Apple Autotetraploids with Enhanced Resistance to Apple Scab (Venturia inaequalis) Due to Genome Duplication-Phenotypic and Genetic Evaluation
Abstract
:1. Introduction
2. Results
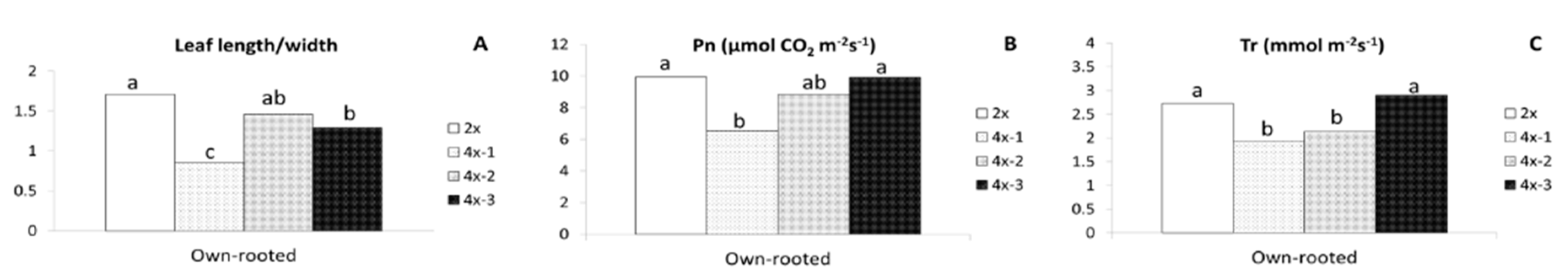
2.1. Morphological Observations
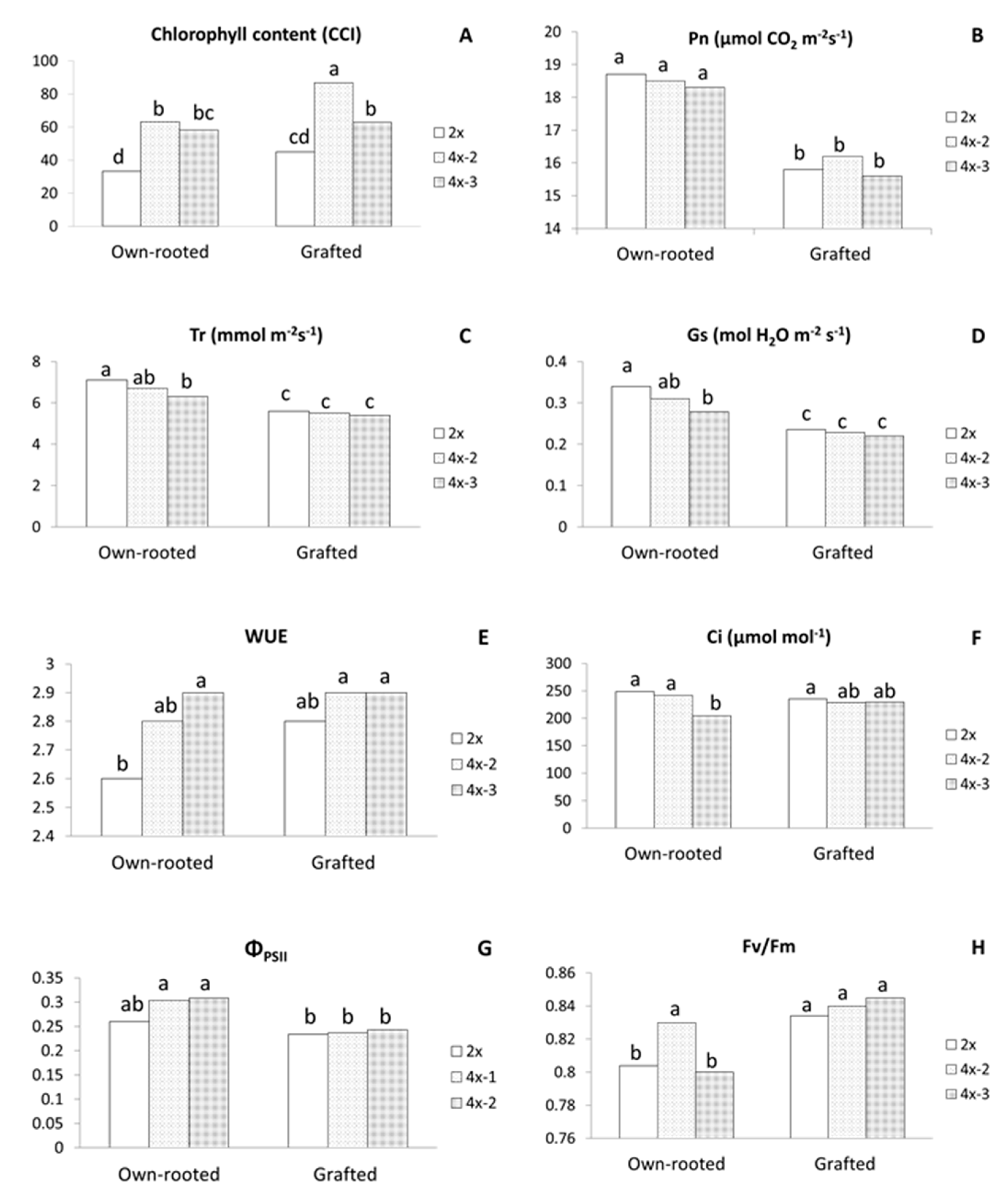
2.2. Physiological Parameters
2.3. Susceptibility to Apple Scab
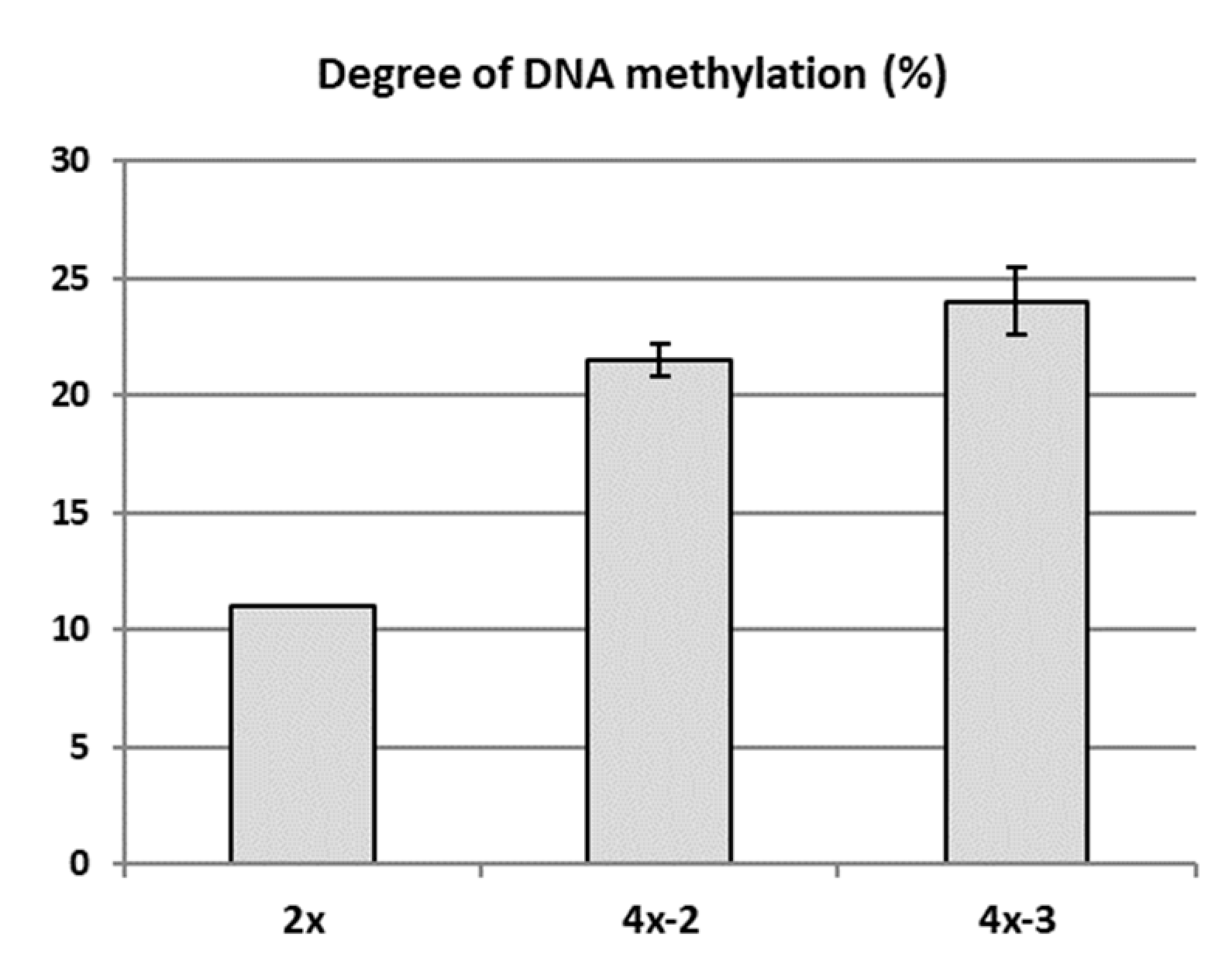
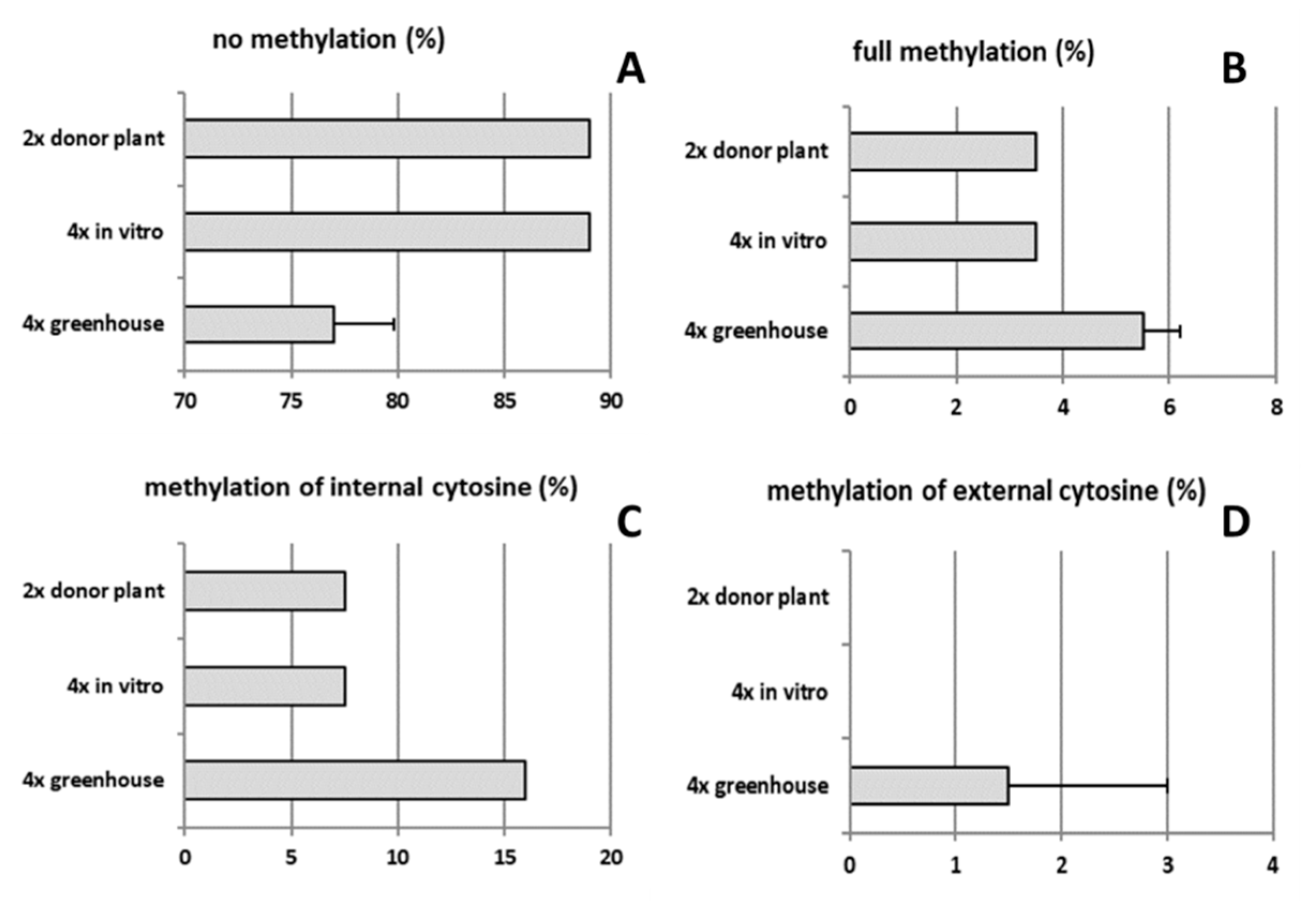
2.4. Analysis of Genetic and Epigenetic Changes in Tetraploids Compared to the Diploid Genotype
2.5. Analysis of Rvi Genes
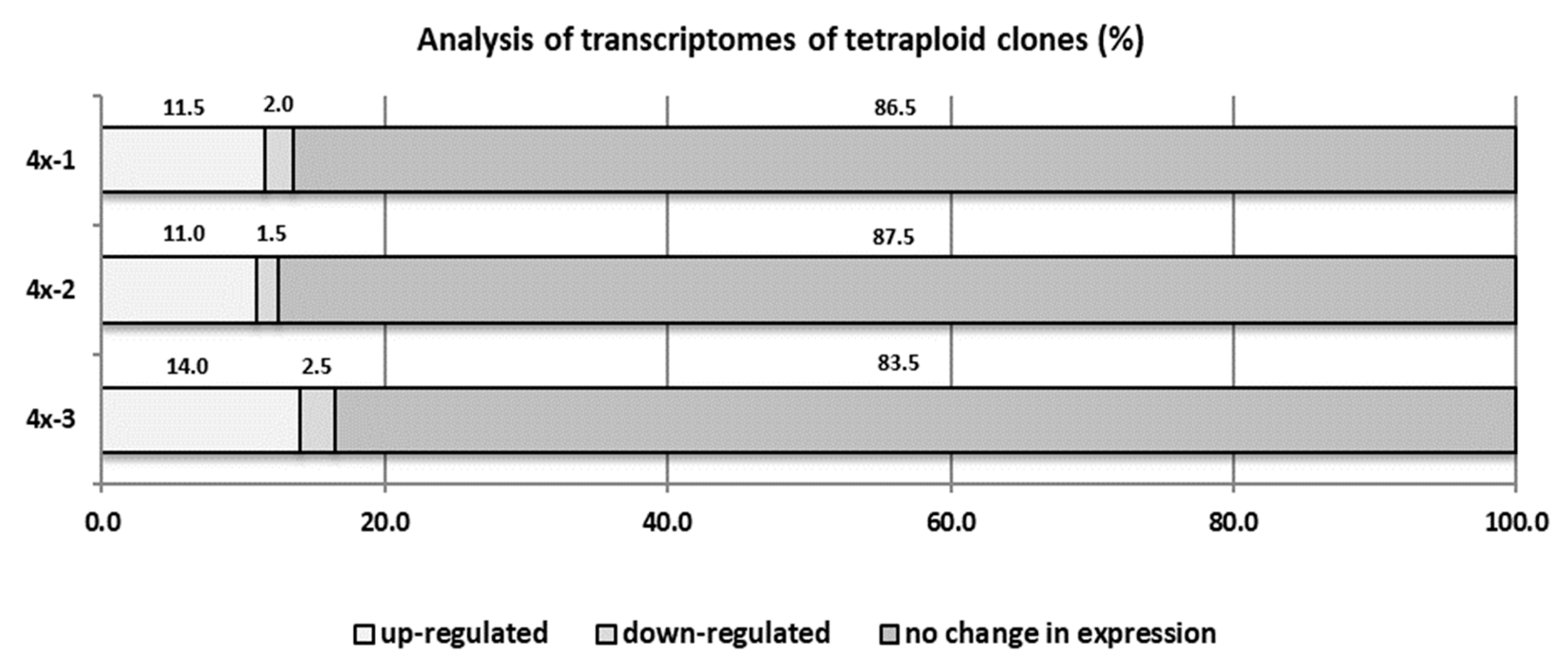
2.6. Comparative Transcriptome Analysis of Diploid and Tetraploid Apple Plants Inoculated with V. inaequalis (cDNA-AFLP (Amplified Fragment Length Polymorphism) Analysis)
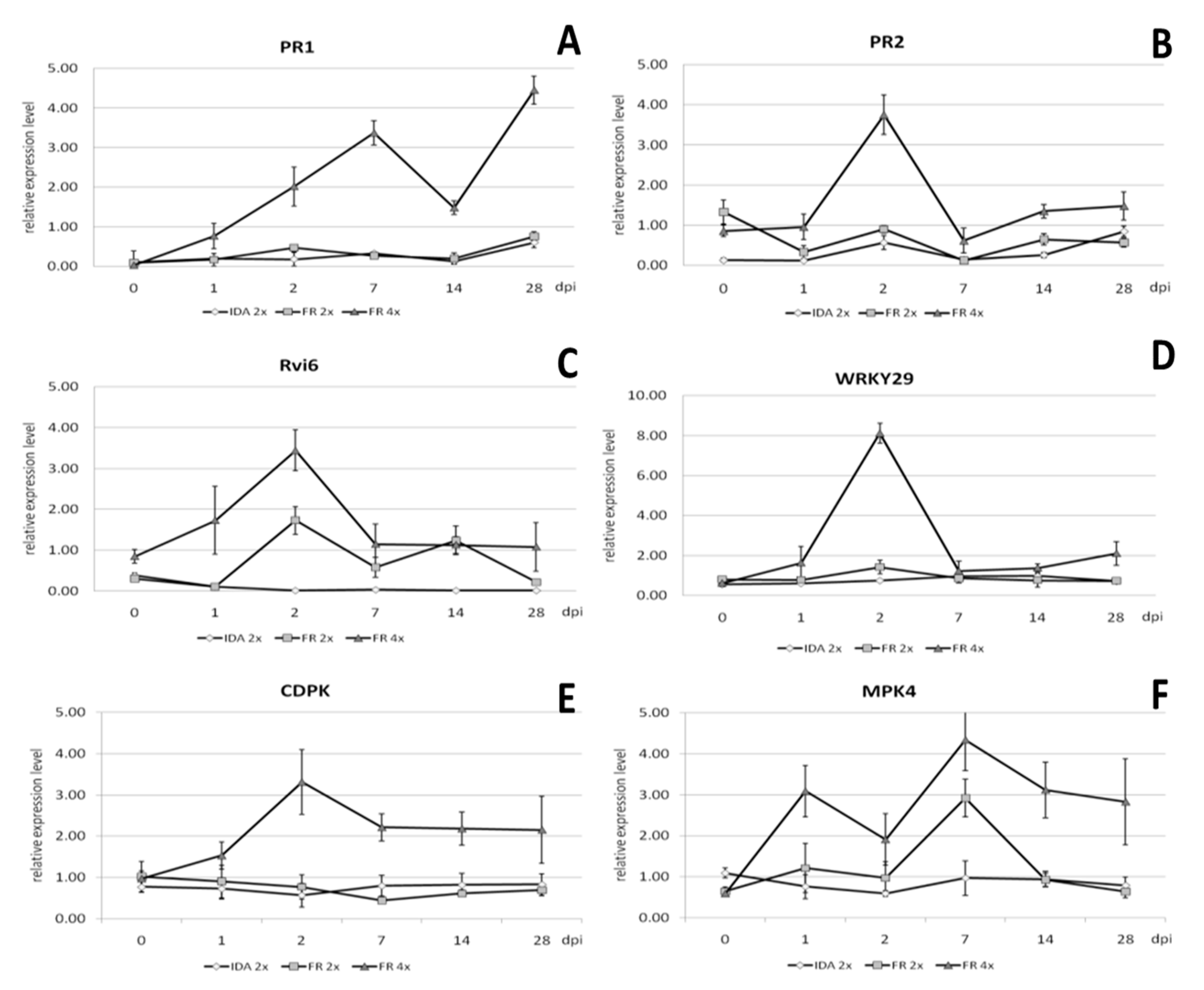
2.7. Gene Expression Analysis
3. Discussion
3.1. Morphological and Physiological Differences between Diploid and Tetraploid Plants
3.2. Poor Vigor of Neotetraploids in the Context of Increased DNA Methylation
3.3. Phenotypic and Genetic Variation among the Sibling Neotetraploids
3.4. Presence of Rvi Genes in Diploid and Tetraploid Plants
3.5. Enhanced Resistance of Neotetraploids to Apple Scab as the Result of Increased Expression Levels of Resistance-Related Genes
3.6. Comparative Transcriptome Analysis of Diploid and Tetraploid Apple Plants Inoculated with V. inaequalis
