Mechanistic Insight into Royal Protein Inhibiting the Gram-Positive Bacteria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling of RJ from the First, Second, and Third Instar Larvae
2.2. Antibacterial Assays of RJ Aqueous Solution, Water-Soluble Fractions, and N-MRJP2
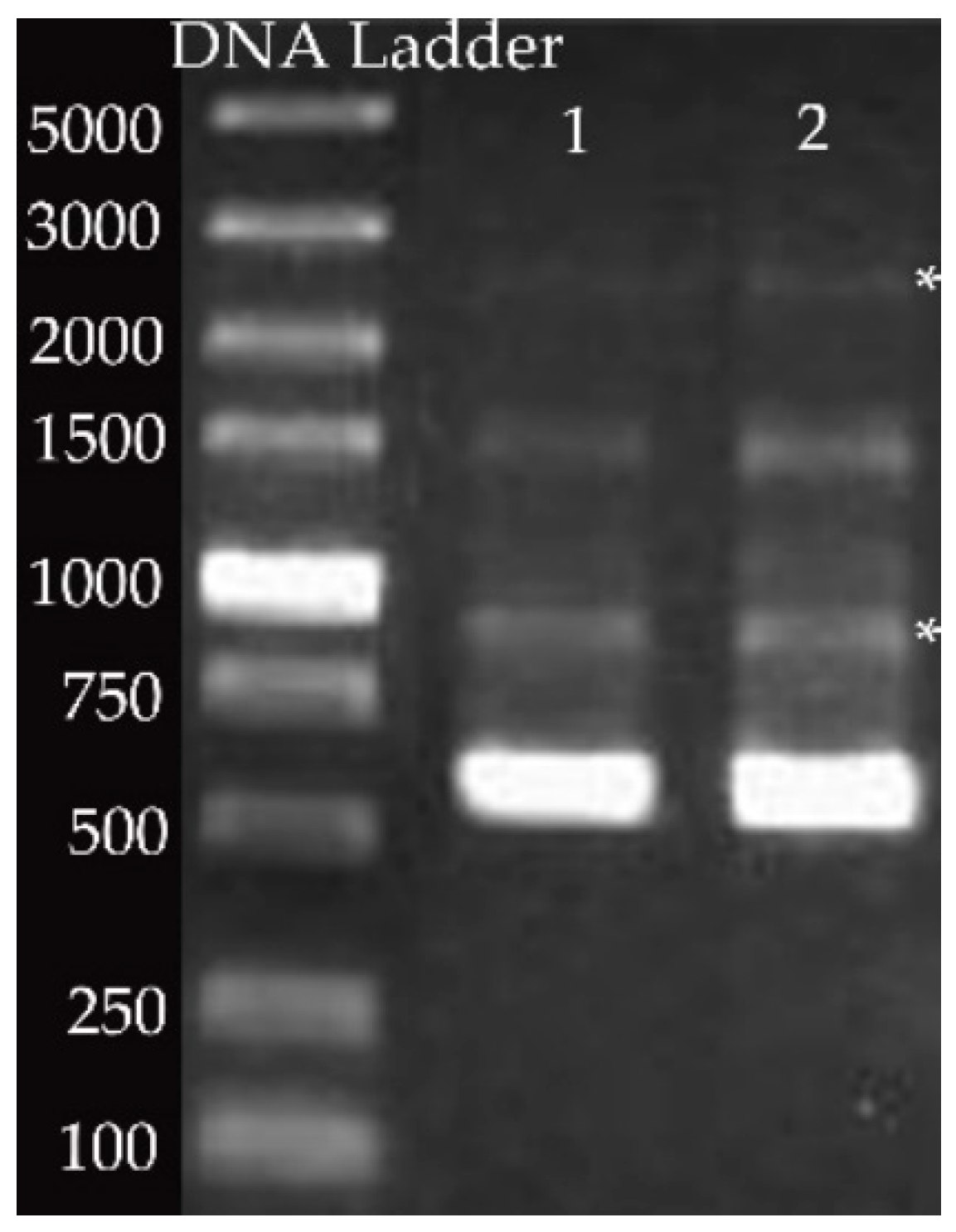
2.3. Genotyping of P. larvae
2.4. Determination of Released UV-Absorbing Material of P. larvae
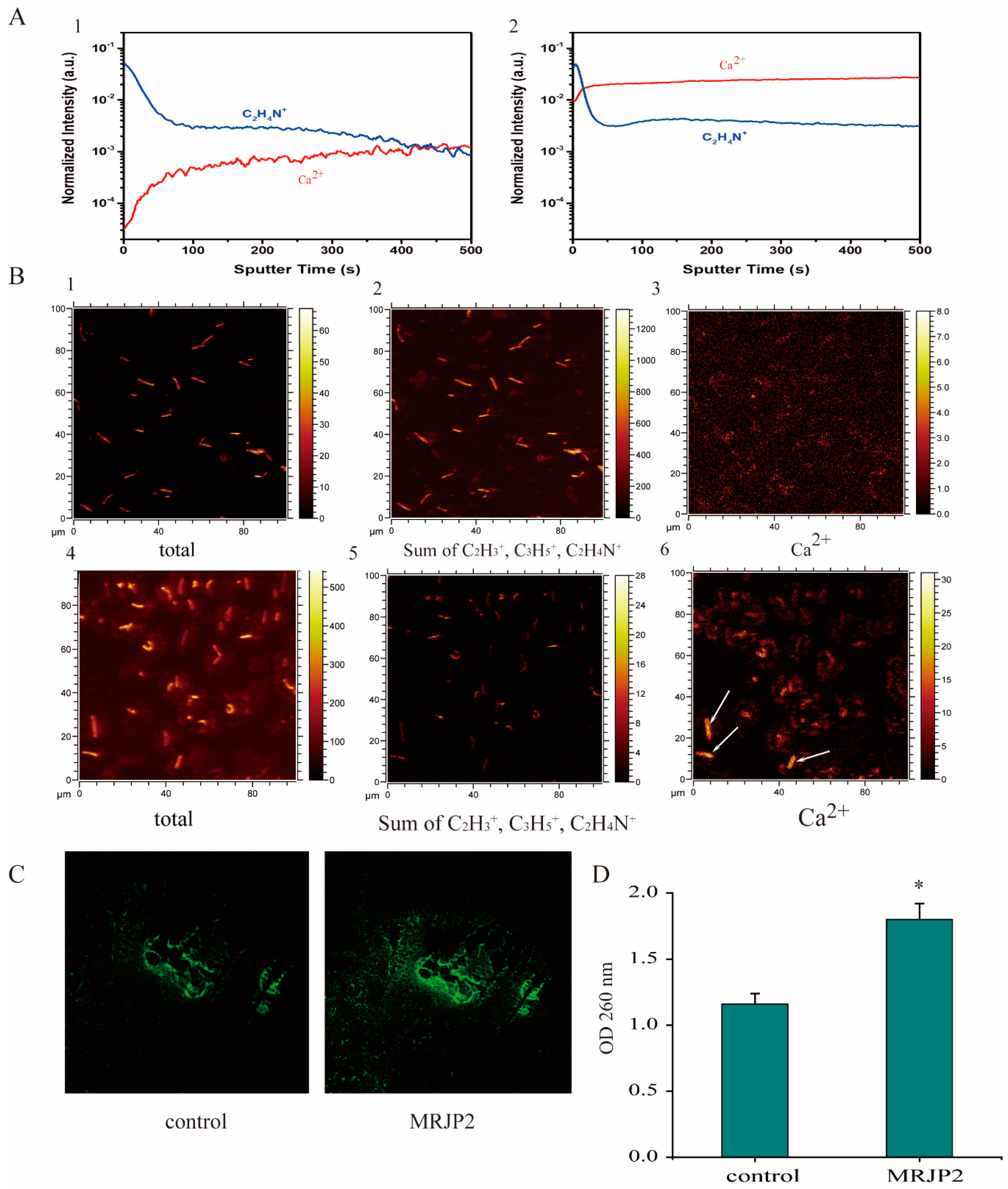
2.5. Intracellular Ca2+ Concentration Evaluation by Laser Confocal Microscopy and Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS)
2.6. Membrane Protein Preparation of P. larvae
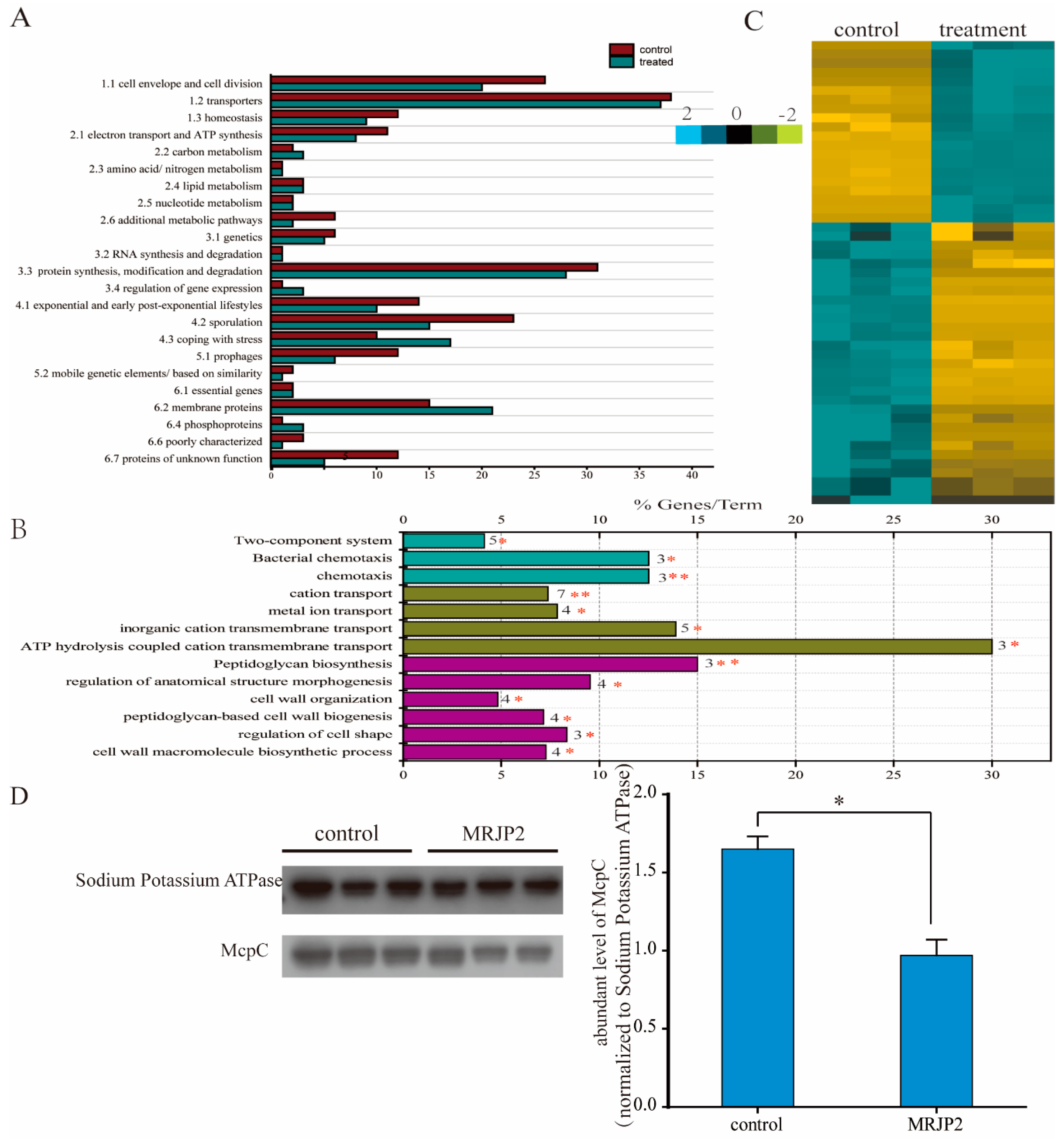
2.7. Membrane Proteome Analysis of P. larvae
2.8. Label-Free Quantification of Abundant Level of Membrane Protein
2.9. Bioinformatics Analysis of Membrane Proteome
2.10. Western Blotting Verification of Protein Related to Chemotaxis
2.11. Statistical Analysis
3. Results
3.1. A Similar Antibacterial Activity of RJ from Queen Cells of Immature Instar Larvae
3.2. Genotype Pattern of P. larvae Using Specific Gel Bands Acquired by Rep-PCR
3.3. N-MRJP2 Treatment Inducing a Cell Membrane Permeability Increase in P. larvae
3.4. Membrane Proteome Dissecting Molecular Basis of N-MRJP2 against P. larvae
4. Discussion
4.1. Immune Regulatory Capacity of RJ to Ensure Larval Development
4.2. N-MRJP2 Has Potential to Kill P. larvae of ERIC II Pattern
4.3. N-MRJP2 Disrupts Cell Membrane Permeability and Cell Wall of P. larvae
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Genersch, E. American Foulbrood in honeybees and its causative agent, Paenibacillus larvae. J. Invertebr. Pathol. 2010, 103 (Suppl. 1), S10–S19. [Google Scholar] [CrossRef]
- Genersch, E.; Forsgren, E.; Pentikainen, J.; Ashiralieva, A.; Rauch, S.; Kilwinski, J.; Fries, I. Reclassification of Paenibacillus larvae subsp pulvifaciens and Paenibacillus larvae subsp larvae as Paenibacillus larvae without subspecies differentiation. Int. J. Syst. Evol. Micr. 2006, 56, 501–511. [Google Scholar] [CrossRef] [Green Version]
