γ-Aminobutyric Acid Suppresses Iron Transportation from Roots to Shoots in Rice Seedlings by Inducing Aerenchyma Formation
Abstract
1. Introduction
2. Results
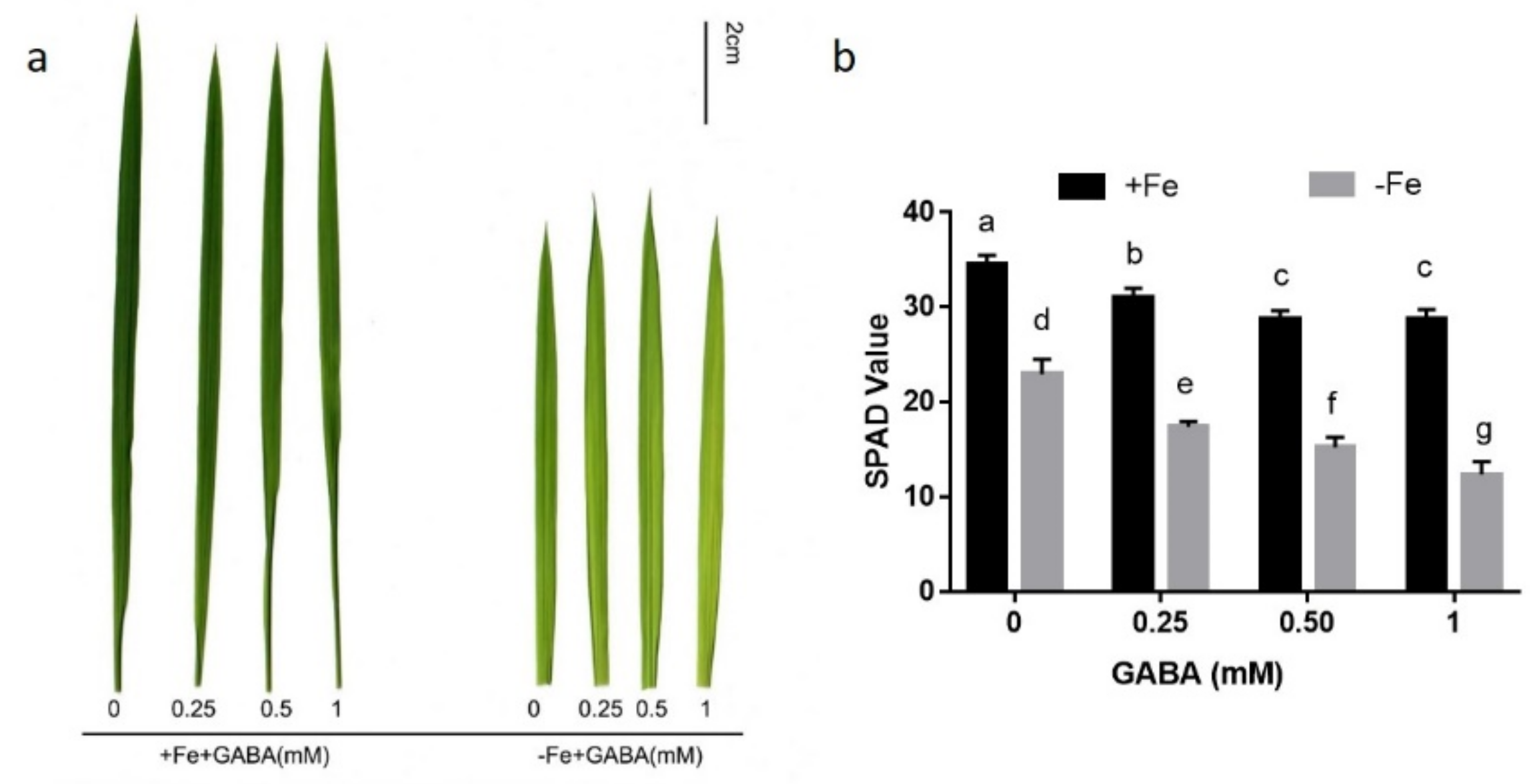
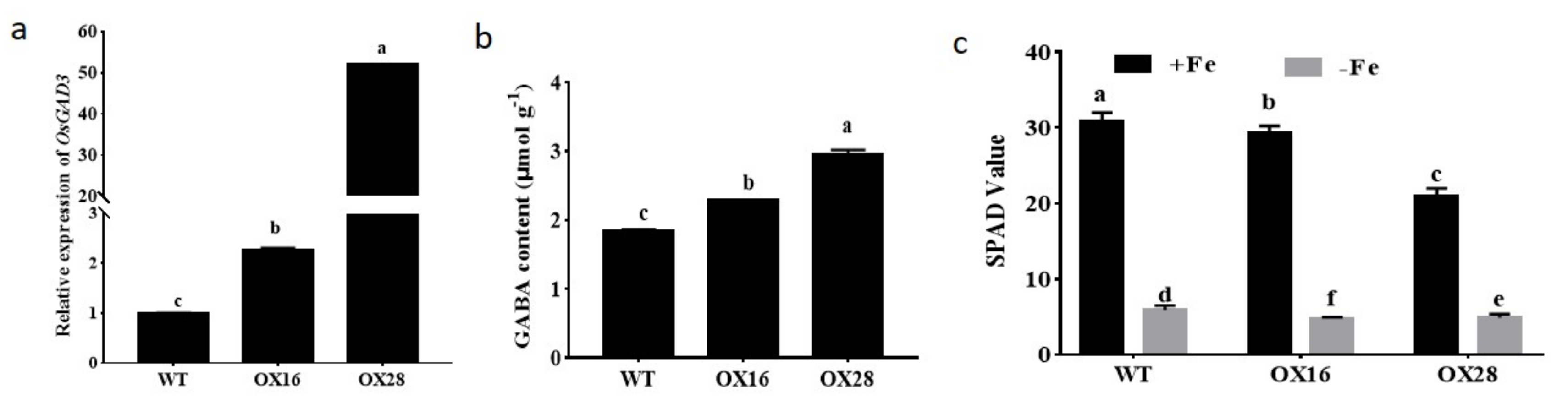
2.1. GABA Participates in the Regulation