Abstract
The tumor microenvironment (TME) has become the focus of interest in cancer research and treatment. It includes the extracellular matrix (ECM) and ECM-modifying enzymes that are secreted by cancer and neighboring cells. The ECM serves both to anchor the tumor cells embedded in it and as a means of communication between the various cellular and non-cellular components of the TME. The cells of the TME modify their surrounding cancer-characteristic ECM. This in turn provides feedback to them via cellular receptors, thereby regulating, together with cytokines and exosomes, differentiation processes as well as tumor progression and spread. Matrix remodeling is accomplished by altering the repertoire of ECM components and by biophysical changes in stiffness and tension caused by ECM-crosslinking and ECM-degrading enzymes, in particular matrix metalloproteinases (MMPs). These can degrade ECM barriers or, by partial proteolysis, release soluble ECM fragments called matrikines, which influence cells inside and outside the TME. This review examines the changes in the ECM of the TME and the interaction between cells and the ECM, with a particular focus on MMPs.
1. Introduction
The tumor microenvironment (TME) describes the conditions within and in the vicinity of a solid tumor mass. It is shaped in an orchestrated manner by the oncogenically transformed cells and their neighboring tissue cells. It comprises cellular and noncellular constituents of macromolecular size and smaller molecules, as well as several biophysical parameters, such as pH, redox status [1], and mechanical tension within the tissue [2,3] (Figure 1). Among the small molecules, aberrant concentrations of redox potential-determining compounds, such as glutathione and reactive oxygen species (ROS) as well as extracellular ATP, characterize the TME [1].
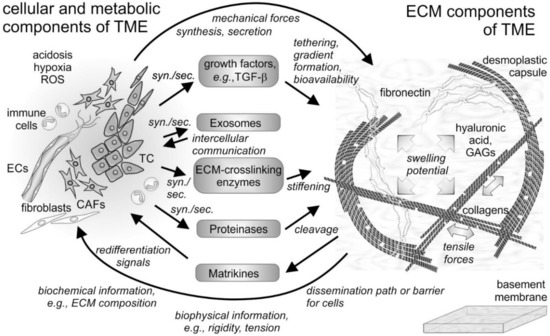
Figure 1.
Cellular and non-cellular components of the tumor microenvironment (TME) and their interplay. In addition to the tumor cells (TCs), fibroblasts and their derivatives, the cancer-associated fibroblasts (CAFs), as well as ingrowing endothelial cells (ECs) and infiltrating immune cells are the cellular components of the tumor microenvironment (TME). The cells synthesize and secrete (syn./sec.) not only the extracellular matrix (ECM) components, but also growth factors, exosomes, and ECM-modifying enzymes, such as proteinases. Both cells and their secretion products interact with the fibrillar and non-fibrillar components of the ECM. Hyaluronic acid and glycosaminoglycan (GAG)-chains of proteoglycans increase the swelling potential of the interstitial space, which is counterbalanced by the tensile force-bearing fibrils of collagens, elastin and fibronectin. Modified by tethered growth factors, by crosslinking and cleaving enzymes, and by contractile forces, the ECM and its fragments with cytokine-like functions (matrikines) influence the cells within the TME in various ways. ROS, reactive oxygen species.
Fibrillar and non-fibrillar proteins and rather amorphous proteoglycans together form the insoluble scaffold of the extracellular matrix [4,5] (Figure 1). The TME is also unique in its composition of soluble extracellular proteins, such as cytokines [6,7] and enzymes. Among the latter, extracellular matrix (ECM)-degrading matrix metalloproteinases (MMPs) and ECM-crosslinking lysyl-oxidases and transglutaminases characteristically contribute to the plasticity and distinction of the TME [8,9,10,11]. They are actively produced and remodeled by cells of the tumor mass and constitute the surroundings, which influence cancer cells and their neighboring cells in a tumor-supportive manner. This review will shed light on the ECM of the TME and will take into account its TME-characteristic remodeling with a special emphasis on the MMPs. Moreover, it will summarize the current knowledge on the interactions of TME-embedded cells, both cancer and resident cells, with the ECM and the mutual effects on each other in maintaining tumor-supportive surroundings and in fostering metastasis.
2. The Extracellular Matrix as a Key Component of the TME
The palpation of, e.g., the mammary gland [3], is a simple procedure to detect volume-demanding and stiffer tumor tissue [2,12]. Biophysical differences between normal tissue and tumor mass are caused by cell growth and the increased deposition of ECM components, known as desmoplasia, which is typically observed in healing wounds and fibrosis [13].
Most of the mass of solid tumors consists of ECM [14]. Having developed from collagen-rich stromal tissue, the TME is rich in collagens, especially if the tumor mass induces a desmoplastic reaction [13,15]. Collagens as the most abundant proteins of the human body crucially contribute to the scaffolding function of the ECM. The almost 30 members of the collagen family share several characteristics: (i) Their three chains consist of the repetitive Gly–X–Y amino acid sequence with X and Y being different amino acids, most frequently proline and hydroxyproline; (ii) They form a characteristic, staggered triple helix with the glycine residues of all triplet sequences in its center; (iii) They self-assemble into supramolecular structures, in which several triple-helical collagen molecules associate forming fibrils, networks, and other highly ordered aggregates [5,14,16]. Fibrils of type I collagen, together with collagen types III and V, bear the tensile forces within normal stromal tissue and in the TME of the tumor mass. They are preferentially deposited in desmoplastic environments, where the resident stromal cells are induced by tumor cells to produce and deposit collagen type I to form the stiff TME or a capsule surrounding the tumor mass [13,15]. Another collagen isoform, type IV collagen, along with collagens XV and XVIII, forms a network-like suprastructure, which is typical of basement membranes (BMs), the specialized sheet-like ECM that separates stromal tissue from other tissues. As it confines cells to their respective tissue type, its breaching by malignant cells is a hallmark of cancer [17].
Spanning the interstitial stroma, collagen fibrils provide an ideal path for cell migration and promote cancer cell dissemination along these fibrils (Figure 2). In contrast, the meshwork of stromal collagen fibrils and the desmoplastic capsule around the tumor mass like the type IV collagen network of basement membranes are extremely dense and impede tumor cell infiltration [18,19]. These ECM barriers are overcome by the cancer cells or their accompanying CAFs by the cleavage of collagen with particular collagenases [20].
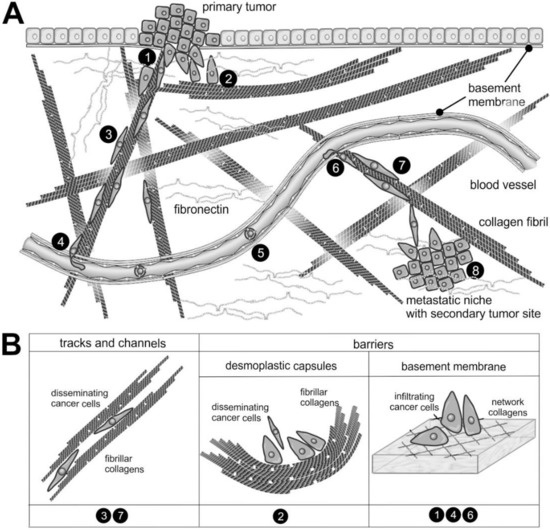
Figure 2.
The supramolecular structure of collagen as a substrate or impediment of cancer cell dissemination: (A) The metastatic cascade of malignant carcinoma cells includes the penetration of the epithelial basement membrane (❶); Solid tumors within the stroma tissue are often surrounded by a desmoplastic collagen capsule, which impedes cancer cell migration (❷); Through the interstitial stroma, cancer cells utilize collagen-rich fibrils to quickly reach a nearby blood vessel (❸); There, they intravasate through the endothelial basement membrane (❹); When blood-borne, they float with the blood stream, mostly sheltered by platelets (❺); To extravasate, they attach to the vessel wall of a distant organ and again breach the endothelial basement membrane (❻); Moving quickly along collagen-rich fibrils (❼), they reach the metastatic niche (❽), where they grow into a metastasis. (B) The different dissemination-supporting and impeding functions of the collagen suprastructures are highlighted for the different steps of the metastatic cascade. The cancer cells experience, particularly by integrins, the ECM as dissemination-supporting tracks and channels, especially if migration and fibrils point into the same direction and if the fibril network is not too dense. In contrast, the dense array of collagen fibrils in desmoplastic capsules and of network-forming collagens within the basement membrane impedes cancer cell progression and requires the use matrix-metalloproteinases. Moreover, the orientation of collagen fibrils within the desmoplastic capsule is mostly perpendicular to the direction of dissemination [28].
In addition to the network-forming collagens, laminins, which form a family of about 20 members, are typical constituents of basement membranes [21,22,23]. Their N termini and the C terminus formed by the globular G domain of the laminin α chain protrude from an α-helical coiled coil [21]. Although laminins are normally exclusively found in BMs, some types of laminins, such as laminin-332, also occur ectopically within the TME [24], but their role in the TME has not yet been fully deciphered.
Collagen fibrils are often found together with elastin and fibulin containing elastic fibrils [25], which, due to their reversible elasticity, allow the resilience of the ECM. Interestingly, most of the body’s elastin is formed pre- and early postnatally but hardly in the adult body [26]. However, some cancer entities stimulate and reinitiate elastin production and deposition, which is known as elastosis [25]. Similarly noteworthy, elastin degradation peptides (EDPs) are released in the TME, which stimulate tumor cell growth and progression via different receptors [27] (Figure 2).
Fibronectin is another scaffold-forming glycoprotein found in BMs as well as in the ECM of the TME [29,30], with distinct splice variants being produced in the TME [31]. Fibronectin consists of two disulfide-linked protein chains with a characteristic modular character of fibronectin repeats of types I, II, and III with about 30, 60, and 90 amino acids, respectively. The type I and II repeats allow the formation of disulfide-crosslinked supramolecular fibronectin networks in the tissue stroma [30]. Fibronectin isoforms with the extra domains ED-A and ED-B are normally expressed under the control of mainly TGF-β during embryonic development and wound repair but also in the hypoxic TME [32,33]. Thus, ED-A- and/or ED-B- fibronectin are employed as a marker to image tumor nodes [34]. Other glycoproteins marking a tumor-modified stroma are tenascins-C and W [35,36,37]. Like the other two members of the tenascin family [38], they are disulfide-crosslinked homotrimers, each consisting of three chains forming an α-helical coiled coil. With their ECM-typical EGF-like and type III fibronectin domains, and a C-terminal fibrinogen globe module, they perform various functions in wound healing and tumor progression [36]. These tenascin isoforms or fragments thereof can also be released from the primary tumor into the blood circulation and precondition distant sites as premetastatic niches [39]. Thus, the blood levels of tenascins are a diagnostic tumor marker [40]. Their abundance in the TME is tested for diagnostic imaging and therapeutic exploitation [41]. Tenascins and other pericellular ECM proteins, such as periostin [42], galectins [43], small integrin-binding ligand N-linked glycoproteins (SIBLINGs), secreted protein acidic and rich in cysteine (SPARC), thrombospondin, angiopoietin-like proteins, certain proteoglycans, and CCN family members are referred to as matricellular proteins [44]. Rather than performing scaffolding functions, they modulate the supramolecular architecture of the collagen and fibronectin network [42] and they regulate cellular behavior within the TME [37]. For example, CCNs regulate the proliferation and migration of ECM scaffold-embedded cells [44,45].
Functionally similarly versatile are glycosaminoglycans (GAGs), and the protein- and sulfate-free hyaluronic acid (HA) made up of N-acetylglucosamine and glucuronic acid [46,47], as well as sulfated GAG-chain containing proteoglycans, such as heparan sulfate and keratan sulfate, and galactosamine-containing chondroitin sulfate and dermatan sulfate [4,5,48,49]. According to their location, proteoglycans are divided into extracellular, membrane-bound, and intracellular proteoglycans. The extracellular group includes the hyaluronic acid-binding hyalectans (e.g., aggrecan, versican with four different splice variants, neurocan, and brevican), the small leucine-rich proteoglycans (SLRPs, e.g., decorin, biglycan, fibromodulin, and lumican), and the BM-located pericellular proteoglycans (e.g., perlecan, agrin, and collagen XVIII). Perlecan, agrin, and the network-forming type VIII collagen bear heparan sulfate GAG chains [48], while the hyalectans and SLRPs are decorated with different numbers of covalently attached chondroitin sulfate, sometimes in combination with additional dermatan sulfate chains (decorin) or keratan sulfate chains (aggrecan, fibromodulin, lumican) [49]. The extracellular proteoglycans non-covalently link up with the scaffold of fibril-forming ECM components in different ways. Type IX collagen decorates the surface of type II collagen fibrils, while type XVIII collagen forms the BM-typical chicken wire network together with type IV collagen. Although alloyed into collagen fibrils, they are also proteoglycans due to their chondroitin sulfate and heparan sulfate GAG-chains, respectively [4,5]. Alternatively, several proteoglycans, such as decorin and other SLRPs, interact via their protein cores with type I collagen-containing fibrils and thus control the suprastructure of the ECM scaffold [46]. Moreover, specific type III repeats of fibronectin, certain G-domains of laminin α-chains, tenascins and thrombospondins harbor heparan sulfate binding sites, thereby allowing protein-carbohydrate binding interactions to connect the “amorphous” proteoglycans with higher suprastructure-forming ECM components [5]. Likewise, the hyalectans have a carbohydrate-binding domain, which allows their binding to hyaluronic acid.
Proteoglycans with their GAG chains also specifically tether growth factors and assist in presenting them to the respective cellular receptors. Such tethering of growth factors and cytokines stabilizes the formation of spatial gradients, which are indispensable for normal development and also contribute to pathologic processes. For example, in tumor angiogenesis, endothelial cells (ECs) follow a VEGF gradient that is stabilized by tethering to heparan sulfate GAG chains [47,48].
3. Fibrillar and Non-Fibrillar ECM Components Orchestrate the Biophysical and Biochemical Properties of the TME
Even if the cellular origin of the ECM and its impact on the cells of the TME were neglected, the different ECM components alone would interplay in a manner that would already explain some of typical features of the TME, e.g., its increased stiffness and tension [2]. Due to its high charge density, GAGs, either as hyaluronic acid or as carbohydrate conjugates of proteoglycans, absorb tremendous amounts of water, causing their immense swelling capacity [50]. However, they do not reach their equilibrium swelling volume due to the limited space of the ECM scaffold and due to the tight connections between the ECM network with interwoven proteoglycans. Thus, the tissue pressure is counteracted by tensile forces, which are taken up by the fibrillar ECM components. This tensed force equilibrium shapes the interstitial stroma tissue and contributes to the cushion function of the ECM [14,15]. The enhanced deposition of hyaluronic acid and proteoglycans in the tumor mass, e.g., in pancreatic duct carcinomas, is accompanied by a desmoplastic increase in collagens [13,51] resulting in increased tissue rigidity and intra-tissue tension [2]. Embedded fibroblasts exert mechanical forces on the ECM scaffold and thus increase its stiffness and tension [52]. At the same time, they also sense changes in the biophysical properties of the TME and adapt their behavior (see below). The increased interstitial pressure of the TME impedes the access of anti-cancer drugs to the tumor mass [1,2,53].
The increased tensile forces on the fibrillar ECM proteins also regulate the bioavailability of growth factors deposited on them, such as TGF-β, one of the key growth factors of the TME [54,55]. Its preform, the latency-associated peptide (LAP) consisting of two non-covalently associated chains, is released after intracellular furin cleavage and tethered by fibrillin to elastic fibrils. When mechanical forces stretch the fibrillin, LAP detaches and becomes accessible to extracellular proteases that release active TGF-β, which then binds to its respective receptors and triggers de- and re-differentiation of cells within the TME [56].
4. The “Give and Take” between the ECM and Cells within the TME
The cellular players within the TME are as complex as the ECM. Formerly, tumor research focused on cancer cells, their oncogenic transformation, metabolic reprogramming, proliferation, dissemination and metastasis. This has provided important knowledge on cancer cells, their diagnostic detection and therapeutic targeting. However, recent decades have also highlighted the important role of resident and infiltrating cells of the TME, which under the influence of the nearby cancer cells, change their properties and metabolism in a tumor-supportive manner. This has broadened the perspective of cancer research and created a more holistic picture of cancer cells and their neighboring cells, among them resident fibroblasts, ingrowing ECs and infiltrating immune cells [57]. Especially the fibroblasts have become important in the context of the TME, as they are the major source of ECM components [58,59]. The ingrowth of ECs and the infiltration of immune cells also significantly contribute to the TME and are in turn influenced by the TME [60], which, however, is beyond the scope of this article and examined elsewhere [61,62,63].
Under the influence of cancer cells, tissue-resident stromal fibroblasts differentiate into cancer-associated fibroblasts (CAFs) [60,64,65]. However, CAFs are a heterogeneous group of cells and may originate from other cell types as well [52,65,66]. Due to their importance in tumor progression, CAFs are now considered as a promising target in cancer treatment [65,67]. Several clinical trials directed against CAF-marker proteins have also been launched to overcome drug resistance in treatment of cancer [68]
Cancer cells secrete growth factors, such as TGF-β [55], that promote the differentiation CAFs from resident fibroblasts [69]. Similar to fibrosis-associated fibroblasts in wound healing, CAFs synthesize, secrete, and deposit TME-typical ECM components, and contract the tissue with their α-smooth muscle actin (αSMA)-reinforced cytoskeleton [70]. The more rigid the ECM of the TME, the more its tension fosters CAF differentiation with YAP/TAZ acting as a sensor for structural and mechanical features of the ECM [71].
The repertoire of cellular adhesion molecules (CAMs) is in turn modulated by the altered ECM of the TME. Among them are hyaluronic acid receptors, membrane-bound proteoglycans, and integrins. They anchor the cells to the ECM scaffold allowing the transmission of cellular forces onto ECM components, and serve as signal relays transmitting cues of the ECM into the cells and vice versa. However, unlike growth factor receptors, most of the CAMs lack kinase domains and signal via associating proteins.
The hyaluronic acid receptors CD44 and RHAMM are closely linked to cancer progression and dissemination. With its known signaling mechanisms [72], CD44 has dual functions in the TME that can both support and attenuate tumor progression, depending on the differential expression, processing and fragmentation of its ligand, hyaluronic acid, in the course of tumor growth [73]. Newly synthesized high molecular weight hyaluronic acid (HMW-HA) is bound by CD44, while at a later disseminating stage of tumor progression, HMW-HA is fragmented by hyaluronidases and the resulting low molecular weight (LMW)-HA fragments. While LMW-HAs do also bind to CD44 and thereby exert important regulatory roles in the TME, as they inhibit CD44 clustering and have an antagonistic effect on HMW-HA activity, LMW-HAs are preferentially recognized by RHAMM [74,75], which is upregulated in disseminating cancer cells and alters the HA signaling of CD44 [74,75].
Furthermore, membrane-bound proteoglycans, such as syndecans, glypicans, and betaglycan, influence interactions between cancer cells and the TME [50]. Notably, the expression of the transmembrane proteoglycan NG2 (CSPG4) is altered on cancer cells and CAFs and may serve as a CAF-marker [49]. The mode of action of membrane-bound proteoglycans is diverse. In addition to tethering growth factors, such as TGF-β, VEGF, and hepatocyte growth factor (HGF), they act as co-receptors too [76,77,78]. Betaglycan is also known as TGF-β type III receptor [5]. Syndecans serve as co-signaling receptors for integrins and modulate their activity [79,80,81]. Soluble SLRPs also interact through their protein core with the extracellular domains of growth factor receptors and integrins, thereby regulating their extracellular ligand binding. For example, decorin, affects the binding of α2β1 integrin to collagen [82], and also the receptors for VEGF, HGF, and epidermal growth factor (EGF), as well as the immunomodulatory Toll-like receptors (TLR)-2 and -4 [76,83,84], the latter preferentially binding biglycan [84]. A similar receptor modulation appears to be effective in the fragmentation of the pericellular proteoglycan perlecan, whose N-terminal fragment endorepellin specifically binds to integrin α2β1 [85,86].
The integrins are undoubtedly the most extensive family among the CAMs in almost all multicellular organisms [87,88,89]. There are 24 different heterodimeric integrins, each consisting of one of 18 α-subunits and 8 β-subunits. The β1-containing integrins α1β1, α2β1, α10β1, and α11β1 recognize collagens, and the β3-containing integrins α3β1, α6β1, α6β4, and α7β1are laminin receptors, whereas fibronectin can be bound by the integrins α5β1, αVβ1, αVβ3, and αVβ6 [87,88]. Like other CAMs, they mechanically anchor the cells in the ECM and also transduce signals between cells and their environment. For this purpose, integrins are associated with the actin cytoskeleton and its motor proteins and form the transmembrane connectors to the ECM scaffold, on which they exert tensile [90]. With their integrins, CAFs, contribute significantly to the interstitial tissue pressure typical of the TME [91], whereby integrin αvβ3 in this context can apparently replace collagen binding β1 integrins [92]. In integrin signaling, significant conformational changes, and clustering into a supramolecular array on the cell surface, as well as the recruitment of several adapter and signal molecules such as kinases and G-proteins are involved [93,94,95,96,97]. This results in the formation of a new cell organelle, the adhesome, a complex supramolecular network of proteins with a highly ordered and hierarchical structure [98,99]. During the oncogenic transformation, the ECM ligand binding activity is regulated by changing various parameters, such as extracellular and intracellular ligands, e.g., the proteoglycan decorin or intracellular kindlins, as well as divalent cations and protons, redox-active compounds, and mechanical forces [82,100,101,102,103,104,105]. Therefore, integrin expression and surface abundance on cancer cells can serve as tumor (suppressor) markers, although this may vary between tumor entities [91,106,107,108]. For example, the expression of α3β1 and α11β1 integrins changes during CAF differentiation, whereupon CAFs interact via these integrins with ectopically expressed laminin-332 and desmoplastically abundant collagen, respectively, and thus support cancer cells [24,109]. Moreover, integrin expression and function is regulated by the TME characteristic TGF-β, and vice versa [110].
Although many studies have been undertaken to define whether certain ECM-proteins, such as fibronectin, and their receptors promote or attenuate tumor progression and spreading, a comprehensive picture of the role of all ECM proteins within the TME has not been fully drawn yet [30]. This is due to the fact, that the ECM of the TME can be different for certain tumor entities [111] and for different stages of tumor progression [30]. Likewise, the role of the ECM receptors is diverse and not entirely defined for each cell type within the TME at any stage of tumor progression and dissemination [72,75,78,112]. However, the ECM contacts of cells in the TME affect inter alia the secretion and deposition, as well as the remodeling of ECM components. Thus, the ECM–CAM axis is a way of signaling between the TME and its cells comparable to paths of paracrine signaling, via growth factors, cytokines and exosomes [69,70,113,114,115], which are all interdependent [116] and additionally integrate other features of the TME, such as the lactacidosis, hypoxia, and altered levels of ROS [117,118,119].