3.7. Summary
4. Materials and Methods
4.1. Plant Material
4.2. Morphological Observation
4.3. Evaluation of Physiological Parameters
4.4. Testing for Susceptibility to Apple Scab
4.5. AFLP Analysis
4.6. Methylation-Sensitive Amplification Polymorphism (MSAP) Analysis
4.7. Determination of Rvi Genes
4.8. Analysis of Transcriptome and Expression of Resistance-Related Genes under V. inaequalis Infection
4.8.1. Transcriptome Analysis (cDNA-AFLP Analysis)
4.8.2. Gene Expression Analysis
4.9. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bus, V.G.; Rikkerink, E.H.; Caffier, V.; Durel, C.E.; Plummer, K.M. Revision of the nomenclature of the differential host-pathogen interactions of Venturia inaequalis and Malus. Annu. Rev. Phytopathol. 2011, 49, 391–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masny, S. Occurrence of Venturia inaequalis races in Poland able to overcome specific apple scab resistance genes. Eur. J. Plant Pathol. 2017, 147, 313–323. [Google Scholar] [CrossRef] [Green Version]
- Regulation (EC) No 1107/2009 of the European Parliament and of the Council of 21 October 2009 Concerning the Placing of Plant Protection Products on the Market. 2020. Available online: https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2009:309:0001:0050:EN:PDF (accessed on 5 January 2021).
