid and its methyl ester induce the formation of Protoporphyrin IX in cultured sensory neurones. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2011, 384, 583–602. [Google Scholar] [CrossRef]
- Brennan, M.J.W.; Cantrill, R.C. δ-Aminolaevulinic acid is a potent agonist for GABA autoreceptors. Nature 1979, 280, 514–515. [Google Scholar] [CrossRef] [PubMed]
- Rud, E.; Gederaas, O.; Høgset, A.; Berg, K. 5-Aminolevulinic Acid, but not 5-Aminolevulinic Acid Esters, is Transported into Adenocarcinoma Cells by System BETA Transporters. Photochem. Photobiol. 2000, 71, 640–647. [Google Scholar] [CrossRef]
- Warren, M.J.; Cooper, J.B.; Wood, S.P.; Shoolingin-Jordan, P.M. Lead poisoning, haem synthesis and 5-aminolaevulinic acid dehydratase. Trends Biochem. Sci. 1998, 23, 217–221. [Google Scholar] [CrossRef]
- Jaffe, E.K. Porphobilinogen synthase: An equilibrium of different assemblies in human health. Prog. Mol. Biol. Transl. Sci. 2020, 169, 85–104. [Google Scholar] [CrossRef]
- Lawrence, S.H.; Selwood, T.; Jaffe, E.K. Diverse Clinical Compounds Alter the Quaternary Structure and Inhibit the Activity of an Essential Enzyme. ChemMedChem 2011, 6, 1067–1073. [Google Scholar] [CrossRef][Green Version]
- Akagi, R.; Kato, N.; Inoue, R.; Anderson, K.E.; Jaffe, E.K.; Sassa, S. δ-Aminolevulinate dehydratase (ALAD) porphyria: The first case in North America with two novel ALAD mutations. Mol. Genet. Metab. 2006, 87, 329–336. [Google Scholar] [CrossRef]
- Lahiji, A.P.; Anderson, K.E.; Chan, A.; Simon, A.; Desnick, R.J.; Ramanujam, V.M.S. 5-Aminolevulinate dehydratase porphyria: Update on hepatic 5-aminolevulinic acid synthase induction and long-term response to hemin. Mol. Genet. Metab. 2020, 131, 418–423. [Google Scholar] [CrossRef]
- Sassa, S.; Kappas, A. Hereditary Tyrosinemia and the Heme Biosynthetic Pathway. Profound inhibition of δ-aminolevulinic acid dehydratase activity by succinylacetone. J. Clin. Investig. 1983, 71, 625–634. [Google Scholar] [CrossRef]
- Bung, N.; Roy, A.; Chen, B.; Das, D.; Pradhan, M.; Yasuda, M.; New, M.I.; Desnick, R.J.; Bulusu, G. Human hydroxymethylbilane synthase: Molecular dynamics of the pyrrole chain elongation identifies step-specific residues that cause AIP. Proc. Natl. Acad. Sci. USA 2018, 115, E4071–E4080. [Google Scholar] [CrossRef] [PubMed]
- Jordan, P.M.; Warren, M.J. Evidence for a dipyrromethane cofactor at the catalytic site of E. coli porphobilinogen deaminase. FEBS Lett. 1987, 225, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Warren, M.J.; Jordan, P.M. Investigation into the nature of substrate binding to the dipyrromethane cofactor of Escherichia coli porphobilinogen deaminase. Biochemistry 1988, 27, 9020–9030. [Google Scholar] [CrossRef]
- Louie, G.V.; Brownlie, P.D.; Lambert, R.; Cooper, J.B.; Blundell, T.L.; Wood, S.P.; Warren, M.J.; Woodcock, S.C.; Jordan, P.M. Structure of porphobilinogen deaminase reveals a flexible multidomain polymerase with a single catalytic site. Nature 1992, 359, 33–39. [Google Scholar] [CrossRef]
- Song, G.; Li, Y.; Cheng, C.; Zhao, Y.; Gao, A.; Zhang, R.; Joachimiak, A.; Shaw, N.; Liu, Z.J. Structural insight into acute intermittent porphyria. FASEB J. 2009, 23, 396–404. [Google Scholar] [CrossRef]
- Helliwell, J.R.; Nieh, Y.-P.; Raftery, J.; Cassetta, A.; Habash, J.; Carr, P.D.; Ursby, T.; Wulff, M.; Thompson, A.W.; Niemann, A.C.; et al. Time-resolved structures of hydroxymethylbilane synthase (Lys59Gln mutant) as it is loaded with substrate in the crystal determined by Laue diffraction. J. Chem. Soc. Faraday Trans. 1998, 94, 2615–2622. [Google Scholar] [CrossRef]
- Hadener, A.; Matzinger, P.K.; Battersby, A.R.; McSweeney, S.; Thompson, A.W.; Hammersley, A.P.; Harrop, S.J.; Cassetta, A.; Deacon, A.; Hunter, W.N.; et al. Determination of the structure of seleno-methionine-labelled hydroxymethylbilane synthase in its active form by multi-wavelength anomalous dispersion. Acta Crystallogr. D Biol. Crystallogr. 1999, 55, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Nieh, Y.P.; Raftery, J.; Weisgerber, S.; Habash, J.; Schotte, F.; Ursby, T.; Wulff, M.; Hädener, A.; Campbell, J.W.; Hao, Q.; et al. Accurate and highly complete synchrotron protein crystal Laue diffraction data using the ESRF CCD and the Daresbury Laue software. J. Synchrotron Radiat. 1999, 6, 995–1006. [Google Scholar] [CrossRef]
- Helliwell, J.R.; Nieh, Y.P.; Habash, J.; Faulder, P.F.; Raftery, J.; Cianci, M.; Wulff, M.; Hadener, A. Time-resolved and static-ensemble structural chemistry of hydroxymethylbilane synthase. Faraday Discuss 2003, 122, 131–144; discussion 171–190. [Google Scholar] [CrossRef]
- Gill, R.; Kolstoe, S.E.; Mohammed, F.; Al D-Bass, A.; Mosely, J.E.; Sarwar, M.; Cooper, J.B.; Wood, S.P.; Shoolingin-Jordan, P.M. Structure of human porphobilinogen deaminase at 2.8 Å: The molecular basis of acute intermittent porphyria. Biochem. J. 2009, 420, 17–25. [Google Scholar] [CrossRef]