The remodeling of the ECM within the TME includes biochemical changes of its composition as well as the biophysical alteration of tissue rigidity and tension [2,3,12]. In addition, cells of the TME also secrete ECM-modifying enzymes, such as covalently crosslinking enzymes that stabilize ECM fibrils and stiffen the TME [15], but also ECM scaffold-cleaving proteases, such as MMPs [31,70,120]. Members of the lysyl-oxidases and of the transglutaminases, especially the isoforms LOXL2 and transglutaminase 2, respectively, connect the ECM proteins via uncleavable linkages and isopeptide bonds [10,121,122]. Thus, they determine not only the TME of the primary tumor site, but also determine the premetastatic niche by modifying the ECM scaffold of distant tissues [3,117,118,119,123]. Proteases secreted into the TME are also a means of communication between cells and the ECM [124]. They specifically cleave ECM components to release soluble ECM fragments with signaling functions for the cells, called matrikines [125]. Furthermore, they degrade proteins of the ECM, thereby weakening its scaffold structure, or locally open the ECM barrier for disseminating cancer cells accompanied by CAFs [19,124]. The expression and function of ADAMs (a disintegrin and a metalloproteinase) in the latter has recently been reviewed [126]. GAG-degrading hydrolases such as hyaluronidases are also involved in ECM remodeling [127]. A dysfunctional imbalance in ECM synthesis and degradation leads to desmoplasia and fibrosis. Among the matrix-degrading enzymes, MMPs play a central role in the entire tumor microenvironment [128].
5. Cancer Progression is Dependent on the Proteolytic Action of MMPs on the ECM of the TME
5.1. The Metastatic Cascade
In such a tumor microenvironment with mutually influencing ECM and cells located therein, the metastatic cascade begins. This comprises several steps by which cancer cells disseminate from the primary tumor, spread throughout the body, and metastasize into distant organs (Figure 2). First, a tumor cell leaves the primary site and enters the bloodstream via the lymph or, with the help of proteolytic enzymes, breaches the vascular wall and thus enters the bloodstream directly. On its route, it breaks through various barriers, such as the EC layer during intra- and extravasation, BMs and other scaffolds of extracellular matrix (ECM) molecules [5].
While developing epithelial neoplasms remain in their original location, the main characteristic of malignancy is that cancer cells destroy the BM that separates the epithelium from the adjacent connective tissue [17]. In particular, the dense network of the BM is impermeable to cells with the exception of immune cells and malignant tumor cells. Such cells secrete numerous different proteases [18,129,130,131]. The synthesis and secretion of MMPs by cancer cells depends on various TME factors, such as special growth factors and tumor-specific variants of fibronectin [132]. Among these proteases, the 28-member family of Zn2+-dependent matrix metalloproteinases (MMPs) plays an important role, and MMPs are therefore of prognostic importance [133,134,135,136,137]. Collagens are particularly resistant to proteolytic attack due to their triple-helicity but can be cleaved by collagenases outperforming gelatinases that can cleave only single strands of denatured collagen [5]. The main interest from the beginning was therefore in the two collagenases MMP-1 and MMP-14 (MT1-MMP), the latter of which is membrane bound, and in the gelatinases MMP-9 and MMP-2 [135,138,139,140,141,142,143,144,145]. Collagenolytic, MMPs partially untwist the collagen triple helix with their hemopexin domain and then cleave the now accessible chains with their catalytic domain [146]. The collagenolytic cleavage destabilizes the comparatively proteolysis-resistant triple helix of the resulting fragments, which partially unwind and thereby become substrates for gelatinases MMP-2 and -9 [20,147,148]. MMP-14 also activates MMP-2 in a complex binding and cleavage mechanism [20,149] with collagen-binding β1 integrins, especially α2β1, which bring collagen to MMP-14 for proteolytic cleavage.
5.2. Structural and Functional Diversity of Matrix Metalloproteinases
Matrix metalloproteinases (MMPs) are of crucial importance for invasive cancer cells to break ECM barriers (Figure 3). Within the metzincin superfamily, which also includes astacins, reprolysins, meprins and ADAMs, MMPs form a family of zinc- and calcium-dependent endopeptidases that can cleave all BM and ECM molecules [150]. Of the 28 MMPs identified in vertebrates, 24 are found in humans, including two MMP-23 isoforms encoded at different loci (MMP-23A and MMP-23B) [151,152,153]. The MMP family members can be divided according to their sequence similarity, domain organization and substrate specificity, into (i) collagenases, (ii) gelatinases, (iii) stromelysins, (iv) matrilysins, (v) transmembrane type I, (vi) transmembrane type II, (vii) glycosylphosphatidylinositol-anchored (GPI-anchored), and (viii) other MMPs (Figure 3) [9,150].
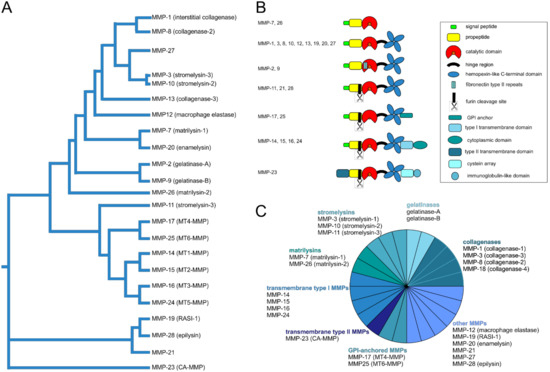
Figure 3.
Phylogenetic and functional relationship of human matrix metalloproteinases (MMPs). In addition to the numbering of the MMPs, which corresponds to the order of their discovery, the MMPs can be ordered according to their sequence similarity [154,155] (A). Regarding their domain organization (B) and substrate specificity they can be assigned to different groups within the MMP family (C): soluble collagenases, gelatinases, stromelysins, matrilysins, membrane-anchored transmembrane type I and type II as well as glycosylphosphatidylinositol (GPI)-anchored MMPs, and other MMPs [152]. All MMPs except MMP-23 have a propeptide with an N-terminal signal sequence. To activate an MMP, this propeptide has to be cleaved off in order to make a zinc ion in the active site of the catalytic domain, which also holds calcium ions, accessible through a cysteine switch. CA-MMP, cysteine array matrix metalloproteinase; RASI-1, rheumatoid arthritis synovium inflamed-1.
In addition to their structural and functional similarities, the individual MMPs are characterized by their distinct domain structure [137,152]: equipped with an N-terminal signal and a propeptide, MMPs have a catalytic metalloproteinase domain that is linked by a hinge region to a hemopexin domain. This central catalytic domain contains a catalytically active zinc ion chelated by three histidine residues in the binding motif HEXXHXXGXXH within the active center of the catalytic domain. Another zinc ion and two to three calcium ions within the catalytic domain have structural roles and account for substrate specificity. In addition to these characteristics common to all MMPs, MMPs have individual structural features. While gelatinases are characterized by three type II fibronectin repeats in their catalytic domain, matrilysins have neither a hinge region nor a hemopexin domain. Furin-dependent MMPs have a furin-like pro-protein convertase recognition sequence at the C-terminus of their propeptide. Unlike the soluble MMPs, membrane-bound MMPs of the membrane type (MT-MMPs) have an additional transmembrane domain and a cytosolic domain that consists of the case of MMP-23 of a cysteine-rich region and an immunoglobulin-like domain, or they are attached to the membrane with a glycosylphosphatidylinositol (GPI) anchor. The transmembrane domain is usually located towards the C-terminus with the only exception of again MMP-23, which instead of a propeptide, has an N-terminal type II transmembrane domain, which contains the sequence ALCLLPA instead of the consensus motif PRCGXPD that keeps MMP-23 it in its latent proMMP form.
proMMPs are activated to MMPs via limited proteolysis by trypsin, other MMPs, plasmin or furin-like convertases. By splitting off the propeptide, the latent proMMP is converted into its active form by means of a cysteine switch, as the removal of the cysteine sulfhydryl group in the sequence PRCGXPD, or PRCGVTD for MMP-28, of the propeptide, opens the catalytic site for the substrate molecule. Alternatively, some MMPs are also allosterically activated by a substrate molecule binding to a so-called MMP exosite outside the catalytic domain, or by chemicals, such as ROS, that affect the binding of the catalytic zinc ion to the cysteine thiol group of the cysteine switch [156].
5.3. MMP-14 Decisively Controls Tumor Progression
Only a few MMPs (MMP-1, -8, -13, and -14) are capable of cleaving triple helical collagen. Membrane-bound MT-MMPs with a C-terminal transmembrane domain can shed membrane proteins and cleave pericellular ECM components. Among them, MMP-14 is the only collagenolytic membrane-bound MMP [141]. It is expressed by various cell types, e.g., ECs and adipocytes [157], and moreover, it is of key importance on cancer cells [158,159,160]. Its expression correlates with their metastatic potential and is an important prognostic marker, e.g., in breast cancer [141,143,161]. In addition, CAFs express MMP-14 and contribute to invasion and metastasis in a murine breast cancer model [162]. Consequently, MMP-14 decisively controls collagen turnover and the breaking of ECM barriers in tumor progression. Its deficiency cannot be compensated for by other MMPs, neither in physiological developmental processes nor in cancer progression. In mice, MMP-14 ablation results in delayed ossification, decreased angiogenesis, severe fibrosis, and early lethality, and MMP-14 silencing with siRNA effectively reduces cancer cell invasion as does the proteolytic removal of the MMP-14 ectodomain from its transmembrane domain [159,160,163]. Conversely, the overexpression of MMP-14 promotes the invasiveness of cancer cells in a collagen-rich environment [158]. The MMP-14-cleavable substrates comprise fibril-forming collagen types I, II and III and other ECM proteins, in particular BM laminins and laminin-332 ectopically expressed in the tumor stroma [141]. In addition, MMP-14 can release membrane-bound cell and matrix receptors such as E-cadherin [164], syndecan-1, and the hyaluronan-receptor CD44 [165] from the cell surface. The outstanding importance of MMP-14 is further underpinned by its ability to activate the zymogen forms of the soluble gelatinases MMP-2 and -9 as well as the collagenolytic MMP-13.
Another yet non-proteolytic activity of MMP-14 that is of particular interest in the context of hypoxic TME is the ability of MMP-14 to activate hypoxia-inducible factors (HIFs) via Munc18-1-interacting protein 3 (Mint3) and factor inhibiting HIF-1 (FIH-1), thus promoting the expression of HIF target genes and the Warburg effect ([166] and references therein).
5.4. Regulation of MMP Expression
MMPs play a central role in the matrix-degrading enzyme activity that is associated with cancer metastasis and secondary tumor development. In most cases, there are no underlying gene amplifications or activating mutations. Instead, the MMP activity is dysregulated [167,168].
To prevent accidental proteolysis, the gene expression of MMPs is strictly controlled at intracellular and extracellular levels by cell–matrix and intercellular interactions and the input of various growth factors, glucocorticoids, cytokines, retinoic acid, interleukins and eicosanoids [169,170]. All of these factors induce the expression of MMPs via, for example, NFκB, MAPK and JAK/STAT signaling ([171] and references therein). There is some degree of co-regulation, as several MMP promoters share structural features [169]. Notably, the expression of MMPs-2, -14, and -28 is less responsive than that of others to cytokines and growth factors [9]. Nonetheless, MMP-14 in particular, with its diverse functions, must be precisely regulated (Figure 4). Accordingly, MMP-14 is subject to epigenetic control by histone modification, chromatin remodeling and DNA methylation-sensitive transcription factors such as SP1 [169,172,173]. The methylation status of MMP-14 and MMP-2 promoters inversely correlates with gene expression and cell migration in vitro, with hypomethylated promoters and undermethylated histone H3 being associated with high expression levels of MMP-14 and MMP-2 [172].
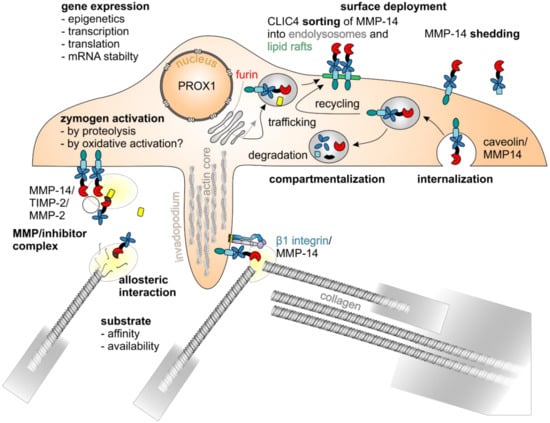
Figure 4.
MMP-14 is regulated by various mechanisms. The enzymatic activity of MMP-14 is regulated by various mechanisms: at the level of gene expression, there are epigenetic [172], transcriptional, and posttranscriptional regulatory mechanisms, such as the cotranslational cleavage of its signal sequence at the endoplasmatic reticulum, the furin-mediated removal of its self-inhibitory prodomain within the Golgi compartment [174], the O-glycosylation of protease-sensitive linker regions, and the phosphorylation as well as palmitoylation of its cytoplasmic domain [175,176], with the transcription factor PROX1 playing an important role [157]. Zymogen activation [156], compartmentalization, surface deployment and internalization [177,178], sorting into lipid rafts [179], homodimerization and interaction with other proteins, such as TIMPs [180], integrin β1 [181], and substrates, as well as shedding [182,183] are closely interlinked.
MMP-14 expression is also tightly regulated at the transcriptional level. The MMP-14 promoter has at least five different transcription start sites and differs from that of other MMPs in that it does not have a TATA box (Goldberg Hogness box), but has a functional, yet unconventional, binding site for the Sp1 transcription factor [184]. Moreover, binding sites for the activating transcription factors HIF-2α, Egr1, SP1, as well as E2F1, -3 and -5 have been found within the MMP-14 promoter, all of which are associated with increased malignancy in the context of various cancers ([168] and references therein). The collagenous microenvironment of the TME and mechanical forces increase the level of the transcription factor Egr1, resulting in an increased expression of MMP-14 [185,186]. Notably, the collagen receptor α2β1 integrin induces MMP-14 expression in fibroblasts and breast cancer cells [187,188]. Although all 23 human MMP gene promoters have an E2F binding site, only MMP-9, -14, and -15 respond to this transcription factor [189]. Crucially important for the regulation of cancer cell invasion, there is a repressive regulatory PROX1 binding site in front of the main transcription start site [157,184,190].
A decreased expression of the serine/threonine kinase D1 correlates with the invasiveness of breast cancer cells and negatively regulates MMPs -2, -7, -9, -10, -11, -13, -14, and -15 [191,192]. In invasive breast cancer, it is epigenetically shut down by DNA methylation, which induces MMP14 and other MMPs [191].
The synthesis of MMPs is also regulated at the post-transcriptional level by stabilization and destabilization, respectively, of their mRNAs with trans-acting RNA-binding proteins and microRNAs ([9] and references therein).
5.5. Regulation of MMP Activity
In addition to MMP activation from inactive zymogen forms, there are diverse other ways of regulating their activity. The proteolytic activity of MMPs as well as ADAMs and ADAMTSs is tightly controlled by the tissue inhibitors of metalloproteinases (TIMPs) that can form 1:1 stoichiometric complexes with MMPs, in which either the N-terminal domain of the TIMP chelates the catalytic zinc ion in the active site of the MMP, thus inactivating it, or in which the C-terminal domain of the TIMP interacts with the hemopexin domain to activate the MMP [193,194,195]. In particular, TIMP-1 is involved in pro-MMP-9/TIMP-1/MMP-3 complex formation to activate MMP-9 [196]. Similarly, TIMP-2 is involved in formation of a proMMP-2/TIMP-2/MMP-14 complex resulting in the activation of proMMP-2 [159]. Two MMP-14 molecules dimerize on the cell surface, whereupon the N-terminal domain of TIMP-2 binds to the catalytic center of one MMP-14, thus facilitating the interaction of the hemopexin domain of proMMP-2 with the C-terminal domain of TIMP-2 to orient the proMMP-2 in such a way that the second MMP-14 can cleave it and release the active MMP-2 [180]. This type of pro-MMP-2 activation occurs particularly in the invadopodia of invading neoplastic cells. Moreover, the homodimerization of two MMP-14 molecules on the cell surface enhances their collagenolytic activity [141]. This is also regulated by interactions with other membrane proteins, such as integrins, CD44, chondroitin/heparin sulfate proteoglycans, and tetraspanins, as well as pericellular MMP-14-inhibiting proteins like TIMP-2, -3, -4, and RECK (reversion-inducing-cysteine-rich protein with Kazal motifs) [141,197]. The invasiveness of cancer cells is likewise regulated by the lifespan of MMP-14, which depends on the pattern of its O-glycosylation, and by the internalization of MMP-14 and the subsequent degradation or recycling to the cell surface [141,198].
In order to enable focalized pericellular ECM proteolysis typical of invading cancer cells, integrin-mediated contacts with the collagen-rich ECM of the tumor stroma bring about a directed transport and fusion of MMP-14-containing vesicles in invadopodia and invasive cell fronts [19,140,143,197]. In particular, caveolin-1 negatively regulates MMP-14 on the cell surface by promoting its internalization [177]. Endocytosed by a flotillin- and Rab5-dependent mechanism and safely stored in intracellular vesicles, such as endolysosomes, activated MMP-14 can be quickly deployed to the cell surface when needed [199,200,201]. The endoplasmic reticulum protein protrudin mediates contact with endosomes containing MMP-14 and the Rab7-binding kinesin adapter protein FYCO1 and facilitates the translocation of MMP-14-containing endosomes to the plasma membrane [202]. The endosomal trafficking of MMP-14 is furthermore regulated by the chloride intracellular channel 4 (CLIC4) which not only binds to the endosomal sorting complex required for transport (ESCRT) but also to proMMP-14 promoting its proteolytic activation in lipid rafts [179].
It has recently been shown that β1 integrin-mediated Src-EGFR signaling regulates MMP-14 phosphorylation and thus the recycling of MMP-14 to sites of invadopodia formation during the invasion of cancer cells [181]. However, it is not yet fully understood, how collagen-binding integrins regulate the expression and activity of MMP-14, and how the topography and biophysical properties of supramolecular collagen affects MMP-14-mediated cancer cell invasion. In fibroblasts migrating through dense collagen, MMP-14 is apparently activated by associating with collagen-binding β1 integrins, especially α2β1, which bring collagen to MMP-14 for proteolytic cleavage [19,197,203]. By this association, proteolytically active MMP-14 may also be kept longer on the cell surface [143]. On the other hand, adhesion- and integrin-mediated signaling via focal adhesion kinase (FAK) and Src can phosphorylate the cytoplasmic tail of MMP-14 and caveolin-1, thereby triggering clathrin- and caveolin-dependent internalization and recycling of MMP-14 [140].
Autocatalytic and non-autocatalytic shedding of the MMP-14 ectodomain from the cell surface yielding either catalytically inactive fragments or soluble catalytically active ectodomains also seems to be a way to regulate its activity and may be important in cancer progression [182,183,204].
MMPs have also been found in mitochondria and in the cell nucleus ([9] and references therein). In hepatocellular carcinoma in particular, MMPs-2 and -14 have been detected inside cell nuclei, although their role there is still unclear [205,206].
5.6. Invasive Cancer Cells Breach ECM Barriers with Invadopodia as Drill Heads
Cancer cells form various adhesome structures to interact with their surrounding matrix. Of particular interest for metastasis are special adhesomes termed invadopodia which, in addition to adhesion-promoting integrins, have matrix-degrading enzymes at their proteolytic tip [28,207,208]. In addition to the basement membrane, the dense arrangement of fibrillar collagens, as in the fibrous capsules of desmoplastic tumors, represents an obstacle to the spread of tumor cells (Figure 2) [209]. Integrins, the expression of which is altered in many tumors, play a key role as the most important ECM receptors in the formation of invadosomes. [207,210]. In particular, collagen-binding β1 integrins, but also β3 integrins are involved [181,211]. When invasive cancer cells encounter such rigid collagen fibrils, they break them down similarly to basement membranes using invadopodia [143,212]. MMP-14 resides in such invadopodia and is essential for breaching the ECM barrier and invading the stroma [137,140,159,207]. Invadopodia are spatially complex structures formed in cancer cells under the influence of the TME, whereby in addition to TGF-β, HGF and epidermal growth factor (EGF), hypoxia in particular stimulates their formation [140,213,214,215,216]. Here, the receptors LPA1R and EGFR cooperate to promote invadopodia formation via Src and PI3K/Akt signaling [217]. Likewise, via EGFR, the prostaglandin PGE2 receptor EP4 promotes the invadopodia-mediated ECM degradation [218]. Conversely, the tumor suppressor nischarin that interacts with multiple signaling molecules, such as Rac1, LIM kinase (LIMK), liver kinase B1 (LKB1), p21 protein (Cdc42/Rac)-activated kinase 1 (PAK1), Rab14 and insulin receptor substrates 1-4, inhibits the formation of invadopodia and the expression of the integrin subunit α2, while it upregulates subunits α1, α4, and α7 [219]. The strongest stimulus for invadopodia formation is, however, the mechanical stiffness of the ECM, and high collagen density stimulates the incorporation of MMP-14 into tumor cell invadopodia [140,220]. It is sensed by integrins in a Rho- and WASP/WAVE-dependent manner and results in a Rac-, PAK1- and cortactin-dependent formation of the invadopodia’s core structure [94,140,214,221,222,223,224]. At moderate ECM stiffness, cells show rapid invadopodia protrusion–retraction cycles with maximum invadopodia-mediated ECM degradation [225]. The dynamics of invadopodia can also be controlled by the endothelin A receptor ETAR via RhoC and cofilin with the help of the scaffold protein β-arrestin1 as a signal-integrating module [216]. F-actin in the core structure is bundled by actinin-1 and -4, with actinin-4 in particular promoting the formation of invadopodia [226]. This core structure of F-actin bundles is further stabilized by recruited signaling molecules such as PAK-1 and -4, thus increasing both the lifespan of the invadopodium and its depth of penetration into the ECM [140,214,227,228]. Other scaffolding proteins stabilize the core structure. FBP17 from the family of F-BAR proteins, which regulate membrane dynamics, is associated with actin-regulatory proteins, such as cortactin, dynamin and ARP2/3 [229]. Similar to cortactin, coronin 1C is required for the formation of invadopodia, but in contrast, it is also involved in the regulation of intracellular endolysosomal transport and the distribution of MMP-14 [230]. By additionally recruiting Tks-4 and -5, invadopodia remain stable for more than 60 min, which distinguishes them from otherwise structurally and functionally similar podosomes of angiogenic ECs that sprout into the tumor mass during tumor-induced angiogenesis [231,232,233]. Invadopodia and podosomes, collectively called invadosomes, both have the proteolytic activity of MMP-14, which enables penetration into the ECM and also activates the soluble gelatinase MMP-2 [140]. In addition to MMP-14, cancer cells have proteinases of the ADAMTS family, which are structurally homologous and likewise membrane-anchored, with which they also can degrade cell migration barriers [234]. Simulations with multiple invadopodia suggest that the distance between individual invadopodia and the secretion rate of soluble MMPs together determine the extent of the ECM degradation. Hence, for invasion cancer cells must fine-tune the inter-invadopodia distance and the secretion rate of soluble MMPs corresponding to the density of the ECM [235].