- Patocchi, A.; Wehrli, A.; Dubuis, P.H.; Auwerkerken, A.; Leida, C.; Cipriani, G.; Paasey, T.; Staples, M.; Didelot, F.; Philion, V.; et al. Ten years of VINQUEST: First insight for breeding new apple cultivars with durable apple scab resistance. Plant Dis. 2020, 104, 2074–2081. [Google Scholar] [CrossRef]
- Khajuria, Y.P.; Kaul, S.; Wani, A.A.; Dhar, M.K. Genetics of resistance in apple against Venturia inaequalis (Wint.) Cke. Tree Genet. Genomes 2018, 14, 16. [Google Scholar] [CrossRef]
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Parisi, L.; Lespinasse, Y.; Guillaumes, J.; Krüger, J. A new race of Venturia inaequalis virulent to apples with resistance due to the Vf gene. Phytopathology 1993, 83, 533–537. [Google Scholar] [CrossRef]
- Aversano, R.; Ercolano, M.R.; Caruso, I.; Fasano, C.; Rosellini, D.; Carputo, D. Molecular tools for exploring polyploid genomes in plants. Int. J. Mol. Sci. 2012, 13, 10316–10335. [Google Scholar] [CrossRef] [Green Version]
- Soltis, P.S.; Liu, X.; Marchant, D.B.; Visger, C.J.; Soltis, D.E. Polyploidy and novelty: Gottlieb’s legacy. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130351. [Google Scholar] [CrossRef] [Green Version]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef]
