Research Trends in the Efficacy of Stem Cell Therapy for Hepatic Diseases Based on MicroRNA Profiling
Abstract
:1. Introduction
2. Main body
2.1. Classifications of Stem Cells
2.2. Stem Cell Therapy in Degenerative Diseases
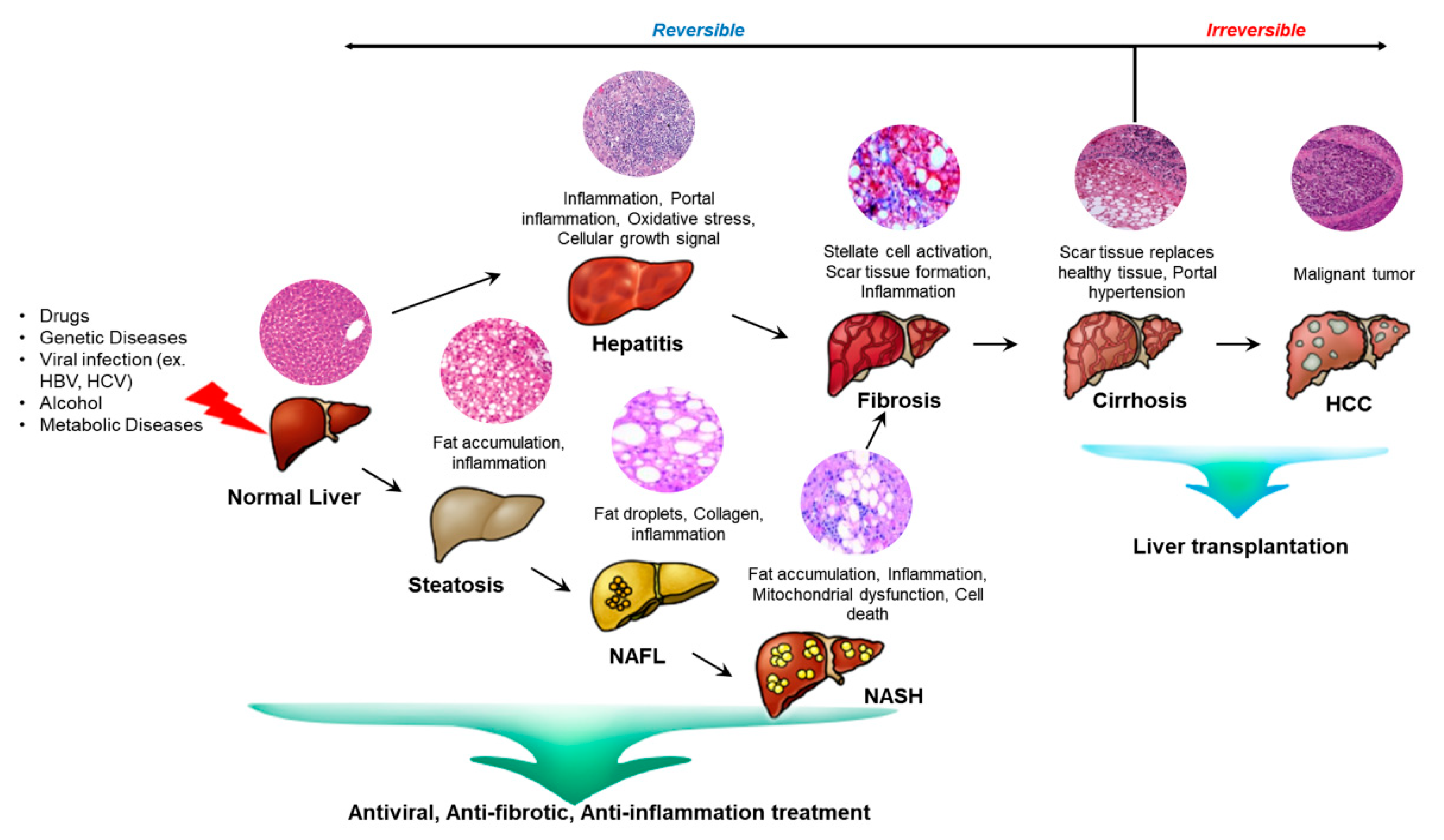
2.3. Liver Diseases
2.4. Stem Cell Therapy for Liver Diseases
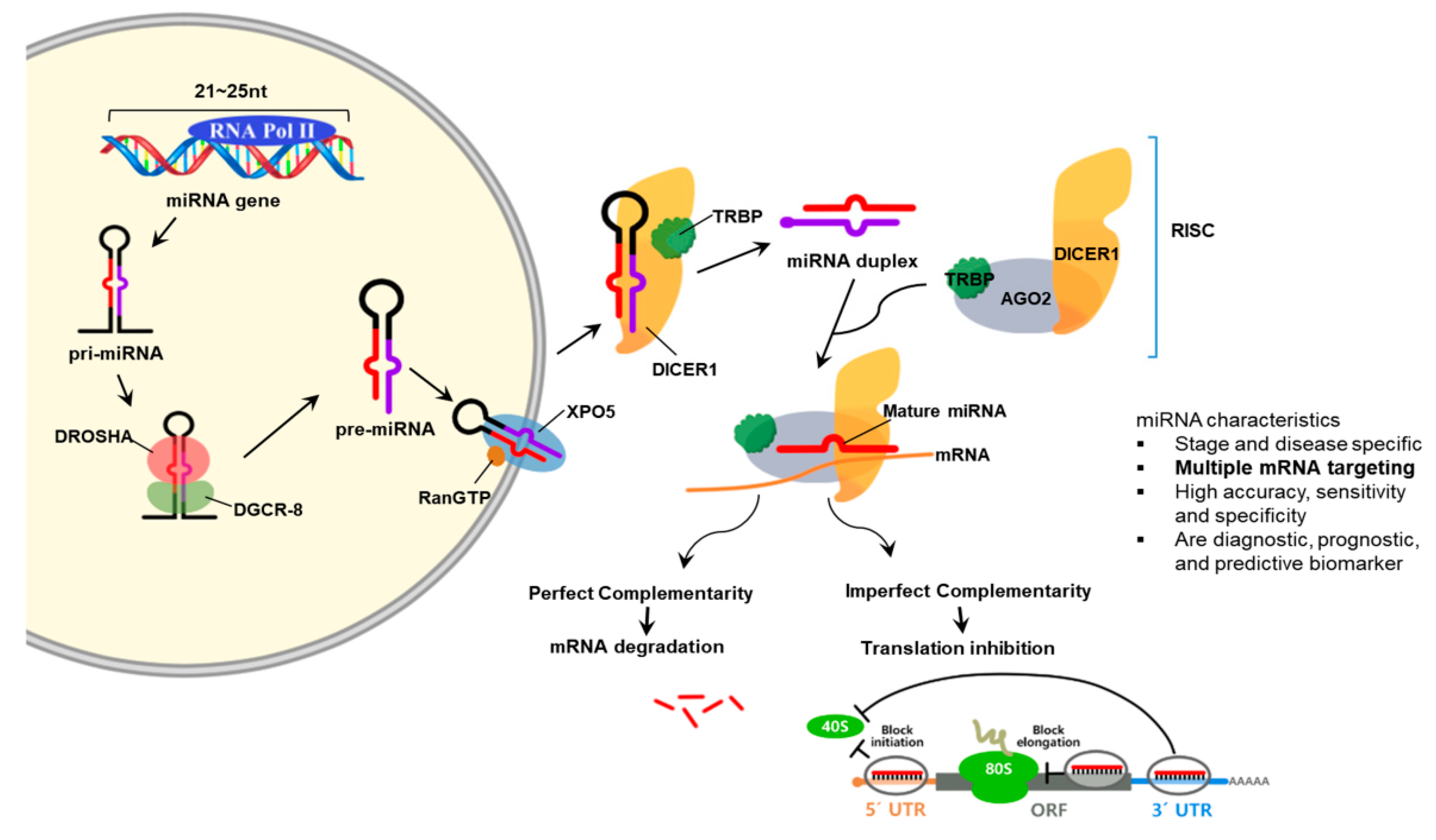
2.5. MicroRNA
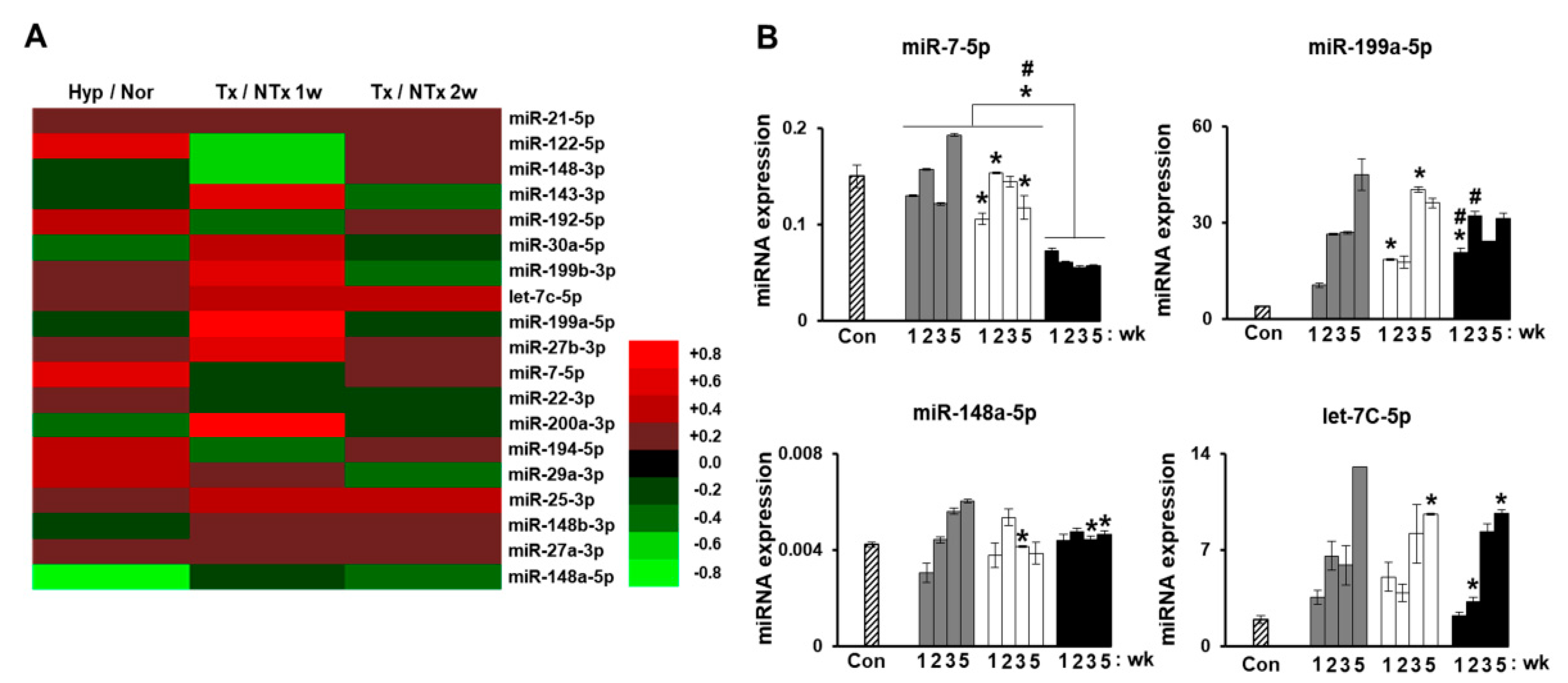
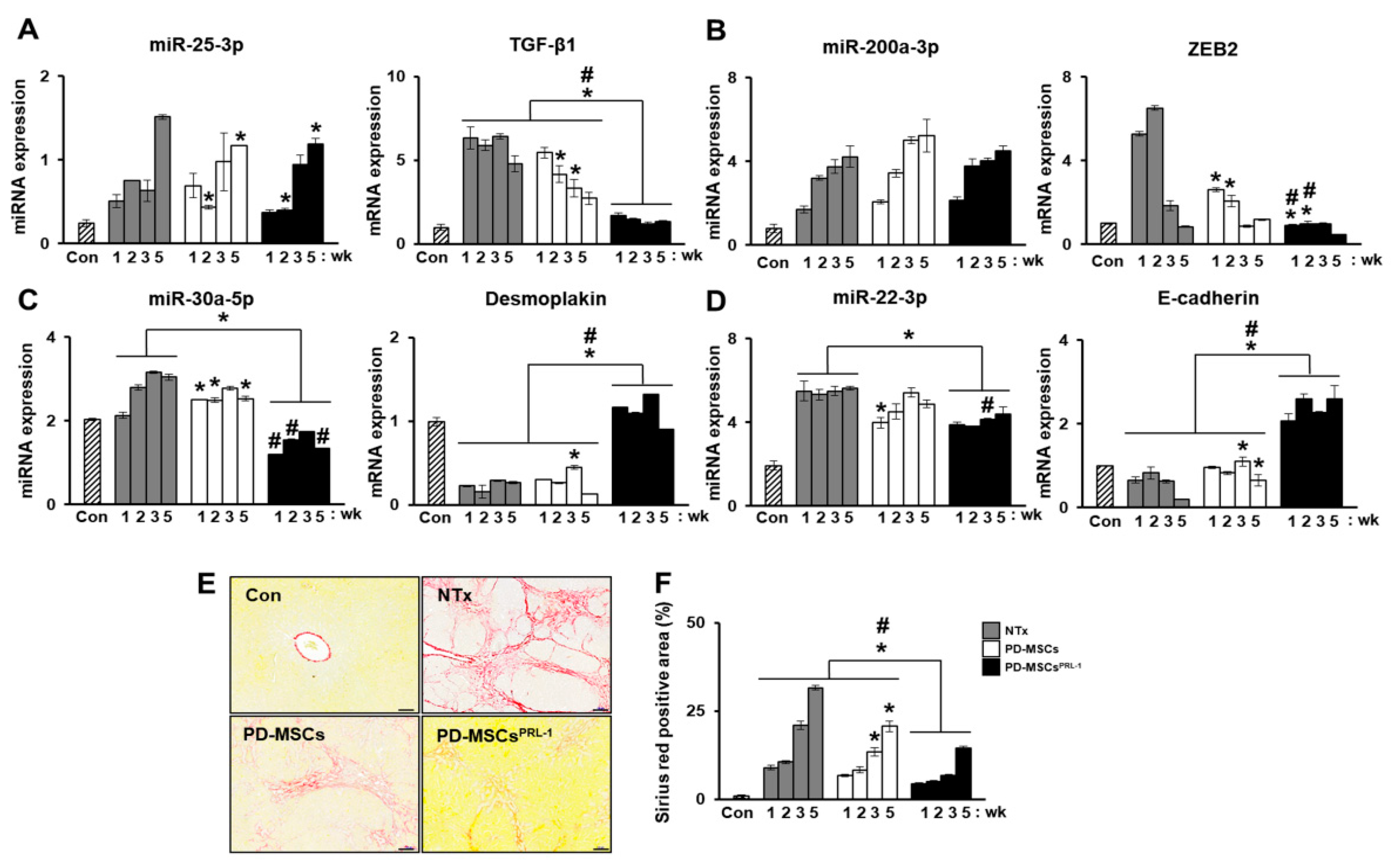
2.6. Monitoring Stem Cell Therapy
2.7. MicroRNA in Degenerative Diseases
2.8. MicroRNAs in Liver Disease
| I. Liver Development | ||
| Function | Target miRNAs | Reference |
| Definitive endoderm formation | miR-10a, miR-24, miR-196a, miR-196b, miR-218, miR-222, miR-338-5p, miR-340-3p, miR-371-3p, miR-371-5p, miR-373, miR-375, miR-520 | [115,116,117,118,119] |
| Epithelial- to-mesenchymal transition (EMT) | miR-30b/c/d/e, miR-218, miR-495 | [120,121,122,123,124,125] |
| Hepatocyte proliferation and differentiation | miR-33, miR-122, miR-148a | [126,127,128,129,130] |
| HSC activation and proliferation | miR-16, miR-19a, miR-27a, miR-27b, miR-29b, miR-146a, miR-150, miR-194, miR-195, miR-335 | [110,131,132,133,134,135,136,137,138,139,140] |
| Liver glucose metabolism | miR-27b, miR-33, miR-96, miR-122, miR185, miR-206, miR-223, miR-370 | [141,142,143,144,145,146,147,148] |
| Liver iron metabolism | miR-485-3p | [149,150] |
| Detoxification | miR-10a, miR-21, miR-24, miR-27b, dre-miR-27b, miR-34a, miR-93, miR-126, miR-130, miR-132, miR-142-3p, miR-148a, miR-185, miR-200c, miR-214, miR-378, miR-699, miR-892a | [151,152,153,154,155,156,157,158,159] |
| II. Liver Regeneration | ||
| Function | Target miRNAs | Reference |
| Regeneration promoters | miR-19a, miR-21, miR-214, miR-106a, miR-20a, miR-20b, miR-93, miR-33, miR-153, miR-298, miR-301b, miR-489, miR-743b, miR-883, miR-126, miR-130a, miR-20a, miR520e | [160] |
| Regeneration inhibitors | let-7a/b/c/d/e/f/i, miR-23a/b, miR-26a/b, miR-29a, miR-30d, miR-33, miR-125b-5p, miR-126, miR-127, miR-145, miR-146a, miR-150, miR-207, miR-223, miR-352, miR-375, miR-378, miR-503, miR-532-3p, miR-663, miR-872 | [160] |
| Disease | Target miRNAs | Reference |
|---|---|---|
| Chronic Hepatitis C (CHC) | miR-21, miR-23b, miR-27a, miR-106, miR-122, miR-134, miR-320c, miR-424-3p, miR-451, miR-629-5p, miR-638, miR-762, miR1207-5p, miR-1225-5p, miR1275, miR-1246 miR-1974 | [102,161,162] |
| Chronic Hepatitis B | miR-16, miR-19b, miR-20a, miR-92a, miR-106a, miR-122, miR-122-3p, miR-125b, miR-146a-5p, miR-194, miR-223, miR-328-3p, miR-572, miR-575, miR-638, miR-744 | [163,164,165,166] |
| NAFLD | miR-16, miR-21, miR-27b-3p, miR-34a, miR-122, miR-122-5p, miR-145, miR-192-5p, miR-451, miR-1290 | [167,168,169] |
| Liver fibrosis | miR-29, miR-29a, miR-34a, miR-199a, miR-200a, miR-200b, miR-571, miR-513-3p, miR-652 | [110,170,171] |
| Liver cirrhosis | miR-29, miR-106b, miR-181b, miR-513-3p, miR-571, miR-652, miR-885-5p | [100,172,173] |
| Hepatocellular Carcinoma (HCC) | miR-15b, miR-18a, miR-21, miR-26a-5p, miR-29b, miR-106b, miR-122, miR-122-5p, miR-122a, miR-130b, miR-141-3p, miR-143, miR-183, miR-192-5p, miR-193a-3p, miR-199a-5p, miR-206, miR-215, miR-369-5p, miR-429, miR-433-5p, miR-483-5p, miR-672, miR-1228-5p | [174,175,176] |
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AFLD | Alcoholic fatty liver diseases |
| AGO | Argonaut |
| ALF | Acute liver failure |
| CHB | Chronic hepatitis B |
| ESCs | Embryonic stem cells |
| HBV | Hepatitis B virus |
| HCC | Hepatocellular carcinoma |
| hESCs | Human embryonic stem cells |
| HSCs | Hematopoietic stem cells |
| IPSCs | Induced pluripotent stem cells |
| MiRNA | microRNA |
| MSCs | Mesenchymal stem cells |
| NAFLD | Nonalcoholic fatty liver disease |
| NASH | Nonalcoholic steatohepatitis |
References
- Trefts, E.; Gannon, M.; Wasserman, D.H. The liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef] [PubMed]
- Bechmann, L.P.; Hannivoort, R.A.; Gerken, G.; Hotamisligil, G.S.; Trauner, M.; Canbay, A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J. Hepatol. 2012, 56, 952–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michalopoulos, G.K.; DeFrances, M.C. Liver regeneration. Science 1997, 276, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Siddiqui, H.; Bhat, M.H. Hepatic progenitor cells in action: Liver regeneration or fibrosis? Am. J. Pathol. 2015, 185, 2342–2350. [Google Scholar] [CrossRef] [PubMed]
- Campana, L.; Iredale, J.P. Regression of liver fibrosis. Semin. Liver Dis. 2017, 37, 1–10. [Google Scholar] [PubMed]
- Jung, Y.K.; Yim, H.J. Reversal of liver cirrhosis: Current evidence and expectations. Korean J. Intern. Med. 2017, 32, 213–228. [Google Scholar] [CrossRef] [Green Version]
- Collaborators, G.B.D.C. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990–2017: A systematic analysis for the global burden of disease study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 245–266. [Google Scholar]
- Gowda, S.; Desai, P.B.; Hull, V.V.; Math, A.A.; Vernekar, S.N.; Kulkarni, S.S. A review on laboratory liver function tests. Pan. Afr. Med. J. 2009, 3, 17. [Google Scholar]
- Mendizabal, M.; Silva, M.O. Liver transplantation in acute liver failure: A challenging scenario. World J. Gastroenterol. 2016, 22, 1523–1531. [Google Scholar] [CrossRef]
- Alfaifi, M.; Eom, Y.W.; Newsome, P.N.; Baik, S.K. Mesenchymal stromal cell therapy for liver diseases. J. Hepatol. 2018, 68, 1272–1285. [Google Scholar] [CrossRef] [Green Version]
- Carpentier, B.; Gautier, A.; Legallais, C. Artificial and bioartificial liver devices: Present and future. Gut 2009, 58, 1690–1702. [Google Scholar] [CrossRef] [PubMed]