5.7. ECM Degradation and Remodeling Releases Bioactive Matrikines
MMPs are not only essential for ECM remodeling by cleaving structural macromolecules, but they also regulate the release and activation of growth factors, chemokines, cytokines, and adhesion molecules [156]. Furthermore, MMPs may affect the redox conditions in the tumor microenvironment and vice versa, as, for example, MMP-2 and MMP-9 are upregulated by NOX/ROS-dependent NFκB activity [236,237], whereas MMP-3 activation induces mitochondrial ROS production and NADPH oxidase 1 (Nox1) [238]. While the loss of contact with the ECM changes the cellular redox equilibrium leading to apoptosis, cancer cells can recalibrate their redox equilibrium in order to survive and metastasize [239]. Associated with ECM degradation, MMPs proteolytically release from many insoluble structural ECM components soluble bioactive fragments with anti-metastatic properties, referred to as matrikines [240,241,242] (Table 1). Oddly enough, there can be a large number of circulating tumor cells while only one to a few metastases develop, which is due to the release of such antimetastatically acting matrikines produced by proteolytic processing of ECM components in the TME [28,58,243].

Table 1.
Matrix proteins of the TME can release bioactive peptides called matrikines, which regulate tumor progression and metastasis, and can be used diagnostically.
The entire TME is significantly influenced by such matrikines which are released by various proteases from insoluble ECM molecules (Table 1). For example, defined fragments of basement membrane collagen types IV, XV, XVIII and XIX, which are split off by infiltrating cancer cells [295], act on the one hand on the cancer cells, and on the other hand, have an angiostatic effect by reducing the sprouting of ECs into the tumor mass [50,240,276,286]. In addition, endostatin can reverse the immunosuppressive environment [102,404], and versicine, a matrikine derived from versican, causes the selective recruitment of certain dendritic cells into the tumor stroma [326]. Furthermore, endorepellin, a fragment of the basement membrane proteoglycan perlecan, can inhibit angiogenesis by interaction with integrin α2β1 on ECs [85,86]. On the other hand, it has been reported that fragments of several matricellular proteins and laminin-332 promote the motility of cancer cells by binding agonistically to the EGF receptor [241,302]. In addition, elastin peptides also act as matrikines and show a broad spectrum of biological activities [300,301,405].
5.8. MMPs Promote Epithelial–Mesenchymal Transition
Signals generated by ECM remodeling and degradation play a crucial role in the EMT process during tumor progression by causing numerous structural and functional changes, such as loss of cell polarity and tight intercellular contacts, the production of mesenchymal proteins, and acquisition of an invasive phenotype [406]. In addition to releasing signal-triggering matrikines and breaking ECM barriers, MMPs can proteolytically cleave members of the protease-activated receptor (PAR) family. In particular, the extracellular N-terminus of PARs, such as PAR-1 and PAR-3, which are expressed by cancer cells and also CAFs, can be canonically cleaved by thrombin and also non-canonically by certain MMPs, such as MMP-1 and MMP-13 [407,408,409]. Canonically, thrombin is secreted by activated monocytes/macrophages in the tumor stroma and activated by the extrinsic coagulation cascade that is triggered by the tissue factor (TF) that is usually expressed on cancer cells [410]. Non-canonically, MMPs proteolytically activate the Gα12/13 of the heterotrimeric G protein and thus Rho signaling. This Rho signaling increases cell contractility and cell movement via the actomyosin machinery and its motor protein myosin II, thus stimulating invasion through the ECM barrier. In addition, activated Rho promotes the EMT of cancer cells that are still connected to one another via cadherins. Cadherins can also be cleaved by MMP-14 [164,407,408,411]. Carcinoma cells that invade as coherent cohorts rather than individual cells are interconnected by homophilically interacting E-cadherin [161]. Its loss through cleavage by MMP-14 leads to their spread and changes in their cell morphology, which are characteristic of EMT [164,412].
6. Translational Assessment and Future Prospect
To monitor tumor progression and staging, the diagnostic palpatory examination of the correlating tissue stiffening can be refined microscopically histologically and with other imaging methods, such as magnetic resonance imaging (MRI), positron emission tomography (PET), single photon emission computer tomography (SPECT), computed tomography (CT); and ultrasound (US) [413]. For example, changes typical of tumor stroma, such as the appearance of the fibronectin splice variants ED-A or ED-B, laminin-332, periostin and tenascin-W, can be histochemically analyzed in tissue biopsies ([28] and references therein). This allows a prognosis to be made and suggests that such tumor markers may be used for drug development. The small leucine-rich proteoglycan lumican, for instance, inhibits and even reverses some of the characteristics of EMT, such as invadopodia formation, suggesting lumican as a marker for staging of several cancers and the development of cancer drugs based on lumican [414,415]. However, the use of such ECM components as antigenic targets to direct anti-tumor agents to the tumor site has not yet been successful beyond the experimental level, and therefore therapeutics that directly target ECM structural components have to be awaited [416,417].
In addition to structural TME components, ECM receptors can be envisioned as target structures for the pharmacological blocking of tumor progression and metastasis [418,419,420]. Moreover, the inhibitors of MMP-activated PARs that increase metastatic invasion of cancer cells have been investigated in clinical studies [407].
ECM-derived peptides, which are released by enzymes of the TME, are also important not only for diagnostic, prognostic and predictive purposes and as possible therapeutic target structures, but also as lead structures for potential antimetastatic drugs. Their use as antimetastatic therapeutics has been examined in detail in a number of excellent reviews [58,83,85,243,307,419,421,422,423,424,425,426]. Since some of the typical tumor stroma proteins and fragments thereof can be detected diagnostically in blood samples, such as laminin-γ2 chains [89,359], easily accessible and reliable diagnostic, prognostic, and predictive tumor markers may be developed from them.
In addition to structural TME components, the enzyme activities within the TME also represent potential approaches for cancer therapy. Pharmacologically targeting ECM-modifying enzymes such as lysyl oxidase-like LOXLs and MMPs was more successful to date, and some strategies even made it to the stage of clinical trials [129,131,144,427]. In a phase II study, lysyl oxidase activity, involved in forming a premetastatic niche in triple-negative breast cancer, was successfully targeted with copper-scavenging tetrathiomolybdate [428,429]. However, then, simtuzumab, a function-blocking antibody against LOXL2 with an antidesmoplastic effect in vitro, did not improve the clinical results in patients with KRAS-mutated colorectal or pancreatic adenocarcinoma [430,431,432,433].
In recent years, not only the implications of MMPs in cancer progression but also novel strategies for improved targeting and delivery as well as for regulation of MMP activity in tumors have been extensively reviewed [425,434,435]. Inhibitors of ECM-modifying enzymes generally have only a limited selectivity and accordingly show undesirable side effects. Notably, therapeutic strategies for MMP targeting are hampered by limited selectivity. For example, the broad-spectrum MMP inhibitor marimastat, although more bioavailable than its analogue batimastat, turned out unsuitable in phase III studies, since it caused pain and inflammation of the musculoskeletal system [436,437]. Moreover, the development of the low molecular weight inhibitor CGS 27023A/MMI270 targeting MMPs -2, -8, and -9 was stopped early in phase II studies for the treatment of non-small cell lung carcinoma because of poorly tolerated joint and muscle pain [438]. Several new MMP inhibitors with improved properties are currently being investigated ([425] and references therein).
Targeting MMPs therapeutically is obvious, as they are significantly involved in all stages of the metastatic cascade. In particular, inhibiting cancer cell-derived MMPs could increase the stability of the endogenous ECM barrier and thus prevent cancer cells from breaking the basement membrane, which in turn would reduce or even suppress metastases [439,440]. In order to precisely identify tumor tissue during tumor surgery, an MMP-cleavable FRET probe that is internalized by cancer cells has been developed [441]. Elevated serum levels of soluble MMP-14 are a marker for poor prognosis and possibly indicate the presence of distant metastases [442]. In addition, the inhibition of MMP-14 as master MMP is interesting from a therapeutic point of view. A monoclonal antibody against MMP-14 selectively blocks proMMP-2 activation which is required for lymphangiogenesis in vitro and ex vivo [443]. In preclinical models, several monoclonal antibodies against MMP-9 [444,445] and MMP-14 [446] initially appeared promising, but the generation of highly selective MMP antibody inhibitors remains a desideratum [447]. Phage display library-derived peptides against the MT loop of MMP-14, such as HS7, which may be useful in the early diagnosis of tumors and for the development of peptide-mediated drugs [448]. Using a yeast display approach, the rather non-specific MMP inhibitor N-TIMP-2 could be converted into a specific inhibitor that blocks MMP-9 activity thousand-fold more than MMP-14 activity [449].
In view of the recently enormously growing understanding of gene regulation in TME, regulatory RNAs appear attractive, since diagnostic, prognostic and predictive biomarkers could be found in both microRNAs and long non-coding RNAs [450,451]. In particular, microRNA-mediated post-transcriptional MMP regulation is of interest for tumor therapy [452]. For example, miR-181a-5p can downregulate MMP-14 and thereby inhibit the migration and angiogenesis of cancer cells [453]. Similarly, miR139-5p and miR203 slow down processes involving the gelatinase MMP-9 and the matrilysin MMP-7, respectively [454,455]. Other microRNAs, such as miR192 and miR140-5p are involved in the upregulation of MMP-2 [456] and ADAMTS5 [457], respectively, suggesting the development and use of respective antagomiRs. It is also conceivable to inhibit oncomiRs, e.g., miR-21 in order to upregulate the expression of MMP-regulating proteins, such as RECK or TIMP-3, to reduce MMP activity [458].
Whatever the methodological approach, attempts to normalize the dysregulated TME and its ECM in a non-tumor-supporting environment [459] and to prevent the tumor-induced formation of premetastatic niches [460,461] are, in any case, goals of prime importance.
Author Contributions
S.N. and J.A.E. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Deutsche Forschungsgemeinschaft (DFG grant SFB1009 project A09).
Acknowledgments
The authors sincerely apologize to the authors of important works not cited here for reasons of space limitations.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Abbreviations
| ADAM(TS) | a disintegrin and a metalloproteinase (with thrombospondin-1 motif) |
| BM | basement membrane |
| CAF | cancer-associated fibroblast |
| CAM | cell adhesion molecule |
| CCN | connective tissue growth factor (CTGF), cysteine-rich protein (Cyr61) and nephroblastoma overexpressed (NOV) |
| EC | endothelial cell |
| ECM | extracellular matrix |
| ED-A/B | extra domain-A/B |
| EGF(R) | epidermal growth factor (receptor) |
| EMT | epithelial–mesenchymal transition |
| GAG | glycosaminoglycan |
| GPI | glycosylphosphatidylinositol |
| HGF | hepatocyte growth factor |
| HIF | hypoxia-inducible factor |
| H/LMW-HA | high/low molecular weight hyaluronic acid |
| LOXL | lysyl oxidase-like |
| LPA1R | lysophosphatidic acid receptor 1 |
| (MT-)MMP | (membrane-type) matrix metalloproteinase |
| NOX | nicotinamide adenine dinucleotide phosphate (NADPH) oxidase |
| PAR | protease-activated receptor |
| RHAMM | receptor for hyaluronic acid-mediated migration |
| ROS | reactive oxygen species |
| SIBLING | small integrin-binding ligand N-linked glycoprotein |
| SLRP | small leucine-rich protein |
| SPARC | secreted protein acidic and rich in cysteine |
| TC | tumor cell |
| TGF-β | transforming growth factor- β |
| TIMP | tissue inhibitors of metalloproteinases |
| TLR | Toll-like receptor |
| TME | tumor microenvironment |
| VEGF | vascular endothelial cell growth factor |
References
- He, Q.; Chen, J.; Yan, J.; Cai, S.; Xiong, H.; Liu, Y.; Peng, D.; Mo, M.; Liu, Z. Tumor microenvironment responsive drug delivery systems. Asian J. Pharm. Sci. 2020, 15, 416–448. [Google Scholar] [CrossRef]
- Emon, B.; Bauer, J.; Jain, Y.; Jung, B.; Saif, T. Biophysics of Tumor Microenvironment and Cancer Metastasis—A Mini Review. Comput. Struct. Biotechnol. J. 2018, 16, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, H.; Sahai, E. Mechanisms and impact of altered tumour mechanics. Nat. Cell Biol. 2018, 20, 766–774. [Google Scholar] [CrossRef] [PubMed]
- Iozzo, R.V.; Schaefer, L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015, 42, 11–55. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, A.D.; Skandalis, S.S.; Gialeli, C.; Karamanos, N.K. Extracellular matrix structure. Adv. Drug Deliv. Rev. 2016, 97, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, J.; Deng, X.; Xiong, F.; Zhang, S.; Gong, Z.; Li, X.; Cao, K.; Deng, H.; He, Y.; et al. The role of microenvironment in tumor angiogenesis. J. Exp. Clin. Cancer Res. 2020, 39, 204. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wei, J.; Sun, J. Roles of TGF-beta signaling pathway in tumor microenvirionment and cancer therapy. Int. Immunopharmacol. 2020, 89, 107101. [Google Scholar] [CrossRef]
- Chitty, J.L.; Setargew, Y.F.I.; Cox, T.R. Targeting the lysyl oxidases in tumour desmoplasia. Biochem. Soc. Trans. 2019, 47, 1661–1678. [Google Scholar] [CrossRef]
- Gonzalez-Avila, G.; Sommer, B.; Mendoza-Posada, D.A.; Ramos, C.; Garcia-Hernandez, A.A.; Falfan-Valencia, R. Matrix metalloproteinases participation in the metastatic process and their diagnostic and therapeutic applications in cancer. Crit. Rev. Oncol. Hematol. 2019, 137, 57–83. [Google Scholar] [CrossRef]
- Amendola, P.G.; Reuten, R.; Erler, J.T. Interplay Between LOX Enzymes and Integrins in the Tumor Microenvironment. Cancers 2019, 11, 729. [Google Scholar] [CrossRef]
- Lentini, A.; Abbruzzese, A.; Provenzano, B.; Tabolacci, C.; Beninati, S. Transglutaminases: Key regulators of cancer metastasis. Amino Acids 2013, 44, 25–32. [Google Scholar] [CrossRef]
- Leight, J.L.; Drain, A.P.; Weaver, V.M. Extracellular Matrix Remodeling and Stiffening Modulate Tumor Phenotype and Treatment Response. Ann. Rev. Cancer Biol. 2017, 1, 313–334. [Google Scholar] [CrossRef]
- Martins Cavaco, A.C.; Damaso, S.; Casimiro, S.; Costa, L. Collagen biology making inroads into prognosis and treatment of cancer progression and metastasis. Cancer Metastasis Rev. 2020, 39, 603–623. [Google Scholar] [CrossRef] [PubMed]
- Bourgot, I.; Primac, I.; Louis, T.; Noel, A.; Maquoi, E. Reciprocal Interplay Between Fibrillar Collagens and Collagen-Binding Integrins: Implications in Cancer Progression and Metastasis. Front. Oncol. 2020, 10, 1488. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Xu, H.; Wang, W.; Li, S.; Li, H.; Li, T.; Zhang, W.; Yu, X.; Liu, L. The role of collagen in cancer: From bench to bedside. J. Transl. Med. 2019, 17, 309. [Google Scholar] [CrossRef] [PubMed]
- Ricard-Blum, S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011, 3, a004978. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Wolf, K.; Friedl, P. Extracellular matrix determinants of proteolytic and non-proteolytic cell migration. Trends Cell Biol. 2011, 21, 736–744. [Google Scholar] [CrossRef]
- Wolf, K.; Te Lindert, M.; Krause, M.; Alexander, S.; Te Riet, J.; Willis, A.L.; Hoffman, R.M.; Figdor, C.G.; Weiss, S.J.; Friedl, P. Physical limits of cell migration: Control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 2013, 201, 1069–1084. [Google Scholar] [CrossRef]
- Overall, C.M. Matrix metalloproteinase substrate binding domains, modules and exosites. Overview and experimental strategies. Methods Mol. Biol. 2001, 151, 79–120. [Google Scholar]
- Aumailley, M. The laminin family. Cell Adh. Migr. 2013, 7, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Halfter, W.; Oertle, P.; Monnier, C.A.; Camenzind, L.; Reyes-Lua, M.; Hu, H.; Candiello, J.; Labilloy, A.; Balasubramani, M.; Henrich, P.B.; et al. New concepts in basement membrane biology. FEBS J. 2015, 282, 4466–4479. [Google Scholar] [CrossRef] [PubMed]
- Hohenester, E.; Yurchenco, P.D. Laminins in basement membrane assembly. Cell Adh. Migr. 2013, 7, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Cavaco, A.C.M.; Rezaei, M.; Caliandro, M.F.; Lima, A.M.; Stehling, M.; Dhayat, S.A.; Haier, J.; Brakebusch, C.; Eble, J.A. The Interaction between Laminin-332 and alpha3beta1 Integrin Determines Differentiation and Maintenance of CAFs, and Supports Invasion of Pancreatic Duct Adenocarcinoma Cells. Cancers 2018, 11, 14. [Google Scholar] [CrossRef]
- Wang, Y.; Song, E.C.; Resnick, M.B. Elastin in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1272, 1–16. [Google Scholar] [CrossRef]
- Muiznieks, L.D.; Weiss, A.S.; Keeley, F.W. Structural disorder and dynamics of elastin. Biochem. Cell. Biol. 2010, 88, 239–250. [Google Scholar] [CrossRef]
- Heinz, A. Elastases and elastokines: Elastin degradation and its significance in health and disease. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 252–273. [Google Scholar] [CrossRef]
- Eble, J.A.; Niland, S. The extracellular matrix in tumor progression and metastasis. Clin. Exp. Metastasis 2019, 36, 171–198. [Google Scholar] [CrossRef]
- White, E.S.; Muro, A.F. Fibronectin splice variants: Understanding their multiple roles in health and disease using engineered mouse models. IUBMB Life 2011, 63, 538–546. [Google Scholar] [CrossRef]
- Lin, T.C.; Yang, C.H.; Cheng, L.H.; Chang, W.T.; Lin, Y.R.; Cheng, H.C. Fibronectin in Cancer: Friend or Foe. Cells 2019, 9, 27. [Google Scholar] [CrossRef]
- Efthymiou, G.; Saint, A.; Ruff, M.; Rekad, Z.; Ciais, D.; Van Obberghen-Schilling, E. Shaping Up the Tumor Microenvironment With Cellular Fibronectin. Front. Oncol. 2020, 10, 641. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lee, Y.J.; Han, H.J. Role of hypoxia-induced fibronectin-integrin beta1 expression in embryonic stem cell proliferation and migration: Involvement of PI3K/Akt and FAK. J. Cell. Physiol. 2011, 226, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Ryu, M.H.; Park, H.M.; Chung, J.; Lee, C.H.; Park, H.R. Hypoxia-inducible factor-1alpha mediates oral squamous cell carcinoma invasion via upregulation of alpha5 integrin and fibronectin. Biochem. Biophys. Res. Commun. 2010, 393, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Sollini, M.; Boni, R.; Traino, A.C.; Lazzeri, E.; Pasqualetti, F.; Modeo, L.; Mariani, G.; Petrini, M.; Erba, P.A. New approaches for imaging and therapy of solid cancer. Q. J. Nucl. Med. Mol. Imaging 2015, 59, 168–183. [Google Scholar]