- Müller, S.; Garcia-Gonzalez, E.; Genersch, E.; Süssmuth, R.D. Involvement of secondary metabolites in the pathogenesis of the American foulbrood of honey bees caused by Paenibacillus larvae. Nat. Prod. Rep. 2015, 32, 765–778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poppinga, L.; Janesch, B.; Funfhaus, A.; Sekot, G.; Garcia-Gonzalez, E.; Hertlein, G.; Hedtke, K.; Schaffer, C.; Genersch, E. Identification and functional analysis of the S-layer protein SplA of Paenibacillus larvae, the causative agent of American Foulbrood of honey bees. PLoS Pathog. 2012, 8, e1002716. [Google Scholar] [CrossRef] [Green Version]
- Funfhaus, A.; Poppinga, L.; Genersch, E. Identification and characterization of two novel toxins expressed by the lethal honey bee pathogen Paenibacillus larvae, the causative agent of American foulbrood. Environ. Microbiol. 2013, 15, 2951–2965. [Google Scholar]
- Rauch, S.; Ashiralieva, A.; Hedtke, K.; Genersch, E. Negative Correlation between Individual-Insect-Level Virulence and Colony-Level Virulence of Paenibacillus larvae, the Etiological Agent of American Foulbrood of Honeybees. Appl. Environ. Microb. 2009, 75, 3344–3347. [Google Scholar] [CrossRef] [Green Version]
- Miyagi, T.; Peng, C.Y.; Chuang, R.Y.; Mussen, E.C.; Spivak, M.S.; Doi, R.H. Verification of oxytetracycline-resistant American foulbrood pathogen Paenibacillus larvae in the United States. J. Invertebr. Pathol. 2000, 75, 95–96. [Google Scholar] [CrossRef]
- Reybroeck, W.; Daeseleire, E.; De Brabander, H.F.; Herman, L. Antimicrobials in beekeeping. Vet. Microbiol. 2012, 158, 1–11. [Google Scholar] [CrossRef]
- Ghorbani-Nezami, S.; LeBlanc, L.; Yost, D.G.; Amy, P.S. Phage Therapy is Effective in Protecting Honeybee Larvae from American Foulbrood Disease. J. Insect Sci. 2015, 15, 84. [Google Scholar] [CrossRef] [Green Version]
- Remolina, S.C.; Hughes, K.A. Evolution and mechanisms of long life and high fertility in queen honey bees. Age 2008, 30, 177–185. [Google Scholar] [CrossRef] [Green Version]
- Fratini, F.; Cilia, G.; Mancini, S.; Felicioli, A. Royal Jelly: An ancient remedy with remarkable antibacterial properties. Microbiol. Res. 2016, 192, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Kohno, K.; Okamoto, I.; Sano, O.; Arai, N.; Iwaki, K.; Ikeda, M.; Kurimoto, M. Royal jelly inhibits the production of proinflammatory cytokines by activated macrophages. Biosci. Biotechnol. Biochem. 2004, 68, 138–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, H.; Kouzuma, Y.; Yonekura, M. Isolation and properties of antioxidative peptides from water-soluble royal jelly protein hydrolysate. Food Sci. Technol. Res. 2005, 11, 222–230. [Google Scholar] [CrossRef] [Green Version]