- Hieu, P.V. Polyploid gene expression and regulation in polysomic polyploids. Am. J. Plant Sci. 2019, 10, 1409–1443. [Google Scholar] [CrossRef] [Green Version]
- Parisod, C.; Holderegger, R.; Brochmann, C. Evolutionary consequences of autopolyploidy. New Phytol. 2010, 186, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ye, C.Y.; Cheng, Z.M.; Tschaplinski, T.J.; Wullschleger, S.D.; Yin, W.; Xia, X.; Tuskan, G. Genomic aspects of research involving polyploid plants. Plant Cell Tissue Organ Cult. 2011, 104, 387–397. [Google Scholar] [CrossRef]
- Oswald, B.P.; Nuismer, S.L. Neopolyploidy and pathogen resistance. Proc. R. Soc. B 2007, 274, 2393–2397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wendel, J.; Lisch, D.; Hu, G.; Mason, A.S. The long and short of doubling down: Polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Curr. Opin. Genet. Dev. 2018, 49, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sedyscheva, G.A.; Gorbacheva, N.G. Estimation of new tetraploid apple forms as donors of diploid gametes for selection on a polyploidy level. Univ. J. Plant Sci. 2013, 1, 49–54. [Google Scholar]
- Jia, S. In Vitro Induction and Identification of Tetraploid in Malus Zumi and Its Evaluation of Stress-Tolerance. Master’s Thesis, Agricultural University of Hebei, Baoding, China, 2009. Available online: http://www.dissertationtopic.net/doc/517919 (accessed on 5 January 2021).