- Degen, M.; Brellier, F.; Kain, R.; Ruiz, C.; Terracciano, L.; Orend, G.; Chiquet-Ehrismann, R. Tenascin-W is a novel marker for activated tumor stroma in low-grade human breast cancer and influences cell behavior. Cancer Res. 2007, 67, 9169–9179. [Google Scholar] [CrossRef]
- Midwood, K.S.; Orend, G. The role of tenascin-C in tissue injury and tumorigenesis. J. Cell Commun. Signal. 2009, 3, 287–310. [Google Scholar] [CrossRef]
- Scherberich, A.; Tucker, R.P.; Degen, M.; Brown-Luedi, M.; Andres, A.C.; Chiquet-Ehrismann, R. Tenascin-W is found in malignant mammary tumors, promotes alpha8 integrin-dependent motility and requires p38MAPK activity for BMP-2 and TNF-alpha induced expression in vitro. Oncogene 2005, 24, 1525–1532. [Google Scholar] [CrossRef]
- Brellier, F.; Tucker, R.P.; Chiquet-Ehrismann, R. Tenascins and their implications in diseases and tissue mechanics. Scand. J. Med. Sci. Sports 2009, 19, 511–519. [Google Scholar] [CrossRef]
- Oskarsson, T.; Acharyya, S.; Zhang, X.H.; Vanharanta, S.; Tavazoie, S.F.; Morris, P.G.; Downey, R.J.; Manova-Todorova, K.; Brogi, E.; Massague, J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat. Med. 2011, 17, 867–874. [Google Scholar] [CrossRef]
- Degen, M.; Brellier, F.; Schenk, S.; Driscoll, R.; Zaman, K.; Stupp, R.; Tornillo, L.; Terracciano, L.; Chiquet-Ehrismann, R.; Ruegg, C.; et al. Tenascin-W, a new marker of cancer stroma, is elevated in sera of colon and breast cancer patients. Int. J. Cancer 2008, 122, 2454–2461. [Google Scholar] [CrossRef]
- Spenle, C.; Saupe, F.; Midwood, K.; Burckel, H.; Noel, G.; Orend, G. Tenascin-C: Exploitation and collateral damage in cancer management. Cell Adh. Migr. 2015, 9, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Kii, I.; Nishiyama, T.; Li, M.; Matsumoto, K.; Saito, M.; Amizuka, N.; Kudo, A. Incorporation of tenascin-C into the extracellular matrix by periostin underlies an extracellular meshwork architecture. J. Biol. Chem. 2010, 285, 2028–2039. [Google Scholar] [CrossRef] [PubMed]
- Girotti, M.R.; Salatino, M.; Dalotto-Moreno, T.; Rabinovich, G.A. Sweetening the hallmarks of cancer: Galectins as multifunctional mediators of tumor progression. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef] [PubMed]
- Thakur, R.; Mishra, D.P. Matrix reloaded: CCN, tenascin and SIBLING group of matricellular proteins in orchestrating cancer hallmark capabilities. Pharmacol. Ther. 2016, 168, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Leask, A. Conjunction junction, what’s the function? CCN proteins as targets in fibrosis and cancers. Am. J. Physiol. Cell Physiol. 2020, 318, C1046–C1054. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Birk, D.E. The regulatory roles of small leucine-rich proteoglycans in extracellular matrix assembly. FEBS J. 2013, 280, 2120–2137. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Li, J.P. Heparan sulfate proteoglycan—A common receptor for diverse cytokines. Cell Signal. 2019, 54, 115–121. [Google Scholar] [CrossRef]
- De Pasquale, V.; Pavone, L.M. Heparan Sulfate Proteoglycan Signaling in Tumor Microenvironment. Int. J. Mol. Sci. 2020, 21, 6588. [Google Scholar] [CrossRef]
- Mellai, M.; Casalone, C.; Corona, C.; Crociara, P.; Favole, A.; Cassoni, P.; Schiffer, D.; Boldorini, R. Chondroitin Sulphate Proteoglycans in the Tumour Microenvironment. Adv. Exp. Med. Biol. 2020, 1272, 73–92. [Google Scholar] [CrossRef]
- Iozzo, R.V.; Sanderson, R.D. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J. Cell. Mol. Med. 2011, 15, 1013–1031. [Google Scholar] [CrossRef]
- Weniger, M.; Honselmann, K.C.; Liss, A.S. The Extracellular Matrix and Pancreatic Cancer: A Complex Relationship. Cancers 2018, 10, 316. [Google Scholar] [CrossRef] [PubMed]
- Otranto, M.; Sarrazy, V.; Bonté, F.; Hinz, B.; Gabbiani, G.; Desmoulière, A. The role of the myofibroblast in tumor stroma remodeling. Cell Adh. Migr. 2012, 6, 203–219. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, F.M.; Vitale, D.L.; Sevic, I.; Alaniz, L. Hyaluronan in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1245, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Batlle, E.; Massague, J. Transforming Growth Factor-beta Signaling in Immunity and Cancer. Immunity 2019, 50, 924–940. [Google Scholar] [CrossRef]
- Ahmadi, A.; Najafi, M.; Farhood, B.; Mortezaee, K. Transforming growth factor-beta signaling: Tumorigenesis and targeting for cancer therapy. J. Cell. Physiol. 2019, 234, 12173–12187. [Google Scholar] [CrossRef]
- Itatani, Y.; Kawada, K.; Sakai, Y. Transforming Growth Factor-beta Signaling Pathway in Colorectal Cancer and Its Tumor Microenvironment. Int. J. Mol. Sci. 2019, 20, 5822. [Google Scholar] [CrossRef]
- Poltavets, V.; Kochetkova, M.; Pitson, S.M.; Samuel, M.S. The Role of the Extracellular Matrix and Its Molecular and Cellular Regulators in Cancer Cell Plasticity. Front. Oncol. 2018, 8, 431. [Google Scholar] [CrossRef]
- Brassart-Pasco, S.; Brezillon, S.; Brassart, B.; Ramont, L.; Oudart, J.B.; Monboisse, J.C. Tumor Microenvironment: Extracellular Matrix Alterations Influence Tumor Progression. Front. Oncol. 2020, 10, 397. [Google Scholar] [CrossRef]
- Najafi, M.; Farhood, B.; Mortezaee, K. Extracellular matrix (ECM) stiffness and degradation as cancer drivers. J. Cell. Biochem. 2018, 120, 2782–2790. [Google Scholar] [CrossRef]
- Bussard, K.M.; Mutkus, L.; Stumpf, K.; Gomez-Manzano, C.; Marini, F.C. Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res. 2016, 18, 84. [Google Scholar] [CrossRef]
- Sobierajska, K.; Ciszewski, W.M.; Sacewicz-Hofman, I.; Niewiarowska, J. Endothelial Cells in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1234, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.; Brehm, C.U.; Gress, T.M.; Buchholz, M.; Alashkar Alhamwe, B.; von Strandmann, E.P.; Slater, E.P.; Bartsch, J.W.; Bauer, C.; Lauth, M. The Immune Microenvironment in Pancreatic Cancer. Int. J. Mol. Sci. 2020, 21, 7307. [Google Scholar] [CrossRef] [PubMed]
- Sadeghalvad, M.; Mohammadi-Motlagh, H.R.; Rezaei, N. Immune microenvironment in different molecular subtypes of ductal breast carcinoma. Breast Cancer Res. Treat. 2020, 3, 1–19. [Google Scholar] [CrossRef]
- Truffi, M.; Sorrentino, L.; Corsi, F. Fibroblasts in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1234, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, D.; Chandra, R.; Karalis, J.; Teke, M.; Aguilera, T.; Maddipati, R.; Wachsmann, M.B.; Ghersi, D.; Siravegna, G.; Zeh, H.J., III; et al. Cancer-Associated Fibroblasts: Versatile Players in the Tumor Microenvironment. Cancers 2020, 12, 2652. [Google Scholar] [CrossRef] [PubMed]
- Mhaidly, R.; Mechta-Grigoriou, F. Fibroblast heterogeneity in tumor micro-environment: Role in immunosuppression and new therapies. Semin. Immunol. 2020, 48, 101417. [Google Scholar] [CrossRef] [PubMed]
- Leask, A. A centralized communication network: Recent insights into the role of the cancer associated fibroblast in the development of drug resistance in tumors. Semin. Cell Dev. Biol. 2020, 101, 111–114. [Google Scholar] [CrossRef]
- Bu, L.; Baba, H.; Yasuda, T.; Uchihara, T.; Ishimoto, T. Functional diversity of cancer-associated fibroblasts in modulating drug resistance. Cancer Sci. 2020, 111, 3468–3477. [Google Scholar] [CrossRef]
- Wang, M.; Zhao, J.; Zhang, L.; Wei, F.; Lian, Y.; Wu, Y.; Gong, Z.; Zhang, S.; Zhou, J.; Cao, K.; et al. Role of tumor microenvironment in tumorigenesis. J. Cancer 2017, 8, 761–773. [Google Scholar] [CrossRef]
- Gal, P.; Varinska, L.; Faber, L.; Novak, S.; Szabo, P.; Mitrengova, P.; Mirossay, A.; Mucaji, P.; Smetana, K. How Signaling Molecules Regulate Tumor Microenvironment: Parallels to Wound Repair. Molecules 2017, 22, 1818. [Google Scholar] [CrossRef]
- Noguchi, S.; Saito, A.; Nagase, T. YAP/TAZ Signaling as a Molecular Link between Fibrosis and Cancer. Int. J. Mol. Sci. 2018, 19, 3674. [Google Scholar] [CrossRef] [PubMed]
- Ouhtit, A.; Rizeq, B.; Saleh, H.A.; Rahman, M.M.; Zayed, H. Novel CD44-downstream signaling pathways mediating breast tumor invasion. Int. J. Biol. Sci. 2018, 14, 1782–1790. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Tolg, C.; Turley, E. Dissecting the Dual Nature of Hyaluronan in the Tumor Microenvironment. Front. Immunol. 2019, 10, 947. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.; Hascall, V.C.; Markwald, R.R.; Ghatak, S. Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer. Front. Immunol. 2015, 6, 201. [Google Scholar] [CrossRef] [PubMed]
- Tolg, C.; McCarthy, J.B.; Yazdani, A.; Turley, E.A. Hyaluronan and RHAMM in wound repair and the “cancerization” of stromal tissues. Biomed. Res. Int. 2014, 2014, 103923. [Google Scholar] [CrossRef] [PubMed]
- Noriega-Guerra, H.; Freitas, V.M. Extracellular Matrix Influencing HGF/c-MET Signaling Pathway: Impact on Cancer Progression. Int. J. Mol. Sci. 2018, 19, 3300. [Google Scholar] [CrossRef]
- Hassan, N.; Greve, B.; Espinoza-Sánchez, N.A.; Götte, M. Cell-surface heparan sulfate proteoglycans as multifunctional integrators of signaling in cancer. Cell Signal. 2020, 77, 109822. [Google Scholar] [CrossRef]
- Kim, S.H.; Turnbull, J.; Guimond, S. Extracellular matrix and cell signalling: The dynamic cooperation of integrin, proteoglycan and growth factor receptor. J. Endocrinol. 2011, 209, 139–151. [Google Scholar] [CrossRef]
- Wang, H.; Leavitt, L.; Ramaswamy, R.; Rapraeger, A.C. Interaction of syndecan and alpha6beta4 integrin cytoplasmic domains: Regulation of ErbB2-mediated integrin activation. J. Biol. Chem. 2010, 285, 13569–13579. [Google Scholar] [CrossRef]
- Morgan, M.R.; Humphries, M.J.; Bass, M.D. Synergistic control of cell adhesion by integrins and syndecans. Nat. Rev. Mol. Cell Biol. 2007, 8, 957–969. [Google Scholar] [CrossRef]
- Vuoriluoto, K.; Jokinen, J.; Kallio, K.; Salmivirta, M.; Heino, J.; Ivaska, J. Syndecan-1 supports integrin alpha2beta1-mediated adhesion to collagen. Exp. Cell Res. 2008, 314, 3369–3381. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, L.R.; Eble, J.A. Decorin regulates endothelial cell-matrix interactions during angiogenesis. Cell Adh. Migr. 2009, 3, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Neill, T.; Schaefer, L.; Iozzo, R.V. Decorin as a multivalent therapeutic agent against cancer. Adv. Drug Deliv. Rev. 2016, 97, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, L.; Tredup, C.; Gubbiotti, M.A.; Iozzo, R.V. Proteoglycan neofunctions: Regulation of inflammation and autophagy in cancer biology. FEBS J. 2017, 284, 10–26. [Google Scholar] [CrossRef]
- Poluzzi, C.; Iozzo, R.V.; Schaefer, L. Endostatin and endorepellin: A common route of action for similar angiostatic cancer avengers. Adv. Drug Deliv. Rev. 2016, 97, 156–173. [Google Scholar] [CrossRef]
- Woodall, B.P.; Nystrom, A.; Iozzo, R.A.; Eble, J.A.; Niland, S.; Krieg, T.; Eckes, B.; Pozzi, A.; Iozzo, R.V. Integrin alpha2beta1 is the required receptor for endorepellin angiostatic activity. J. Biol. Chem. 2008, 283, 2335–2343. [Google Scholar] [CrossRef]
- Barczyk, M.; Carracedo, S.; Gullberg, D. Integrins. Cell Tissue Res. 2010, 339, 269–280. [Google Scholar] [CrossRef]
- Humphries, J.D.; Byron, A.; Humphries, M.J. Integrin ligands at a glance. J. Cell Sci. 2006, 119, 3901–3903. [Google Scholar] [CrossRef]
- Zheng, Y.; Leftheris, K. Insights into Protein-Ligand Interactions in Integrin Complexes: Advances in Structure Determinations. J. Med. Chem. 2020, 63, 5675–5696. [Google Scholar] [CrossRef]
- Jang, I.; Beningo, K.A. Integrins, CAFs and Mechanical Forces in the Progression of Cancer. Cancers 2019, 11, 721. [Google Scholar] [CrossRef]
- DiPersio, C.M.; Van De Water, L. Integrin Regulation of CAF Differentiation and Function. Cancers 2019, 11, 715. [Google Scholar] [CrossRef] [PubMed]
- Liden, A.; Karlsen, T.V.; Guss, B.; Reed, R.K.; Rubin, K. Integrin alphaV beta3 can substitute for collagen-binding beta1 -integrins in vivo to maintain a homeostatic interstitial fluid pressure. Exp. Physiol. 2018, 103, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Arnaout, M.A.; Goodman, S.L.; Xiong, J.P. Structure and mechanics of integrin-based cell adhesion. Curr. Opin. Cell Biol. 2007, 19, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Humphries, J.D.; Chastney, M.R.; Askari, J.A.; Humphries, M.J. Signal transduction via integrin adhesion complexes. Curr. Opin. Cell Biol. 2019, 56, 14–21. [Google Scholar] [CrossRef]
- Luo, B.H.; Carman, C.V.; Springer, T.A. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 2007, 25, 619–647. [Google Scholar] [CrossRef]
- Wang, W.; Luo, B.H. Structural basis of integrin transmembrane activation. J. Cell. Biochem. 2010, 109, 447–452. [Google Scholar] [CrossRef]
- Lima, A.M.; Wegner, S.V.; Martins Cavaco, A.C.; Estevão-Costa, M.I.; Sanz-Soler, R.; Niland, S.; Nosov, G.; Klingauf, J.; Spatz, J.P.; Eble, J.A. The spatial molecular pattern of integrin recognition sites and their immobilization to colloidal nanobeads determine α2β1 integrin-dependent platelet activation. Biomaterials 2018, 167, 107–120. [Google Scholar] [CrossRef]
- Kanchanawong, P.; Shtengel, G.; Pasapera, A.M.; Ramko, E.B.; Davidson, M.W.; Hess, H.F.; Waterman, C.M. Nanoscale architecture of integrin-based cell adhesions. Nature 2010, 468, 580–584. [Google Scholar] [CrossRef]
- Zaidel-Bar, R.; Geiger, B. The switchable integrin adhesome. J. Cell Sci. 2010, 123, 1385–1388. [Google Scholar] [CrossRef]
- Li, J.; Springer, T.A. Integrin extension enables ultrasensitive regulation by cytoskeletal force. Proc. Natl. Acad. Sci. USA 2017, 114, 4685–4690. [Google Scholar] [CrossRef]
- Li, J.; Su, Y.; Xia, W.; Qin, Y.; Humphries, M.J.; Vestweber, D.; Cabanas, C.; Lu, C.; Springer, T.A. Conformational equilibria and intrinsic affinities define integrin activation. EMBO J. 2017, 36, 629–645. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Costell, M.; Fassler, R. Integrin activation by talin, kindlin and mechanical forces. Nat. Cell Biol. 2019, 21, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, I.; Eble, J.A.; Hanschmann, E.M. Thiol switches in membrane proteins—Extracellular redox regulation in cell biology. Biol. Chem. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Chen, J. The regulation of integrin function by divalent cations. Cell Adh. Migr. 2012, 6, 20–29. [Google Scholar] [CrossRef]
- Bergerhausen, L.; Grosche, J.; Meißner, J.; Hecker, C.; Caliandro, M.F.; Westerhausen, C.; Kamenac, A.; Rezaei, M.; Mörgelin, M.; Poschmann, G.; et al. Extracellular Redox Regulation of α7β Integrin-Mediated Cell Migration Is Signaled via a Dominant Thiol-Switch. Antioxidants 2020, 9, 227. [Google Scholar] [CrossRef]
- Ng, L.; Poon, R.T.; Pang, R. Biomarkers for predicting future metastasis of human gastrointestinal tumors. Cell. Mol. Life Sci. 2013, 70, 3631–3656. [Google Scholar] [CrossRef]
- Waerzeggers, Y.; Monfared, P.; Viel, T.; Faust, A.; Kopka, K.; Schafers, M.; Tavitian, B.; Winkeler, A.; Jacobs, A. Specific biomarkers of receptors, pathways of inhibition and targeted therapies: Pre-clinical developments. Br. J. Radiol. 2011, 84, S168–S178. [Google Scholar] [CrossRef]
- Arias-Mejias, S.M.; Warda, K.Y.; Quattrocchi, E.; Alonso-Quinones, H.; Sominidi-Damodaran, S.; Meves, A. The role of integrins in melanoma: A review. Int. J. Dermatol. 2020, 59, 525–534. [Google Scholar] [CrossRef]
- Zeltz, C.; Alam, J.; Liu, H.; Erusappan, P.M.; Hoschuetzky, H.; Molven, A.; Parajuli, H.; Cukierman, E.; Costea, D.-E.; Lu, N.; et al. α11β1 Integrin is Induced in a Subset of Cancer-Associated Fibroblasts in Desmoplastic Tumor Stroma and Mediates In Vitro Cell Migration. Cancers 2019, 11, 765. [Google Scholar] [CrossRef]
- Margadant, C.; Sonnenberg, A. Integrin-TGF-beta crosstalk in fibrosis, cancer and wound healing. EMBO Rep. 2010, 11, 97–105. [Google Scholar] [CrossRef]
- Sala, M.; Ros, M.; Saltel, F. A Complex and Evolutive Character: Two Face Aspects of ECM in Tumor Progression. Front. Oncol. 2020, 10, 1620. [Google Scholar] [CrossRef] [PubMed]
- Su, C.Y.; Li, J.Q.; Zhang, L.L.; Wang, H.; Wang, F.H.; Tao, Y.W.; Wang, Y.Q.; Guo, Q.R.; Li, J.J.; Liu, Y.; et al. The Biological Functions and Clinical Applications of Integrins in Cancers. Front. Pharmacol. 2020, 11, 579068. [Google Scholar] [CrossRef]
- Webber, J.; Steadman, R.; Mason, M.D.; Tabi, Z.; Clayton, A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010, 70, 9621–9630. [Google Scholar] [CrossRef] [PubMed]
- Weidle, U.H.; Birzele, F.; Kollmorgen, G.; Ruger, R. The Multiple Roles of Exosomes in Metastasis. Cancer Genom. Proteom. 2017, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; Wang, X.; Gong, Z.; Yu, M.; Wu, H.; Zhang, D. Exosome-mediated metabolic reprogramming: The emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct. Target Ther. 2020, 5, 242. [Google Scholar] [CrossRef]
- Yang, J.; Bahcecioglu, G.; Zorlutuna, P. The Extracellular Matrix and Vesicles Modulate the Breast Tumor Microenvironment. Bioengineering 2020, 7, 124. [Google Scholar] [CrossRef]
- Korenchan, D.E.; Flavell, R.R. Spatiotemporal pH Heterogeneity as a Promoter of Cancer Progression and Therapeutic Resistance. Cancers 2019, 11, 1026. [Google Scholar] [CrossRef]
- Pillai, S.R.; Damaghi, M.; Marunaka, Y.; Spugnini, E.P.; Fais, S.; Gillies, R.J. Causes, consequences, and therapy of tumors acidosis. Cancer Metastasis Rev. 2019, 38, 205–222. [Google Scholar] [CrossRef]
- Vaupel, P.; Multhoff, G. Accomplices of the Hypoxic Tumor Microenvironment Compromising Antitumor Immunity: Adenosine, Lactate, Acidosis, Vascular Endothelial Growth Factor, Potassium Ions, and Phosphatidylserine. Front. Immunol. 2017, 8, 1887. [Google Scholar] [CrossRef]
- Miles, F.L.; Sikes, R.A. Insidious changes in stromal matrix fuel cancer progression. Mol. Cancer Res. 2014, 12, 297–312. [Google Scholar] [CrossRef]
- Huang, L.; Xu, A.M.; Liu, W. Transglutaminase 2 in cancer. Am. J. Cancer Res. 2015, 5, 2756–2776. [Google Scholar] [PubMed]
- Kim, S.Y. New Insights into Development of Transglutaminase 2 Inhibitors as Pharmaceutical Lead Compounds. Med. Sci. 2018, 6, 87. [Google Scholar] [CrossRef] [PubMed]
- Hoye, A.M.; Erler, J.T. Structural ECM components in the premetastatic and metastatic niche. Am. J. Physiol. Cell Physiol. 2016, 310, C955–C967. [Google Scholar] [CrossRef] [PubMed]
- Ji, K.; Mayernik, L.; Moin, K.; Sloane, B.F. Acidosis and proteolysis in the tumor microenvironment. Cancer Metastasis Rev. 2019, 38, 103–112. [Google Scholar] [CrossRef]
- Wells, J.M.; Gaggar, A.; Blalock, J.E. MMP generated matrikines. Matrix Biol. 2015, 44–46, 122–129. [Google Scholar] [CrossRef]
- Mochizuki, S.; Ao, T.; Sugiura, T.; Yonemura, K.; Shiraishi, T.; Kajiwara, Y.; Okamoto, K.; Shinto, E.; Okada, Y.; Ueno, H. Expression and Function of a Disintegrin and Metalloproteinases in Cancer-Associated Fibroblasts of Colorectal Cancer. Digestion 2020, 101, 18–24. [Google Scholar] [CrossRef]
- Stern, R.; Jedrzejas, M.J. Hyaluronidases: Their genomics, structures, and mechanisms of action. Chem. Rev. 2006, 106, 818–839. [Google Scholar] [CrossRef]
- Quintero-Fabian, S.; Arreola, R.; Becerril-Villanueva, E.; Torres-Romero, J.C.; Arana-Argaez, V.; Lara-Riegos, J.; Ramirez-Camacho, M.A.; Alvarez-Sanchez, M.E. Role of Matrix Metalloproteinases in Angiogenesis and Cancer. Front. Oncol. 2019, 9, 1370. [Google Scholar] [CrossRef]