- Roberts, A.; Gill, R.; Hussey, R.J.; Mikolajek, H.; Erskine, P.T.; Cooper, J.B.; Wood, S.P.; Chrystal, E.J.T.; Shoolingin-Jordan, P.M. Insights into the mechanism of pyrrole polymerization catalysed by porphobilinogen deaminase: High-resolution X-ray studies of the Arabidopsis thaliana enzyme. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 471–485. [Google Scholar] [CrossRef]
- Azim, N.; Deery, E.; Warren, M.J.; Wolfenden, B.A.; Erskine, P.; Cooper, J.B.; Coker, A.; Wood, S.P.; Akhtar, M. Structural evidence for the partially oxidized dipyrromethene and dipyrromethanone forms of the cofactor of porphobilinogen deaminase: Structures of the Bacillus megaterium enzyme at near-atomic resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 744–751. [Google Scholar] [CrossRef]
- Guo, J.; Erskine, P.; Coker, A.R.; Wood, S.P.; Cooper, J.B. Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium. Acta Crystallogr. F Struct. Biol. Commun. 2017, 73, 612–620. [Google Scholar] [CrossRef] [PubMed]
- Pluta, P.; Roversi, P.; Bernardo-Seisdedos, G.; Rojas, A.L.; Cooper, J.B.; Gu, S.; Pickersgill, R.W.; Millet, O. Structural basis of pyrrole polymerization in human porphobilinogen deaminase. Biochim. Biophys. Acta BBA Gen. Subj. 2018, 1862, 1948–1955. [Google Scholar] [CrossRef] [PubMed]
- Bustad, H.J.; Kallio, J.P.; Laitaoja, M.; Toska, K.; Kursula, I.; Martinez, A.; Jänis, J. Structural characterisation of porphobilinogen deaminase mutants associated with the acute intermittent porphyria reveals that Arg173 is crucial for the polypyrrole elongation mechanism. iScience 2021. (in revision). [Google Scholar]
- Jordan, P.M.; Woodcock, S.C. Mutagenesis of arginine residues in the catalytic cleft of Escherichia coli porphobilinogen deaminase that affects dipyrromethane cofactor assembly and tetrapyrrole chain initiation and elongation. Biochem. J. 1991, 280, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.E.; Collins, S. Open-Label Study of Hemin for Acute Porphyria: Clinical Practice Implications. Am. J. Med. 2006, 119, 526.e19–526.e25. [Google Scholar] [CrossRef] [PubMed]
- Bonkowsky, H.L.; Tschudy, D.P.; Collins, A.; Doherty, J.; Bossenmaier, I.; Cardinal, R.; Watson, C.J. Repression of the Overproduction of Porphyrin Precursors in Acute Intermittent Porphyria by Intravenous Infusions of Hematin. Proc. Natl. Acad. Sci. USA 1971, 68, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Tenhunen, R.; Tokola, O.; Lindén, I.-B. Haem arginate: A new stable haem compound. J. Pharm. Pharmacol. 1987, 39, 780–786. [Google Scholar] [CrossRef]
- May, B.K.; Dogra, S.C.; Sadlon, T.J.; Bhasker, C.R.; Cox, T.C.; Bottomley, S.S. Molecular Regulation of Heme Biosynthesis in Higher Vertebrates. Prog. Nucleic Acid Res. Mol. Biol. 1995, 51, 1–51. [Google Scholar] [CrossRef]
- Watson, C.J.; Pierach, C.A.; Bossenmaier, I.; Cardinal, R. Postulated deficiency of hepatic heme and repair by hematin infusions in the “inducible” hepatic porphyrias. Proc. Natl. Acad. Sci. USA 1977, 74, 2118–2120. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.E.; Bloomer, J.R.; Bonkovsky, H.L.; Kushner, J.P.; Pierach, C.A.; Pimstone, N.R.; Desnick, R.J. Recommendations for the Diagnosis and Treatment of the Acute Porphyrias. Ann. Intern. Med. 2005, 142, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Willandt, B.; Langendonk, J.G.; Biermann, K.; Meersseman, W.; D’Heygere, F.; George, C.; Verslype, C.; Monbaliu, D.; Cassiman, D. Liver Fibrosis Associated with Iron Accumulation Due to Long-Term Heme-Arginate Treatment in Acute Intermittent Porphyria: A Case Series. JIMD Rep. 2016, 25, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, C.; Lenglet, H.; Yu, A.; Delaby, C.; Benecke, A.; Lefebvre, T.; Letteron, P.; Paradis, V.; Wahlin, S.; Sandberg, S.; et al. Recurrent attacks of acute hepatic porphyria: Major role of the chronic inflammatory response in the liver. J. Intern. Med. 2018, 284, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Handschin, C.; Lin, J.; Rhee, J.; Peyer, A.-K.; Chin, S.; Wu, P.-H.; Meyer, U.A.; Spiegelman, B.M. Nutritional Regulation of Hepatic Heme Biosynthesis and Porphyria through PGC-1α. Cell 2005, 122, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Brunet, A.; Bonni, A.; Zigmond, M.J.; Lin, M.Z.; Juo, P.; Hu, L.S.; Anderson, M.J.; Arden, K.C.; Blenis, J.; Greenberg, M.E. Akt Promotes Cell Survival by Phosphorylating and Inhibiting a Forkhead Transcription Factor. Cell 1999, 96, 857–868. [Google Scholar] [CrossRef]
- Nakae, J.; Kitamura, T.; Ogawa, W.; Kasuga, M.; Accili, D. Insulin regulation of gene expression through the forkhead transcription factor Foxo1 (Fkhr) requires kinases distinct from Akt. Biochemistry 2001, 40, 11768–11776. [Google Scholar] [CrossRef]
- Anderson, K.E.; Spitz, I.M.; Bardin, C.W.; Kappas, A. A Gonadotropin Releasing Hormone Analogue Prevents Cyclical Attacks of Porphyria. Arch. Intern. Med. 1990, 150, 1469–1474. [Google Scholar] [CrossRef]
- Innala, E.; Bäckström, T.; Bixo, M.; Andersson, C. Evaluation of gonadotropin-releasing hormone agonist treatment for prevention of menstrual-related attacks in acute porphyria. Acta Obstet. Gynecol. Scand. 2010, 89, 95–100. [Google Scholar] [CrossRef]