- Xue, H.; Zhang, F.; Zhang, Z.H.; Fu, J.F.; Wang, F.; Zhang, B.; Ma, Y. Differences in salt tolerance between diploid and autotetraploid apple seedlings exposed to salt stress. Sci. Hortic. 2015, 190, 24–30. [Google Scholar] [CrossRef]
- Xue, H.; Zhang, B.; Tian, J.R.; Chen, M.M.; Zhang, Y.Y.; Zhang, Z.H.; Ma, Y. Comparison of the morphology, growth and development of diploid and autotetraploid ‘Hanfu’ apple trees. Sci. Hortic. 2017, 225, 277–285. [Google Scholar] [CrossRef]
- Chen, M.; Wang, F.; Zhang, Z.; Fu, J.; Ma, Y. Characterization of fungi resistance in two autotetraploid apple cultivars. Sci. Hortic. 2017, 220, 27–35. [Google Scholar] [CrossRef]
- Hias, N.; De Dauw, K.; Davey, M.W.; Leus, L.; Van Labeke, M.C.; Van Huylenbroeck, J.; Keulemans, J. Influence of ploidy level on the physiological response of apple to water deficit. Acta Hortic. 2017, 1177, 333–338. [Google Scholar] [CrossRef]
- Hias, N.; Svara, A.; Keulemans, J.W. Effect of polyploidisation on the response of apple (Malus × domestica Borkh.) to Venturia inaequalis infection. Eur. J. Plant Pathol. 2018, 151, 515–526. [Google Scholar] [CrossRef]
- Podwyszyńska, M.; Markiewicz, M.; Klamkowski, K.; Broniarek, A.; Wojtania, A. The genetic background of the phenotypic variability observed in apple autotetraploids. Acta Hortic. 2020, in press. [Google Scholar]
- Podwyszyńska, M.; Sowik, I.; Puławska, J. In vitro testing of apple tetraploids for resistance to fire blight. Acta Hortic. 2020, 1282, 343–350. [Google Scholar] [CrossRef]
- Podwyszyńska, M.; Sowik, I.; Machlańska, A.; Kruczyńska, D.; Dyki, B. In Vitro tetraploid induction of Malus × domestica Borkh. Using leaf or shoot explants. Sci. Hortic. 2017, 226, 379–388. [Google Scholar] [CrossRef]
- Podwyszyńska, M.; Cieślińska, M. Rooting shoots of apple varieties and their tetraploids obtained by the in vitro technique. Acta Sci. Pol.-Hortorum Cultus 2018, 17, 49–64. [Google Scholar] [CrossRef]
- Soledade, M.; Pedras, C.; Yaya, E.E. Plant chemical defenses: Are all constitutive antimicrobial metabolites phytoanticipins? Nat. Prod. Commun. 2015, 10, 2009–2018. [Google Scholar]
- Czolpinska, M.; Rurek, M. Plant Glycine-Rich Proteins in Stress Response: An Emerging, Still Prospective Story. Front. Plant Sci. 2018, 9, 302. [Google Scholar] [CrossRef]
- Markiewicz, M.; Michalczuk, L.; Neumüller, M. Hypersensitive reaction of plum (Prunus domestica) in response to Plum pox virus infection: Changes in gene expression and identification of potential molecular markers. Sci. Hortic. 2019, 247, 430–435. [Google Scholar] [CrossRef]
- Markiewicz, M.; Michalczuk, L. Molecular response of resistant and susceptible apple genotypes to Erwinia amylovora infection. Eur. J. Plant Pathol. 2015, 143, 515–526. [Google Scholar] [CrossRef]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212, 29–37. [Google Scholar] [CrossRef]
- Sarowar, S.; Alam, S.T.; Makandar, R.; Lee, H.; Trick, H.N.; Dong, Y.; Shah, J. Targeting the pattern-triggered immunity pathway to enhance resistance to Fusarium graminearum. Mol. Plant Pathol. 2019, 20, 626–640. [Google Scholar] [CrossRef] [Green Version]
- Shi, S.; Li, S.; Asim, M.; Mao, J.; Xu, D.; Ullah, Z.; Liu, G.; Wang, Q.; Liu, H. The Arabidopsis calcium-dependent protein kinases (CDPKs) and their roles in plant growth regulation and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 1900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulz, P.; Herde, M.; Romeis, T. Calcium-dependent protein kinases: Hubs in plant stress signaling and development. Plant Physiol. 2013, 163, 523–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Schneider, J.D.; Lin, C.; Geng, S.; Ma, T.; Lawrence, S.R.; Dufresne, C.P.; Harmon, A.C.; Chen, S. MPK4 phosphorylation dynamics and interacting proteins in plant immunity. J. Proteome Res. 2019, 18, 826–840. [Google Scholar] [CrossRef] [PubMed]
- Podwyszyńska, M.; Gabryszewska, E.; Dyki, B.; Stępowska, A.; Kowalski, A.; Jasiński, A. Phenotypic and genome size changes (variation) in synthetic tetraploids of daylily (Hemerocallis) in relation to their diploid counterparts. Euphytica 2015, 203, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Xue, H.; Zhang, L.; Zhang, F.; Ou, C.; Wang, F.; Zhang, Z. Involvement of auxin and brassinosteroid in dwarfism of autotetraploid apple (Malus × domestica). Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Rodríguez, A.M.; Grajal-Martín, M.J. Physiological behaviour of mangos with different ploidy levels. Acta Hortic. 2013, 992, 155–158. [Google Scholar] [CrossRef]
- Korban, S.S.; Wannarat, W.; Rayburn, C.M.; Tatum, T.C.; Rayburn, A.L. Genome size and nucleotypic variation in Malus germplasm. Genome 2009, 52, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Podwyszyńska, M.; Kruczynska, D.; Machlanska, A.; Dyki, B.; Sowik, I. Nuclear DNA content and ploidy level of apple cultivars including Polish ones in relation to some morphological traits. Acta Biol. Crac. Ser. Bot. 2016, 58, 81–93. [Google Scholar] [CrossRef]