- Eatemadi, A.; Aiyelabegan, H.T.; Negahdari, B.; Mazlomi, M.A.; Daraee, H.; Daraee, N.; Eatemadi, R.; Sadroddiny, E. Role of protease and protease inhibitors in cancer pathogenesis and treatment. Biomed. Pharmacother. 2017, 86, 221–231. [Google Scholar] [CrossRef]
- Crotti, S.; Piccoli, M.; Rizzolio, F.; Giordano, A.; Nitti, D.; Agostini, M. Extracellular Matrix and Colorectal Cancer: How Surrounding Microenvironment Affects Cancer Cell Behavior? J. Cell. Physiol. 2017, 232, 967–975. [Google Scholar] [CrossRef]
- Stefanidakis, M.; Koivunen, E. Cell-surface association between matrix metalloproteinases and integrins: Role of the complexes in leukocyte migration and cancer progression. Blood 2006, 108, 1441–1450. [Google Scholar] [CrossRef] [PubMed]
- Saad, S.; Gottlieb, D.J.; Bradstock, K.F.; Overall, C.M.; Bendall, L.J. Cancer cell-associated fibronectin induces release of matrix metalloproteinase-2 from normal fibroblasts. Cancer Res. 2002, 62, 283–289. [Google Scholar] [PubMed]
- Nagase, H.; Visse, R.; Murphy, G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res. 2006, 69, 562–573. [Google Scholar] [CrossRef] [PubMed]
- Jacob, A.; Prekeris, R. The regulation of MMP targeting to invadopodia during cancer metastasis. Front. Cell Dev. Biol. 2015, 3, 4. [Google Scholar] [CrossRef]
- Ren, F.; Tang, R.; Zhang, X.; Madushi, W.M.; Luo, D.; Dang, Y.; Li, Z.; Wei, K.; Chen, G. Overexpression of MMP Family Members Functions as Prognostic Biomarker for Breast Cancer Patients: A Systematic Review and Meta-Analysis. PLoS ONE 2015, 10, e0135544. [Google Scholar] [CrossRef]
- Vandooren, J.; Van den Steen, P.E.; Opdenakker, G. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): The next decade. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 222–272. [Google Scholar] [CrossRef]
- Gonzalez-Avila, G.; Sommer, B.; García-Hernández, A.A.; Ramos, C. Matrix Metalloproteinases’ Role in Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1245, 97–131. [Google Scholar] [CrossRef]
- Huang, H. Matrix Metalloproteinase-9 (MMP-9) as a Cancer Biomarker and MMP-9 Biosensors: Recent Advances. Sensors 2018, 18, 3249. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, S.; Guo, J.; Zhou, L.; You, L.; Zhang, T.; Zhao, Y. Insights into the distinct roles of MMP-11 in tumor biology and future therapeutics (Review). Int. J. Oncol. 2016, 48, 1783–1793. [Google Scholar] [CrossRef]
- Castro-Castro, A.; Marchesin, V.; Monteiro, P.; Lodillinsky, C.; Rossé, C.; Chavrier, P. Cellular and Molecular Mechanisms of MT1-MMP-Dependent Cancer Cell Invasion. Annu. Rev. Cell Dev. Biol. 2016, 32, 555–576. [Google Scholar] [CrossRef]
- Pahwa, S.; Stawikowski, M.J.; Fields, G.B. Monitoring and Inhibiting MT1-MMP during Cancer Initiation and Progression. Cancers 2014, 6, 416–435. [Google Scholar] [CrossRef] [PubMed]
- Alcantara, M.B.; Dass, C.R. Regulation of MT1-MMP and MMP-2 by the serpin PEDF: A promising new target for metastatic cancer. Cell. Physiol. Biochem. 2013, 31, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Poincloux, R.; Lizarraga, F.; Chavrier, P. Matrix invasion by tumour cells: A focus on MT1-MMP trafficking to invadopodia. J. Cell Sci. 2009, 122, 3015–3024. [Google Scholar] [CrossRef] [PubMed]
- Radisky, E.S.; Radisky, D.C. Matrix metalloproteinases as breast cancer drivers and therapeutic targets. Front. Biosci. Landmark 2015, 20, 1144–1163. [Google Scholar] [CrossRef] [PubMed]
- Parvanescu, V.; Georgescu, M.; Georgescu, I.; Surlin, V.; Patrascu, S.; Picleanu, A.M.; Georgescu, E. The Role of Matrix Metalloproteinase-9 (MMP-9) as a Prognostic Factor in Epithelial and Lymphatic Neoplasia. Chirurgia 2015, 110, 506–510. [Google Scholar]
- Fields, G.B. Biophysical studies of matrix metalloproteinase/triple-helix complexes. Adv. Protein Chem. Struct. Biol. 2014, 97, 37–48. [Google Scholar] [CrossRef]
- Tam, E.M.; Moore, T.R.; Butler, G.S.; Overall, C.M. Characterization of the distinct collagen binding, helicase and cleavage mechanisms of matrix metalloproteinase 2 and 14 (gelatinase A and MT1-MMP): The differential roles of the MMP hemopexin c domains and the MMP-2 fibronectin type II modules in collagen triple helicase activities. J. Biol. Chem. 2004, 279, 43336–43344. [Google Scholar] [CrossRef]
- Farina, A.R.; Mackay, A.R. Gelatinase B/MMP-9 in Tumour Pathogenesis and Progression. Cancers 2014, 6, 240–296. [Google Scholar] [CrossRef]
- Sato, H.; Takino, T. Coordinate action of membrane-type matrix metalloproteinase-1 (MT1-MMP) and MMP-2 enhances pericellular proteolysis and invasion. Cancer Sci. 2010, 101, 843–847. [Google Scholar] [CrossRef]
- Murphy, G.; Nagase, H. Progress in matrix metalloproteinase research. Mol. Aspects Med. 2008, 29, 290–308. [Google Scholar] [CrossRef]
- Jackson, B.C.; Nebert, D.W.; Vasiliou, V. Update of human and mouse matrix metalloproteinase families. Hum. Genom. 2010, 4, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Cui, N.; Hu, M.; Khalil, R.A. Biochemical and Biological Attributes of Matrix Metalloproteinases. Prog. Mol. Biol. Transl. Sci. 2017, 147, 1–73. [Google Scholar] [CrossRef] [PubMed]
- Mittal, R.; Patel, A.P.; Debs, L.H.; Nguyen, D.; Patel, K.; Grati, M.; Mittal, J.; Yan, D.; Chapagain, P.; Liu, X.Z. Intricate Functions of Matrix Metalloproteinases in Physiological and Pathological Conditions. J. Cell. Physiol. 2016, 231, 2599–2621. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Mandal, M.; Chakraborti, T.; Mandal, A.; Chakraborti, S. Structure and evolutionary aspects of matrix metalloproteinases: A brief overview. Mol. Cell. Biochem. 2003, 253, 31–40. [Google Scholar] [CrossRef]
- Massova, I.; Kotra, L.P.; Fridman, R.; Mobashery, S. Matrix metalloproteinases: Structures, evolution, and diversification. FASEB J. 1998, 12, 1075–1095. [Google Scholar] [CrossRef]
- Loffek, S.; Schilling, O.; Franzke, C.W. Series “matrix metalloproteinases in lung health and disease”: Biological role of matrix metalloproteinases: A critical balance. Eur. Respir. J. 2011, 38, 191–208. [Google Scholar] [CrossRef]
- Gramolelli, S.; Cheng, J.; Martinez-Corral, I.; Vähä-Koskela, M.; Elbasani, E.; Kaivanto, E.; Rantanen, V.; Tuohinto, K.; Hautaniemi, S.; Bower, M.; et al. PROX1 is a transcriptional regulator of MMP14. Sci. Rep. 2018, 8, 9531. [Google Scholar] [CrossRef]
- Hotary, K.; Allen, E.; Punturieri, A.; Yana, I.; Weiss, S.J. Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J. Cell Biol. 2000, 149, 1309–1323. [Google Scholar] [CrossRef]
- Itoh, Y.; Seiki, M. MT1-MMP: A potent modifier of pericellular microenvironment. J. Cell. Physiol. 2006, 206, 1–8. [Google Scholar] [CrossRef]
- Sabeh, F.; Ota, I.; Holmbeck, K.; Birkedal-Hansen, H.; Soloway, P.; Balbin, M.; Lopez-Otin, C.; Shapiro, S.; Inada, M.; Krane, S.; et al. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J. Cell Biol. 2004, 167, 769–781. [Google Scholar] [CrossRef]
- Wolf, K.; Wu, Y.I.; Liu, Y.; Geiger, J.; Tam, E.; Overall, C.; Stack, M.S.; Friedl, P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat. Cell Biol. 2007, 9, 893–904. [Google Scholar] [CrossRef]
- Szabova, L.; Chrysovergis, K.; Yamada, S.S.; Holmbeck, K. MT1-MMP is required for efficient tumor dissemination in experimental metastatic disease. Oncogene 2008, 27, 3274–3281. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, Z.; Apte, S.S.; Soininen, R.; Cao, R.; Baaklini, G.Y.; Rauser, R.W.; Wang, J.; Cao, Y.; Tryggvason, K. Impaired endochondral ossification and angiogenesis in mice deficient in membrane-type matrix metalloproteinase I. Proc. Natl. Acad. Sci. USA 2000, 97, 4052–4057. [Google Scholar] [CrossRef] [PubMed]
- David, J.M.; Rajasekaran, A.K. Dishonorable discharge: The oncogenic roles of cleaved E-cadherin fragments. Cancer Res. 2012, 72, 2917–2923. [Google Scholar] [CrossRef] [PubMed]
- Kajita, M.; Itoh, Y.; Chiba, T.; Mori, H.; Okada, A.; Kinoh, H.; Seiki, M. Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J. Cell Biol. 2001, 153, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Seiki, M. Integrated functions of membrane-type 1 matrix metalloproteinase in regulating cancer malignancy: Beyond a proteinase. Cancer Sci. 2017, 108, 1095–1100. [Google Scholar] [CrossRef] [PubMed]
- Egeblad, M.; Werb, Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2002, 2, 161–174. [Google Scholar] [CrossRef]
- Gonzalez-Molina, J.; Gramolelli, S.; Liao, Z.; Carlson, J.W.; Ojala, P.M.; Lehti, K. MMP14 in Sarcoma: A Regulator of Tumor Microenvironment Communication in Connective Tissues. Cells 2019, 8, 991. [Google Scholar] [CrossRef]
- Yan, C.; Boyd, D.D. Regulation of matrix metalloproteinase gene expression. J. Cell Physiol. 2007, 211, 19–26. [Google Scholar] [CrossRef]
- Chatterjee, K.; Jana, S.; Choudhary, P.; Swarnakar, S. Triumph and tumult of matrix metalloproteinases and their crosstalk with eicosanoids in cancer. Cancer. Metastasis Rev. 2018, 37, 279–288. [Google Scholar] [CrossRef]
- Kim, Y.S.; Joh, T.H. Matrix metalloproteinases, new insights into the understanding of neurodegenerative disorders. Biomol. Ther. 2012, 20, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Chernov, A.V.; Sounni, N.E.; Remacle, A.G.; Strongin, A.Y. Epigenetic control of the invasion-promoting MT1-MMP/MMP-2/TIMP-2 axis in cancer cells. J. Biol. Chem. 2009, 284, 12727–12734. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; He, Z.; Wang, C.; Zhou, Y.; Li, F.; Pu, W.; Zhang, X.; Feng, X.; Zhang, M.; Yecheng, X.; et al. Epigenetic silencing of ZNF132 mediated by methylation-sensitive Sp1 binding promotes cancer progression in esophageal squamous cell carcinoma. Cell Death Dis. 2018, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Remacle, A.G.; Rozanov, D.V.; Fugere, M.; Day, R.; Strongin, A.Y. Furin regulates the intracellular activation and the uptake rate of cell surface-associated MT1-MMP. Oncogene 2006, 25, 5648–5655. [Google Scholar] [CrossRef]
- Williams, K.C.; Coppolino, M.G. Phosphorylation of membrane type 1-matrix metalloproteinase (MT1-MMP) and its vesicle-associated membrane protein 7 (VAMP7)-dependent trafficking facilitate cell invasion and migration. J. Biol. Chem. 2011, 286, 43405–43416. [Google Scholar] [CrossRef]
- Remacle, A.G.; Chekanov, A.V.; Golubkov, V.S.; Savinov, A.Y.; Rozanov, D.V.; Strongin, A.Y. O-glycosylation regulates autolysis of cellular membrane type-1 matrix metalloproteinase (MT1-MMP). J. Biol. Chem. 2006, 281, 16897–16905. [Google Scholar] [CrossRef]
- Kruglikov, I.L.; Joffin, N.; Scherer, P.E. The MMP14-caveolin axis and its potential relevance for lipoedema. Nat. Rev. Endocrinol. 2020, 16, 669–674. [Google Scholar] [CrossRef]
- Kim, H.N.; Chung, H.S. Caveolin-1 inhibits membrane-type 1 matrix metalloproteinase activity. BMB Rep. 2008, 41, 858–862. [Google Scholar] [CrossRef]
- Hsu, K.S.; Otsu, W.; Li, Y.; Wang, H.C.; Chen, S.; Tsang, S.H.; Chuang, J.Z.; Sung, C.H. CLIC4 regulates late endosomal trafficking and matrix degradation activity of MMP14 at focal adhesions in RPE cells. Sci. Rep. 2019, 9, 12247. [Google Scholar] [CrossRef]
- Itoh, Y. Membrane-type matrix metalloproteinases: Their functions and regulations. Matrix Biol. 2015, 44-46, 207–223. [Google Scholar] [CrossRef]
- Grafinger, O.R.; Gorshtein, G.; Stirling, T.; Brasher, M.I.; Coppolino, M.G. beta1 integrin-mediated signaling regulates MT1-MMP phosphorylation to promote tumor cell invasion. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef] [PubMed]
- Tobar, N.; Avalos, M.C.; Mendez, N.; Smith, P.C.; Bernabeu, C.; Quintanilla, M.; Martinez, J. Soluble MMP-14 produced by bone marrow-derived stromal cells sheds epithelial endoglin modulating the migratory properties of human breast cancer cells. Carcinogenesis 2014, 35, 1770–1779. [Google Scholar] [CrossRef] [PubMed]
- Osenkowski, P.; Toth, M.; Fridman, R. Processing, shedding, and endocytosis of membrane type 1-matrix metalloproteinase (MT1-MMP). J. Cell. Physiol. 2004, 200, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Lohi, J.; Lehti, K.; Westermarck, J.; Kähäri, V.M.; Keski-Oja, J. Regulation of membrane-type matrix metalloproteinase-1 expression by growth factors and phorbol 12-myristate 13-acetate. Eur. J. Biochem. 1996, 239, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Nakamura, T.; Suzuki, Y.; Imizu, T.; Matsumoto, K. 3-D collagen-dependent cell surface expression of MT1-MMP and MMP-2 activation regardless of integrin β1 function and matrix stiffness. Biochem. Biophys. Res. Commun. 2011, 412, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Haas, T.L.; Stitelman, D.; Davis, S.J.; Apte, S.S.; Madri, J.A. Egr-1 mediates extracellular matrix-driven transcription of membrane type 1 matrix metalloproteinase in endothelium. J. Biol. Chem. 1999, 274, 22679–22685. [Google Scholar] [CrossRef]
- Borrirukwanit, K.; Pavasant, P.; Blick, T.; Lafleur, M.A.; Thompson, E.W. High threshold of β1 integrin inhibition required to block collagen I-induced membrane type-1 matrix metalloproteinase (MT1-MMP) activation of matrix metalloproteinase 2 (MMP-2). Cancer Cell Int. 2014, 14, 99. [Google Scholar] [CrossRef]
- Zigrino, P.; Drescher, C.; Mauch, C. Collagen-induced proMMP-2 activation by MT1-MMP in human dermal fibroblasts and the possible role of alpha2beta1 integrins. Eur. J. Cell Biol. 2001, 80, 68–77. [Google Scholar] [CrossRef]
- Johnson, J.L.; Pillai, S.; Pernazza, D.; Sebti, S.M.; Lawrence, N.J.; Chellappan, S.P. Regulation of matrix metalloproteinase genes by E2F transcription factors: Rb-Raf-1 interaction as a novel target for metastatic disease. Cancer Res. 2012, 72, 516–526. [Google Scholar] [CrossRef]
- Elsir, T.; Smits, A.; Lindström, M.S.; Nistér, M. Transcription factor PROX1: Its role in development and cancer. Cancer Metastasis Rev. 2012, 31, 793–805. [Google Scholar] [CrossRef]
- Eiseler, T.; Döppler, H.; Yan, I.K.; Goodison, S.; Storz, P. Protein kinase D1 regulates matrix metalloproteinase expression and inhibits breast cancer cell invasion. Breast Cancer Res. 2009, 11, R13. [Google Scholar] [CrossRef] [PubMed]
- Onishi, Y.; Kawamoto, T.; Kishimoto, K.; Hara, H.; Fukase, N.; Toda, M.; Harada, R.; Kurosaka, M.; Akisue, T. PKD1 negatively regulates cell invasion, migration and proliferation ability of human osteosarcoma. Int. J. Oncol. 2012, 40, 1839–1848. [Google Scholar] [CrossRef] [PubMed]
- Jackson, H.W.; Defamie, V.; Waterhouse, P.; Khokha, R. TIMPs: Versatile extracellular regulators in cancer. Nat. Rev. Cancer 2017, 17, 38–53. [Google Scholar] [CrossRef] [PubMed]
- Murphy, G. Tissue inhibitors of metalloproteinases. Genome Biol. 2011, 12, 233. [Google Scholar] [CrossRef] [PubMed]
- Strongin, A.Y.; Collier, I.; Bannikov, G.; Marmer, B.L.; Grant, G.A.; Goldberg, G.I. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J. Biol. Chem. 1995, 270, 5331–5338. [Google Scholar] [CrossRef] [PubMed]
- Ogata, Y.; Itoh, Y.; Nagase, H. Steps involved in activation of the pro-matrix metalloproteinase 9 (progelatinase B)-tissue inhibitor of metalloproteinases-1 complex by 4-aminophenylmercuric acetate and proteinases. J. Biol. Chem. 1995, 270, 18506–18511. [Google Scholar] [CrossRef]
- Mori, H.; Lo, A.T.; Inman, J.L.; Alcaraz, J.; Ghajar, C.M.; Mott, J.D.; Nelson, C.M.; Chen, C.S.; Zhang, H.; Bascom, J.L.; et al. Transmembrane/cytoplasmic, rather than catalytic, domains of Mmp14 signal to MAPK activation and mammary branching morphogenesis via binding to integrin β1. Development 2013, 140, 343–352. [Google Scholar] [CrossRef]
- Nguyen, A.T.; Chia, J.; Ros, M.; Hui, K.M.; Saltel, F.; Bard, F. Organelle Specific O-Glycosylation Drives MMP14 Activation, Tumor Growth, and Metastasis. Cancer Cell 2017, 32, 639–653. [Google Scholar] [CrossRef]
- Planchon, D.; Rios Morris, E.; Genest, M.; Comunale, F.; Vacher, S.; Bieche, I.; Denisov, E.V.; Tashireva, L.A.; Perelmuter, V.M.; Linder, S.; et al. MT1-MMP targeting to endolysosomes is mediated by upregulation of flotillins. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef]
- Infante, E.; Castagnino, A.; Ferrari, R.; Monteiro, P.; Agüera-González, S.; Paul-Gilloteaux, P.; Domingues, M.J.; Maiuri, P.; Raab, M.; Shanahan, C.M.; et al. LINC complex-Lis1 interplay controls MT1-MMP matrix digest-on-demand response for confined tumor cell migration. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Ferrari, R.; Infante, E.; Chavrier, P. Nucleus-Invadopodia Duo During Cancer Invasion. Trends Cell Biol. 2019, 29, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.M.; Wenzel, E.M.; Wang, L.; Antoine, S.; Chavrier, P.; Stenmark, H.; Raiborg, C. Protrudin-mediated ER-endosome contact sites promote MT1-MMP exocytosis and cell invasion. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef] [PubMed]
- Ellerbroek, S.M.; Wu, Y.I.; Overall, C.M.; Stack, M.S. Functional interplay between type I collagen and cell surface matrix metalloproteinase activity. J. Biol. Chem. 2001, 276, 24833–24842. [Google Scholar] [CrossRef] [PubMed]
- Laudański, P.; Swiatecka, J.; Kozłowski, L.; Leśniewska, M.; Wojtukiewicz, M.; Wołczyński, S. Increased serum level of membrane type 1-matrix metalloproteinase (MT1-MMP/MMP-14) in patients with breast cancer. Folia Histochem. Cytobiol. 2010, 48, 101–103. [Google Scholar] [CrossRef] [PubMed]
- Ip, Y.C.; Cheung, S.T.; Fan, S.T. Atypical localization of membrane type 1-matrix metalloproteinase in the nucleus is associated with aggressive features of hepatocellular carcinoma. Mol. Carcinog. 2007, 46, 225–230. [Google Scholar] [CrossRef]
- Mannello, F.; Medda, V. Nuclear localization of matrix metalloproteinases. Prog. Histochem. Cytochem. 2012, 47, 27–58. [Google Scholar] [CrossRef] [PubMed]
- Augoff, K.; Hryniewicz-Jankowska, A.; Tabola, R. Invadopodia: Clearing the way for cancer cell invasion. Ann. Transl. Med. 2020, 8, 902. [Google Scholar] [CrossRef]
- Branch, K.M.; Hoshino, D.; Weaver, A.M. Adhesion rings surround invadopodia and promote maturation. Biol. Open 2012, 1, 711–722. [Google Scholar] [CrossRef]
- Malik, R.; Luong, T.; Cao, X.; Han, B.; Shah, N.; Franco-Barraza, J.; Han, L.; Shenoy, V.B.; Lelkes, P.I.; Cukierman, E. Rigidity controls human desmoplastic matrix anisotropy to enable pancreatic cancer cell spread via extracellular signal-regulated kinase 2. Matrix Biol. 2019, 81, 50–69. [Google Scholar] [CrossRef]
- Peláez, R.; Pariente, A.; Pérez-Sala, Á.; Larrayoz, I.M. Integrins: Moonlighting Proteins in Invadosome Formation. Cancers 2019, 11, 615. [Google Scholar] [CrossRef]
- Pelaez, R.; Morales, X.; Salvo, E.; Garasa, S.; Ortiz de Solorzano, C.; Martinez, A.; Larrayoz, I.M.; Rouzaut, A. beta3 integrin expression is required for invadopodia-mediated ECM degradation in lung carcinoma cells. PLoS ONE 2017, 12, e0181579. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y. MT1-MMP: A key regulator of cell migration in tissue. IUBMB Life 2006, 58, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, S.; Leon, R.P.; Gribbin, K.P.; Zhang, Y.; Navarro, J.; Smith, R.; Devlin, K.; Wang, L.G.; Gibbs, S.L.; Korkola, J.; et al. Crosstalk between invadopodia and the extracellular matrix. Eur. J. Cell Biol. 2020, 99, 151122. [Google Scholar] [CrossRef] [PubMed]
- Eddy, R.J.; Weidmann, M.D.; Sharma, V.P.; Condeelis, J.S. Tumor Cell Invadopodia: Invasive Protrusions that Orchestrate Metastasis. Trends Cell Biol. 2017, 27, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Revach, O.Y.; Geiger, B. The interplay between the proteolytic, invasive, and adhesive domains of invadopodia and their roles in cancer invasion. Cell Adh. Migr. 2014, 8, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Bagnato, A.; Rosano, L. Endothelin-1 receptor drives invadopodia: Exploiting how beta-arrestin-1 guides the way. Small GTPases 2018, 9, 394–398. [Google Scholar] [CrossRef]