- Singal, A.K.; Parker, C.; Bowden, C.; Thapar, M.; Liu, L.; McGuire, B.M. Liver transplantation in the management of porphyria. Hepatology 2014, 60, 1082–1089. [Google Scholar] [CrossRef]
- Dar, F.S.; Asai, K.; Haque, A.R.; Cherian, T.; Rela, M.; Heaton, N. Liver transplantation for acute intermittent porphyria: A viable treatment? Hepatobiliary Pancreat. Dis. Int. 2010, 9, 93–96. [Google Scholar] [PubMed]
- Soonawalla, Z.F.; Orug, T.; Badminton, M.N.; Elder, G.H.; Rhodes, J.M.; Bramhall, S.R.; Elias, E. Liver transplantation as a cure for acute intermittent porphyria. Lancet 2004, 363, 705–706. [Google Scholar] [CrossRef]
- Dowman, J.K.; Gunson, B.K.; Mirza, D.F.; Bramhall, S.R.; Badminton, M.N.; Newsome, P.N. Liver transplantation for acute intermittent porphyria is complicated by a high rate of hepatic artery thrombosis. Liver Transplant. 2011, 18, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Malinzak, E.B.; Knudsen, N.W.; Udani, A.D.; Vikraman, D.; Sudan, D.L.; Miller, T.E. Perioperative Challenges in Liver Transplantation for a Patient with Acute Intermittent Porphyria. J. Cardiothorac. Vasc. Anesth. 2018, 32, 2716–2720. [Google Scholar] [CrossRef]
- Tchernitchko, D.; Tavernier, Q.; Lamoril, J.; Schmitt, C.; Talbi, N.; Lyoumi, S.; Robreau, A.-M.; Karim, Z.; Gouya, L.; Thervet, E.; et al. A Variant of Peptide Transporter 2 Predicts the Severity of Porphyria-Associated Kidney Disease. J. Am. Soc. Nephrol. 2017, 28, 1924–1932. [Google Scholar] [CrossRef]
- Sardh, E.; Andersson, D.E.; Henrichson, A.; Harper, P. Porphyrin precursors and porphyrins in three patients with acute intermittent porphyria and end-stage renal disease under different therapy regimes. Cell. Mol. Biol. 2009, 55, 66–71. [Google Scholar]
- Lazareth, H.; Talbi, N.; Kamar, N.; Levi, C.; Moulin, B.; Caillard, S.; Frimat, L.; Chemouny, J.; Chatelet, V.; Vachey, C.; et al. Kidney transplantation improves the clinical outcomes of Acute Intermittent Porphyria. Mol. Genet. Metab. 2020. [Google Scholar] [CrossRef]
- Ferreira, G.D.S.A.; De Oliveira, L.C.; Ulisses, L.R.D.S.; Watanabe, A.L.C.; Medeiros, I.N.; Cardoso, H.S.S.; Alves, I.C.D.C.; De Almeida, T.M.; De Lima, L.V.; Fontoura, R.P.; et al. Combined Liver and Kidney Transplant in Acute Intermittent Porphyria: A Case Report. Am. J. Case Rep. 2020, 21, e927832. [Google Scholar] [CrossRef]
- Wahlin, S.; Harper, P.; Sardh, E.; Andersson, C.; Andersson, D.E.; Ericzon, B.-G. Combined liver and kidney transplantation in acute intermittent porphyria. Transpl. Int. 2010, 23, e18–e21. [Google Scholar] [CrossRef]
- Sardh, E.; Harper, P.; Balwani, M.; Stein, P.; Rees, D.; Bissell, D.M.; Desnick, R.; Parker, C.; Phillips, J.; Bonkovsky, H.L.; et al. Phase 1 Trial of an RNA Interference Therapy for Acute Intermittent Porphyria. N. Engl. J. Med. 2019, 380, 549–558. [Google Scholar] [CrossRef]
- Chan, A.; Liebow, A.; Yasuda, M.; Gan, L.; Racie, T.; Maier, M.; Kuchimanchi, S.; Foster, D.; Milstein, S.; Charisse, K.; et al. Preclinical Development of a Subcutaneous ALAS1 RNAi Therapeutic for Treatment of Hepatic Porphyrias Using Circulating RNA Quantification. Mol. Ther. Nucleic Acids 2015, 4, e263. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, M.; Gan, L.; Chen, B.; Kadirvel, S.; Yu, C.; Phillips, J.D.; New, M.I.; Liebow, A.; Fitzgerald, K.; Querbes, W.; et al. RNAi-mediated silencing of hepatic Alas1 effectively prevents and treats the induced acute attacks in acute intermittent porphyria mice. Proc. Natl. Acad. Sci. USA 2014, 111, 7777–7782. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.J. Givosiran: First Approval. Drugs 2020, 80, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Balwani, M.; Sardh, E.; Ventura, P.; Peiró, P.A.; Rees, D.C.; Stölzel, U.; Bissell, D.M.; Bonkovsky, H.L.; Windyga, J.; Anderson, K.E.; et al. Phase 3 Trial of RNAi Therapeutic Givosiran for Acute Intermittent Porphyria. N. Engl. J. Med. 2020, 382, 2289–2301. [Google Scholar] [CrossRef] [PubMed]
- ENVISION: A Study to Evaluate the Efficacy and Safety of Givosiran (ALN-AS1) in Patients with Acute Hepatic Porphyrias (AHP). Available online: https://clinicaltrials.gov/ct2/show/NCT03338816 (accessed on 18 November 2020).
- Johansson, A.; Möller, C.; Fogh, J.; Harper, P. Biochemical Characterization of Porphobilinogen Deaminase–Deficient Mice During Phenobarbital Induction of Heme Synthesis and the Effect of Enzyme Replacement. Mol. Med. 2003, 9, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Sardh, E.; Rejkjaer, L.; Andersson, D.E.H.; Harper, P. Safety, Pharmacokinetics and Pharmocodynamics of Recombinant Human Porphobilinogen Deaminase in Healthy Subjects and Asymptomatic Carriers of the Acute Intermittent Porphyria Gene Who Have Increased Porphyrin Precursor Excretion. Clin. Pharmacokinet. 2007, 46, 335–349. [Google Scholar] [CrossRef]
- Andersson, C.; Peterson, J.; Anderson, K.E.; Deybach, J.C.; Kauppinen, R.; Skadberg, Ø.; Andersson, D.E.; Bonkovsky, H.L.; Fogh, J.; Rejkjaer, L. Randomized clinical trial of recombinant human porphobilinogen deaminase (rhPBGD) in acute attacks of porphyria. In Proceedings of the Porphyrins and Porphyrias International Meeting, Rotterdam, The Netherlands, 29 April–3 May 2007; pp. 152–216. [Google Scholar]