- Karvellas, C.J.; Subramanian, R.M. Current evidence for extracorporeal liver support systems in acute liver failure and acute-on-chronic liver failure. Crit. Care Clin. 2016, 32, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Shekkeris, A.S.; Jaiswal, P.K.; Khan, W.S. Clinical applications of mesenchymal stem cells in the treatment of fracture non-union and bone defects. Curr. Stem Cell Res. Ther. 2012, 7, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.B.; Chen, E.Q.; Tang, H. Stem cell transplantation for the treatment of end-stage liver disease. World J. Hepatol. 2018, 10, 907–910. [Google Scholar] [CrossRef]
- Bacakova, L.; Zarubova, J.; Travnickova, M.; Musilkova, J.; Pajorova, J.; Slepicka, P.; Kasalkova, N.S.; Svorcik, V.; Kolska, Z.; Motarjemi, H.; et al. Stem cells: Their source, potency and use in regenerative therapies with focus on adipose-derived stem cells—A review. Biotechnol. Adv. 2018, 36, 1111–1126. [Google Scholar] [CrossRef]
- Clevers, H.; Loh, K.M.; Nusse, R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 2014, 346, 1248012. [Google Scholar] [CrossRef]
- Navone, S.; Cristini, S.; Canzi, L.; Parati, E.A.; Invernici, G. Stem cell patents: An innovative approach to anti-cancer drug discovery. Recent Pat. Anticancer Drug Discov. 2010, 5, 14–21. [Google Scholar] [CrossRef]
- Sugaya, K.; Vaidya, M. Stem cell therapies for neurodegenerative diseases. Adv. Exp. Med. Biol. 2018, 1056, 61–84. [Google Scholar]
- Uhl, P.; Fricker, G.; Haberkorn, U.; Mier, W. Current status in the therapy of liver diseases. Int. J. Mol. Sci. 2014, 15, 7500–7512. [Google Scholar] [CrossRef] [Green Version]
- Kim, G.; Eom, Y.W.; Baik, S.K.; Shin, Y.; Lim, Y.L.; Kim, M.Y.; Kwon, S.O.; Chang, S.J. Therapeutic effects of mesenchymal stem cells for patients with chronic liver diseases: Systematic review and meta-analysis. J. Korean Med. Sci. 2015, 30, 1405–1415. [Google Scholar] [CrossRef] [Green Version]
- Fu, S.; Wu, D.; Jiang, W.; Li, J.; Long, J.; Jia, C.; Zhou, T. Molecular biomarkers in drug-induced liver injury: Challenges and future perspectives. Front. Pharmacol. 2019, 10, 1667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohr, A.M.; Mott, J.L. Overview of microrna biology. Semin. Liver Dis. 2015, 35, 3–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Giau, V.; An, S.S. Emergence of exosomal mirnas as a diagnostic biomarker for alzheimer’s disease. J. Neurol. Sci. 2016, 360, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.; Teodoro, M.; Rugolo, C.A.; Alibrando, C.; Giambo, F.; Briguglio, G.; Fenga, C. Micrornas alteration as early biomarkers for cancer and neurodegenerative diseases: New challenges in pesticides exposure. Toxicol. Rep. 2020, 7, 759–767. [Google Scholar] [CrossRef]
- Sharkis, S.J. Canadian stem cell scientists take the prize. Cell 2005, 122, 817–819. [Google Scholar] [CrossRef] [Green Version]
- Laplane, L.; Solary, E. Towards a classification of stem cells. Elife 2019, 8. [Google Scholar] [CrossRef]
- De Kretser, D. Totipotent, pluripotent or unipotent stem cells: A complex regulatory enigma and fascinating biology. J. Law Med. 2007, 15, 212–218. [Google Scholar]
- Estrov, Z. Stem cells and somatic cells: Reprogramming and plasticity. Clin. Lymphoma Myeloma 2009, 9 (Suppl. S3), S319–S328. [Google Scholar] [CrossRef]
- Damdimopoulou, P.; Rodin, S.; Stenfelt, S.; Antonsson, L.; Tryggvason, K.; Hovatta, O. Human embryonic stem cells. Best Pract. Res. Clin. Obstet. Gynaecol. 2016, 31, 2–12. [Google Scholar] [CrossRef]
- Ohnuki, M.; Takahashi, K. Present and future challenges of induced pluripotent stem cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140367. [Google Scholar] [CrossRef]
- Lin, Y.; Gil, C.H.; Yoder, M.C. Differentiation, evaluation, and application of human induced pluripotent stem cell-derived endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 2014–2025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansel, M.C.; Davila, J.C.; Vosough, M.; Gramignoli, R.; Skvorak, K.J.; Dorko, K.; Marongiu, F.; Blake, W.; Strom, S.C. The use of induced pluripotent stem cells for the study and treatment of liver diseases. Curr. Protoc. Toxicol. 2016, 67, 14 13 11–14 13 27. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, A.; Khanlarkhani, N.; Baazm, M.; Mohammadzadeh, F.; Najafi, A.; Mehdinejadiani, S.; Sargolzaei Aval, F. Multipotent stem cell and current application. Acta Med. Iran. 2017, 55, 6–23. [Google Scholar] [PubMed]
- Mishra, P.J.; Banerjee, D. Activation and differentiation of mesenchymal stem cells. Methods Mol. Biol. 2017, 1554, 201–209. [Google Scholar]
- Hoffman, A.M.; Dow, S.W. Concise review: Stem cell trials using companion animal disease models. Stem Cells 2016, 34, 1709–1729. [Google Scholar] [CrossRef]
- Duscher, D.; Barrera, J.; Wong, V.W.; Maan, Z.N.; Whittam, A.J.; Januszyk, M.; Gurtner, G.C. Stem cells in wound healing: The future of regenerative medicine? A mini-review. Gerontology 2016, 62, 216–225. [Google Scholar] [CrossRef]
- Kiss, J.; Urban, V.S.; Dudics, V.; Vas, V.; Uher, F. Mesenchymal stem cells and the immune system—Immunosuppression without drugs? Orv. Hetil. 2008, 149, 339–346. [Google Scholar] [CrossRef]
- Uccelli, A.; Moretta, L.; Pistoia, V. Mesenchymal stem cells in health and disease. Nat. Rev. Immunol. 2008, 8, 726–736. [Google Scholar] [CrossRef]
- El Agha, E.; Kramann, R.; Schneider, R.K.; Li, X.; Seeger, W.; Humphreys, B.D.; Bellusci, S. Mesenchymal stem cells in fibrotic disease. Cell Stem Cell 2017, 21, 166–177. [Google Scholar] [CrossRef]