- Harper, K.; Lavoie, R.R.; Charbonneau, M.; Brochu-Gaudreau, K.; Dubois, C.M. The hypoxic tumor microenvironment promotes invadopodia formation and metastasis through LPA1 receptor and EGFR cooperation. Mol. Cancer Res. 2018, 16, 1601–1613. [Google Scholar] [CrossRef]
- Tonisen, F.; Perrin, L.; Bayarmagnai, B.; van den Dries, K.; Cambi, A.; Gligorijevic, B. EP4 receptor promotes invadopodia and invasion in human breast cancer. Eur. J. Cell Biol. 2017. [Google Scholar] [CrossRef]
- Maziveyi, M.; Dong, S.; Baranwal, S.; Alahari, S.K. Nischarin regulates focal adhesion and Invadopodia formation in breast cancer cells. Mol. Cancer. 2018, 17, 21. [Google Scholar] [CrossRef]
- Artym, V.V.; Swatkoski, S.; Matsumoto, K.; Campbell, C.B.; Petrie, R.J.; Dimitriadis, E.K.; Li, X.; Mueller, S.C.; Bugge, T.H.; Gucek, M.; et al. Dense fibrillar collagen is a potent inducer of invadopodia via a specific signaling network. J. Cell Biol. 2015, 208, 331–350. [Google Scholar] [CrossRef]
- Alekhina, O.; Burstein, E.; Billadeau, D.D. Cellular functions of WASP family proteins at a glance. J. Cell Sci. 2017, 130, 2235–2241. [Google Scholar] [CrossRef] [PubMed]
- Frugtniet, B.; Jiang, W.G.; Martin, T.A. Role of the WASP and WAVE family proteins in breast cancer invasion and metastasis. Breast Cancer 2015, 7, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Parekh, A.; Weaver, A.M. Regulation of invadopodia by mechanical signaling. Exp. Cell Res. 2016, 343, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Jeannot, P.; Besson, A. Cortactin function in invadopodia. Small GTPases 2017, 1–15. [Google Scholar] [CrossRef]
- Pourfarhangi, K.E.; Bergman, A.; Gligorijevic, B. ECM Cross-Linking Regulates Invadopodia Dynamics. Biophys. J. 2018, 114, 1455–1466. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Ito, Y.; Miura, N.; Nagamura, Y.; Nakabo, A.; Fukami, K.; Honda, K.; Sakai, R. Actinin-1 and actinin-4 play essential but distinct roles in invadopodia formation by carcinoma cells. Eur. J. Cell Biol. 2017, 96, 685–694. [Google Scholar] [CrossRef]
- Linder, S. The matrix corroded: Podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol. 2007, 17, 107–117. [Google Scholar] [CrossRef]
- Nicholas, N.S.; Pipili, A.; Lesjak, M.S.; Wells, C.M. Differential role for PAK1 and PAK4 during the invadopodia lifecycle. Small GTPases 2017, 10, 1–7. [Google Scholar] [CrossRef]
- Suman, P.; Mishra, S.; Chander, H. High expression of FBP17 in invasive breast cancer cells promotes invadopodia formation. Med. Oncol. 2018, 35, 71. [Google Scholar] [CrossRef]
- Castagnino, A.; Castro-Castro, A.; Irondelle, M.; Guichard, A.; Lodillinsky, C.; Fuhrmann, L.; Vacher, S.; Aguera-Gonzalez, S.; Zagryazhskaya-Masson, A.; Romao, M.; et al. Coronin 1C promotes triple-negative breast cancer invasiveness through regulation of MT1-MMP traffic and invadopodia function. Oncogene 2018. [Google Scholar] [CrossRef]
- Seano, G.; Primo, L. Podosomes and invadopodia: Tools to breach vascular basement membrane. Cell Cycle 2015, 14, 1370–1374. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Deryugina, E.I.; Quigley, J.P. Tumor angiogenesis: MMP-mediated induction of intravasation- and metastasis-sustaining neovasculature. Matrix Biol. 2015, 44-46, 94–112. [Google Scholar] [CrossRef] [PubMed]
- Genis, L.; Galvez, B.G.; Gonzalo, P.; Arroyo, A.G. MT1-MMP: Universal or particular player in angiogenesis? Cancer Metastasis Rev. 2006, 25, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Binder, M.J.; McCoombe, S.; Williams, E.D.; McCulloch, D.R.; Ward, A.C. The extracellular matrix in cancer progression: Role of hyalectan proteoglycans and ADAMTS enzymes. Cancer Lett. 2017, 385, 55–64. [Google Scholar] [CrossRef]
- Kumar, S.; Das, A.; Barai, A.; Sen, S. MMP Secretion Rate and Inter-invadopodia Spacing Collectively Govern Cancer Invasiveness. Biophys. J. 2018, 114, 650–662. [Google Scholar] [CrossRef]
- Binker, M.G.; Binker-Cosen, A.A.; Gaisano, H.Y.; de Cosen, R.H.; Cosen-Binker, L.I. TGF-β1 increases invasiveness of SW1990 cells through Rac1/ROS/NF-κB/IL-6/MMP-2. Biochem. Biophys. Res. Commun. 2011, 405, 140–145. [Google Scholar] [CrossRef]
- Fan, Z.; Duan, X.; Cai, H.; Wang, L.; Li, M.; Qu, J.; Li, W.; Wang, Y.; Wang, J. Curcumin inhibits the invasion of lung cancer cells by modulating the PKCα/Nox-2/ROS/ATF-2/MMP-9 signaling pathway. Oncol. Rep. 2015, 34, 691–698. [Google Scholar] [CrossRef]
- Choi, D.H.; Kim, J.H.; Seo, J.H.; Lee, J.; Choi, W.S.; Kim, Y.S. Matrix metalloproteinase-3 causes dopaminergic neuronal death through Nox1-regenerated oxidative stress. PLoS ONE 2014, 9, e115954. [Google Scholar] [CrossRef]
- Hawk, M.A.; Schafer, Z.T. Mechanisms of redox metabolism and cancer cell survival during extracellular matrix detachment. J. Biol. Chem. 2018, 293, 7531–7537. [Google Scholar] [CrossRef]
- Monboisse, J.C.; Oudart, J.B.; Ramont, L.; Brassart-Pasco, S.; Maquart, F.X. Matrikines from basement membrane collagens: A new anti-cancer strategy. Biochim. Biophys. Acta 2014, 1840, 2589–2598. [Google Scholar] [CrossRef]
- Tran, K.T.; Lamb, P.; Deng, J.S. Matrikines and matricryptins: Implications for cutaneous cancers and skin repair. J. Dermatol. Sci. 2005, 40, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Hornebeck, W.; Maquart, F.X. Proteolyzed matrix as a template for the regulation of tumor progression. Biomed. Pharmacother. 2003, 57, 223–230. [Google Scholar] [CrossRef]
- Ricard-Blum, S.; Vallet, S.D. Fragments generated upon extracellular matrix remodeling: Biological regulators and potential drugs. Matrix Biol. 2019, 75-76, 170–189. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Fleury, C.; Jalalvand, F.; Riesbeck, K. Human pathogens utilize host extracellular matrix proteins laminin and collagen for adhesion and invasion of the host. FEMS Microbiol. Rev. 2012, 36, 1122–1180. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, D.; Camardella, L.; Ulivi, V.; Guasco, G.; Manduca, P. Trimer carboxyl propeptide of collagen I produced by mature osteoblasts is chemotactic for endothelial cells. J. Biol. Chem. 2000, 275, 32658–32663. [Google Scholar] [CrossRef]
- Palmieri, D.; Astigiano, S.; Barbieri, O.; Ferrari, N.; Marchisio, S.; Ulivi, V.; Volta, C.; Manduca, P. Procollagen I COOH-terminal fragment induces VEGF-A and CXCR4 expression in breast carcinoma cells. Exp. Cell Res. 2008, 314, 2289–2298. [Google Scholar] [CrossRef]
- Visigalli, D.; Palmieri, D.; Strangio, A.; Astigiano, S.; Barbieri, O.; Casartelli, G.; Zicca, A.; Manduca, P. The carboxyl terminal trimer of procollagen I induces pro-metastatic changes and vascularization in breast cancer cells xenografts. BMC Cancer 2009, 9, 59. [Google Scholar] [CrossRef]
- Fernandes, R.J.; Hirohata, S.; Engle, J.M.; Colige, A.; Cohn, D.H.; Eyre, D.R.; Apte, S.S. Procollagen II amino propeptide processing by ADAMTS-3. Insights on dermatosparaxis. J. Biol. Chem. 2001, 276, 31502–31509. [Google Scholar] [CrossRef]
- Wang, Z.; Bryan, J.; Franz, C.; Havlioglu, N.; Sandell, L.J. Type IIB procollagen NH(2)-propeptide induces death of tumor cells via interaction with integrins alpha(V)beta(3) and alpha(V)beta(5). J. Biol. Chem. 2010, 285, 20806–20817. [Google Scholar] [CrossRef]
- Hayashi, S.; Wang, Z.; Bryan, J.; Kobayashi, C.; Faccio, R.; Sandell, L.J. The type II collagen N-propeptide, PIIBNP, inhibits cell survival and bone resorption of osteoclasts via integrin-mediated signaling. Bone 2011, 49, 644–652. [Google Scholar] [CrossRef]
- Lipton, A.; Leitzel, K.; Ali, S.M.; Polimera, H.V.; Nagabhairu, V.; Marks, E.; Richardson, A.E.; Krecko, L.; Ali, A.; Koestler, W.; et al. High turnover of extracellular matrix reflected by specific protein fragments measured in serum is associated with poor outcomes in two metastatic breast cancer cohorts. Int. J. Cancer 2018, 143, 3027–3034. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.J.; Nedergaard, A.F.; Sun, S.; Veidal, S.S.; Larsen, L.; Zheng, Q.; Suetta, C.; Henriksen, K.; Christiansen, C.; Karsdal, M.A.; et al. The neo-epitope specific PRO-C3 ELISA measures true formation of type III collagen associated with liver and muscle parameters. Am. J. Transl. Res. 2013, 5, 303–315. [Google Scholar] [PubMed]
- Leitinger, B.; Hohenester, E. Mammalian collagen receptors. Matrix Biol. 2007, 26, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Leeming, D.; He, Y.; Veidal, S.; Nguyen, Q.; Larsen, D.; Koizumi, M.; Segovia-Silvestre, T.; Zhang, C.; Zheng, Q.; Sun, S.; et al. A novel marker for assessment of liver matrix remodeling: An enzyme-linked immunosorbent assay (ELISA) detecting a MMP generated type I collagen neo-epitope (C1M). Biomarkers 2011, 16, 616–628. [Google Scholar] [CrossRef]
- Abdul Roda, M.; Fernstrand, A.M.; Redegeld, F.A.; Blalock, J.E.; Gaggar, A.; Folkerts, G. The matrikine PGP as a potential biomarker in COPD. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 308, L1095–L1101. [Google Scholar] [CrossRef]
- Patel, D.F.; Snelgrove, R.J. The multifaceted roles of the matrikine Pro-Gly-Pro in pulmonary health and disease. Eur. Respir. Rev. 2018, 27. [Google Scholar] [CrossRef]
- Provenzano, P.P.; Vanderby, R., Jr. Collagen fibril morphology and organization: Implications for force transmission in ligament and tendon. Matrix Biol. 2006, 25, 71–84. [Google Scholar] [CrossRef]
- Veidal, S.S.; Vassiliadis, E.; Barascuk, N.; Zhang, C.; Segovia-Silvestre, T.; Klickstein, L.; Larsen, M.R.; Qvist, P.; Christiansen, C.; Vainer, B.; et al. Matrix metalloproteinase-9-mediated type III collagen degradation as a novel serological biochemical marker for liver fibrogenesis. Liver Int. 2010, 30, 1293–1304. [Google Scholar] [CrossRef]
- Iyengar, P.; Espina, V.; Williams, T.W.; Lin, Y.; Berry, D.; Jelicks, L.A.; Lee, H.; Temple, K.; Graves, R.; Pollard, J.; et al. Adipocyte-derived collagen VI affects early mammary tumor progression in vivo, demonstrating a critical interaction in the tumor/stroma microenvironment. J. Clin. Investig. 2005, 115, 1163–1176. [Google Scholar] [CrossRef]
- Motrescu, E.R.; Blaise, S.; Etique, N.; Messaddeq, N.; Chenard, M.P.; Stoll, I.; Tomasetto, C.; Rio, M.C. Matrix metalloproteinase-11/stromelysin-3 exhibits collagenolytic function against collagen VI under normal and malignant conditions. Oncogene 2008, 27, 6347–6355. [Google Scholar] [CrossRef]
- Park, J.; Scherer, P.E. Adipocyte-derived endotrophin promotes malignant tumor progression. J. Clin. Investig. 2012, 122, 4243–4256. [Google Scholar] [CrossRef] [PubMed]
- Willumsen, N.; Jorgensen, L.N.; Karsdal, M.A. Vastatin (the NC1 domain of human type VIII collagen a1 chain) is linked to stromal reactivity and elevated in serum from patients with colorectal cancer. Cancer Biol. Ther. 2019, 20, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, J.; Woo, Y.M.; Shen, Z.; Yao, H.; Cai, Y.; Lin, M.C.; Poon, W.S. Enhanced expression of Vastatin inhibits angiogenesis and prolongs survival in murine orthotopic glioblastoma model. BMC Cancer 2017, 17, 126. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Yao, C.; Wang, Z.; Yue, L.; Fang, Z.; Yao, H.; Lin, F.; Zhao, H.; Sun, Y.J.; Bian, X.W.; et al. Vastatin, an Endogenous Antiangiogenesis Polypeptide That Is Lost in Hepatocellular Carcinoma, Effectively Inhibits Tumor Metastasis. Mol. Ther. 2016, 24, 1358–1368. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Yao, Z.Y.; Xin, L.; Zhang, Q.; Li, T.P.; Gan, R.B. NC1 domain of human type VIII collagen (alpha 1) inhibits bovine aortic endothelial cell proliferation and causes cell apoptosis. Biochem. Biophys. Res. Commun. 2001, 289, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Boudko, S.P.; Danylevych, N.; Hudson, B.G.; Pedchenko, V.K. Basement membrane collagen IV: Isolation of functional domains. Methods Cell Biol. 2018, 143, 171–185. [Google Scholar] [CrossRef]
- Heljasvaara, R.; Aikio, M.; Ruotsalainen, H.; Pihlajaniemi, T. Collagen XVIII in tissue homeostasis and dysregulation—Lessons learned from model organisms and human patients. Matrix Biol. 2017, 57–58, 55–75. [Google Scholar] [CrossRef]
- Mak, K.M.; Mei, R. Basement Membrane Type IV Collagen and Laminin: An Overview of Their Biology and Value as Fibrosis Biomarkers of Liver Disease. Anat. Rec. 2017, 300, 1371–1390. [Google Scholar] [CrossRef]
- Rebustini, I.T.; Myers, C.; Lassiter, K.S.; Surmak, A.; Szabova, L.; Holmbeck, K.; Pedchenko, V.; Hudson, B.G.; Hoffman, M.P. MT2-MMP-dependent release of collagen IV NC1 domains regulates submandibular gland branching morphogenesis. Dev. Cell 2009, 17, 482–493. [Google Scholar] [CrossRef]
- Sugiyama, A.; Mitsui, A.; Okada, M.; Yamawaki, H. Cathepsin S degrades arresten and canstatin in infarcted area after myocardial infarction in rats. J. Vet. Med. Sci. 2019, 81, 522–531. [Google Scholar] [CrossRef]
- Hamano, Y.; Zeisberg, M.; Sugimoto, H.; Lively, J.C.; Maeshima, Y.; Yang, C.; Hynes, R.O.; Werb, Z.; Sudhakar, A.; Kalluri, R. Physiological levels of tumstatin, a fragment of collagen IV alpha3 chain, are generated by MMP-9 proteolysis and suppress angiogenesis via alphaV beta3 integrin. Cancer Cell 2003, 3, 589–601. [Google Scholar] [CrossRef]
- Nyberg, P.; Xie, L.; Sugimoto, H.; Colorado, P.; Sund, M.; Holthaus, K.; Sudhakar, A.; Salo, T.; Kalluri, R. Characterization of the anti-angiogenic properties of arresten, an alpha1beta1 integrin-dependent collagen-derived tumor suppressor. Exp. Cell Res. 2008, 314, 3292–3305. [Google Scholar] [CrossRef] [PubMed]
- Kamphaus, G.D.; Colorado, P.C.; Panka, D.J.; Hopfer, H.; Ramchandran, R.; Torre, A.; Maeshima, Y.; Mier, J.W.; Sukhatme, V.P.; Kalluri, R. Canstatin, a novel matrix-derived inhibitor of angiogenesis and tumor growth. J. Biol. Chem. 2000, 275, 1209–1215. [Google Scholar] [CrossRef]
- Hamano, Y.; Kalluri, R. Tumstatin, the NC1 domain of alpha3 chain of type IV collagen, is an endogenous inhibitor of pathological angiogenesis and suppresses tumor growth. Biochem. Biophys. Res. Commun. 2005, 333, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Karagiannis, E.D.; Popel, A.S. Identification of novel short peptides derived from the alpha 4, alpha 5, and alpha 6 fibrils of type IV collagen with anti-angiogenic properties. Biochem. Biophys. Res. Commun. 2007, 354, 434–439. [Google Scholar] [CrossRef]
- Brassart-Pasco, S.; Senechal, K.; Thevenard, J.; Ramont, L.; Devy, J.; Di Stefano, L.; Dupont-Deshorgue, A.; Brezillon, S.; Feru, J.; Jazeron, J.F.; et al. Tetrastatin, the NC1 domain of the alpha4(IV) collagen chain: A novel potent anti-tumor matrikine. PLoS ONE 2012, 7, e29587. [Google Scholar] [CrossRef]
- Weckmann, M.; Moir, L.M.; Heckman, C.A.; Black, J.L.; Oliver, B.G.; Burgess, J.K. Lamstatin—A novel inhibitor of lymphangiogenesis derived from collagen IV. J. Cell. Mol. Med. 2012, 16, 3062–3073. [Google Scholar] [CrossRef]
- Magnon, C.; Galaup, A.; Mullan, B.; Rouffiac, V.; Bouquet, C.; Bidart, J.M.; Griscelli, F.; Opolon, P.; Perricaudet, M. Canstatin acts on endothelial and tumor cells via mitochondrial damage initiated through interaction with alphavbeta3 and alphavbeta5 integrins. Cancer Res. 2005, 65, 4353–4361. [Google Scholar] [CrossRef]
- Torres, P.H.; Sousa, G.L.; Pascutti, P.G. Structural analysis of the N-terminal fragment of the antiangiogenic protein endostatin: A molecular dynamics study. Proteins 2011, 79, 2684–2692. [Google Scholar] [CrossRef]
- Mundel, T.M.; Yliniemi, A.M.; Maeshima, Y.; Sugimoto, H.; Kieran, M.; Kalluri, R. Type IV collagen alpha6 chain-derived noncollagenous domain 1 (alpha6(IV)NC1) inhibits angiogenesis and tumor growth. Int. J. Cancer 2008, 122, 1738–1744. [Google Scholar] [CrossRef]
- Petitclerc, E.; Boutaud, A.; Prestayko, A.; Xu, J.; Sado, Y.; Ninomiya, Y.; Sarras, M.P., Jr.; Hudson, B.G.; Brooks, P.C. New functions for non-collagenous domains of human collagen type IV. Novel integrin ligands inhibiting angiogenesis and tumor growth in vivo. J. Biol. Chem. 2000, 275, 8051–8061. [Google Scholar] [CrossRef] [PubMed]
- Veidal, S.S.; Karsdal, M.A.; Nawrocki, A.; Larsen, M.R.; Dai, Y.; Zheng, Q.; Hägglund, P.; Vainer, B.; Skjøt-Arkil, H.; Leeming, D.J. Assessment of proteolytic degradation of the basement membrane: A fragment of type IV collagen as a biochemical marker for liver fibrosis. Fibrogenes. Tissue Repair 2011, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Heljasvaara, R.; Nyberg, P.; Luostarinen, J.; Parikka, M.; Heikkilä, P.; Rehn, M.; Sorsa, T.; Salo, T.; Pihlajaniemi, T. Generation of biologically active endostatin fragments from human collagen XVIII by distinct matrix metalloproteases. Exp. Cell Res. 2005, 307, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Veillard, F.; Saidi, A.; Burden, R.E.; Scott, C.J.; Gillet, L.; Lecaille, F.; Lalmanach, G. Cysteine cathepsins S and L modulate anti-angiogenic activities of human endostatin. J. Biol. Chem. 2011, 286, 37158–37167. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Moses, M.A.; Wiederschain, D.; Arbiser, J.L.; Folkman, J. The generation of endostatin is mediated by elastase. Cancer Res. 1999, 59, 6052–6056. [Google Scholar] [PubMed]
- Folkman, J. Antiangiogenesis in cancer therapy--endostatin and its mechanisms of action. Exp. Cell Res. 2006, 312, 594–607. [Google Scholar] [CrossRef]
- Shi, H.; Huang, Y.; Zhou, H.; Song, X.; Yuan, S.; Fu, Y.; Luo, Y. Nucleolin is a receptor that mediates antiangiogenic and antitumor activity of endostatin. Blood 2007, 110, 2899–2906. [Google Scholar] [CrossRef]
- Wickström, S.A.; Alitalo, K.; Keski-Oja, J. Endostatin associates with integrin alpha5beta1 and caveolin-1, and activates Src via a tyrosyl phosphatase-dependent pathway in human endothelial cells. Cancer Res. 2002, 62, 5580–5589. [Google Scholar]
- Kim, Y.M.; Hwang, S.; Kim, Y.M.; Pyun, B.J.; Kim, T.Y.; Lee, S.T.; Gho, Y.S.; Kwon, Y.G. Endostatin blocks vascular endothelial growth factor-mediated signaling via direct interaction with KDR/Flk-1. J. Biol. Chem. 2002, 277, 27872–27879. [Google Scholar] [CrossRef]
- Karumanchi, S.A.; Jha, V.; Ramchandran, R.; Karihaloo, A.; Tsiokas, L.; Chan, B.; Dhanabal, M.; Hanai, J.I.; Venkataraman, G.; Shriver, Z.; et al. Cell surface glypicans are low-affinity endostatin receptors. Mol. Cell. 2001, 7, 811–822. [Google Scholar] [CrossRef]
- Lin, H.C.; Chang, J.H.; Jain, S.; Gabison, E.E.; Kure, T.; Kato, T.; Fukai, N.; Azar, D.T. Matrilysin cleavage of corneal collagen type XVIII NC1 domain and generation of a 28-kDa fragment. Investig. Ophthalmol. Vis. Sci. 2001, 42, 2517–2524. [Google Scholar]
- Chang, J.H.; Javier, J.A.; Chang, G.Y.; Oliveira, H.B.; Azar, D.T. Functional characterization of neostatins, the MMP-derived, enzymatic cleavage products of type XVIII collagen. FEBS Lett. 2005, 579, 3601–3606. [Google Scholar] [CrossRef] [PubMed]
- Arai, M.; Yada, T.; Suzuki, S.; Kimata, K. Isolation and characterization of type IX collagen-proteoglycan from the Swarm rat chondrosarcoma. Biochim. Biophys. Acta 1992, 1117, 60–70. [Google Scholar] [CrossRef]
- Oudart, J.B.; Brassart-Pasco, S.; Vautrin, A.; Sellier, C.; Machado, C.; Dupont-Deshorgue, A.; Brassart, B.; Baud, S.; Dauchez, M.; Monboisse, J.C.; et al. Plasmin releases the anti-tumor peptide from the NC1 domain of collagen XIX. Oncotarget 2015, 6, 3656–3668. [Google Scholar] [CrossRef]
- Ramont, L.; Brassart-Pasco, S.; Thevenard, J.; Deshorgue, A.; Venteo, L.; Laronze, J.Y.; Pluot, M.; Monboisse, J.C.; Maquart, F.X. The NC1 domain of type XIX collagen inhibits in vivo melanoma growth. Mol. Cancer Ther. 2007, 6, 506–514. [Google Scholar] [CrossRef]