- Johansson, A.; Möller, C.; Gellerfors, P.; Harper, P. Non-viral mediated gene transfer of porphobilinogen deaminase into mammalian cells. Scand. J. Clin. Lab. Investig. 2002, 62, 105–113. [Google Scholar] [CrossRef]
- Johansson, A.; Nowak, G.; Möller, C.; Blomberg, P.; Harper, P. Adenoviral-mediated expression of porphobilinogen deaminase in liver restores the metabolic defect in a mouse model of acute intermittent porphyria. Mol. Ther. 2004, 10, 337–343. [Google Scholar] [CrossRef]
- Unzu, C.; Sampedro, A.; Mauleón, I.; González-Aparicio, M.; De Salamanca, R.E.; Prieto, J.; Aragón, T.; Fontanellas, A. Helper-dependent adenoviral liver gene therapy protects against induced attacks and corrects protein folding stress in acute intermittent porphyria mice. Hum. Mol. Genet. 2013, 22, 2929–2940. [Google Scholar] [CrossRef]
- Unzu, C.; Sampedro, A.; Mauleón, I.; Alegre, M.; Beattie, S.G.; De Salamanca, R.E.; Snapper, J.; Twisk, J.; Petry, H.; González-Aseguinolaza, G.; et al. Sustained Enzymatic Correction by rAAV-Mediated Liver Gene Therapy Protects Against Induced Motor Neuropathy in Acute Porphyria Mice. Mol. Ther. 2011, 19, 243–250. [Google Scholar] [CrossRef]
- Yasuda, M.; Bishop, D.F.; Fowkes, M.; Cheng, S.H.; Gan, L.; Desnick, R.J. AAV8-mediated Gene Therapy Prevents Induced Biochemical Attacks of Acute Intermittent Porphyria and Improves Neuromotor Function. Mol. Ther. 2010, 18, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Pañeda, A.; López-Franco, E.; Kaeppel, C.; Unzu, C.; Gil-Royo, A.G.; D’Avola, D.; Beattie, S.G.; Olague, C.; Ferrero, R.; Sampedro, A.; et al. Safety and Liver Transduction Efficacy of rAAV5-cohPBGD in Nonhuman Primates: A Potential Therapy for Acute Intermittent Porphyria. Hum. Gene Ther. 2013, 24, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Brunetti-Pierri, N.; Newsome, P.N. AAV-mediated liver-directed gene therapy for Acute Intermittent Porphyria: It is safe but is it effective? J. Hepatol. 2016, 65, 666–667. [Google Scholar] [CrossRef] [PubMed]
- D’Avola, D.; López-Franco, E.; Sangro, B.; Pañeda, A.; Grossios, N.; Gil-Farina, I.; Benito, A.; Twisk, J.; Paz, M.; Ruiz, J.; et al. Phase I open label liver-directed gene therapy clinical trial for acute intermittent porphyria. J. Hepatol. 2016, 65, 776–783. [Google Scholar] [CrossRef]
- Serrano-Mendioroz, I.; Sampedro, A.; Alegre, M.; De Salamanca, R.E.; Berraondo, P.; Fontanellas, A. An Inducible Promoter Responsive to Different Porphyrinogenic Stimuli Improves Gene Therapy Vectors for Acute Intermittent Porphyria. Hum. Gene Ther. 2018, 29, 480–491. [Google Scholar] [CrossRef]
- Serrano-Mendioroz, I.; Sampedro, A.; Serna, N.; De Salamanca, R.E.; Sanz-Parra, A.; Corrales, F.; Berraondo, P.; Millet, O.; Fontanellas, A. Bioengineered PBGD variant improves the therapeutic index of gene therapy vectors for acute intermittent porphyria. Hum. Mol. Genet. 2018, 27, 3688–3696. [Google Scholar] [CrossRef]
- Jiang, L.; Berraondo, P.; Jericó, D.; Guey, L.T.; Sampedro, A.; Frassetto, A.; Benenato, K.E.; Burke, K.; Santamaría, E.; Alegre, M.; et al. Systemic messenger RNA as an etiological treatment for acute intermittent porphyria. Nat. Med. 2018, 24, 1899–1909. [Google Scholar] [CrossRef]
- Jorns, C.; Ellis, E.C.; Nowak, G.; Fischler, B.; Nemeth, A.; Strom, S.C.; Ericzon, B.G. Hepatocyte transplantation for inherited metabolic diseases of the liver. J. Intern. Med. 2012, 272, 201–223. [Google Scholar] [CrossRef]
- Yin, Z.; Wahlin, S.; Ellis, E.C.S.; Harper, P.; Ericzon, B.-G.; Nowak, G. Hepatocyte Transplantation Ameliorates the Metabolic Abnormality in a Mouse Model of Acute Intermittent Porphyria. Cell Transplant. 2014, 23, 1153–1162. [Google Scholar] [CrossRef]
- Cantz, T.; Sharma, A.D.; Ott, M. Concise Review: Cell Therapies for Hereditary Metabolic Liver Diseases-Concepts, Clinical Results, and Future Developments. Stem Cells 2015, 33, 1055–1062. [Google Scholar] [CrossRef]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In vivo aspects of protein folding and quality control. Science 2016, 353, aac4354. [Google Scholar] [CrossRef] [PubMed]
- Clark, P.L. Protein folding in the cell: Reshaping the folding funnel. Trends Biochem. Sci. 2004, 29, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Halskau, Ø.; Perez-Jimenez, R.; Ibarra-Molero, B.; Underhaug, J.; Muñoz, V.; Martinez, A.; Sanchez-Ruiz, J.M. Large-scale modulation of thermodynamic protein folding barriers linked to electrostatics. Proc. Natl. Acad. Sci. USA 2008, 105, 8625–8630. [Google Scholar] [CrossRef]
- Fuxreiter, M. Fold or not to fold upon binding—Does it really matter? Curr. Opin. Struct. Biol. 2019, 54, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Breinig, S.; Kervinen, J.; Stith, L.; Wasson, A.S.; Fairman, R.; Wlodawer, A.; Zdanov, A.; Jaffe, E.K. Control of tetrapyrrole biosynthesis by alternate quaternary forms of porphobilinogen synthase. Nat. Struct. Mol. Biol. 2003, 10, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Murzin, A.G. Biochemistry: Metamorphic Proteins. Science 2008, 320, 1725–1726. [Google Scholar] [CrossRef]