- Berardis, S.; Dwisthi Sattwika, P.; Najimi, M.; Sokal, E.M. Use of mesenchymal stem cells to treat liver fibrosis: Current situation and future prospects. World J. Gastroenterol. 2015, 21, 742–758. [Google Scholar] [CrossRef]
- Rockey, D.C.; Bell, P.D.; Hill, J.A. Fibrosis—A common pathway to organ injury and failure. N. Engl. J. Med. 2015, 372, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A.; Ramalingam, T.R. Mechanisms of fibrosis: Therapeutic translation for fibrotic disease. Nat. Med. 2012, 18, 1028–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corless, J.K.; Middleton, H.M., 3rd. Normal liver function. A basis for understanding hepatic disease. Arch. Intern. Med. 1983, 143, 2291–2294. [Google Scholar] [CrossRef] [PubMed]
- Connolly, M.K.; Bedrosian, A.S.; Mallen-St Clair, J.; Mitchell, A.P.; Ibrahim, J.; Stroud, A.; Pachter, H.L.; Bar-Sagi, D.; Frey, A.B.; Miller, G. In liver fibrosis, dendritic cells govern hepatic inflammation in mice via tnf-alpha. J. Clin. Investig. 2009, 119, 3213–3225. [Google Scholar]
- Poley, J.R.; Nowicki, M.J. Other hereditary diseases and the liver. Baillieres Clin. Gastroenterol. 1998, 12, 369–407. [Google Scholar] [CrossRef]
- Keeffe, E.B. Hepatitis a and b superimposed on chronic liver disease: Vaccine-preventable diseases. Trans. Am. Clin. Climatol. Assoc. 2006, 117, 227–237, discussion 237–228. [Google Scholar]
- Sedhom, D.; D’Souza, M.; John, E.; Rustgi, V. Viral hepatitis and acute liver failure: Still a problem. Clin. Liver Dis. 2018, 22, 289–300. [Google Scholar] [CrossRef]
- Zhu, J.Z.; Yi, H.W.; Huang, W.; Pang, T.; Zhou, H.P.; Wu, X.D. Fatty liver diseases, mechanisms, and potential therapeutic plant medicines. Chin. J. Nat. Med. 2020, 18, 161–168. [Google Scholar] [CrossRef]
- Hashimoto, E.; Taniai, M.; Tokushige, K. Characteristics and diagnosis of nafld/nash. J. Gastroenterol. Hepatol. 2013, 28 (Suppl. S4), 64–70. [Google Scholar] [CrossRef] [Green Version]
- Johnston, M.P.; Patel, J.; Byrne, C.D. Causes of mortality in non-alcoholic fatty liver disease (nafld) and alcohol related fatty liver disease (afld). Curr. Pharm. Des. 2020, 26, 1079–1092. [Google Scholar] [CrossRef]
- Sanchez-Valle, V.; Chavez-Tapia, N.C.; Uribe, M.; Mendez-Sanchez, N. Role of oxidative stress and molecular changes in liver fibrosis: A review. Curr. Med. Chem. 2012, 19, 4850–4860. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L.; Roll, F.J.; Boyles, J.; Bissell, D.M. Hepatic lipocytes: The principal collagen-producing cells of normal rat liver. Proc. Natl. Acad. Sci. USA 1985, 82, 8681–8685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aydin, M.M.; Akcali, K.C. Liver fibrosis. Turk. J. Gastroenterol. 2018, 29, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Lee, U.E.; Friedman, S.L. Mechanisms of hepatic fibrogenesis. Best Pract. Res. Clin. Gastroenterol. 2011, 25, 195–206. [Google Scholar] [CrossRef]
- Zhou, W.C.; Zhang, Q.B.; Qiao, L. Pathogenesis of liver cirrhosis. World J. Gastroenterol. 2014, 20, 7312–7324. [Google Scholar] [CrossRef]
- Altamirano-Barrera, A.; Barranco-Fragoso, B.; Mendez-Sanchez, N. Management strategies for liver fibrosis. Ann. Hepatol. 2017, 16, 48–56. [Google Scholar] [CrossRef]
- Pais, R.; Barritt, A.S.t.; Calmus, Y.; Scatton, O.; Runge, T.; Lebray, P.; Poynard, T.; Ratziu, V.; Conti, F. Nafld and liver transplantation: Current burden and expected challenges. J. Hepatol. 2016, 65, 1245–1257. [Google Scholar] [CrossRef] [Green Version]
- De Miguel, M.P.; Prieto, I.; Moratilla, A.; Arias, J.; Aller, M.A. Mesenchymal stem cells for liver regeneration in liver failure: From experimental models to clinical trials. Stem Cells Int. 2019, 2019, 3945672. [Google Scholar] [CrossRef]
- Zhao, L.; Chen, S.; Shi, X.; Cao, H.; Li, L. A pooled analysis of mesenchymal stem cell-based therapy for liver disease. Stem Cell Res. Ther. 2018, 9, 72. [Google Scholar] [CrossRef]
- Watanabe, Y.; Tsuchiya, A.; Seino, S.; Kawata, Y.; Kojima, Y.; Ikarashi, S.; Starkey Lewis, P.J.; Lu, W.Y.; Kikuta, J.; Kawai, H.; et al. Mesenchymal stem cells and induced bone marrow-derived macrophages synergistically improve liver fibrosis in mice. Stem Cells Transl. Med. 2019, 8, 271–284. [Google Scholar] [CrossRef] [Green Version]
- Huang, B.; Cheng, X.; Wang, H.; Huang, W.; la Ga Hu, Z.; Wang, D.; Zhang, K.; Zhang, H.; Xue, Z.; Da, Y.; et al. Mesenchymal stem cells and their secreted molecules predominantly ameliorate fulminant hepatic failure and chronic liver fibrosis in mice respectively. J. Transl. Med. 2016, 14, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kordelas, L.; Rebmann, V.; Ludwig, A.K.; Radtke, S.; Ruesing, J.; Doeppner, T.R.; Epple, M.; Horn, P.A.; Beelen, D.W.; Giebel, B. Msc-derived exosomes: A novel tool to treat therapy-refractory graft-versus-host disease. Leukemia 2014, 28, 970–973. [Google Scholar] [CrossRef] [PubMed]
- Ramos, T.L.; Sánchez-Abarca, L.I.; Muntión, S.; Preciado, S.; Puig, N.; López-Ruano, G.; Hernández-Hernández, Á.; Redondo, A.; Ortega, R.; Rodríguez, C.; et al. Msc surface markers (cd44, cd73, and cd90) can identify human msc-derived extracellular vesicles by conventional flow cytometry. Cell Commun. Signal. 2016, 14, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antinozzi, C.; Sgro, P.; Di Luigi, L. Advantages of phosphodiesterase type 5 inhibitors in the management of glucose metabolism disorders: A clinical and translational issue. Int. J. Endocrinol. 2020, 2020, 7078108. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.X.; He, H.L.; Pan, S.W.; Chen, Y.L.; Zhang, M.L.; Zhu, S.; Gao, Z.L.; Peng, L.; Li, J.G. Combination treatments of plasma exchange and umbilical cord-derived mesenchymal stem cell transplantation for patients with hepatitis b virus-related acute-on-chronic liver failure: A clinical trial in china. Stem Cells Int. 2019, 2019, 4130757. [Google Scholar] [CrossRef]