of Iron Homeostasis
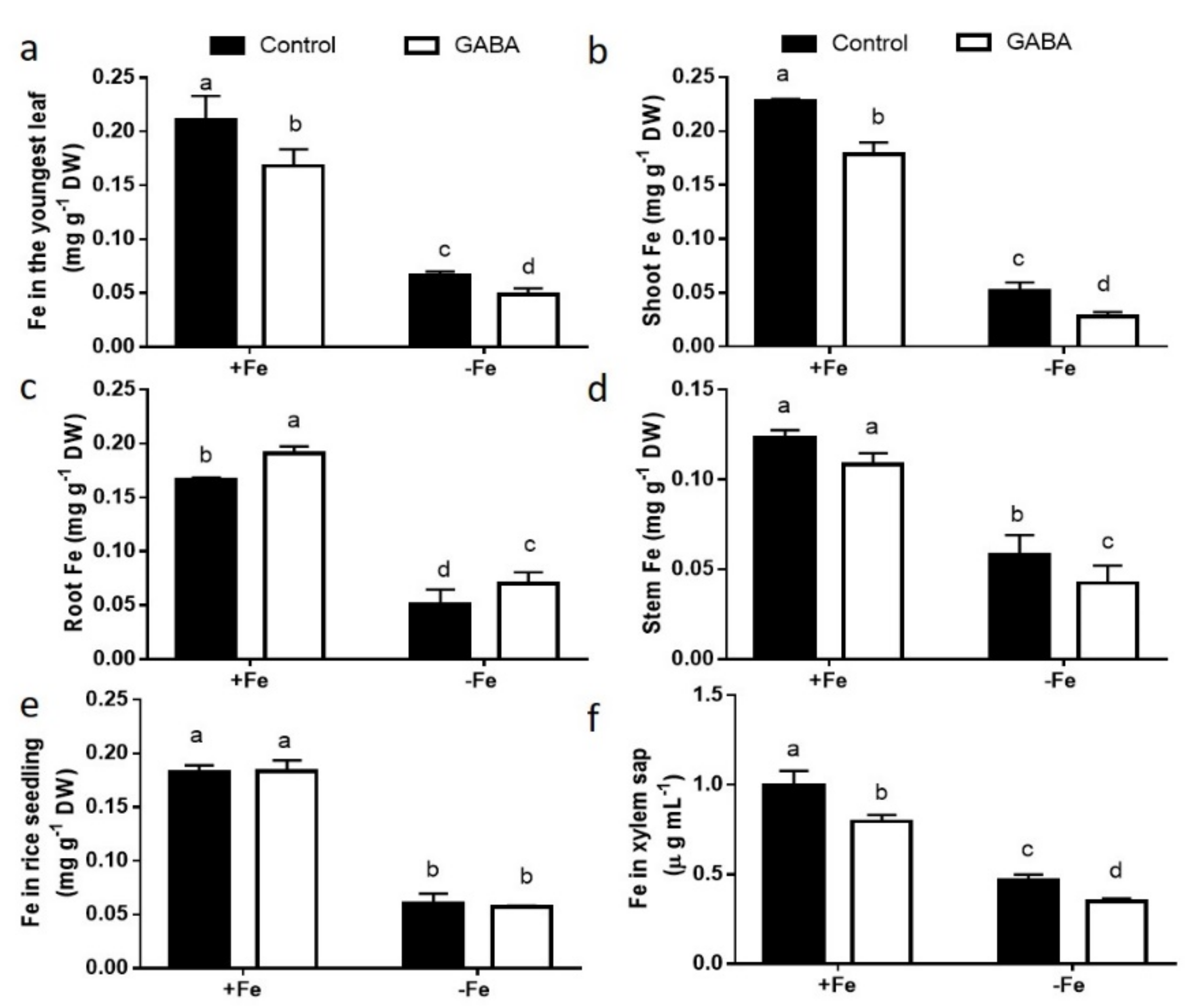
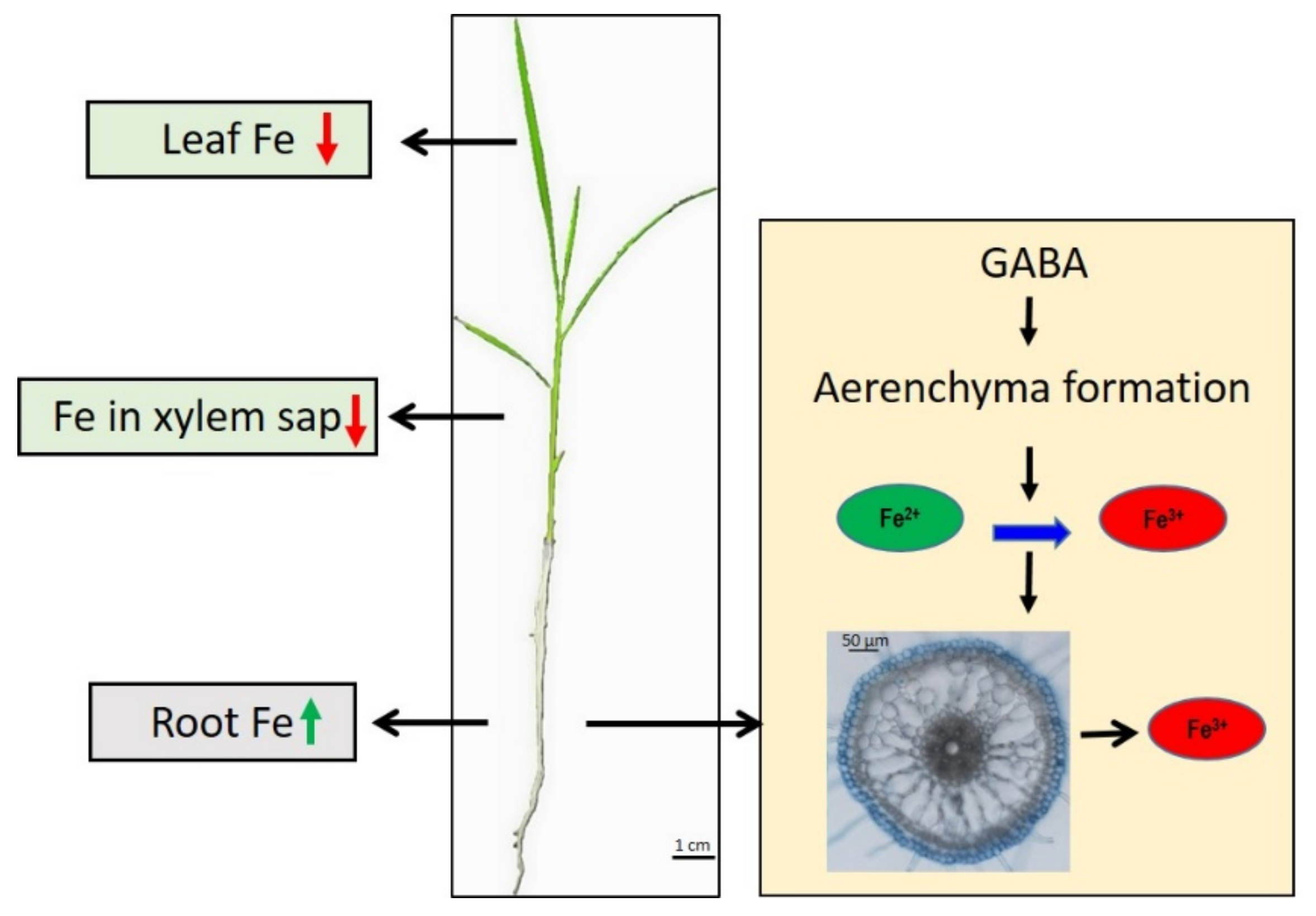
2.2. GABA Treatment Suppressed Fe