- Bassett, C.L.; Glenn, D.M.; Forsline, P.L.; Wisniewski, M.E.; Farrell, R.E. Characterizing water use efficiency and water deficit responses in apple (Malus × domestica Borkh. and Malus sieversii Ledeb.) M. Roem. Hortic. Sci. 2011, 46, 1079–1084. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Xue, H.; Lu, X.; Zhang, B.; Wang, F.; Ma, Y.; Zhang, Z. Autotetraploidization enhances drought stress tolerance in two apple cultivars. Trees 2015, 29, 1773–1780. [Google Scholar] [CrossRef]
- Foster, T.M.; Mc Atee, P.A.; Waite, C.N.; Boldingh, H.L.; Mc Ghie, T. Apple dwarfing rootstocks exhibit an imbalance in carbohydrate allocation and reduced cell growth and metabolism. Hortic. Res. 2017, 4, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; An, H.S.; Wang, Y.; Zhang, X.Z.; Han, Z.H. Low expression of PIN gene family members is involved in triggering the dwarfing effect in M9 interstem but not in M9 rootstock apple trees. Acta Physiol. Plant. 2015, 37, 104. [Google Scholar] [CrossRef]
- Jensen, P.J.; Makalowska, I.; Altman, N.; Fazio, G.; Praul, C.; Maximova, S.N.; McNellis, T.W. Rootstock-regulated gene expression patterns in apple tree scions. Tree Genet. Genomes 2010, 6, 57–72. [Google Scholar] [CrossRef]
- He, P.; Cheng, L.; Li, H.; Wang, H.; Li, L. A comparative analysis of DNA methylation in diploid and tetraploid apple (Malus × domestica Borkh.). Czech J. Genet. Plant Breed. 2017, 53, 63–68. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zhang, W.; Chen, G.; Wang, J. DNA methylation alteration is a major consequence of genome doubling in autotetraploid Brassica rapa. Arch. Biol. Sci. 2017, 69, 689–697. [Google Scholar] [CrossRef]
- Lavania, U.C.; Srivastava, S.; Lavania, S.; Basu, S.; Misra, N.K.; Mukai, Y. Autopolyploidy differentially influences body size in plants, but facilitates enhanced accumulation of secondary metabolites, causing increased cytosine methylation. Plant J. 2012, 71, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [Green Version]
- Shilpa, K.; Sunkad, G.; Srinivasu, K.; Marri, S.; Padmashree, K.; Jadhav, D.R.; Sahrawat, K.L.; Mallikarjuna, N. Biochemical composition and disease resistance in newly synthesized amphidiploid and autotetraploid peanuts. Food Nutr. Sci. 2013, 4, 169–176. [Google Scholar] [CrossRef] [Green Version]
- Šedivá, J.; Mrázková, M.; Zahumenická, P.; Cusimamani, E.F.; Zahradník, D. Identification of Phytophthora tolerance in the Anemone sylvestris tetraploid. Sci. Hortic. 2019, 256, 108579. [Google Scholar] [CrossRef]
- Korbin, M.; Keller-Przybyłkowicz, S.; Żurawicz, E. Application of molecular markers in evaluation of selected apple genotypes for apple scab resistance genes. Biotechnologia 2008, 2, 153–161. [Google Scholar]
- Gygax, M.; Gianfranceschi, L.; Liebhard, R.; Kellerhals, M.; Gessler, C.; Patocchi, A. Molecular markers linked to the apple scab resistance gene Vbj derived from Malus baccata jackii. Theor. Appl. Genet. 2004, 109, 1702–1709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Żurawicz, E.; Lewandowski, M.; Broniarek-Niemiec, A.; Rutkowski, K. Preliminary results on the production value of new scab-resistant apple cultivars bred at the Research Institute of Pomology and Floriculture (RIPF), Skierniewice, Poland. Acta Hortic. 2004, 663, 879–882. [Google Scholar] [CrossRef]
- Chizzali, C.; Gusberti, M.; Schouten, H.J.; Gessler, C.; Broggini, G.A. Cisgenic Rvi6 scab-resistant apple lines show no differences in Rvi6 transcription when compared with conventionally bred cultivars. Planta 2016, 243, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Jiang, Y.; Rahman, M.H.; Deyholos, M.K.; Kav, N.N. Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol. 2009, 9, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choudhary, D.K.; Prakash, A.; Johri, B.N. Induced systemic resistance (ISR) in plants: Mechanism of action. Indian J. Microbiol. 2007, 47, 289–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, J. Plants under attack: Systemic signals in defense. Curr. Opin. Plant Biol. 2009, 12, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.; Guo, J.; Qiao, Q.; Guo, X.; Ma, Y. The WRKY transcription factor PlWRKY65 enhances the resistance of Paeonia lactiflora (herbaceous peony) to Alternaria tenuissima. Hortic. Res. 2020, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Aldon, D.; Mbengue, M.; Mazars, C.; Galaud, J.P. Calcium signaling in plant biotic interactions. Int. J. Mol. Sci. 2018, 19, 665. [Google Scholar] [CrossRef] [Green Version]
- Genenncher, B.; Wirthmueller, L.; Roth, C.; Klenke, M.; Ma, L.; Sharon, A.; Wiermer, M. Nucleoporin-regulated MAP kinase signaling in immunity to a necrotrophic fungal pathogen. Plant Physiol. 2016, 172, 1293–1305. [Google Scholar] [CrossRef] [Green Version]