- Liang, J.; Zhang, H.; Zhao, C.; Wang, D.; Ma, X.; Zhao, S.; Wang, S.; Niu, L.; Sun, L. Effects of allogeneic mesenchymal stem cell transplantation in the treatment of liver cirrhosis caused by autoimmune diseases. Int. J. Rheum. Dis. 2017, 20, 1219–1226. [Google Scholar] [CrossRef]
- Suk, K.T.; Yoon, J.H.; Kim, M.Y.; Kim, C.W.; Kim, J.K.; Park, H.; Hwang, S.G.; Kim, D.J.; Lee, B.S.; Lee, S.H.; et al. Transplantation with autologous bone marrow-derived mesenchymal stem cells for alcoholic cirrhosis: Phase 2 trial. Hepatology 2016, 64, 2185–2197. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microrna biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Catalanotto, C.; Cogoni, C.; Zardo, G. Microrna in control of gene expression: An overview of nuclear functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [Green Version]
- Cannell, I.G.; Kong, Y.W.; Bushell, M. How do micrornas regulate gene expression? Biochem. Soc. Trans. 2008, 36, 1224–1231. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Hara, H.; Dai, Y.; Mou, L.; Cooper, D.K.; Wu, C.; Cai, Z. Circulating organ-specific micrornas serve as biomarkers in organ-specific diseases: Implications for organ allo- and xeno-transplantation. Int. J. Mol. Sci. 2016, 17, 1232. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, M.; Babic, A. (Eds.) The European Blood and Marrow Transplantation Textbook for Nurses: Under the Auspices of Ebmt; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Shafiq, M.; Jung, Y.; Kim, S.H. Insight on stem cell preconditioning and instructive biomaterials to enhance cell adhesion, retention, and engraftment for tissue repair. Biomaterials 2016, 90, 85–115. [Google Scholar] [CrossRef] [PubMed]
- Nowicki, M.; Szemraj, J.; Wierzbowska, A.; Misiewicz, M.; Malachowski, R.; Pluta, A.; Grzybowska-Izydorczyk, O.; Robak, T.; Szmigielska-Kaplon, A. Mirna-15a, mirna-16, mirna-126, mirna-146a, and mirna-223 expressions in autologous hematopoietic stem cell transplantation and their impact on engraftment. Eur. J. Haematol. 2018, 100, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Accomasso, L.; Gallina, C.; Turinetto, V.; Giachino, C. Stem cell tracking with nanoparticles for regenerative medicine purposes: An overview. Stem Cells Int. 2016, 2016, 7920358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, S.Y.; Ricles, L.M.; Suggs, L.J.; Emelianov, S.Y. In vivo ultrasound and photoacoustic monitoring of mesenchymal stem cells labeled with gold nanotracers. PLoS ONE 2012, 7, e37267. [Google Scholar] [CrossRef] [Green Version]
- Boisselier, E.; Astruc, D. Gold nanoparticles in nanomedicine: Preparations, imaging, diagnostics, therapies and toxicity. Chem. Soc. Rev. 2009, 38, 1759–1782. [Google Scholar] [CrossRef]
- Ricles, L.M.; Nam, S.Y.; Sokolov, K.; Emelianov, S.Y.; Suggs, L.J. Function of mesenchymal stem cells following loading of gold nanotracers. Int. J. Nanomed. 2011, 6, 407–416. [Google Scholar] [CrossRef] [Green Version]
- Wei, M.; Li, S.; Le, W. Nanomaterials modulate stem cell differentiation: Biological interaction and underlying mechanisms. J. Nanobiotechnol. 2017, 15, 75. [Google Scholar] [CrossRef] [Green Version]
- Bang, C.; Thum, T. Exosomes: New players in cell-cell communication. Int. J. Biochem. Cell Biol. 2012, 44, 2060–2064. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [Green Version]
- Barile, L.; Vassalli, G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol. Ther. 2017, 174, 63–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Console, L.; Scalise, M.; Indiveri, C. Exosomes in inflammation and role as biomarkers. Clin. Chim. Acta 2019, 488, 165–171. [Google Scholar] [CrossRef]
- Wu, J.; Qu, Z.; Fei, Z.W.; Wu, J.H.; Jiang, C.P. Role of stem cell-derived exosomes in cancer. Oncol. Lett. 2017, 13, 2855–2866. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Ji, X.; Lv, R.; Pei, J.J.; Du, Y.; Shen, C.; Hou, X. Targetting exosomes as a new biomarker and therapeutic approach for alzheimer’s disease. Clin. Interv. Aging 2020, 15, 195–205. [Google Scholar] [CrossRef] [Green Version]
- Han, S.; Kang, B.; Jang, E.; Ki, J.; Kim, E.; Jeong, M.Y.; Huh, Y.M.; Son, H.Y.; Haam, S. Convenient monitoring system of intracellular microrna expression during adipogenesis via mechanical stimulus-induced exocytosis of lipovesicular mirna beacon. Adv. Healthc. Mater. 2018, 7, 1701019. [Google Scholar] [CrossRef] [PubMed]
- Gothelf, Y.; Kaspi, H.; Abramov, N.; Aricha, R. Mirna profiling of nurown(r): Mesenchymal stem cells secreting neurotrophic factors. Stem Cell Res. Ther. 2017, 8, 249. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Fu, B.; Sun, X.; Li, D.; Huang, Q.; Zhao, W.; Chen, X. Differentially expressed micrornas in bone marrow mesenchymal stem cell-derived microvesicles in young and older rats and their effect on tumor growth factor-beta1-mediated epithelial-mesenchymal transition in hk2 cells. Stem Cell Res. Ther. 2015, 6, 185. [Google Scholar] [CrossRef] [Green Version]
- Sekar, D.; Saravanan, S.; Karikalan, K.; Thirugnanasambantham, K.; Lalitha, P.; Islam, V.I. Role of microrna 21 in mesenchymal stem cell (msc) differentiation: A powerful biomarker in mscs derived cells. Curr. Pharm. Biotechnol. 2015, 16, 43–48. [Google Scholar] [CrossRef]