- Jaffe, E.K. Morpheeins—A new structural paradigm for allosteric regulation. Trends Biochem. Sci. 2005, 30, 490–497. [Google Scholar] [CrossRef]
- Gendoo, D.M.; Harrison, P.M. Discordant and chameleon sequences: Their distribution and implications for amyloidogenicity. Protein Sci. 2011, 20, 567–579. [Google Scholar] [CrossRef]
- Dishman, A.F.; Volkman, B.F. Unfolding the Mysteries of Protein Metamorphosis. ACS Chem. Biol. 2018, 13, 1438–1446. [Google Scholar] [CrossRef]
- Bustad, H.J.; Toska, K.; Schmitt, C.; Vorland, M.; Skjærven, L.; Kallio, J.P.; Simonin, S.; Letteron, P.; Underhaug, J.; Sandberg, S.; et al. A Pharmacological Chaperone Therapy for Acute Intermittent Porphyria. Mol. Ther. 2020, 28, 677–689. [Google Scholar] [CrossRef]
- Chen, C.H.; Astrin, K.H.; Lee, G.; Anderson, K.E.; Desnick, R.J. Acute intermittent porphyria: Identification and expression of exonic mutations in the hydroxymethylbilane synthase gene. An initiation codon missense mutation in the housekeeping transcript causes “variant acute intermittent porphyria” with normal expression of the erythroid-specific enzyme. J. Clin. Investig. 1994, 94, 1927–1937. [Google Scholar] [CrossRef] [PubMed]
- Ulbrichová, D.; Hrdinka, M.; Saudek, V.; Martasek, P. Acute intermittent porphyria—Impact of mutations found in the hydroxymethylbilane synthase gene on biochemical and enzymatic protein properties. FEBS J. 2009, 276, 2106–2115. [Google Scholar] [CrossRef] [PubMed]
- Ulbrichová, D.; Schneider-Yin, X.; Mamet, R.; Saudek, V.; Martasek, P.; Minder, E.I.; Schoenfeld, N. Correlation between biochemical findings, structural and enzymatic abnormalities in mutated HMBS identified in six Israeli families with acute intermittent porphyria. Blood Cells, Mol. Dis. 2009, 42, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Liguori, L.; Monticelli, M.; Allocca, M.; Mele, B.H.; Lukas, J.; Cubellis, M.V.; Andreotti, G. Pharmacological Chaperones: A Therapeutic Approach for Diseases Caused by Destabilizing Missense Mutations. Int. J. Mol. Sci. 2020, 21, 489. [Google Scholar] [CrossRef] [PubMed]
- Nussinov, R.; Tsai, C.-J.; Jang, H. Protein ensembles link genotype to phenotype. PLoS Comput. Biol. 2019, 15, e1006648. [Google Scholar] [CrossRef] [PubMed]
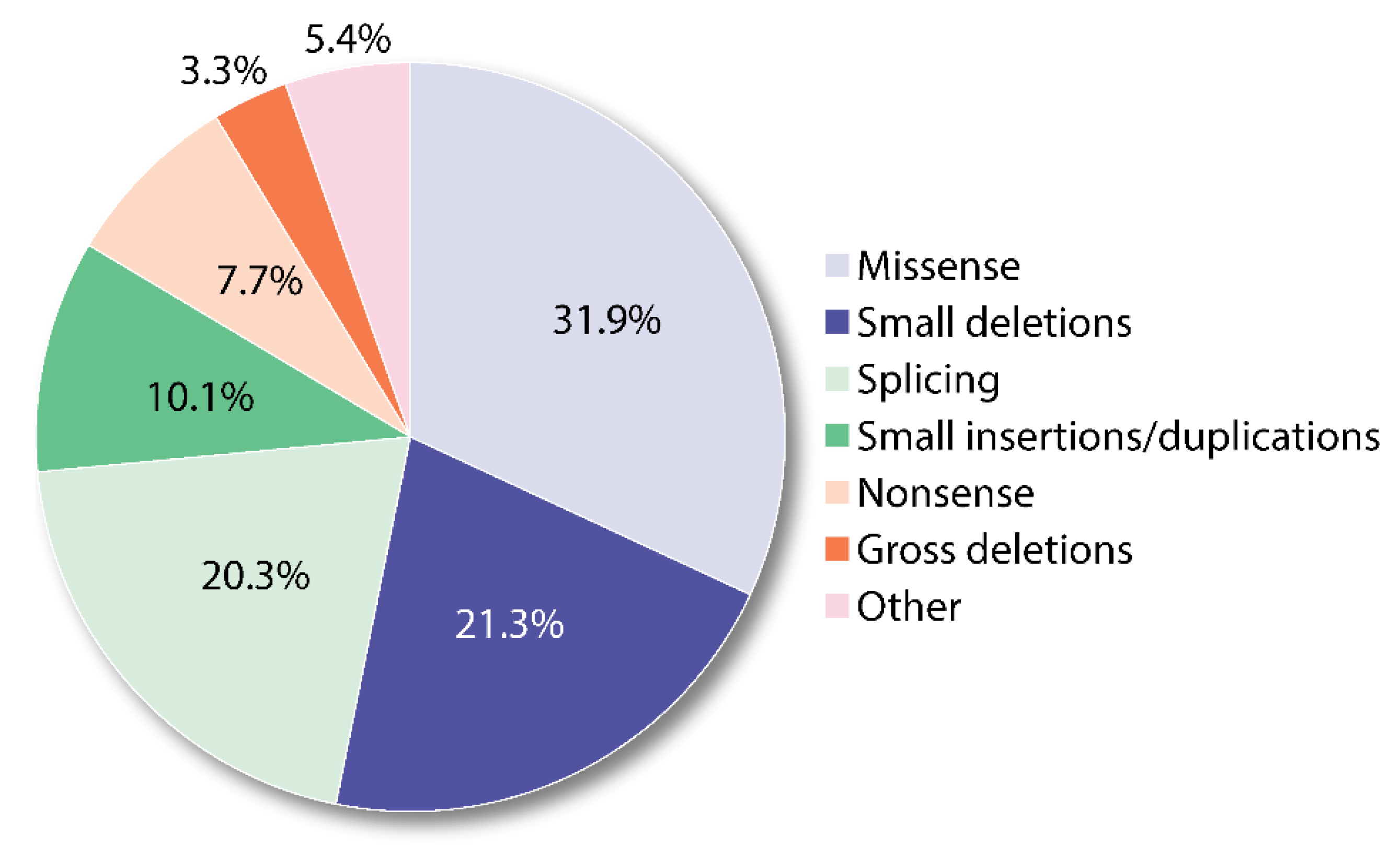
- Chen, B.; Whatley, S.; Badminton, M.; Aarsand, A.K.; Anderson, K.E.; Bissell, D.M.; Bonkovsky, H.L.; Cappellini, M.D.; Floderus, Y.; Friesema, E.C.H.; et al. International Porphyria Molecular Diagnostic Collaborative: An evidence-based database of verified pathogenic and benign variants for the porphyrias. Genet. Med. 2019, 21, 2605–2613. [Google Scholar] [CrossRef]
- De Siervi, A.; Rossetti, M.V.; Parera, V.E.; Astrin, K.H.; Aizencang, G.I.; Glass, I.A.; Batlle, A.M.; Desnick, R.J. Identification and characterization of hydroxymethylbilane synthase mutations causing acute intermittent porphyria: Evidence for an ancestral founder of the common G111R mutation. Am. J. Med. Genet. 1999, 86, 366–375. [Google Scholar] [CrossRef]
- De Siervi, A.; Weiss Cadiz, D.E.; Parera, V.E.; del C. Batlle, A.M.; Rossetti, M.V. Identification and characterization of two novel mutations that produce acute intermittent porphyria: A 3-base deletion (841-843delGGA) and a missense mutation (T35M). Hum. Mutat. 2000, 16, 373. [Google Scholar] [CrossRef]
- Delfau, M.H.; Picat, C.; de Rooij, F.W.; Hamer, K.; Bogard, M.; Wilson, J.H.; Deybach, J.C.; Nordmann, Y.; Grandchamp, B. Two different point G to A mutations in exon 10 of the porphobilinogen deaminase gene are responsible for acute intermittent porphyria. J. Clin. Investig. 1990, 86, 1511–1516. [Google Scholar] [CrossRef]
- Gu, X.F.; de Rooij, F.; Voortman, G.; Te Velde, K.; Deybach, J.C.; Nordmann, Y.; Grandchamp, B. Detection of eleven mutations causing acute intermittent porphyria using denaturing gradient gel electrophoresis. Hum. Genet. 1994, 93, 47–52. [Google Scholar] [CrossRef]
- Kauppinen, R.; Mustajoki, S.; Pihlaja, H.; Peltonen, L.; Mustajoki, P. Acute intermittent porphyria in Finland: 19 mutations in the porphobilinogen deaminase gene. Hum. Mol. Genet. 1995, 4, 215–222. [Google Scholar] [CrossRef]
- Mgone, C.S.; Lanyon, W.G.; Moore, M.R.; Connor, J.M. Detection of seven point mutations in the porphobilinogen deaminase gene in patients with acute intermittent porphyria, by direct sequencing of in vitro amplified cDNA. Hum. Genet. 1992, 90, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Moran-Jimenez, M.J.; Borrero-Corte, M.J.; Jara-Rubio, F.; Garcia-Pastor, I.; Diaz-Diaz, S.; Castelbon-Fernandez, F.J.; Enriquez-de-Salamanca, R.; Mendez, M. Molecular Analysis of 55 Spanish Patients with Acute Intermittent Porphyria. Genes (Basel) 2020, 11, 924. [Google Scholar] [CrossRef] [PubMed]