- Asano, T.; Nguyen, T.H.N.; Yasuda, M.; Sidiq, Y.; Nishimura, K.; Nakashita, H.; Nishiuchi, T. Arabidopsis MAPKKK δ-1 is required for full immunity against bacterial and fungal infection. J. Exp. Bot. 2020, 71, 2085–2097. [Google Scholar] [CrossRef]
- Giuntoli, B.; Perata, P. Group VII ethylene response factors in Arabidopsis: Regulation and physiological roles. Plant Physiol. 2018, 176, 1143–1155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.G.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Mindrebo, J.T.; Nartey, C.M.; Seto, Y.; Burkart, M.D.; Noel, J.P. Unveiling the functional diversity of the alpha/beta hydrolase superfamily in the plant kingdom. Curr. Opin. Struct. Biol. 2016, 41, 233–246. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.M.; Song, H.R.; Han, S.-K.; Han, M.; Kim, C.Y.; Park, J.; Lee, Y.-H.; Jeon, J.-S.; Noh, Y.-S.; Noh, B. HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J. 2012, 71, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [Green Version]
- Koh, S.; Wiles, A.M.; Sharp, J.S.; Naider, F.R.; Becker, J.M.; Stacey, G. An oligopeptide transporter gene family in Arabidopsis. Plant Physiol. 2002, 128, 21–29. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, D.; Kang, Y.; Wu, J.X.; Yao, F.; Pan, C.; Yan, Z.; Song, C.; Chen, L. Structure of the mechanosensitive OSCA channels. Nat. Struct. Mol. Biol. 2018, 25, 850–858. [Google Scholar] [CrossRef]
- Pauwels, L.; Barbero, F.G.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Perez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [Green Version]
- van den Burg, H.A.; Tsitsigiannis, D.I.; Rowland, O.; Lo, J.; Rallapalli, G.; Maclean, D.; Takken, F.L.W.; Jones, J.D.G. The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell 2008, 20, 697–719. [Google Scholar] [CrossRef] [Green Version]
- Ji, R.; Wang, Y.; Wang, X.; Liu, Y.; Shen, X.; Feng, H. Proteomic analysis of the interaction between Plasmodiophora brassicae and Chinese cabbage (Brassica rapa L. ssp. pekinensis) at the initial infection stage. Sci. Hortic. 2018, 233, 386–393. [Google Scholar]
- Janick, J. The PRI apple breeding program. HortScience 2006, 41, 8–10. [Google Scholar] [CrossRef] [Green Version]
- Vos, P.; Hogers, R.; Blecker, M.; Reijans, M.; Lee, T.V.D.; Hornes, M.; Frijters, A.; Pot, L.; Peleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, L.Z.; Xu, C.G.; Maroof, S.; Zhang, G. Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol. Gen. Genet. 1999, 261, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Peraza-Echeverria, S.; Herrera-Valencia, V.A.; James-Kay, A. Detection of DNA methylation changes in micropropagated banana plants using methylation-sensitive amplification polymorphism (MSAP). Plant Sci. 2001, 161, 359–367. [Google Scholar] [CrossRef]
- Fulneček, J.; Kowařik, A. How to interpret methylation sensitive amplified polymorphism (MSAP) profiles? BMC Genet. 2014, 15, 2. [Google Scholar] [CrossRef] [Green Version]
- Money, T.; Reader, S.; Qu, L.J.; Dunford, R.P.; Moore, G. AFLP-based mRNA fingerprinting. Nucleic Acids Res. 1996, 24, 2616–2617. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Cong, P.; Tian, Y.; Zhu, Y. Using RNA-seq data to select reference genes for normalizing gene expression in apple roots. PLoS ONE 2017, 12, e0185288. [Google Scholar] [CrossRef]
- Larionov, A.; Krause, A.; Miller, W. A standard curve based method for relative real time PCR data processing. BMC Bioinform. 2005, 6, 62. [Google Scholar] [CrossRef] [Green Version]



| Trait | Diploid | Tetraploid * | p |
|---|---|---|---|
| Shoot length (cm) | 54.1 a | 14.5 b | 0.000 |
| Stem diameter measured at the middle of the shoot (mm) | 4.9 a | 4.1 b | 0.005 |
| Number of nodes | 27.0 a | 16.7 b | 0.000 |
| Leaf area (cm2) | 39.1a | 31.6 a | 0.100 |
| Leaf length (cm) | 9.7 a | 7.7 b | 0.005 |
| Leaf width (cm) | 5.7 a | 5.9 a | 0.503 |
| Leaf length/width | 1.71 a | 1.30 b | 0.000 |
| Stomata length (µm) | 28.3 b | 37.8 a | 0.000 |
| Chlorophyll content index (CCI) | 32.0 b | 38.4 a | 0.000 |
| Net photosynthesis rate (Pn) (µmol CO2 m−2 s−1) | 9.94 a | 8.68 a | 0.276 |
| Transpiration rate (Tr) (mmol m−2 s−1) | 2.73 a | 2.45 a | 0.132 |
| Water-use efficiency (WUE) | 3.63 a | 3.59 a | 0.916 |
| Quantum efficiency of open photosystem II centres (Fv/Fm) | 0.797 a | 0.768 b | 0.024 |
| Photosynthetic quantum yield (ΦPSII) | 0.154 b | 0.200 a | 0.019 |
| Genotype | Fungal Lesion (Scale 0–5) | |||
|---|---|---|---|---|
| 2017 | 2018 | 2019 | 2020 | |
| ‘Idared’ (reference) | 4.0 a | 3.3 a | - | 2.0 a |
| ‘Free Redstar’ 2x | 1.5 b | 3.7 a | 4.3 a | 2.7 a |
| 4x-1 | 0.0 c | 0.0 b | 0.0 b | 0.0 b |
| 4x-2 | 0.0 c | 0.0 b | 0.1 b | 0.0 b |
| 4x-3 | 0.0 c | 0.0 b | 0.1 b | 0.0 b |
| 4x-4 | 0.0 c | 0.0 b | 0.0 b | 0.0 b |
| Rvi Genes | Genotypes | ||||
|---|---|---|---|---|---|
| 4x-1 | 4x-2 | 4x-3 | 2x | Idared 2x (Reference) | |