- Kagawa, T.; Shirai, Y.; Oda, S.; Yokoi, T. Identification of specific microrna biomarkers in early stages of hepatocellular injury, cholestasis, and steatosis in rats. Toxicol. Sci. 2018, 166, 228–239. [Google Scholar] [CrossRef]
- Gilad, S.; Meiri, E.; Yogev, Y.; Benjamin, S.; Lebanony, D.; Yerushalmi, N.; Benjamin, H.; Kushnir, M.; Cholakh, H.; Melamed, N.; et al. Serum micrornas are promising novel biomarkers. PLoS ONE 2008, 3, e3148. [Google Scholar] [CrossRef] [Green Version]
- Laterza, O.F.; Lim, L.; Garrett-Engele, P.W.; Vlasakova, K.; Muniappa, N.; Tanaka, W.K.; Johnson, J.M.; Sina, J.F.; Fare, T.L.; Sistare, F.D.; et al. Plasma micrornas as sensitive and specific biomarkers of tissue injury. Clin. Chem. 2009, 55, 1977–1983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of micrornas in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Q.; Zhao, Y.; Zhou, X.; Luan, J.; Cui, Y.; Han, J. Comparison of the extraction and determination of serum exosome and mirna in serum and the detection of mir-27a-3p in serum exosome of als patients. Intract. Rare Dis. Res. 2018, 7, 13–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, M.M.; Yang, H.; Zhang, L.; Shu, W.; Blair, D.G.; Morrisey, E.E. The bone morphogenic protein antagonist gremlin regulates proximal-distal patterning of the lung. Dev. Dyn. 2001, 222, 667–680. [Google Scholar] [CrossRef] [PubMed]
- Divi, S.N.; Markova, D.Z.; Fang, T.; Guzek, R.; Kurd, M.F.; Rihn, J.A.; Hilibrand, A.S.; Anderson, D.G.; Vaccaro, A.R.; Schroeder, G.D.; et al. Circulating mir-155-5p as a novel biomarker of lumbar degenerative disc disease. Spine 2020, 45, E499–E507. [Google Scholar] [CrossRef]
- Ma, J.; Lin, X.; Chen, C.; Li, S.; Zhang, S.; Chen, Z.; Li, D.; Zhao, F.; Yang, C.; Yin, C.; et al. Circulating mir-181c-5p and mir-497-5p are potential biomarkers for prognosis and diagnosis of osteoporosis. J. Clin. Endocrinol. Metab. 2020, 105, dgz300. [Google Scholar] [CrossRef]
- Kumar, S.; Reddy, P.H. Are circulating micrornas peripheral biomarkers for alzheimer’s disease? Biochim. Biophys. Acta 2016, 1862, 1617–1627. [Google Scholar] [CrossRef]
- Shi, J.; Liu, H.; Wang, H.; Kong, X. Microrna expression signature in degenerative aortic stenosis. Biomed. Res. Int. 2016, 2016, 4682172. [Google Scholar] [CrossRef] [Green Version]
- Gui, J.; Tian, Y.; Wen, X.; Zhang, W.; Zhang, P.; Gao, J.; Run, W.; Tian, L.; Jia, X.; Gao, Y. Serum microrna characterization identifies mir-885-5p as a potential marker for detecting liver pathologies. Clin. Sci. 2011, 120, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhang, Z.; Dai, F.; Shi, B.; Chen, L.; Zhang, X.; Zang, G.; Zhang, J.; Chen, X.; Qian, F.; et al. Comparison of circulating, hepatocyte specific messenger rna and microrna as biomarkers for chronic hepatitis b and c. PLoS ONE 2014, 9, e92112. [Google Scholar] [CrossRef]
- Zhang, S.; Ouyang, X.; Jiang, X.; Gu, D.; Lin, Y.; Kong, S.K.; Xie, W. Dysregulated serum microrna expression profile and potential biomarkers in hepatitis c virus-infected patients. Int. J. Med. Sci. 2015, 12, 590–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukasiak, S.; Schiller, C.; Oehlschlaeger, P.; Schmidtke, G.; Krause, P.; Legler, D.F.; Autschbach, F.; Schirmacher, P.; Breuhahn, K.; Groettrup, M. Proinflammatory cytokines cause fat10 upregulation in cancers of liver and colon. Oncogene 2008, 27, 6068–6074. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.Q.; Meng, H.; Wang, N.; Liang, L.N.; Liu, L.N.; Lu, S.M.; Luan, Y. Serum microrna 143 and microrna 215 as potential biomarkers for the diagnosis of chronic hepatitis and hepatocellular carcinoma. Diagn. Pathol. 2014, 9, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.; Ge, G.; Pan, T.; Wen, D.; Gan, J. Serum mirna panel as potential biomarkers for chronic hepatitis b with persistently normal alanine aminotransferase. Clin. Chim. Acta 2015, 451, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Rottiers, V.; Naar, A.M. Micrornas in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012, 13, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Cermelli, S.; Ruggieri, A.; Marrero, J.A.; Ioannou, G.N.; Beretta, L. Circulating micrornas in patients with chronic hepatitis c and non-alcoholic fatty liver disease. PLoS ONE 2011, 6, e23937. [Google Scholar] [CrossRef] [Green Version]
- Pirola, C.J.; Fernandez Gianotti, T.; Castano, G.O.; Mallardi, P.; San Martino, J.; Mora Gonzalez Lopez Ledesma, M.; Flichman, D.; Mirshahi, F.; Sanyal, A.J.; Sookoian, S. Circulating microrna signature in non-alcoholic fatty liver disease: From serum non-coding rnas to liver histology and disease pathogenesis. Gut 2015, 64, 800–812. [Google Scholar] [CrossRef] [Green Version]
- Saleh, H.A.; Abu-Rashed, A.H. Liver biopsy remains the gold standard for evaluation of chronic hepatitis and fibrosis. J. Gastrointest. Liver Dis. 2007, 16, 425–426. [Google Scholar]
- Roderburg, C.; Urban, G.W.; Bettermann, K.; Vucur, M.; Zimmermann, H.; Schmidt, S.; Janssen, J.; Koppe, C.; Knolle, P.; Castoldi, M.; et al. Micro-rna profiling reveals a role for mir-29 in human and murine liver fibrosis. Hepatology 2011, 53, 209–218. [Google Scholar] [CrossRef]
- Lopez-Riera, M.; Conde, I.; Quintas, G.; Pedrola, L.; Zaragoza, A.; Perez-Rojas, J.; Salcedo, M.; Benlloch, S.; Castell, J.V.; Jover, R. Non-invasive prediction of nafld severity: A comprehensive, independent validation of previously postulated serum microrna biomarkers. Sci. Rep. 2018, 8, 10606. [Google Scholar] [CrossRef]
- Bala, S.; Marcos, M.; Szabo, G. Emerging role of micrornas in liver diseases. World J. Gastroenterol. 2009, 15, 5633–5640. [Google Scholar] [CrossRef] [PubMed]