- Mustajoki, S.; Pihlaja, H.; Ahola, H.; Petersen, N.E.; Mustajoki, P.; Kauppinen, R. Three splicing defects, an insertion, and two missense mutations responsible for acute intermittent porphyria. Hum. Genet. 1998, 102, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Ramdall, R.B.; Cunha, L.; Astrin, K.H.; Katz, D.R.; Anderson, K.E.; Glucksman, M.; Bottomley, S.S.; Desnick, R.J. Acute intermittent porphyria: Novel missense mutations in the human hydroxymethylbilane synthase gene. Genet. Med. 2000, 2, 290–295. [Google Scholar] [CrossRef][Green Version]
- Solis, C.; Lopez-Echaniz, I.; Sefarty-Graneda, D.; Astrin, K.H.; Desnick, R.J. Identification and expression of mutations in the hydroxymethylbilane synthase gene causing acute intermittent porphyria (AIP). Mol. Med. 1999, 5, 664–671. [Google Scholar] [CrossRef]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Hartl, F.U. Molecular Chaperone Functions in Protein Folding and Proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef]
- Klaips, C.L.; Jayaraj, G.G.; Hartl, F.U. Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 2018, 217, 51–63. [Google Scholar] [CrossRef]
- Sinnige, T.; Yu, A.; Morimoto, R.I. Challenging Proteostasis: Role of the Chaperone Network to Control Aggregation-Prone Proteins in Human Disease. Adv. Exp. Med. Biol. 2020, 1243, 53–68. [Google Scholar] [CrossRef]
- Jayaraj, G.G.; Hipp, M.S.; Hartl, F.U. Functional Modules of the Proteostasis Network. Cold Spring Harb. Perspect. Biol. 2020, 12, a033951. [Google Scholar] [CrossRef]
- Saibil, H.R. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 2013, 14, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Sontag, E.M.; Samant, R.S.; Frydman, J. Mechanisms and Functions of Spatial Protein Quality Control. Annu. Rev. Biochem. 2017, 86, 97–122. [Google Scholar] [CrossRef] [PubMed]
- Lamark, T.; Svenning, S.; Johansen, T. Regulation of selective autophagy: The p62/SQSTM1 paradigm. Essays Biochem. 2017, 61, 609–624. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef]
- Gregersen, N.; Bross, P.; Vang, S.; Christensen, J.H. Protein Misfolding and Human Disease. Annu. Rev. Genom. Hum. Genet. 2006, 7, 103–124. [Google Scholar] [CrossRef]
- Kocaturk, N.M.; Gozuacik, D. Crosstalk Between Mammalian Autophagy and the Ubiquitin-Proteasome System. Front. Cell Dev. Biol. 2018, 6, 128. [Google Scholar] [CrossRef]
- Mukherjee, A.; Morales-Scheihing, D.; Butler, P.C.; Soto, C. Type 2 diabetes as a protein misfolding disease. Trends Mol. Med. 2015, 21, 439–449. [Google Scholar] [CrossRef]
- Sade, D.; Shaham-Niv, S.; Arnon, Z.A.; Tavassoly, O.; Gazit, E. Seeding of proteins into amyloid structures by metabolite assemblies may clarify certain unexplained epidemiological associations. Open Biol. 2018, 8, 170229. [Google Scholar] [CrossRef]
- Maitra, D.; Cunha, J.B.; Elenbaas, J.S.; Bonkovsky, H.L.; Shavit, J.A.; Omary, M.B. Porphyrin-Induced Protein Oxidation and Aggregation as a Mechanism of Porphyria-Associated Cell Injury. Cell. Mol. Gastroenterol. Hepatol. 2019, 8, 535–548. [Google Scholar] [CrossRef]
- Aminaka, M.; Kondo, M.; Takata, A.; Yamauchi, H.; Ikeda, M.; Yoshida, K. Oxidative stress in porphyria and carriers. Nippon Eiseigaku Zasshi 2008, 63, 628–635. [Google Scholar] [CrossRef]
- Gidalevitz, T.; Wang, N.; Deravaj, T.; Alexander-Floyd, J.; Morimoto, R.I. Natural genetic variation determines susceptibility to aggregation or toxicity in a C. elegans model for polyglutamine disease. BMC Biol. 2013, 11, 100. [Google Scholar] [CrossRef] [PubMed]
- Seemann, S.; Ernst, M.; Cimmaruta, C.; Struckmann, S.; Cozma, C.; Koczan, D.; Knospe, A.-M.; Haake, L.R.; Citro, V.; Bräuer, A.U.; et al. Proteostasis regulators modulate proteasomal activity and gene expression to attenuate multiple phenotypes in Fabry disease. Biochem. J. 2020, 477, 359–380. [Google Scholar] [CrossRef] [PubMed]
- Ong, D.S.T.; Kelly, J.W. Chemical and/or biological therapeutic strategies to ameliorate protein misfolding diseases. Curr. Opin. Cell Biol. 2011, 23, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Swallows, C.L.; Zhang, C.; Lu, J.; Xiao, H.; Brady, R.O.; Zhuang, Z. Celastrol increases glucocerebrosidase activity in Gaucher disease by modulating molecular chaperones. Proc. Natl. Acad. Sci. USA 2014, 111, 249–254. [Google Scholar] [CrossRef]
- Sarodaya, N.; Suresh, B.; Kim, K.-S.; Ramakrishna, S. Protein Degradation and the Pathologic Basis of Phenylketonuria and Hereditary Tyrosinemia. Int. J. Mol. Sci. 2020, 21, 4996. [Google Scholar] [CrossRef] [PubMed]
- Blouin, J.-M.; Bernardo-Seisdedos, G.; Sasso, E.; Esteve, J.; Ged, C.; Lalanne, M.; Sanz-Parra, A.; Urquiza, P.; De Verneuil, H.; Millet, O.; et al. Missense UROS mutations causing congenital erythropoietic porphyria reduce UROS homeostasis that can be rescued by proteasome inhibition. Hum. Mol. Genet. 2017, 26, 1565–1576. [Google Scholar] [CrossRef]
- Blouin, J.-M.; Duchartre, Y.; Costet, P.; Lalanne, M.; Ged, C.; Lain, A.; Millet, O.; De Verneuil, H.; Richard, E. Therapeutic potential of proteasome inhibitors in congenital erythropoietic porphyria. Proc. Natl. Acad. Sci. USA 2013, 110, 18238–18243. [Google Scholar] [CrossRef]
- Ernst, R.; Claessen, J.H.L.; Mueller, B.; Sanyal, S.; Spooner, E.; Van Der Veen, A.G.; Kirak, O.; Schlieker, C.D.; Weihofen, W.A.; Ploegh, H.L. Enzymatic Blockade of the Ubiquitin-Proteasome Pathway. PLoS Biol. 2011, 8, e1000605. [Google Scholar] [CrossRef]