- Kamakura, M.; Mitani, N.; Fukuda, T.; Fukushima, M. Antifatigue effect of fresh royal jelly in mice. J. Nutr. Sci. Vitaminol. 2001, 47, 394–401. [Google Scholar] [CrossRef] [Green Version]
- Fan, P.; Han, B.; Feng, M.; Fang, Y.; Zhang, L.; Hu, H.; Hao, Y.; Qi, Y.; Zhang, X.; Li, J. Functional and Proteomic Investigations Reveal Major Royal Jelly Protein 1 Associated with Anti-hypertension Activity in Mouse Vascular Smooth Muscle Cells. Sci. Rep. 2016, 6, 30230. [Google Scholar] [CrossRef] [Green Version]
- Kimura, Y.; Ushijima, T.; Maeda, M.; Hama, Y.; Kimura, M.; Okihara, K.; Sugimoto, H.; Yamada, H. Tumor antigen occurs in N-glycan of royal jelly glycoproteins: Honeybee cells synthesize T-antigen unit in N-glycan moiety. Biosci. Biotechnol. Biochem. 2006, 70, 2583–2587. [Google Scholar] [CrossRef] [Green Version]
- Pourmoradian, S.; Mahdavi, R.; Mobasseri, M.; Faramarzi, E.; Mobasseri, M. Effects of royal jelly supplementation on glycemic control and oxidative stress factors in type 2 diabetic female: A randomized clinical trial. Chin. J. Integr. Med. 2014, 20, 347–352. [Google Scholar] [CrossRef]
- Zamani, Z.; Reisi, P.; Alaei, H.; Pilehvarian, A.A. Effect of Royal Jelly on spatial learning and memory in rat model of streptozotocin-induced sporadic Alzheimer’s disease. Adv. Biomed. Res. 2012, 1, 26. [Google Scholar]
- Hattori, N.; Ohta, S.; Sakamoto, T.; Mishima, S.; Furukawa, S. Royal Jelly Facilitates Restoration of the Cognitive Ability in Trimethyltin-Intoxicated Mice. Evid. Based Complement. Alternat. Med. 2011, 2011, 165968. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Liu, F.; Wan, J.B.; Lai, C.Q.; Shen, L.R. Effect of Major Royal Jelly Proteins on Spatial Memory in Aged Rats: Metabolomics Analysis in Urine. J. Agric. Food Chem. 2017, 65, 3151–3159. [Google Scholar] [CrossRef]
- Ramadan, M.F.; Al-Ghamdi, A. Bioactive compounds and health-promoting properties of royal jelly: A review. J. Funct. Foods 2012, 4, 39–52. [Google Scholar] [CrossRef]