- Fathi-Kazerooni, M.; Kazemnejad, S.; Khanjani, S.; Saltanatpour, Z.; Tavoosidana, G. Down-regulation of mir-122 after transplantation of mesenchymal stem cells in acute liver failure in mice model. Biologicals 2019, 58, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.; Ge, J.; Xiu, L.; Zhao, Z.; Duan, X.; Tian, L.; Xie, J.; Yang, L.; Li, L. Hur mediates motility of human bone marrow-derived mesenchymal stem cells triggered by sphingosine 1-phosphate in liver fibrosis. J. Mol. Med. 2017, 95, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Jun, J.H.; Park, S.Y.; Yang, S.W.; Bae, S.H.; Kim, G.J. Dynamic regulation of mirna expression by functionally enhanced placental mesenchymal stem cells promoteshepatic regeneration in a rat model with bile duct ligation. Int. J. Mol. Sci. 2019, 20, 5299. [Google Scholar] [CrossRef] [Green Version]
- Fu, S.; Fei, Q.; Jiang, H.; Chuai, S.; Shi, S.; Xiong, W.; Jiang, L.; Lu, C.; Atadja, P.; Li, E.; et al. Involvement of histone acetylation of sox17 and foxa2 promoters during mouse definitive endoderm differentiation revealed by microrna profiling. PLoS ONE 2011, 6, e27965. [Google Scholar] [CrossRef] [Green Version]
- Tzur, G.; Levy, A.; Meiri, E.; Barad, O.; Spector, Y.; Bentwich, Z.; Mizrahi, L.; Katzenellenbogen, M.; Ben-Shushan, E.; Reubinoff, B.E.; et al. Microrna expression patterns and function in endodermal differentiation of human embryonic stem cells. PLoS ONE 2008, 3, e3726. [Google Scholar] [CrossRef]
- Hinton, A.; Afrikanova, I.; Wilson, M.; King, C.C.; Maurer, B.; Yeo, G.W.; Hayek, A.; Pasquinelli, A.E. A distinct microrna signature for definitive endoderm derived from human embryonic stem cells. Stem Cells Dev. 2010, 19, 797–807. [Google Scholar] [CrossRef] [Green Version]
- Kim, N.; Kim, H.; Jung, I.; Kim, Y.; Kim, D.; Han, Y.M. Expression profiles of mirnas in human embryonic stem cells during hepatocyte differentiation. Hepatol. Res. 2011, 41, 170–183. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, H.; Liu, J.; Tu, X.; Zang, Y.; Zhu, J.; Chen, J.; Dong, L.; Zhang, J. Mir-30 inhibits tgf-beta1-induced epithelial-to-mesenchymal transition in hepatocyte by targeting snail1. Biochem. Biophys. Res. Commun. 2012, 417, 1100–1105. [Google Scholar] [CrossRef]
- Kao, C.J.; Martiniez, A.; Shi, X.B.; Yang, J.; Evans, C.P.; Dobi, A.; deVere White, R.W.; Kung, H.J. Mir-30 as a tumor suppressor connects egf/src signal to erg and emt. Oncogene 2014, 33, 2495–2503. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.; Hou, J.; Alder, O.; Ye, X.; Lee, S.; Cullum, R.; Chu, A.; Zhao, Y.; Warner, S.M.; Knight, D.A.; et al. Genome-wide microrna and messenger rna profiling in rodent liver development implicates mir302b and mir20a in repressing transforming growth factor-beta signaling. Hepatology 2013, 57, 2491–2501. [Google Scholar] [CrossRef] [PubMed]
- Margagliotti, S.; Clotman, F.; Pierreux, C.E.; Beaudry, J.B.; Jacquemin, P.; Rousseau, G.G.; Lemaigre, F.P. The onecut transcription factors hnf-6/oc-1 and oc-2 regulate early liver expansion by controlling hepatoblast migration. Dev. Biol. 2007, 311, 579–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanhorenbeeck, V.; Jenny, M.; Cornut, J.F.; Gradwohl, G.; Lemaigre, F.P.; Rousseau, G.G.; Jacquemin, P. Role of the onecut transcription factors in pancreas morphogenesis and in pancreatic and enteric endocrine differentiation. Dev. Biol. 2007, 305, 685–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simion, A.; Laudadio, I.; Prevot, P.P.; Raynaud, P.; Lemaigre, F.P.; Jacquemin, P. Mir-495 and mir-218 regulate the expression of the onecut transcription factors hnf-6 and oc-2. Biochem. Biophys. Res. Commun. 2010, 391, 293–298. [Google Scholar] [CrossRef]
- Laudadio, I.; Manfroid, I.; Achouri, Y.; Schmidt, D.; Wilson, M.D.; Cordi, S.; Thorrez, L.; Knoops, L.; Jacquemin, P.; Schuit, F.; et al. A feedback loop between the liver-enriched transcription factor network and mir-122 controls hepatocyte differentiation. Gastroenterology 2012, 142, 119–129. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.G.; Qiu, R.L.; Wu, Y.H.; Li, Z.X.; Xie, P.; Zhang, J.; Zhou, J.J.; Zeng, L.X.; Tang, J.; Maharjan, A.; et al. Overexpression of mir-122 promotes the hepatic differentiation and maturation of mouse escs through a mir-122/foxa1/hnf4a-positive feedback loop. Liver Int. 2014, 34, 281–295. [Google Scholar] [CrossRef]
- Xu, H.; He, J.H.; Xiao, Z.D.; Zhang, Q.Q.; Chen, Y.Q.; Zhou, H.; Qu, L.H. Liver-enriched transcription factors regulate microrna-122 that targets cutl1 during liver development. Hepatology 2010, 52, 1431–1442. [Google Scholar] [CrossRef]
- Gailhouste, L.; Gomez-Santos, L.; Hagiwara, K.; Hatada, I.; Kitagawa, N.; Kawaharada, K.; Thirion, M.; Kosaka, N.; Takahashi, R.U.; Shibata, T.; et al. Mir-148a plays a pivotal role in the liver by promoting the hepatospecific phenotype and suppressing the invasiveness of transformed cells. Hepatology 2013, 58, 1153–1165. [Google Scholar] [CrossRef]
- Allen, R.M.; Marquart, T.J.; Albert, C.J.; Suchy, F.J.; Wang, D.Q.; Ananthanarayanan, M.; Ford, D.A.; Baldan, A. Mir-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol. Med. 2012, 4, 882–895. [Google Scholar] [CrossRef]
- Lakner, A.M.; Steuerwald, N.M.; Walling, T.L.; Ghosh, S.; Li, T.; McKillop, I.H.; Russo, M.W.; Bonkovsky, H.L.; Schrum, L.W. Inhibitory effects of microrna 19b in hepatic stellate cell-mediated fibrogenesis. Hepatology 2012, 56, 300–310. [Google Scholar] [CrossRef] [Green Version]