- Pey, A.L.; Ying, M.; Cremades, N.; Velázquez-Campoy, A.; Scherer, T.; Thöny, B.; Sancho, J.; Martinez, A. Identification of pharmacological chaperones as potential therapeutic agents to treat phenylketonuria. J. Clin. Investig. 2008, 118, 2858–2867. [Google Scholar] [CrossRef]
- Morello, J.-P.; Salahpour, A.; Laperrière, A.; Bernier, V.; Arthus, M.-F.; Lonergan, M.; Petäjä-Repo, U.; Angers, S.; Morin, D.; Bichet, D.G.; et al. Pharmacological chaperones rescue cell-surface expression and function of misfolded V2 vasopressin receptor mutants. J. Clin. Investig. 2000, 105, 887–895. [Google Scholar] [CrossRef]
- Tao, Y.-X.; Conn, P.M. Pharmacoperones as Novel Therapeutics for Diverse Protein Conformational Diseases. Physiol. Rev. 2018, 98, 697–725. [Google Scholar] [CrossRef] [PubMed]
- Brehme, M.; Sverchkova, A.; Voisine, C. Proteostasis network deregulation signatures as biomarkers for pharmacological disease intervention. Curr. Opin. Syst. Biol. 2019, 15, 74–81. [Google Scholar] [CrossRef]
- Wang, M.; Kaufman, R.J. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 2016, 529, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.N.; Neupane, K.; Rezajooei, N.; Cortez, L.M.; Sim, V.L.; Woodside, M.T. Pharmacological chaperone reshapes the energy landscape for folding and aggregation of the prion protein. Nat. Commun. 2016, 7, 12058. [Google Scholar] [CrossRef]
- Dong, C.; Garen, C.R.; Mercier, P.; Petersen, N.O.; Woodside, M.T. Characterizing the inhibition of α-synuclein oligomerization by a pharmacological chaperone that prevents prion formation by the protein PrP. Protein Sci. 2019, 28, 1690–1702. [Google Scholar] [CrossRef]
- Losada Díaz, J.C.; Cepeda Del Castillo, J.; Rodríguez-López, E.A.; Alméciga-Díaz, C.J. Advances in the Development of Pharmacological Chaperones for the Mucopolysaccharidoses. Int. J. Mol. Sci. 2019, 21, 232. [Google Scholar] [CrossRef]
- Yue, W.W. From structural biology to designing therapy for inborn errors of metabolism. J. Inherit. Metab. Dis. 2016, 39, 489–498. [Google Scholar] [CrossRef]
- Martinez, A.; Calvo, A.C.; Teigen, K.; Pey, A.L. Rescuing Proteins of Low Kinetic Stability by Chaperones and Natural Ligands: Phenylketonuria, a Case Study. Prog. Mol. Biol. Trans. Sci. 2008, 89–134. [Google Scholar] [CrossRef]
- Wittung-Stafshede, P. Role of Cofactors in Protein Folding. Acc. Chem. Res. 2002, 35, 201–208. [Google Scholar] [CrossRef]
- Skjærven, L.; Reuter, N.; Martinez, A. Dynamics, flexibility and ligand-induced conformational changes in biological macromolecules: A computational approach. Future Med. Chem. 2011, 3, 2079–2100. [Google Scholar] [CrossRef]
- Teilum, K.; Olsen, J.G.; Kragelund, B.B. Functional aspects of protein flexibility. Cell. Mol. Life Sci. 2009, 66, 2231–2247. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, E.K. Wrangling Shape-Shifting Morpheeins to Tackle Disease and Approach Drug Discovery. Front. Mol. Biosci. 2020, 7, 582966. [Google Scholar] [CrossRef] [PubMed]
- Boyd, R.E.; Lee, G.; Rybczynski, P.; Benjamin, E.R.; Khanna, R.; Wustman, B.A.; Valenzano, K.J. Pharmacological Chaperones as Therapeutics for Lysosomal Storage Diseases. J. Med. Chem. 2013, 56, 2705–2725. [Google Scholar] [CrossRef] [PubMed]
- Guce, A.I.; Clark, N.E.; Salgado, E.N.; Ivanen, D.R.; Kulminskaya, A.A.; Brumer, H.; Garman, S.C. Catalytic Mechanism of Human α-Galactosidase. J. Biol. Chem. 2010, 285, 3625–3632. [Google Scholar] [CrossRef] [PubMed]
- Khanna, R.; Benjamin, E.R.; Pellegrino, L.; Schilling, A.; Rigat, B.A.; Soska, R.; Nafar, H.; Ranes, B.E.; Feng, J.; Lun, Y.; et al. The pharmacological chaperone isofagomine increases the activity of the Gaucher disease L444P mutant form of β-glucosidase. FEBS J. 2010, 277, 1618–1638. [Google Scholar] [CrossRef]
- Fan, J.-Q. A counterintuitive approach to treat enzyme deficiencies: Use of enzyme inhibitors for restoring mutant enzyme activity. Biol. Chem. 2008, 389, 1–11. [Google Scholar] [CrossRef]
- Yu, Y.; Mena-Barragán, T.; Higaki, K.; Johnson, J.L.; Drury, J.E.; Lieberman, R.L.; Nakasone, N.; Ninomiya, H.; Tsukimura, T.; Sakuraba, H.; et al. Molecular Basis of 1-Deoxygalactonojirimycin Arylthiourea Binding to Human α-Galactosidase A: Pharmacological Chaperoning Efficacy on Fabry Disease Mutants. ACS Chem. Biol. 2014, 9, 1460–1469. [Google Scholar] [CrossRef]
- Jung, O.; Patnaik, S.; Marugan, J.; Sidransky, E.; Westbroek, W. Progress and potential of non-inhibitory small molecule chaperones for the treatment of Gaucher disease and its implications for Parkinson disease. Expert Rev. Proteom. 2016, 13, 471–479. [Google Scholar] [CrossRef]
- Nussinov, R.; Tsai, C.-J. Allostery in Disease and in Drug Discovery. Cell 2013, 153, 293–305. [Google Scholar] [CrossRef]
- Janovick, J.A.; Maya-Núñez, G.; Ulloa-Aguirre, A.; Huhtaniemi, I.T.; Dias, J.A.; Verbost, P.; Conn, P.M. Increased plasma membrane expression of human follicle-stimulating hormone receptor by a small molecule thienopyr(im)idine. Mol. Cell. Endocrinol. 2009, 298, 84–88. [Google Scholar] [CrossRef]