- Bilikova, K.; Huang, S.C.; Lin, I.P.; Simuth, J.; Peng, C.C. Structure and antimicrobial activity relationship of royalisin, an antimicrobial peptide from royal jelly of Apis mellifera. Peptides 2015, 68, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.C.; Finola, M.S.; Marioli, J.M. Bioassay Directed Identification of Royal Jelly’s Active Compounds against the Growth of Bacteria Capable of Infecting Cutaneous Wounds. Adv. Microbiol. 2013, 3, 7. [Google Scholar] [CrossRef] [Green Version]
- Fontana, R.; Mendes, M.A.; Souza, B.M.d.; Konno, K.; César, L.M.M.; Malaspina, O.; Palma, M.S. Jelleines: A family of antimicrobial peptides from the Royal Jelly of honeybees (Apis mellifera). Peptides 2004, 25, 919–928. [Google Scholar] [CrossRef]
- Sagona, S.; Turchi, B.; Fratini, F.; Giusti, M.; Torracca, B.; Nuvoloni, R.; Cerri, D.; Felicioli, A. Preliminary evaluation of glucose oxidase and its products in vitro antimicrobial activities on Paenibacillus larvae ATCC9545 vegetative form. Bull. Insectol. 2015, 68, 233–237. [Google Scholar]
- Kim, B.Y.; Jin, B.R. Apolipophorin III from honeybees (Apis cerana) exhibits antibacterial activity. Comp. Biochem. Phys. B 2015, 182, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Brudzynski, K.; Sjaarda, C.; Lannigan, R. MRJP1-containing glycoproteins isolated from honey, a novel antibacterial drug candidate with broad spectrum activity against multi-drug resistant clinical isolates. Front. Microbiol. 2015, 6, 711. [Google Scholar] [CrossRef] [Green Version]
- Feng, M.; Fang, Y.; Han, B.; Xu, X.; Fan, P.; Hao, Y.; Qi, Y.; Hu, H.; Huo, X.; Meng, L.; et al. In-Depth N-Glycosylation Reveals Species-Specific Modifications and Functions of the Royal Jelly Protein from Western (Apis mellifera) and Eastern Honeybees (Apis cerana). J. Proteome Res. 2015, 14, 5327–5340. [Google Scholar] [CrossRef]
- Lin, N.; Li, J.M.; Shao, R.M.; Zhang, H. Site-Specific Analysis of N-Linked Glycosylation Heterogeneity from Royal Jelly Glycoproteins. J. Agric. Food Chem. 2019, 67, 9411–9422. [Google Scholar] [CrossRef]
- Santos, K.S.; Delazari dos Santos, L.; Anita Mendes, M.; Monson de Souza, B.; Malaspina, O.; Palma, M.S. Profiling the proteome complement of the secretion from hypopharyngeal gland of Africanized nurse-honeybees (Apis mellifera L.). Insect Biochem. Mol. Biol. 2005, 35, 85–91. [Google Scholar] [CrossRef]
- Lercker, G.; Capella, P.; Conte, L.; Ruini, F.; Giordani, G. Components of royal jelly: I. Identification of the organic acids. Lipids 1981, 16, 912–919. [Google Scholar] [CrossRef]