- Sekiya, Y.; Ogawa, T.; Iizuka, M.; Yoshizato, K.; Ikeda, K.; Kawada, N. Down-regulation of cyclin e1 expression by microrna-195 accounts for interferon-beta-induced inhibition of hepatic stellate cell proliferation. J. Cell. Physiol. 2011, 226, 2535–2542. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Zhang, J.; Huang, G.; Qian, J.; Wang, X.; Mei, S. Over-expressed microrna-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 2009, 583, 759–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekiya, Y.; Ogawa, T.; Yoshizato, K.; Ikeda, K.; Kawada, N. Suppression of hepatic stellate cell activation by microrna-29b. Biochem. Biophys. Res. Commun. 2011, 412, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Lin, Z.; Dong, P.; Lu, Z.; Gao, S.; Chen, X.; Wu, C.; Yu, F. Activation of hepatic stellate cells is suppressed by microrna-150. Int. J. Mol. Med. 2013, 32, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venugopal, S.K.; Jiang, J.; Kim, T.H.; Li, Y.; Wang, S.S.; Torok, N.J.; Wu, J.; Zern, M.A. Liver fibrosis causes downregulation of mirna-150 and mirna-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 298, G101–G106. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Wu, C.Q.; Zhang, Z.Q.; Yao, D.K.; Zhu, L. Loss of expression of mir-335 is implicated in hepatic stellate cell migration and activation. Exp. Cell Res. 2011, 317, 1714–1725. [Google Scholar] [CrossRef]
- Kwiecinski, M.; Noetel, A.; Elfimova, N.; Trebicka, J.; Schievenbusch, S.; Strack, I.; Molnar, L.; von Brandenstein, M.; Tox, U.; Nischt, R.; et al. Hepatocyte growth factor (hgf) inhibits collagen i and iv synthesis in hepatic stellate cells by mirna-29 induction. PLoS ONE 2011, 6, e24568. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.J.; Pan, Q.; Li, D.G.; Sun, H.; Liu, B.W. Mir-15b and mir-16 are implicated in activation of the rat hepatic stellate cell: An essential role for apoptosis. J. Hepatol. 2009, 50, 766–778. [Google Scholar] [CrossRef]
- He, Y.; Huang, C.; Sun, X.; Long, X.R.; Lv, X.W.; Li, J. Microrna-146a modulates tgf-beta1-induced hepatic stellate cell proliferation by targeting smad4. Cell. Signal. 2012, 24, 1923–1930. [Google Scholar] [CrossRef]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. Mir-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [Green Version]
- Iliopoulos, D.; Drosatos, K.; Hiyama, Y.; Goldberg, I.J.; Zannis, V.I. Microrna-370 controls the expression of microrna-122 and cpt1alpha and affects lipid metabolism. J. Lipid Res. 2010, 51, 1513–1523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rayner, K.J.; Suarez, Y.; Davalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernandez-Hernando, C. Mir-33 contributes to the regulation of cholesterol homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davalos, A.; Goedeke, L.; Smibert, P.; Ramirez, C.M.; Warrier, N.P.; Andreo, U.; Cirera-Salinas, D.; Rayner, K.; Suresh, U.; Pastor-Pareja, J.C.; et al. Mir-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 9232–9237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Jia, X.J.; Jiang, H.J.; Du, Y.; Yang, F.; Si, S.Y.; Hong, B. Micrornas 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol. Cell. Biol. 2013, 33, 1956–1964. [Google Scholar] [CrossRef] [Green Version]
- Vickers, K.C.; Shoucri, B.M.; Levin, M.G.; Wu, H.; Pearson, D.S.; Osei-Hwedieh, D.; Collins, F.S.; Remaley, A.T.; Sethupathy, P. Microrna-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology 2013, 57, 533–542. [Google Scholar] [CrossRef] [Green Version]
- Zhong, D.; Huang, G.; Zhang, Y.; Zeng, Y.; Xu, Z.; Zhao, Y.; He, X.; He, F. Microrna-1 and microrna-206 suppress lxralpha-induced lipogenesis in hepatocytes. Cell. Signal. 2013, 25, 1429–1437. [Google Scholar] [CrossRef]
- Baselga-Escudero, L.; Blade, C.; Ribas-Latre, A.; Casanova, E.; Salvado, M.J.; Arola, L.; Arola-Arnal, A. Grape seed proanthocyanidins repress the hepatic lipid regulators mir-33 and mir-122 in rats. Mol. Nutr. Food Res. 2012, 56, 1636–1646. [Google Scholar] [CrossRef]
- Castoldi, M.; Vujic Spasic, M.; Altamura, S.; Elmen, J.; Lindow, M.; Kiss, J.; Stolte, J.; Sparla, R.; D’Alessandro, L.A.; Klingmuller, U.; et al. The liver-specific microrna mir-122 controls systemic iron homeostasis in mice. J. Clin. Investig. 2011, 121, 1386–1396. [Google Scholar] [CrossRef]
- Sangokoya, C.; Doss, J.F.; Chi, J.T. Iron-responsive mir-485-3p regulates cellular iron homeostasis by targeting ferroportin. PLoS Genet. 2013, 9, e1003408. [Google Scholar] [CrossRef]
- Guengerich, F.P. Cytochrome p450 and chemical toxicology. Chem. Res. Toxicol. 2008, 21, 70–83. [Google Scholar] [CrossRef]
- Rieger, J.K.; Klein, K.; Winter, S.; Zanger, U.M. Expression variability of absorption, distribution, metabolism, excretion-related micrornas in human liver: Influence of nongenetic factors and association with gene expression. Drug Metab. Dispos. 2013, 41, 1752–1762. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.S. Opioid metabolism. Mayo Clin. Proc. 2009, 84, 613–624. [Google Scholar] [CrossRef] [Green Version]
- Takagi, S.; Nakajima, M.; Mohri, T.; Yokoi, T. Post-transcriptional regulation of human pregnane x receptor by micro-rna affects the expression of cytochrome p450 3a4. J. Biol. Chem. 2008, 283, 9674–9680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Ma, J.; Fang, Q.; Li, Y. Transcription alterations of micrornas, cytochrome p4501a1 and 3a65, and ahr and pxr in the liver of zebrafish exposed to crude microcystins. Toxicon 2013, 73, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y.; Nakajima, M.; Mohri, T.; Takamiya, M.; Aoki, Y.; Fukami, T.; Yokoi, T. Aryl hydrocarbon receptor nuclear translocator in human liver is regulated by mir-24. Toxicol. Appl. Pharmacol. 2012, 260, 222–231. [Google Scholar] [CrossRef] [Green Version]