- Oudart, J.B.; Doué, M.; Vautrin, A.; Brassart, B.; Sellier, C.; Dupont-Deshorgue, A.; Monboisse, J.C.; Maquart, F.X.; Brassart-Pasco, S.; Ramont, L. The anti-tumor NC1 domain of collagen XIX inhibits the FAK/ PI3K/Akt/mTOR signaling pathway through αvβ3 integrin interaction. Oncotarget 2016, 7, 1516–1528. [Google Scholar] [CrossRef]
- Nagase, H.; Fields, G.B. Human matrix metalloproteinase specificity studies using collagen sequence-based synthetic peptides. Biopolymers 1996, 40, 399–416. [Google Scholar] [CrossRef]
- Halper, J.; Kjaer, M. Basic components of connective tissues and extracellular matrix: Elastin, fibrillin, fibulins, fibrinogen, fibronectin, laminin, tenascins and thrombospondins. Adv. Exp. Med. Biol. 2014, 802, 31–47. [Google Scholar] [CrossRef]
- Mithieux, S.M.; Weiss, A.S. Elastin. Adv. Protein Chem. 2005, 70, 437–461. [Google Scholar] [CrossRef]
- Scandolera, A.; Odoul, L.; Salesse, S.; Guillot, A.; Blaise, S.; Kawecki, C.; Maurice, P.; El Btaouri, H.; Romier-Crouzet, B.; Martiny, L.; et al. The Elastin Receptor Complex: A Unique Matricellular Receptor with High Anti-tumoral Potential. Front. Pharmacol. 2016, 7, 32. [Google Scholar] [CrossRef]
- Duca, L.; Floquet, N.; Alix, A.J.; Haye, B.; Debelle, L. Elastin as a matrikine. Crit. Rev. Oncol. Hematol. 2004, 49, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Grahovac, J.; Wells, A. Matrikine and matricellular regulators of EGF receptor signaling on cancer cell migration and invasion. Lab. Investig. 2014, 94, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Cain, S.A.; Mularczyk, E.J.; Singh, M.; Massam-Wu, T.; Kielty, C.M. ADAMTS-10 and -6 differentially regulate cell-cell junctions and focal adhesions. Sci. Rep. 2016, 6, 35956. [Google Scholar] [CrossRef] [PubMed]
- Bax, D.V.; Mahalingam, Y.; Cain, S.; Mellody, K.; Freeman, L.; Younger, K.; Shuttleworth, C.A.; Humphries, M.J.; Couchman, J.R.; Kielty, C.M. Cell adhesion to fibrillin-1: Identification of an Arg-Gly-Asp-dependent synergy region and a heparin-binding site that regulates focal adhesion formation. J. Cell Sci. 2007, 120, 1383–1392. [Google Scholar] [CrossRef] [PubMed]
- Jovanovic, J.; Iqbal, S.; Jensen, S.; Mardon, H.; Handford, P. Fibrillin-integrin interactions in health and disease. Biochem. Soc. Trans. 2008, 36, 257–262. [Google Scholar] [CrossRef]
- Han, Z.; Lu, Z.R. Targeting Fibronectin for Cancer Imaging and Therapy. J. Mater. Chem. B 2017, 5, 639–654. [Google Scholar] [CrossRef]
- Kumra, H.; Reinhardt, D.P. Fibronectin-targeted drug delivery in cancer. Adv. Drug Deliv. Rev. 2016, 97, 101–110. [Google Scholar] [CrossRef]
- Mezzenga, R.; Mitsi, M. The Molecular Dance of Fibronectin: Conformational Flexibility Leads to Functional Versatility. Biomacromolecules 2018. [Google Scholar] [CrossRef]
- Zollinger, A.J.; Smith, M.L. Fibronectin, the extracellular glue. Matrix Biol. 2017, 60-61, 27–37. [Google Scholar] [CrossRef]
- Joshi, R.; Goihberg, E.; Ren, W.; Pilichowska, M.; Mathew, P. Proteolytic fragments of fibronectin function as matrikines driving the chemotactic affinity of prostate cancer cells to human bone marrow mesenchymal stromal cells via the alpha5beta1 integrin. Cell Adh. Migr. 2017, 11, 305–315. [Google Scholar] [CrossRef][Green Version]
- Yi, M.; Ruoslahti, E. A fibronectin fragment inhibits tumor growth, angiogenesis, and metastasis. Proc. Natl. Acad. Sci. USA 2001, 98, 620–624. [Google Scholar] [CrossRef] [PubMed]
- Bossard, C.; Van den Berghe, L.; Laurell, H.; Castano, C.; Cerutti, M.; Prats, A.C.; Prats, H. Antiangiogenic properties of fibstatin, an extracellular FGF-2-binding polypeptide. Cancer Res. 2004, 64, 7507–7512. [Google Scholar] [CrossRef] [PubMed]
- Topalovski, M.; Brekken, R.A. Matrix control of pancreatic cancer: New insights into fibronectin signaling. Cancer Lett. 2016, 381, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Bachman, H.; Nicosia, J.; Dysart, M.; Barker, T.H. Utilizing Fibronectin Integrin-Binding Specificity to Control Cellular Responses. Adv. Wound Care 2015, 4, 501–511. [Google Scholar] [CrossRef]
- Wang, K.; Seo, B.R.; Fischbach, C.; Gourdon, D. Fibronectin Mechanobiology Regulates Tumorigenesis. Cell. Mol. Bioeng. 2016, 9, 1–11. [Google Scholar] [CrossRef]
- Prasad, A.; Clark, R.A. Fibronectin interaction with growth factors in the context of general ways extracellular matrix molecules regulate growth factor signaling. G. Ital. Dermatol. Venereol. 2018, 153, 361–374. [Google Scholar] [CrossRef]
- White, E.S.; Baralle, F.E.; Muro, A.F. New insights into form and function of fibronectin splice variants. J. Pathol. 2008, 216, 1–14. [Google Scholar] [CrossRef]
- Faron, G.; Balepa, L.; Parra, J.; Fils, J.F.; Gucciardo, L. The fetal fibronectin test: 25 years after its development, what is the evidence regarding its clinical utility? A systematic review and meta-analysis. J. Matern. Fetal Neonatal Med. 2018, 33, 1–31. [Google Scholar] [CrossRef]
- Sawicka, K.M.; Seeliger, M.; Musaev, T.; Macri, L.K.; Clark, R.A. Fibronectin Interaction and Enhancement of Growth Factors: Importance for Wound Healing. Adv. Wound Care 2015, 4, 469–478. [Google Scholar] [CrossRef]
- Wang, Y.; Ni, H. Fibronectin maintains the balance between hemostasis and thrombosis. Cell. Mol. Life Sci. 2016, 73, 3265–3277. [Google Scholar] [CrossRef]
- Mercuri, F.A.; Maciewicz, R.A.; Tart, J.; Last, K.; Fosang, A.J. Mutations in the interglobular domain of aggrecan alter matrix metalloproteinase and aggrecanase cleavage patterns. Evidence that matrix metalloproteinase cleavage interferes with aggrecanase activity. J. Biol. Chem. 2000, 275, 33038–33045. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, H.; Leung, T.C.; Hoffman, S.; Balsamo, J.; Lilien, J. Coordinate regulation of cadherin and integrin function by the chondroitin sulfate proteoglycan neurocan. J. Cell Biol. 2000, 149, 1275–1288. [Google Scholar] [CrossRef]
- Mohan, V.; Wyatt, E.V.; Gotthard, I.; Phend, K.D.; Diestel, S.; Duncan, B.W.; Weinberg, R.J.; Tripathy, A.; Maness, P.F. Neurocan Inhibits Semaphorin 3F Induced Dendritic Spine Remodeling Through NrCAM in Cortical Neurons. Front. Cell. Neurosci. 2018, 12, 346. [Google Scholar] [CrossRef] [PubMed]
- Viapiano, M.S.; Hockfield, S.; Matthews, R.T. BEHAB/brevican requires ADAMTS-mediated proteolytic cleavage to promote glioma invasion. J. Neurooncol. 2008, 88, 261–272. [Google Scholar] [CrossRef]
- Demircan, K.; Topcu, V.; Takigawa, T.; Akyol, S.; Yonezawa, T.; Ozturk, G.; Ugurcu, V.; Hasgul, R.; Yigitoglu, M.R.; Akyol, O.; et al. ADAMTS4 and ADAMTS5 knockout mice are protected from versican but not aggrecan or brevican proteolysis during spinal cord injury. Biomed. Res. Int. 2014, 2014, 693746. [Google Scholar] [CrossRef] [PubMed]
- Hope, C.; Emmerich, P.B.; Papadas, A.; Pagenkopf, A.; Matkowskyj, K.A.; Van De Hey, D.R.; Payne, S.N.; Clipson, L.; Callander, N.S.; Hematti, P.; et al. Versican-Derived Matrikines Regulate Batf3-Dendritic Cell Differentiation and Promote T Cell Infiltration in Colorectal Cancer. J. Immunol. 2017, 199, 1933–1941. [Google Scholar] [CrossRef] [PubMed]
- Papadas, A.; Asimakopoulos, F. Versican in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1272, 55–72. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, L.; Zheng, P.S.; Yang, B.B. beta 1-Integrin-mediated glioma cell adhesion and free radical-induced apoptosis are regulated by binding to a C-terminal domain of PG-M/versican. J. Biol. Chem. 2002, 277, 12294–12301. [Google Scholar] [CrossRef]
- Overall, C.M. Molecular determinants of metalloproteinase substrate specificity: Matrix metalloproteinase substrate binding domains, modules, and exosites. Mol. Biotechnol. 2002, 22, 51–86. [Google Scholar] [CrossRef]
- Iozzo, R.V.; Moscatello, D.K.; McQuillan, D.J.; Eichstetter, I. Decorin is a biological ligand for the epidermal growth factor receptor. J. Biol. Chem. 1999, 274, 4489–4492. [Google Scholar] [CrossRef]
- Moreth, K.; Iozzo, R.V.; Schaefer, L. Small leucine-rich proteoglycans orchestrate receptor crosstalk during inflammation. Cell Cycle 2012, 11, 2084–2091. [Google Scholar] [CrossRef] [PubMed]
- Goldoni, S.; Humphries, A.; Nystrom, A.; Sattar, S.; Owens, R.T.; McQuillan, D.J.; Ireton, K.; Iozzo, R.V. Decorin is a novel antagonistic ligand of the Met receptor. J. Cell Biol. 2009, 185, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Khan, G.A.; Girish, G.V.; Lala, N.; Di Guglielmo, G.M.; Lala, P.K. Decorin is a novel VEGFR-2-binding antagonist for the human extravillous trophoblast. Mol. Endocrinol. 2011, 25, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- Hausser, H.; Wedekind, P.; Sperber, T.; Peters, R.; Hasilik, A.; Kresse, H. Isolation and cellular localization of the decorin endocytosis receptor. Eur. J. Cell Biol. 1996, 71, 325–331. [Google Scholar]
- Nastase, M.V.; Young, M.F.; Schaefer, L. Biglycan: A multivalent proteoglycan providing structure and signals. J. Histochem. Cytochem. 2012, 60, 963–975. [Google Scholar] [CrossRef]
- Grindel, B.; Li, Q.; Arnold, R.; Petros, J.; Zayzafoon, M.; Muldoon, M.; Stave, J.; Chung, L.W.; Farach-Carson, M.C. Perlecan/HSPG2 and matrilysin/MMP-7 as indices of tissue invasion: Tissue localization and circulating perlecan fragments in a cohort of 288 radical prostatectomy patients. Oncotarget 2016, 7, 10433–10447. [Google Scholar] [CrossRef]
- Cailhier, J.F.; Sirois, I.; Laplante, P.; Lepage, S.; Raymond, M.A.; Brassard, N.; Prat, A.; Iozzo, R.V.; Pshezhetsky, A.V.; Hébert, M.J. Caspase-3 activation triggers extracellular cathepsin L release and endorepellin proteolysis. J. Biol. Chem. 2008, 283, 27220–27229. [Google Scholar] [CrossRef]
- Gubbiotti, M.A.; Neill, T.; Iozzo, R.V. A current view of perlecan in physiology and pathology: A mosaic of functions. Matrix Biol. 2017, 57-58, 285–298. [Google Scholar] [CrossRef]
- Goyal, A.; Pal, N.; Concannon, M.; Paul, M.; Doran, M.; Poluzzi, C.; Sekiguchi, K.; Whitelock, J.M.; Neill, T.; Iozzo, R.V. Endorepellin, the angiostatic module of perlecan, interacts with both the α2β1 integrin and vascular endothelial growth factor receptor 2 (VEGFR2): A dual receptor antagonism. J. Biol. Chem. 2011, 286, 25947–25962. [Google Scholar] [CrossRef]
- Gonzalez, E.M.; Reed, C.C.; Bix, G.; Fu, J.; Zhang, Y.; Gopalakrishnan, B.; Greenspan, D.S.; Iozzo, R.V. BMP-1/Tolloid-like metalloproteases process endorepellin, the angiostatic C-terminal fragment of perlecan. J. Biol. Chem. 2005, 280, 7080–7087. [Google Scholar] [CrossRef]
- Titz, B.; Dietrich, S.; Sadowski, T.; Beck, C.; Petersen, A.; Sedlacek, R. Activity of MMP-19 inhibits capillary-like formation due to processing of nidogen-1. Cell. Mol. Life Sci. 2004, 61, 1826–1833. [Google Scholar] [CrossRef] [PubMed]
- Sage, J.; Leblanc-Noblesse, E.; Nizard, C.; Sasaki, T.; Schnebert, S.; Perrier, E.; Kurfurst, R.; Brömme, D.; Lalmanach, G.; Lecaille, F. Cleavage of nidogen-1 by cathepsin S impairs its binding to basement membrane partners. PLoS ONE 2012, 7, e43494. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.D.; Kaushal, G.P.; Shah, S.V. Meprin A, the major matrix degrading enzyme in renal tubules, produces a novel nidogen fragment in vitro and in vivo. Kidney Int. 1998, 53, 1673–1680. [Google Scholar] [CrossRef]
- Beck, K.; Brodsky, B. Supercoiled protein motifs: The collagen triple-helix and the alpha-helical coiled coil. J. Struct. Biol. 1998, 122, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Kikkawa, Y.; Hozumi, K.; Katagiri, F.; Nomizu, M.; Kleinman, H.K.; Koblinski, J.E. Laminin-111-derived peptides and cancer. Cell Adh. Migr. 2013, 7, 150–256. [Google Scholar] [CrossRef] [PubMed]
- Adair-Kirk, T.L.; Atkinson, J.J.; Broekelmann, T.J.; Doi, M.; Tryggvason, K.; Miner, J.H.; Mecham, R.P.; Senior, R.M. A site on laminin alpha 5, AQARSAASKVKVSMKF, induces inflammatory cell production of matrix metalloproteinase-9 and chemotaxis. J. Immunol. 2003, 171, 398–406. [Google Scholar] [CrossRef]
- Woods, A.; Longley, R.L.; Tumova, S.; Couchman, J.R. Syndecan-4 binding to the high affinity heparin-binding domain of fibronectin drives focal adhesion formation in fibroblasts. Arch. Biochem. Biophys. 2000, 374, 66–72. [Google Scholar] [CrossRef]
- Carulli, S.; Beck, K.; Dayan, G.; Boulesteix, S.; Lortat-Jacob, H.; Rousselle, P. Cell surface proteoglycans syndecan-1 and -4 bind overlapping but distinct sites in laminin alpha3 LG45 protein domain. J. Biol. Chem. 2012, 287, 12204–12216. [Google Scholar] [CrossRef]
- Bachy, S.; Letourneur, F.; Rousselle, P. Syndecan-1 interaction with the LG4/5 domain in laminin-332 is essential for keratinocyte migration. J. Cell. Physiol. 2008, 214, 238–249. [Google Scholar] [CrossRef]
- Ramovs, V.; Te Molder, L.; Sonnenberg, A. The opposing roles of laminin-binding integrins in cancer. Matrix Biol. 2017, 57-58, 213–243. [Google Scholar] [CrossRef]
- Rousselle, P.; Beck, K. Laminin 332 processing impacts cellular behavior. Cell Adh. Migr. 2013, 7, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Tsuruta, D.; Kobayashi, H.; Imanishi, H.; Sugawara, K.; Ishii, M.; Jones, J.C. Laminin-332-integrin interaction: A target for cancer therapy? Curr. Med. Chem. 2008, 15, 1968–1975. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Sekiguchi, K. Molecular Basis of Laminin-Integrin Interactions. Curr. Top. Membr. 2015, 76, 197–229. [Google Scholar] [CrossRef]
- Eble, J.A.; Wucherpfennig, K.W.; Gauthier, L.; Dersch, P.; Krukonis, E.; Isberg, R.R.; Hemler, M.E. Recombinant soluble human alpha 3 beta 1 integrin: Purification, processing, regulation, and specific binding to laminin-5 and invasin in a mutually exclusive manner. Biochemistry 1998, 37, 10945–10955. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.G.; An, H.J.; Kang, S.; Choi, Y.P.; Gao, M.Q.; Park, H.; Cho, N.H. Laminin-332-rich tumor microenvironment for tumor invasion in the interface zone of breast cancer. Am. J. Pathol. 2011, 178, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Marinkovich, M.P. Tumour microenvironment: Laminin 332 in squamous-cell carcinoma. Nat. Rev. Cancer. 2007, 7, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, W.; Wei, J.; Zhou, D.; Zhao, X.; Song, W.; Sun, Q.; Huang, P.; Zheng, S. Overexpression of beta3 Chains of Laminin-332 is Associated With Clinicopathologic Features and Decreased Survival in Patients With Pancreatic Adenocarcinoma. Appl. Immunohistochem. Mol. Morphol. 2015, 23, 516–521. [Google Scholar] [CrossRef]
- Guess, C.M.; Lafleur, B.J.; Weidow, B.L.; Quaranta, V. A decreased ratio of laminin-332 beta3 to gamma2 subunit mRNA is associated with poor prognosis in colon cancer. Cancer Epidemiol. Biomark. Prev. 2009, 18, 1584–1590. [Google Scholar] [CrossRef]
- Katayama, M.; Funakoshi, A.; Sumii, T.; Sanzen, N.; Sekiguchi, K. Laminin gamma2-chain fragment circulating level increases in patients with metastatic pancreatic ductal cell adenocarcinomas. Cancer Lett. 2005, 225, 167–176. [Google Scholar] [CrossRef]
- Hibino, S.; Shibuya, M.; Engbring, J.A.; Mochizuki, M.; Nomizu, M.; Kleinman, H.K. Identification of an active site on the laminin alpha5 chain globular domain that binds to CD44 and inhibits malignancy. Cancer Res. 2004, 64, 4810–4816. [Google Scholar] [CrossRef]
- Kusuma, N.; Anderson, R.L.; Pouliot, N. Laminin α5-derived peptides modulate the properties of metastatic breast tumour cells. Clin. Exp. Metastasis 2011, 28, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Su, J.L.; Chiou, J.; Tang, C.H.; Zhao, M.; Tsai, C.H.; Chen, P.S.; Chang, Y.W.; Chien, M.H.; Peng, C.Y.; Hsiao, M.; et al. CYR61 regulates BMP-2-dependent osteoblast differentiation through the {alpha}v{beta}3 integrin/integrin-linked kinase/ERK pathway. J. Biol. Chem. 2010, 285, 31325–31336. [Google Scholar] [CrossRef] [PubMed]
- Crockett, J.C.; Schütze, N.; Tosh, D.; Jatzke, S.; Duthie, A.; Jakob, F.; Rogers, M.J. The matricellular protein CYR61 inhibits osteoclastogenesis by a mechanism independent of alphavbeta3 and alphavbeta5. Endocrinology 2007, 148, 5761–5768. [Google Scholar] [CrossRef]
- Chen, C.C.; Young, J.L.; Monzon, R.I.; Chen, N.; Todorovic, V.; Lau, L.F. Cytotoxicity of TNFalpha is regulated by integrin-mediated matrix signaling. EMBO J. 2007, 26, 1257–1267. [Google Scholar] [CrossRef] [PubMed]
- Kaasboll, O.J.; Gadicherla, A.K.; Wang, J.H.; Monsen, V.T.; Hagelin, E.M.V.; Dong, M.Q.; Attramadal, H. Connective tissue growth factor (CCN2) is a matricellular preproprotein controlled by proteolytic activation. J. Biol. Chem. 2018, 293, 17953–17970. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.C.; Chang, A.C.; Tsai, C.H.; Huang, Y.L.; Gan, L.; Chen, C.K.; Liu, S.C.; Huang, T.Y.; Fong, Y.C.; Tang, C.H. CCN2 promotes drug resistance in osteosarcoma by enhancing ABCG2 expression. J. Cell. Physiol. 2019, 234, 9297–9307. [Google Scholar] [CrossRef]
- Babic, A.M.; Chen, C.C.; Lau, L.F. Fisp12/mouse connective tissue growth factor mediates endothelial cell adhesion and migration through integrin alphavbeta3, promotes endothelial cell survival, and induces angiogenesis in vivo. Mol. Cell. Biol. 1999, 19, 2958–2966. [Google Scholar] [CrossRef]
- Hutchenreuther, J.; Vincent, K.; Norley, C.; Racanelli, M.; Gruber, S.B.; Johnson, T.M.; Fullen, D.R.; Raskin, L.; Perbal, B.; Holdsworth, D.W.; et al. Activation of cancer-associated fibroblasts is required for tumor neovascularization in a murine model of melanoma. Matrix Biol. 2018. [Google Scholar] [CrossRef]
- Qiao, L.; Liang, N.; Zhang, J.; Xie, J.; Liu, F.; Xu, D.; Yu, X.; Tian, Y. Advanced research on vasculogenic mimicry in cancer. J. Cell. Mol. Med. 2015, 19, 315–326. [Google Scholar] [CrossRef]
- Seftor, R.E.; Seftor, E.A.; Kirschmann, D.A.; Hendrix, M.J. Targeting the tumor microenvironment with chemically modified tetracyclines: Inhibition of laminin 5 gamma2 chain promigratory fragments and vasculogenic mimicry. Mol. Cancer. Ther. 2002, 1, 1173–1179. [Google Scholar]
- Velez, D.O.; Tsui, B.; Goshia, T.; Chute, C.L.; Han, A.; Carter, H.; Fraley, S.I. 3D collagen architecture induces a conserved migratory and transcriptional response linked to vasculogenic mimicry. Nat. Commun. 2017, 8, 1651. [Google Scholar] [CrossRef]
- Adams, J.C.; Chiquet-Ehrismann, R.; Tucker, R.P. The evolution of tenascins and fibronectin. Cell Adh. Migr. 2015, 9, 22–33. [Google Scholar] [CrossRef]
- Chiquet-Ehrismann, R.; Hagios, C.; Matsumoto, K. The tenascin gene family. Perspect. Dev. Neurobiol. 1994, 2, 3–7. [Google Scholar]
- Yoshida, T.; Akatsuka, T.; Imanaka-Yoshida, K. Tenascin-C and integrins in cancer. Cell Adh. Migr. 2015, 9, 96–104. [Google Scholar] [CrossRef]
- Martina, E.; Chiquet-Ehrismann, R.; Brellier, F. Tenascin-W: An extracellular matrix protein associated with osteogenesis and cancer. Int. J. Biochem. Cell. Biol. 2010, 42, 1412–1415. [Google Scholar] [CrossRef]
- Brellier, F.; Martina, E.; Degen, M.; Heuze-Vourc’h, N.; Petit, A.; Kryza, T.; Courty, Y.; Terracciano, L.; Ruiz, C.; Chiquet-Ehrismann, R. Tenascin-W is a better cancer biomarker than tenascin-C for most human solid tumors. BMC Clin. Pathol. 2012, 12, 14. [Google Scholar] [CrossRef]
- Martina, E.; Degen, M.; Ruegg, C.; Merlo, A.; Lino, M.M.; Chiquet-Ehrismann, R.; Brellier, F. Tenascin-W is a specific marker of glioma-associated blood vessels and stimulates angiogenesis in vitro. FASEB J. 2010, 24, 778–787. [Google Scholar] [CrossRef]