- Zheng, H.Q.; Hu, F.L.; Dietemann, V. Changes in composition of royal jelly harvested at different times: Consequences for quality standards. Apidologie 2011, 42, 39–47. [Google Scholar] [CrossRef] [Green Version]
- Jie, H.; Li, P.M.; Zhao, G.J.; Feng, X.L.; Zeng, D.J.; Zhang, C.L.; Lei, M.Y.; Yu, M.; Chen, Q. Amino acid composition of royal jelly harvested at different times after larval transfer. Genet. Mol. Res. 2016, 15, gmr.15038306. [Google Scholar] [CrossRef]
- Liu, J.R.; Yang, Y.C.; Shi, L.S.; Peng, C.C. Antioxidant properties of royal jelly associated with larval age and time of harvest. J. Agric. Food Chem. 2008, 56, 11447–11452. [Google Scholar] [CrossRef]
- Reddy, K.V.R.; Yedery, R.D.; Aranha, C. Antimicrobial peptides: Premises and promises. Int. J. Antimicrob. Agents 2004, 24, 536–547. [Google Scholar] [CrossRef]
- Tian, W.L.; Li, M.; Guo, H.Y.; Peng, W.J.; Xue, X.F.; Hu, Y.F.; Liu, Y.; Zhao, Y.Z.; Fang, X.M.; Wang, K.; et al. Architecture of the native major royal jelly protein 1 oligomer. Nat. Commun. 2018, 9, 3373. [Google Scholar] [CrossRef]
- Tokhtaeva, E.; Mareninova, O.A.; Gukovskaya, A.S.; Vagin, O. Analysis of N- and O-Glycosylation of Lysosomal Glycoproteins. Methods Mol. Biol. 2017, 1594, 35–42. [Google Scholar]
- Genersch, E.; Otten, C. The use of repetitive element PCR fingerprinting (rep-PCR) for genetic subtyping of German field isolates of Paenibacillus larvae subsp larvae. Apidologie 2003, 34, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Septama, A.W.; Xiao, J.; Panichayupakaranant, P. A synergistic effect of artocarpanone from Artocarpus heterophyllus L. (Moraceae) on the antibacterial activity of selected antibiotics and cell membrane permeability. J. Intercult. Ethnopharmacol. 2017, 6, 186–191. [Google Scholar] [CrossRef]
- Carson, C.F.; Mee, B.J.; Riley, T.V. Mechanism of action of Melaleuca alternifolia (tea tree) oil on Staphylococcus aureus determined by time-kill, lysis, leakage, and salt tolerance assays and electron microscopy. Antimicrob. Agents Chemother. 2002, 46, 1914–1920. [Google Scholar] [CrossRef] [Green Version]