- Takagi, S.; Nakajima, M.; Kida, K.; Yamaura, Y.; Fukami, T.; Yokoi, T. Micrornas regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J. Biol. Chem. 2010, 285, 4415–4422. [Google Scholar] [CrossRef] [Green Version]
- Hayes, J.D.; Strange, R.C. Glutathione s-transferase polymorphisms and their biological consequences. Pharmacology 2000, 61, 154–166. [Google Scholar] [CrossRef]
- Maes, O.C.; An, J.; Sarojini, H.; Wang, E. Murine micrornas implicated in liver functions and aging process. Mech. Ageing Dev. 2008, 129, 534–541. [Google Scholar] [CrossRef]
- Yi, P.S.; Zhang, M.; Xu, M.Q. Role of microrna in liver regeneration. Hepatobiliary Pancreat. Dis. Int. 2016, 15, 141–146. [Google Scholar] [CrossRef]
- Murakami, Y.; Toyoda, H.; Tanahashi, T.; Tanaka, J.; Kumada, T.; Yoshioka, Y.; Kosaka, N.; Ochiya, T.; Taguchi, Y.H. Comprehensive mirna expression analysis in peripheral blood can diagnose liver disease. PLoS ONE 2012, 7, e48366. [Google Scholar] [CrossRef] [Green Version]
- El-Hefny, M.; Fouad, S.; Hussein, T.; Abdel-Hameed, R.; Effat, H.; Mohamed, H.; Abdel Wahab, A.H. Circulating micrornas as predictive biomarkers for liver disease progression of chronic hepatitis c (genotype-4) egyptian patients. J. Med. Virol. 2019, 91, 93–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Li, Q.Y.; Guo, Z.Z.; Guan, Y.; Du, J.; Lu, Y.Y.; Hu, Y.Y.; Liu, P.; Huang, S.; Su, S.B. Serum levels of micrornas can specifically predict liver injury of chronic hepatitis b. World J. Gastroenterol. 2012, 18, 5188–5196. [Google Scholar] [PubMed]
- Ji, F.; Yang, B.; Peng, X.; Ding, H.; You, H.; Tien, P. Circulating micrornas in hepatitis b virus-infected patients. J. Viral Hepat. 2011, 18, e242–e251. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Peng, S.F.; Fu, L.; Fu, X.Y.; Wu, D.X.; Liu, B.J.; Tan, D.M.; Ouyang, Y. Serum levels of mirna in patients with hepatitis b virus-associated acute-on-chronic liver failure. Hepatobiliary Pancreat. Dis. Int. 2018, 17, 126–132. [Google Scholar] [CrossRef]
- Chen, S.; Chen, H.; Gao, S.; Qiu, S.; Zhou, H.; Yu, M.; Tu, J. Differential expression of plasma microrna-125b in hepatitis b virus-related liver diseases and diagnostic potential for hepatitis b virus-induced hepatocellular carcinoma. Hepatol. Res. 2017, 47, 312–320. [Google Scholar] [CrossRef]
- Yamada, H.; Suzuki, K.; Ichino, N.; Ando, Y.; Sawada, A.; Osakabe, K.; Sugimoto, K.; Ohashi, K.; Teradaira, R.; Inoue, T.; et al. Associations between circulating micrornas (mir-21, mir-34a, mir-122 and mir-451) and non-alcoholic fatty liver. Clin. Chim. Acta 2013, 424, 99–103. [Google Scholar] [CrossRef]
- Yamada, H.; Ohashi, K.; Suzuki, K.; Munetsuna, E.; Ando, Y.; Yamazaki, M.; Ishikawa, H.; Ichino, N.; Teradaira, R.; Hashimoto, S. Longitudinal study of circulating mir-122 in a rat model of non-alcoholic fatty liver disease. Clin. Chim. Acta 2015, 446, 267–271. [Google Scholar] [CrossRef]
- Tan, Y.; Ge, G.; Pan, T.; Wen, D.; Gan, J. A pilot study of serum micrornas panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS ONE 2014, 9, e105192. [Google Scholar] [CrossRef] [Green Version]
- Murakami, Y.; Toyoda, H.; Tanaka, M.; Kuroda, M.; Harada, Y.; Matsuda, F.; Tajima, A.; Kosaka, N.; Ochiya, T.; Shimotohno, K. The progression of liver fibrosis is related with overexpression of the mir-199 and 200 families. PLoS ONE 2011, 6, e16081. [Google Scholar] [CrossRef]
- Teng, K.Y.; Ghoshal, K. Role of noncoding rnas as biomarker and therapeutic targets for liver fibrosis. Gene Expr. 2015, 16, 155–162. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.J.; Zhu, J.M.; Wu, H.; Fan, J.; Zhou, J.; Hu, J.; Yu, Q.; Liu, T.T.; Yang, L.; Wu, C.L.; et al. Circulating micrornas as a fingerprint for liver cirrhosis. PLoS ONE 2013, 8, e66577. [Google Scholar] [CrossRef] [PubMed]
- Szabo, G.; Bala, S. Micrornas in liver disease. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 542–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Chen, J.; Chen, X.; Tang, J.; Guo, H.; Wang, X.; Qian, J.; Luo, G.; He, F.; Lu, X.; et al. Serum mirnas as predictive and preventive biomarker for pre-clinical hepatocellular carcinoma. Cancer Lett. 2016, 373, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.T.; Liu, S.M.; Ma, H.; Yang, Y.; Zhang, X.; Sun, H.; Zhang, X.; Xu, J.; Wang, J. Systematic review and meta-analysis: Circulating mirnas for diagnosis of hepatocellular carcinoma. J. Cell. Physiol. 2016, 231, 328–335. [Google Scholar] [CrossRef]
- Morishita, A.; Masaki, T. Micrornas as possible biomarkers for hepatocellular carcinoma. Hepatol. Res. 2018, 48, 499–501. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kweon, M.; Kim, J.Y.; Jun, J.H.; Kim, G.J. Research Trends in the Efficacy of Stem Cell Therapy for Hepatic Diseases Based on MicroRNA Profiling. Int. J. Mol. Sci. 2021, 22, 239. https://doi.org/10.3390/ijms22010239
Kweon M, Kim JY, Jun JH, Kim GJ. Research Trends in the Efficacy of Stem Cell Therapy for Hepatic Diseases Based on MicroRNA Profiling. International Journal of Molecular Sciences. 2021; 22(1):239. https://doi.org/10.3390/ijms22010239
Chicago/Turabian StyleKweon, Minyeoung, Jae Yeon Kim, Ji Hye Jun, and Gi Jin Kim. 2021. "Research Trends in the Efficacy of Stem Cell Therapy for Hepatic Diseases Based on MicroRNA Profiling" International Journal of Molecular Sciences 22, no. 1: 239. https://doi.org/10.3390/ijms22010239