- Castello, L.M.; Raineri, D.; Salmi, L.; Clemente, N.; Vaschetto, R.; Quaglia, M.; Garzaro, M.; Gentilli, S.; Navalesi, P.; Cantaluppi, V.; et al. Osteopontin at the Crossroads of Inflammation and Tumor Progression. Mediators Inflamm. 2017, 2017, 4049098. [Google Scholar] [CrossRef]
- Kruger, T.E.; Miller, A.H.; Godwin, A.K.; Wang, J. Bone sialoprotein and osteopontin in bone metastasis of osteotropic cancers. Crit. Rev. Oncol. Hematol. 2014, 89, 330–341. [Google Scholar] [CrossRef]
- Gao, Y.A.; Agnihotri, R.; Vary, C.P.; Liaw, L. Expression and characterization of recombinant osteopontin peptides representing matrix metalloproteinase proteolytic fragments. Matrix Biol. 2004, 23, 457–466. [Google Scholar] [CrossRef]
- Ye, Q.H.; Qin, L.X.; Forgues, M.; He, P.; Kim, J.W.; Peng, A.C.; Simon, R.; Li, Y.; Robles, A.I.; Chen, Y.; et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat. Med. 2003, 9, 416–423. [Google Scholar] [CrossRef]
- Gonzalez-Gonzalez, L.; Alonso, J. Periostin: A Matricellular Protein With Multiple Functions in Cancer Development and Progression. Front. Oncol. 2018, 8, 225. [Google Scholar] [CrossRef]
- Gillan, L.; Matei, D.; Fishman, D.A.; Gerbin, C.S.; Karlan, B.Y.; Chang, D.D. Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res. 2002, 62, 5358–5364. [Google Scholar]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef]
- Micke, P.; Ostman, A. Tumour-stroma interaction: Cancer-associated fibroblasts as novel targets in anti-cancer therapy? Lung Cancer 2004, 45 (Suppl. S2), S163–S175. [Google Scholar] [CrossRef]
- Leprini, A.; Querze, G.; Zardi, L. Tenascin isoforms: Possible targets for diagnosis and therapy of cancer and mechanisms regulating their expression. Perspect. Dev. Neurobiol. 1994, 2, 117–123. [Google Scholar]
- Nicolo, G.; Salvi, S.; Oliveri, G.; Borsi, L.; Castellani, P.; Zardi, L. Expression of tenascin and of the ED-B containing oncofetal fibronectin isoform in human cancer. Cell. Differ. Dev. 1990, 32, 401–408. [Google Scholar] [CrossRef]
- Gritsenko, P.G.; Ilina, O.; Friedl, P. Interstitial guidance of cancer invasion. J. Pathol. 2012, 226, 185–199. [Google Scholar] [CrossRef]
- Kakizaki, Y.; Makino, N.; Tozawa, T.; Honda, T.; Matsuda, A.; Ikeda, Y.; Ito, M.; Saito, Y.; Kimura, W.; Ueno, Y. Stromal Fibrosis and Expression of Matricellular Proteins Correlate With Histological Grade of Intraductal Papillary Mucinous Neoplasm of the Pancreas. Pancreas 2016, 45, 1145–1152. [Google Scholar] [CrossRef][Green Version]
- Viloria, K.; Hill, N.J. Embracing the complexity of matricellular proteins: The functional and clinical significance of splice variation. Biomol. Concepts 2016, 7, 117–132. [Google Scholar] [CrossRef]
- Naschberger, E.; Liebl, A.; Schellerer, V.S.; Schütz, M.; Britzen-Laurent, N.; Kölbel, P.; Schaal, U.; Haep, L.; Regensburger, D.; Wittmann, T.; et al. Matricellular protein SPARCL1 regulates tumor microenvironment-dependent endothelial cell heterogeneity in colorectal carcinoma. J. Clin. Investig. 2016, 126, 4187–4204. [Google Scholar] [CrossRef] [PubMed]
- Kamili, N.A.; Arthur, C.M.; Gerner-Smidt, C.; Tafesse, E.; Blenda, A.; Dias-Baruffi, M.; Stowell, S.R. Key regulators of galectin-glycan interactions. Proteomics 2016, 16, 3111–3125. [Google Scholar] [CrossRef] [PubMed]
- Thijssen, V.L.; Rabinovich, G.A.; Griffioen, A.W. Vascular galectins: Regulators of tumor progression and targets for cancer therapy. Cytokine Growth Factor Rev. 2013, 24, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Huergo, S.P.; Blidner, A.G.; Rabinovich, G.A. Galectins: Emerging regulatory checkpoints linking tumor immunity and angiogenesis. Curr. Opin. Immunol. 2017, 45, 8–15. [Google Scholar] [CrossRef]
- Bellahcène, A.; Castronovo, V.; Ogbureke, K.U.; Fisher, L.W.; Fedarko, N.S. Small integrin-binding ligand N-linked glycoproteins (SIBLINGs): Multifunctional proteins in cancer. Nat. Rev. Cancer. 2008, 8, 212–226. [Google Scholar] [CrossRef]
- Barbouri, D.; Afratis, N.; Gialeli, C.; Vynios, D.H.; Theocharis, A.D.; Karamanos, N.K. Syndecans as modulators and potential pharmacological targets in cancer progression. Front. Oncol. 2014, 4, 4. [Google Scholar] [CrossRef]
- Rapraeger, A.C. Synstatin: A selective inhibitor of the syndecan-1-coupled IGF1R-αvβ3 integrin complex in tumorigenesis and angiogenesis. FEBS J. 2013, 280, 2207–2215. [Google Scholar] [CrossRef]
- Rapraeger, A.C.; Ell, B.J.; Roy, M.; Li, X.; Morrison, O.R.; Thomas, G.M.; Beauvais, D.M. Vascular endothelial-cadherin stimulates syndecan-1-coupled insulin-like growth factor-1 receptor and cross-talk between αVβ3 integrin and vascular endothelial growth factor receptor 2 at the onset of endothelial cell dissemination during angiogenesis. FEBS J. 2013, 280, 2194–2206. [Google Scholar] [CrossRef]
- Wang, H.; Jin, H.; Rapraeger, A.C. Syndecan-1 and Syndecan-4 Capture Epidermal Growth Factor Receptor Family Members and the α3β1 Integrin Via Binding Sites in Their Ectodomains: Novel synstatins prevent kinase capture and inhibit α6β4-integrin-dependent epithelial cell motility. J. Biol. Chem. 2015, 290, 26103–26113. [Google Scholar] [CrossRef]
- Beauvais, D.M.; Ell, B.J.; McWhorter, A.R.; Rapraeger, A.C. Syndecan-1 regulates alphavbeta3 and alphavbeta5 integrin activation during angiogenesis and is blocked by synstatin, a novel peptide inhibitor. J. Exp. Med. 2009, 206, 691–705. [Google Scholar] [CrossRef]
- Jung, O.; Trapp-Stamborski, V.; Purushothaman, A.; Jin, H.; Wang, H.; Sanderson, R.D.; Rapraeger, A.C. Heparanase-induced shedding of syndecan-1/CD138 in myeloma and endothelial cells activates VEGFR2 and an invasive phenotype: Prevention by novel synstatins. Oncogenesis 2016, 5, e202. [Google Scholar] [CrossRef] [PubMed]
- Stephan, A.; Mateos, J.M.; Kozlov, S.V.; Cinelli, P.; Kistler, A.D.; Hettwer, S.; Rülicke, T.; Streit, P.; Kunz, B.; Sonderegger, P. Neurotrypsin cleaves agrin locally at the synapse. FASEB J. 2008, 22, 1861–1873. [Google Scholar] [CrossRef] [PubMed]
- Monzon, M.E.; Fregien, N.; Schmid, N.; Falcon, N.S.; Campos, M.; Casalino-Matsuda, S.M.; Forteza, R.M. Reactive oxygen species and hyaluronidase 2 regulate airway epithelial hyaluronan fragmentation. J. Biol. Chem. 2010, 285, 26126–26134. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Nie, W.; Xie, Q.; Chen, G.; Li, X.; Jia, Y.; Yin, B.; Qu, X.; Li, Y.; Liang, J. Endostatin reverses immunosuppression of the tumor microenvironment in lung carcinoma. Oncol. Lett. 2018, 15, 1874–1880. [Google Scholar] [CrossRef]
- Da Silva, J.; Lameiras, P.; Beljebbar, A.; Berquand, A.; Villemin, M.; Ramont, L.; Dukic, S.; Nuzillard, J.M.; Molinari, M.; Gautier, M.; et al. Structural characterization and in vivo pro-tumor properties of a highly conserved matrikine. Oncotarget 2018, 9, 17839–17857. [Google Scholar] [CrossRef]
- Tzanakakis, G.; Kavasi, R.M.; Voudouri, K.; Berdiaki, A.; Spyridaki, I.; Tsatsakis, A.; Nikitovic, D. Role of the extracellular matrix in cancer-associated epithelial to mesenchymal transition phenomenon. Dev. Dyn. 2018, 247, 368–381. [Google Scholar] [CrossRef]
- Covic, L.; Kuliopulos, A. Protease-Activated Receptor 1 as Therapeutic Target in Breast, Lung, and Ovarian Cancer: Pepducin Approach. Int. J. Mol. Sci. 2018, 19, 2237. [Google Scholar] [CrossRef]
- Liu, X.; Yu, J.; Song, S.; Yue, X.; Li, Q. Protease-activated receptor-1 (PAR-1): A promising molecular target for cancer. Oncotarget 2017, 8, 107334–107345. [Google Scholar] [CrossRef]
- Arakaki, A.K.S.; Pan, W.A.; Trejo, J. GPCRs in Cancer: Protease-Activated Receptors, Endocytic Adaptors and Signaling. Int. J. Mol. Sci. 2018, 19, 1886. [Google Scholar] [CrossRef]
- Zara, M.; Canobbio, I.; Visconte, C.; Canino, J.; Torti, M.; Guidetti, G.F. Molecular mechanisms of platelet activation and aggregation induced by breast cancer cells. Cell Signal. 2018, 48, 45–53. [Google Scholar] [CrossRef]
- Mendonsa, A.M.; Na, T.Y.; Gumbiner, B.M. E-cadherin in contact inhibition and cancer. Oncogene 2018, 37, 4769–4780. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Ramamonjisoa, N.; Ackerstaff, E. Characterization of the Tumor Microenvironment and Tumor-Stroma Interaction by Non-invasive Preclinical Imaging. Front Oncol. 2017, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Coulson-Thomas, V.J.; Coulson-Thomas, Y.M.; Gesteira, T.F.; Andrade de Paula, C.A.; Carneiro, C.R.; Ortiz, V.; Toma, L.; Kao, W.W.; Nader, H.B. Lumican expression, localization and antitumor activity in prostate cancer. Exp. Cell Res. 2013, 319, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Karamanou, K.; Franchi, M.; Vynios, D.; Brezillon, S. Epithelial-to-mesenchymal transition and invadopodia markers in breast cancer: Lumican a key regulator. Semin. Cancer. Biol. 2020, 62, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Ronca, R.; Sozzani, S.; Presta, M.; Alessi, P. Delivering cytokines at tumor site: The immunocytokine-conjugated anti-EDB-fibronectin antibody case. Immunobiology 2009, 214, 800–810. [Google Scholar] [CrossRef] [PubMed]
- Hirata, E.; Sahai, E. Tumor Microenvironment and Differential Responses to Therapy. Cold Spring Harb. Perspect. Med. 2017, 7. [Google Scholar] [CrossRef]
- Arosio, D.; Casagrande, C. Advancement in integrin facilitated drug delivery. Adv. Drug Deliv. Rev. 2016, 97, 111–143. [Google Scholar] [CrossRef]
- Rodríguez-Cabello, J.C.; Arias, F.J.; Rodrigo, M.A.; Girotti, A. Elastin-like polypeptides in drug delivery. Adv. Drug Deliv. Rev. 2016, 97, 85–100. [Google Scholar] [CrossRef]
- Alexander, S.; Friedl, P. Cancer invasion and resistance: Interconnected processes of disease progression and therapy failure. Trends Mol. Med. 2012, 18, 13–26. [Google Scholar] [CrossRef]
- An, B.; Lin, Y.S.; Brodsky, B. Collagen interactions: Drug design and delivery. Adv. Drug Deliv. Rev. 2016, 97, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Sawyer, A.J.; Kyriakides, T.R. Matricellular proteins in drug delivery: Therapeutic targets, active agents, and therapeutic localization. Adv. Drug Deliv. Rev. 2016, 97, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Jiang, D.; Noble, P.W. Hyaluronan as a therapeutic target in human diseases. Adv. Drug Deliv. Rev. 2016, 97, 186–203. [Google Scholar] [CrossRef] [PubMed]
- Dosio, F.; Arpicco, S.; Stella, B.; Fattal, E. Hyaluronic acid for anticancer drug and nucleic acid delivery. Adv. Drug Deliv. Rev. 2016, 97, 204–236. [Google Scholar] [CrossRef]
- Piperigkou, Z.; Manou, D.; Karamanou, K.; Theocharis, A.D. Strategies to Target Matrix Metalloproteinases as Therapeutic Approach in Cancer. Methods Mol. Biol. 2018, 1731, 325–348. [Google Scholar] [CrossRef]
- Au, J.L.; Yeung, B.Z.; Wientjes, M.G.; Lu, Z.; Wientjes, M.G. Delivery of cancer therapeutics to extracellular and intracellular targets: Determinants, barriers, challenges and opportunities. Adv. Drug Deliv. Rev. 2016, 97, 280–301. [Google Scholar] [CrossRef]
- Barker, H.E.; Cox, T.R.; Erler, J.T. The rationale for targeting the LOX family in cancer. Nat. Rev. Cancer. 2012, 12, 540–552. [Google Scholar] [CrossRef]
- Chan, N.; Willis, A.; Kornhauser, N.; Ward, M.M.; Lee, S.B.; Nackos, E.; Seo, B.R.; Chuang, E.; Cigler, T.; Moore, A.; et al. Influencing the Tumor Microenvironment: A Phase II Study of Copper Depletion Using Tetrathiomolybdate in Patients with Breast Cancer at High Risk for Recurrence and in Preclinical Models of Lung Metastases. Clin. Cancer Res. 2017, 23, 666–676. [Google Scholar] [CrossRef]
- Chan, N.; Willis, A.; Kornhauser, N.; Ward, M.M.; Lee, S.B.; Nackos, E.; Seo, B.R.; Chuang, E.; Cigler, T.; Moore, A.; et al. Correction: Influencing the Tumor Microenvironment: A Phase II Study of Copper Depletion Using Tetrathiomolybdate in Patients with Breast Cancer at High Risk for Recurrence and in Preclinical Models of Lung Metastases. Clin. Cancer Res. 2020, 26, 5051. [Google Scholar] [CrossRef]
- Hecht, J.R.; Benson, A.B., III; Vyushkov, D.; Yang, Y.; Bendell, J.; Verma, U. A Phase II, Randomized, Double-Blind, Placebo-Controlled Study of Simtuzumab in Combination with FOLFIRI for the Second-Line Treatment of Metastatic KRAS Mutant Colorectal Adenocarcinoma. Oncologist 2017, 22, 243. [Google Scholar] [CrossRef]
- Benson, A.B., III; Wainberg, Z.A.; Hecht, J.R.; Vyushkov, D.; Dong, H.; Bendell, J.; Kudrik, F. A Phase II Randomized, Double-Blind, Placebo-Controlled Study of Simtuzumab or Placebo in Combination with Gemcitabine for the First-Line Treatment of Pancreatic Adenocarcinoma. Oncologist 2017, 22, 241. [Google Scholar] [CrossRef] [PubMed]
- Barry-Hamilton, V.; Spangler, R.; Marshall, D.; McCauley, S.; Rodriguez, H.M.; Oyasu, M.; Mikels, A.; Vaysberg, M.; Ghermazien, H.; Wai, C.; et al. Allosteric inhibition of lysyl oxidase-like-2 impedes the development of a pathologic microenvironment. Nat. Med. 2010, 16, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, H.M.; Vaysberg, M.; Mikels, A.; McCauley, S.; Velayo, A.C.; Garcia, C.; Smith, V. Modulation of lysyl oxidase-like 2 enzymatic activity by an allosteric antibody inhibitor. J. Biol. Chem. 2010, 285, 20964–20974. [Google Scholar] [CrossRef] [PubMed]
- Fields, G.B. The Rebirth of Matrix Metalloproteinase Inhibitors: Moving Beyond the Dogma. Cells 2019, 8, 984. [Google Scholar] [CrossRef] [PubMed]
- Levin, M.; Udi, Y.; Solomonov, I.; Sagi, I. Next generation matrix metalloproteinase inhibitors—Novel strategies bring new prospects. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1927–1939. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, H.S.; McCann, P.P. Matrix metalloproteinase inhibition as a novel anticancer strategy: A review with special focus on batimastat and marimastat. Pharmacol. Ther. 1997, 75, 69–75. [Google Scholar] [CrossRef]
- Cathcart, J.; Pulkoski-Gross, A.; Cao, J. Targeting Matrix Metalloproteinases in Cancer: Bringing New Life to Old Ideas. Genes Dis. 2015, 2, 26–34. [Google Scholar] [CrossRef]
- Rao, B.G. Recent developments in the design of specific Matrix Metalloproteinase inhibitors aided by structural and computational studies. Curr. Pharm. Des. 2005, 11, 295–322. [Google Scholar] [CrossRef]
- Chang, T.T.; Thakar, D.; Weaver, V.M. Force-dependent breaching of the basement membrane. Matrix Biol. 2017, 57-58, 178–189. [Google Scholar] [CrossRef]
- Vandooren, J.; Opdenakker, G.; Loadman, P.M.; Edwards, D.R. Proteases in cancer drug delivery. Adv. Drug Deliv. Rev. 2016, 97, 144–155. [Google Scholar] [CrossRef]
- Zhan, Y.; Ling, S.; Huang, H.; Zhang, Y.; Chen, G.; Huang, S.; Li, C.; Guo, W.; Wang, Q. Rapid and Nondestructive Histopathological Analysis for Intraoperative Cancer Diagnosis Using an Enzyme-Activated NIR-II Nanoprobe. Angew. Chem. Int. Ed. Engl. 2020, 59, 1–7. [Google Scholar] [CrossRef]
- Kasurinen, A.; Tervahartiala, T.; Laitinen, A.; Kokkola, A.; Sorsa, T.; Böckelman, C.; Haglund, C. High serum MMP-14 predicts worse survival in gastric cancer. PLoS ONE 2018, 13, e0208800. [Google Scholar] [CrossRef] [PubMed]
- Ingvarsen, S.; Porse, A.; Erpicum, C.; Maertens, L.; Jürgensen, H.J.; Madsen, D.H.; Melander, M.C.; Gardsvoll, H.; Hoyer-Hansen, G.; Noel, A.; et al. Targeting a single function of the multifunctional matrix metalloprotease MT1-MMP Impact on Lymphangiogenesis. J. Biol. Chem. 2013, 288, 10195–10204. [Google Scholar] [CrossRef] [PubMed]
- Paemen, L.; Martens, E.; Masure, S.; Opdenakker, G. Monoclonal antibodies specific for natural human neutrophil gelatinase B used for affinity purification, quantitation by two-site ELISA and inhibition of enzymatic activity. Eur. J. Biochem. 1995, 234, 759–765. [Google Scholar] [CrossRef]
- Martens, E.; Leyssen, A.; Van Aelst, I.; Fiten, P.; Piccard, H.; Hu, J.; Descamps, F.J.; Van den Steen, P.E.; Proost, P.; Van Damme, J.; et al. A monoclonal antibody inhibits gelatinase B/MMP-9 by selective binding to part of the catalytic domain and not to the fibronectin or zinc binding domains. Biochim. Biophys. Acta 2007, 1770, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Devy, L.; Huang, L.; Naa, L.; Yanamandra, N.; Pieters, H.; Frans, N.; Chang, E.; Tao, Q.; Vanhove, M.; Lejeune, A.; et al. Selective inhibition of matrix metalloproteinase-14 blocks tumor growth, invasion, and angiogenesis. Cancer Res. 2009, 69, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Nam, D.H.; Ge, X. Generation of Highly Selective MMP Antibody Inhibitors. Methods Mol. Biol. 2018, 1731, 307–324. [Google Scholar] [CrossRef]
- Li, X.; Ma, Z.; Wang, H.; Ren, L.; Zhang, D.; Liang, W.; Zhang, G.; Zhang, J.; Yu, D.; Fang, X. Screening, Identification, and Characterization of an Affinity Peptide Specific to MT1-MMP and Its Application in Tumor Imaging. Bioconjug. Chem. 2019, 30, 1507–1517. [Google Scholar] [CrossRef]
- Shirian, J.; Arkadash, V.; Cohen, I.; Sapir, T.; Radisky, E.S.; Papo, N.; Shifman, J.M. Converting a broad matrix metalloproteinase family inhibitor into a specific inhibitor of MMP-9 and MMP-14. FEBS Lett. 2018, 592, 1122–1134. [Google Scholar] [CrossRef]
- Wen, X.Q.; Qian, X.L.; Sun, H.K.; Zheng, L.L.; Zhu, W.Q.; Li, T.Y.; Hu, J.P. MicroRNAs: Multifaceted Regulators of Colorectal Cancer Metastasis and Clinical Applications. Onco Targets Ther. 2020, 13, 10851–10866. [Google Scholar] [CrossRef]
- D’Angelo, E.; Agostini, M. Long non-coding RNA and extracellular matrix: The hidden players in cancer-stroma cross-talk. Noncoding RNA Res. 2018, 3, 174–177. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, H. Role of microRNA-mediated MMP regulation in the treatment and diagnosis of malignant tumors. Cancer Biol. Ther. 2013, 14, 796–805. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kuscu, C.; Banach, A.; Zhang, Q.; Pulkoski-Gross, A.; Kim, D.; Liu, J.; Roth, E.; Li, E.; Shroyer, K.R.; et al. miR-181a-5p Inhibits Cancer Cell Migration and Angiogenesis via Downregulation of Matrix Metalloproteinase-14. Cancer Res. 2015, 75, 2674–2685. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Wu, Y.; Wang, J.; He, J.; Han, X. Sevoflurane inhibits the migration and invasion of colorectal cancer cells through regulating ERK/MMP-9 pathway by up-regulating miR-203. Eur. J. Pharmacol. 2019, 850, 43–52. [Google Scholar] [CrossRef]
- Zhang, L.; Dong, Y.; Zhu, N.; Tsoi, H.; Zhao, Z.; Wu, C.W.; Wang, K.; Zheng, S.; Ng, S.S.; Chan, F.K.; et al. microRNA-139-5p exerts tumor suppressor function by targeting NOTCH1 in colorectal cancer. Mol. Cancer 2014, 13, 124. [Google Scholar] [CrossRef]
- Cai, H.K.; Chen, X.; Tang, Y.H.; Deng, Y.C. MicroRNA-194 modulates epithelial-mesenchymal transition in human colorectal cancer metastasis. Onco Targets Ther. 2017, 10, 1269–1278. [Google Scholar] [CrossRef]
- Yu, L.; Lu, Y.; Han, X.; Zhao, W.; Li, J.; Mao, J.; Wang, B.; Shen, J.; Fan, S.; Wang, L.; et al. microRNA -140-5p inhibits colorectal cancer invasion and metastasis by targeting ADAMTS5 and IGFBP5. Stem. Cell Res. Ther. 2016, 7, 180. [Google Scholar] [CrossRef]
- Gabriely, G.; Wurdinger, T.; Kesari, S.; Esau, C.C.; Burchard, J.; Linsley, P.S.; Krichevsky, A.M. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol. Cell. Biol. 2008, 28, 5369–5380. [Google Scholar] [CrossRef]
- Hinderer, S.; Layland, S.L.; Schenke-Layland, K. ECM and ECM-like materials—Biomaterials for applications in regenerative medicine and cancer therapy. Adv. Drug Deliv. Rev. 2016, 97, 260–269. [Google Scholar] [CrossRef]
- Phan, T.G.; Croucher, P.I. The dormant cancer cell life cycle. Nat. Rev. Cancer. 2020, 20, 398–411. [Google Scholar] [CrossRef]
- Celià-Terrassa, T.; Kang, Y. Metastatic niche functions and therapeutic opportunities. Nat. Cell Biol. 2018, 20, 868–877. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).