- Tran, T.T.; MiIler, W.R.; Shamoo, Y.; Arias, C.A. Targeting cell membrane adaptation as a novel antimicrobial strategy. Curr. Opin. Microbiol. 2016, 33, 91–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, L.; Abhyankar, W.; Ouwerling, N.; Dekker, H.L.; van Veen, H.; van der Wel, N.N.; Roseboom, W.; de Koning, L.J.; Brul, S.; de Koster, C.G. Bacillus subtilis Spore Inner Membrane Proteome. J. Proteome Res. 2016, 15, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Moller, S.; Croning, M.D.R.; Apweiler, R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 2001, 17, 646–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kall, L.; Krogh, A.; Sonnhammer, E.L.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- Bagos, P.G.; Tslrigos, K.D.; Liakopoulos, T.D.; Hamodrakas, S.J. Prediction of Lipoprotein Signal Peptides in Gram-Positive Bacteria with a Hidden Markov Model. J. Proteome Res. 2008, 7, 5082–5093. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.Z.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [Green Version]
- Michna, R.H.; Commichau, F.M.; Todter, D.; Zschiedrich, C.P.; Stulke, J. SubtiWiki-a database for the model organism Bacillus subtilis that links pathway, interaction and expression information. Nucleic Acids Res. 2014, 42, D692–D698. [Google Scholar] [CrossRef] [Green Version]
- Brodsgaard, C.J.; Ritter, W.; Hansen, H. Response of in vitro reared honey bee larvae to various doses of Paenibacillus larvae larvae spores. Apidologie 1998, 29, 569–578. [Google Scholar] [CrossRef] [Green Version]
- Wilson-Rich, N.; Dres, S.T.; Starks, P.T. The ontogeny of immunity: Development of innate immune strength in the honey bee (Apis mellifera). J. Insect Physiol. 2008, 54, 1392–1399. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alreshoodi, F.M.; Sultanbawa, Y. Antimicrobial activity of royal jelly. Anti-Infect. Agents 2015, 13, 50–59. [Google Scholar] [CrossRef]
- Evans, J.; Aronstein, K.; Chen, Y.; Hetru, C.; Imler, J.L.; Jiang, H.; Kanost, M.; Thompson, G.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Zhang, S.; Yang, K.; Zhu, L.; Lin, D. Toxicity of perfluorooctane sulfonate and perfluorooctanoic acid to Escherichia coli: Membrane disruption, oxidative stress, and DNA damage induced cell inactivation and/or death. Environ. Pollut. 2016, 214, 806–815. [Google Scholar] [CrossRef]
- Shen, L.; Liu, D.; Li, M.; Jin, F.; Din, M.; Parnell, L.D.; Lai, C.Q. Mechanism of action of recombinant acc-royalisin from royal jelly of Asian honeybee against gram-positive bacteria. PLoS ONE 2012, 7, e47194. [Google Scholar] [CrossRef]