- Porto, C.; Ferrara, M.C.; Meli, M.; Acampora, E.; Avolio, V.; Rosa, M.; Cobucci-Ponzano, B.; Colombo, G.; Moracci, M.; Andria, G.; et al. Pharmacological Enhancement of α-Glucosidase by the Allosteric Chaperone N-acetylcysteine. Mol. Ther. 2012, 20, 2201–2211. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.L.; Génisson, Y.; Ballereau, S.; Dehoux, C. Second-Generation Pharmacological Chaperones: Beyond Inhibitors. Molecules 2020, 25, 3145. [Google Scholar] [CrossRef] [PubMed]
- Conn, P.M.; Spicer, T.P.; Scampavia, L.; Janovick, J.A. Assay strategies for identification of therapeutic leads that target protein trafficking. Trends Pharmacol. Sci. 2015, 36, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Goldin, E.; Zheng, W.; Motabar, O.; Southall, N.; Choi, J.H.; Marugan, J.; Austin, C.P.; Sidransky, E. High Throughput Screening for Small Molecule Therapy for Gaucher Disease Using Patient Tissue as the Source of Mutant Glucocerebrosidase. PLoS ONE 2012, 7, e29861. [Google Scholar] [CrossRef] [PubMed]
- Underhaug, J.; Aubi, O.; Martinez, A. Phenylalanine Hydroxylase Misfolding and Pharmacological Chaperones. Curr. Top. Med. Chem. 2012, 12, 2534–2545. [Google Scholar] [CrossRef]
- Lucas, T.G.; Gomes, C.M.; Henriques, B.J. Thermal Shift and Stability Assays of Disease-Related Misfolded Proteins Using Differential Scanning Fluorimetry. Protein Misfolding Dis. 2018, 1873, 255–264. [Google Scholar] [CrossRef]
- Støve, S.I.; Flydal, M.I.; Hausvik, E.; Underhaug, J.; Martinez, A. Differential scanning fluorimetry in the screening and validation of pharmacological chaperones for soluble and membrane proteins. In Protein Homeostasis Diseases; Pey, A.L., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 329–341, Chapter 15. [Google Scholar] [CrossRef]
- Irwin, J.J.; Shoichet, B.K. Docking Screens for Novel Ligands Conferring New Biology. J. Med. Chem. 2016, 59, 4103–4120. [Google Scholar] [CrossRef]
- Lavecchia, A.; Di Giovanni, C. Virtual Screening Strategies in Drug Discovery: A Critical Review. Curr. Med. Chem. 2013, 20, 2839–2860. [Google Scholar] [CrossRef]
- Urquiza, P.; Laín, A.; Sanz-Parra, A.; Moreno, J.; Bernardo-Seisdedos, G.; Dubus, P.; González, E.; Gutiérrez-De-Juan, V.; García, S.; Eraña, H.; et al. Repurposing ciclopirox as a pharmacological chaperone in a model of congenital erythropoietic porphyria. Sci. Transl. Med. 2018, 10, eaat7467. [Google Scholar] [CrossRef]
- Mele, B.H.; Citro, V.; Andreotti, G.; Cubellis, M.V. Drug repositioning can accelerate discovery of pharmacological chaperones. Orphanet J. Rare Dis. 2015, 10, 1–3. [Google Scholar] [CrossRef]
- Surade, S.; Blundell, T.L. Structural Biology and Drug Discovery of Difficult Targets: The Limits of Ligandability. Chem. Biol. 2012, 19, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Maveyraud, L.; Mourey, L. Protein X-ray Crystallography and Drug Discovery. Molecules 2020, 25, 1030. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, B.; Das, D.; Bung, N.; Roy, A.; Bulusu, G. Network analysis of hydroxymethylbilane synthase dynamics. J. Mol. Graph. Model. 2020, 99, 107641. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Huynh, D.T.; Yeates, T.O. A 3.8 Å resolution cryo-EM structure of a small protein bound to an imaging scaffold. Nat. Commun. 2019, 10, 1864. [Google Scholar] [CrossRef]
- Murata, K.; Wolf, M. Cryo-electron microscopy for structural analysis of dynamic biological macromolecules. Biochim. Biophys. Acta BBA Gen. Subj. 2018, 1862, 324–334. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bustad, H.J.; Kallio, J.P.; Vorland, M.; Fiorentino, V.; Sandberg, S.; Schmitt, C.; Aarsand, A.K.; Martinez, A. Acute Intermittent Porphyria: An Overview of Therapy Developments and Future Perspectives Focusing on Stabilisation of HMBS and Proteostasis Regulators. Int. J. Mol. Sci. 2021, 22, 675. https://doi.org/10.3390/ijms22020675
Bustad HJ, Kallio JP, Vorland M, Fiorentino V, Sandberg S, Schmitt C, Aarsand AK, Martinez A. Acute Intermittent Porphyria: An Overview of Therapy Developments and Future Perspectives Focusing on Stabilisation of HMBS and Proteostasis Regulators. International Journal of Molecular Sciences. 2021; 22(2):675. https://doi.org/10.3390/ijms22020675
Chicago/Turabian StyleBustad, Helene J., Juha P. Kallio, Marta Vorland, Valeria Fiorentino, Sverre Sandberg, Caroline Schmitt, Aasne K. Aarsand, and Aurora Martinez. 2021. "Acute Intermittent Porphyria: An Overview of Therapy Developments and Future Perspectives Focusing on Stabilisation of HMBS and Proteostasis Regulators" International Journal of Molecular Sciences 22, no. 2: 675. https://doi.org/10.3390/ijms22020675
APA StyleBustad, H. J., Kallio, J. P., Vorland, M., Fiorentino, V., Sandberg, S., Schmitt, C., Aarsand, A. K., & Martinez, A. (2021). Acute Intermittent Porphyria: An Overview of Therapy Developments and Future Perspectives Focusing on Stabilisation of HMBS and Proteostasis Regulators. International Journal of Molecular Sciences, 22(2), 675. https://doi.org/10.3390/ijms22020675