| Rvi5 | + | + | + | + | − |
| Rvi6 | + | + | + | + | − |
| Rvi7 | − | − | − | − | − |
| Rvi8 | + | + | + | + | − |
| Rvi11 | + | + | + | + | − |
| Rvi14 | + | + | + | + | − |
| Rvi15 | − | − | − | − | − |
| Rvi17 | + | + | + | + | − |
| EST ID | Basic Local Alignment Search Tool (BLAST) Annotation | Gen Bank Access Code | E Value | Functional Classification |
|---|---|---|---|---|
| Homology to Genes from NIH GenBank | ||||
| M12 | No homology | |||
| M14 | Chlorophyll a-b binding protein 6 (Malus × domestica) | XP_028961659.1 | 8e−83 | other (photosynthesis) |
| M15 | Chlorophyll a-b binding protein 6 (Prunus × avium) | XP_021832771.1 | 3e−88 | other (photosynthesis) |
| M17 | Hypothetical protein DVH24_023262 (Malus × domestica) | RXI09118.1 | 2e−88 | unknown |
| M18 | Putative F-box/LRR-repeat protein (Brassica × rapa) | XP_009131094.1 | 2e−18 | protein modification |
| M19 | oligopeptide transporter 3-like isoform X2 (Malus × domestica) | XP_028963015.1 | 2e−05 | cellular transport |
| M21 | Hypothetical protein (Malus × domestica) | RXI09118 | 2e−87 | unknown |
| M22 | No homology | |||
| M23 | Chlorophyll a-b binding protein (Pyrus × bretschneideri) | KJ008954.1 | 3e−178 | other (photosynthesis) |
| M24 | Chlorophyll a-b binding protein 6 (Prunus × persica) | XM_007201177.2 | 2e−179 | other (photosynthesis) |
| M25 | Uncharacterized protein (Malus × domestica) | XM_008372153.3 | 6e−88 | unknown |
| M26 | EFR3 protein (Juglans × regia) | XM_018960757.1 | 1e−14 | defense and host-pathogen interaction |
| M2A | CSC1-like protein HYP1 isoform X1 (Malus × domestica) | XM_008343386.3 | 3e−143 | cellular transport, signal transduction |
| M2 | Ethylene-responsive transcription factor RAP2-7 isoform X2 (Malus × domestica) | XP_028944297.1 | 5e−17 | defense and host-pathogen interaction, signal transduction |
| M3A | CSC1-like protein HYP1 isoform X1 (Malus × domestica) | XP_008341608.1 | 2e−51 | cellular transport signal transduction |
| M49 | Adenylosuccinatesynthetase 2 (Malus × domestica) | XM_008384161.3 | 2e−89 | other (purine synthesis) |
| M51 | No homology | |||
| M53A | Ninja-family protein mc410-like (Malus × domestica) | XM_029110229.1 | 3e−106 | signal transduction |
| M54 | No homology | |||
| M61 | Chlorophyll a-b binding protein 151 (Malus × domestica) | XM_008381765.3 | 1e−46 | other (photosynthesis) |
| M63 | Adenylosuccinatesynthetase 2 (Pyrus × bretschneideri) | XM_009380296.2 | 2e−94 | other (purine synthesis) |
| M64 | Histone deacetylase 19-like protein (Malus × domestica) | XM_008350733.3 | 1e−09 | gene expression regulation |
| M65 | No homology | |||
| M66 | Adenylosuccinatesynthetase 2 (Malus × domestica) | XM_008374895.3 | 2e−84 | other (purine synthesis) |
| M70 | Transcription factor MYC2-like (Malus × domestica) | XM_008343741.3 | 3e−133 | gene expression regulation |
| M71 | S-formylglutathione hydrolase (Nicotiana × attenuata) | XM_019391580.1 | 1e−08 | other (cell detoxification) |
| M75 | Alpha/beta hydrolase (Malus × domestica) | WP_105934285.1 | 1e−33 | defense and host-pathogen interaction |
| M77 | Histone H3.3 (Malus × domestica) | XM_008345103.3 | 1e−48 | gene expression regulation |
| M79 | Far upstream element-binding protein 1-like isoform X2 (Malus × domestica) | XP_029106921.1 | 6e−32 | gene expression regulation |
| M9 | LHC-I protein complex (Nicotiana × tabacum) | X64198.1 | 4e−112 | other (photosynthesis) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Podwyszyńska, M.; Markiewicz, M.; Broniarek-Niemiec, A.; Matysiak, B.; Marasek-Ciolakowska, A. Apple Autotetraploids with Enhanced Resistance to Apple Scab (Venturia inaequalis) Due to Genome Duplication-Phenotypic and Genetic Evaluation. Int. J. Mol. Sci. 2021, 22, 527. https://doi.org/10.3390/ijms22020527
Podwyszyńska M, Markiewicz M, Broniarek-Niemiec A, Matysiak B, Marasek-Ciolakowska A. Apple Autotetraploids with Enhanced Resistance to Apple Scab (Venturia inaequalis) Due to Genome Duplication-Phenotypic and Genetic Evaluation. International Journal of Molecular Sciences. 2021; 22(2):527. https://doi.org/10.3390/ijms22020527
Chicago/Turabian StylePodwyszyńska, Małgorzata, Monika Markiewicz, Agata Broniarek-Niemiec, Bożena Matysiak, and Agnieszka Marasek-Ciolakowska. 2021. "Apple Autotetraploids with Enhanced Resistance to Apple Scab (Venturia inaequalis) Due to Genome Duplication-Phenotypic and Genetic Evaluation" International Journal of Molecular Sciences 22, no. 2: 527. https://doi.org/10.3390/ijms22020527
APA StylePodwyszyńska, M., Markiewicz, M., Broniarek-Niemiec, A., Matysiak, B., & Marasek-Ciolakowska, A. (2021). Apple Autotetraploids with Enhanced Resistance to Apple Scab (Venturia inaequalis) Due to Genome Duplication-Phenotypic and Genetic Evaluation. International Journal of Molecular Sciences, 22(2), 527. https://doi.org/10.3390/ijms22020527