- Mungai, P.T.; Waypa, G.B.; Jairaman, A.; Prakriya, M.; Dokic, D.; Ball, M.K.; Schumacker, P.T. Hypoxia Triggers AMPK Activation through Reactive Oxygen Species-Mediated Activation of Calcium Release-Activated Calcium Channels. Mol. Cell. Biol. 2011, 31, 3531–3545. [Google Scholar] [CrossRef] [Green Version]
- Marino, M.; Hoffmann, T.; Schmid, R.; Mobitz, H.; Jahn, D. Changes in protein synthesis during the adaptation of Bacillus subtilis to anaerobic growth conditions. Microbiol. SGM 2000, 146, 97–105. [Google Scholar] [CrossRef] [Green Version]
- Garrity, L.F.; Schiel, S.L.; Merrill, R.; Reizer, J.; Saier, M.H., Jr.; Ordal, G.W. Unique regulation of carbohydrate chemotaxis in Bacillus subtilis by the phosphoenolpyruvate-dependent phosphotransferase system and the methyl-accepting chemotaxis protein McpC. J. Bacteriol. 1998, 180, 4475–4480. [Google Scholar] [CrossRef] [Green Version]
- Kocaoglu, O.; Calvo, R.A.; Sham, L.T.; Cozy, L.M.; Lanning, B.R.; Francis, S.; Winkler, M.E.; Kearns, D.B.; Carlson, E.E. Selective Penicillin-Binding Protein Imaging Probes Reveal Substructure in Bacterial Cell Division. ACS Chem. Biol. 2012, 7, 1746–1753. [Google Scholar] [CrossRef] [Green Version]
- McPherson, D.C.; Driks, A.; Popham, D.L. Two class A high-molecular-weight penicillin-binding proteins of Bacillus subtilis play redundant roles in sporulation. J. Bacteriol. 2001, 183, 6046–6053. [Google Scholar] [CrossRef] [Green Version]
- Popham, D.L.; Gilmore, M.E.; Setlow, P. Roles of low-molecular-weight penicillin-binding proteins in Bacillus subtilis spore peptidoglycan synthesis and spore properties. J. Bacteriol. 1999, 181, 126–132. [Google Scholar] [CrossRef] [Green Version]
- Yue, D.; Nordhoff, M.; Wieler, L.H.; Genersch, E. Fluorescence in situ hybridization (FISH) analysis of the interactions between honeybee larvae and Paenibacillus larvae, the causative agent of American foulbrood of honeybees (Apis mellifera). Environ. Microbiol. 2008, 10, 1612–1620. [Google Scholar] [CrossRef]
- Melo-Braga, M.N.; Verano-Braga, T.; Leon, I.R.; Antonacci, D.; Nogueira, F.C.S.; Thelen, J.J.; Larsen, M.R.; Palmisano, G. Modulation of Protein Phosphorylation, N-Glycosylation and Lys-Acetylation in Grape (Vitis vinifera) Mesocarp and Exocarp Owing to Lobesia botrana Infection. Mol. Cell. Proteom. 2012, 11, 945–956. [Google Scholar] [CrossRef] [Green Version]
- Muresan, C.I.; Schierhorn, A.; Buttstedt, A. The Fate of Major Royal Jelly Proteins during Proteolytic Digestion in the Human Gastrointestinal Tract. J. Agric. Food Chem. 2018, 66, 4164–4170. [Google Scholar] [CrossRef]
- Varki, A. Biological roles of oligosaccharides: All of the theories are correct. Glycobiology 1993, 3, 97–130. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef]
- Ellegaard, K.M.; Engel, P. Genomic diversity landscape of the honey bee gut microbiota. Nat. Commun. 2019, 10, 446. [Google Scholar] [CrossRef]
- O’Riordan, N.; Kane, M.; Joshi, L.; Hickey, R.M. Structural and functional characteristics of bovine milk protein glycosylation. Glycobiology 2014, 24, 220–236. [Google Scholar] [CrossRef] [Green Version]
- Burt, S. Essential oils: Their antibacterial properties and potential applications in foods—A review. Int. J. Food Microbiol. 2004, 94, 223–253. [Google Scholar] [CrossRef]
- Trombetta, D.; Castelli, F.; Sarpietro, M.G.; Venuti, V.; Cristani, M.; Daniele, C.; Saija, A.; Mazzanti, G.; Bisignano, G. Mechanisms of antibacterial action of three monoterpenes. Antimicrob. Agents Chemother. 2005, 49, 2474–2478. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, M.; Fang, Y.; Ma, C.; Duan, X.; Zhang, Y.; Han, B.; Hu, H.; Meng, L.; Wang, F.; Li, J. Mechanistic Insight into Royal Protein Inhibiting the Gram-Positive Bacteria. Biomolecules 2021, 11, 64. https://doi.org/10.3390/biom11010064
Feng M, Fang Y, Ma C, Duan X, Zhang Y, Han B, Hu H, Meng L, Wang F, Li J. Mechanistic Insight into Royal Protein Inhibiting the Gram-Positive Bacteria. Biomolecules. 2021; 11(1):64. https://doi.org/10.3390/biom11010064
Chicago/Turabian StyleFeng, Mao, Yu Fang, Chuan Ma, Xiangyuan Duan, Yanyan Zhang, Bin Han, Han Hu, Lifeng Meng, Fuyi Wang, and Jianke Li. 2021. "Mechanistic Insight into Royal Protein Inhibiting the Gram-Positive Bacteria" Biomolecules 11, no. 1: 64. https://doi.org/10.3390/biom11010064
APA StyleFeng, M., Fang, Y., Ma, C., Duan, X., Zhang, Y., Han, B., Hu, H., Meng, L., Wang, F., & Li, J. (2021). Mechanistic Insight into Royal Protein Inhibiting the Gram-Positive Bacteria. Biomolecules, 11(1), 64. https://doi.org/10.3390/biom11010064