Transportation from Roots to Shoots
2.3. GABA Regulated Epidermis–Pericycle Fe Translocation in Roots
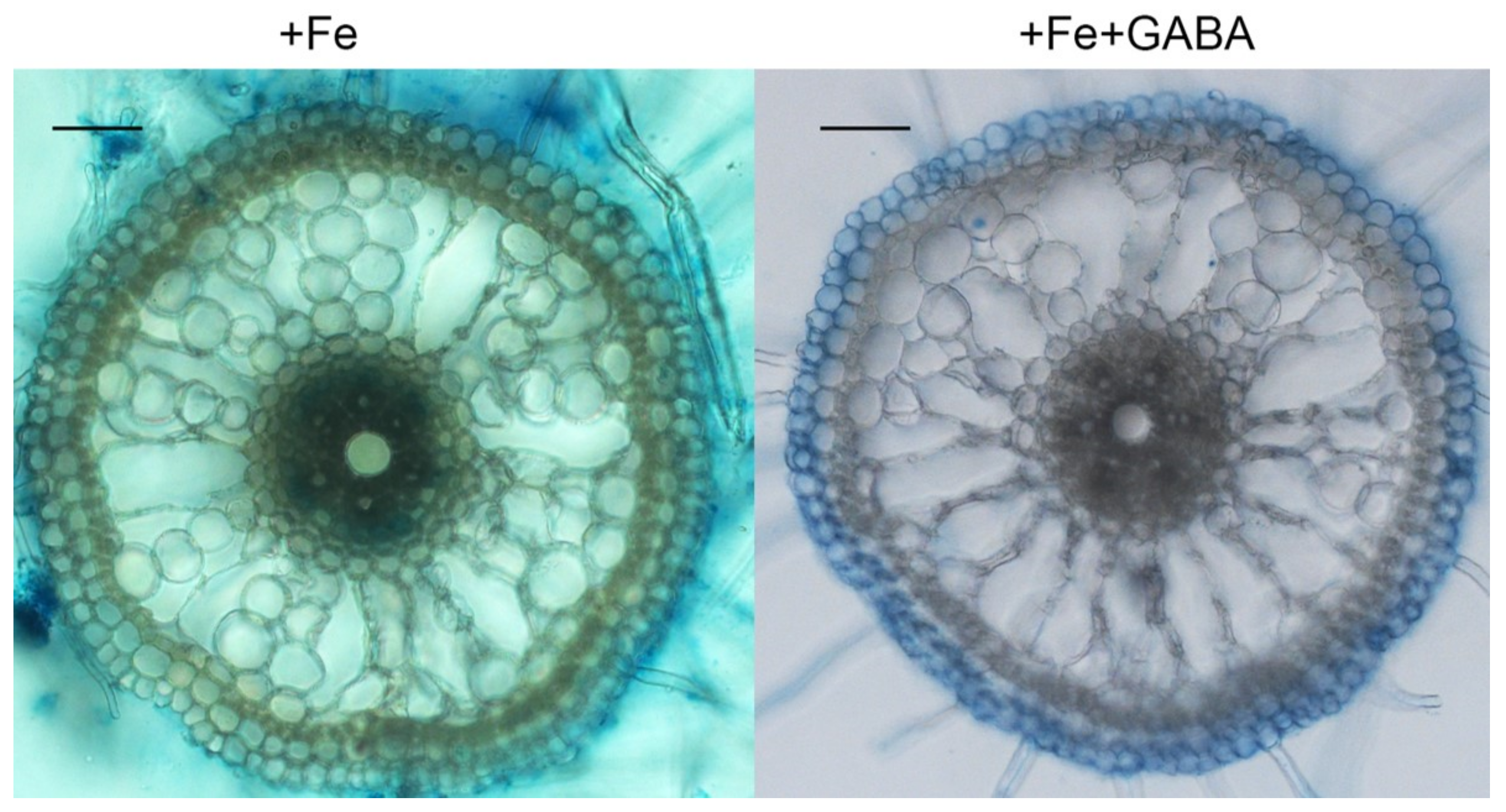
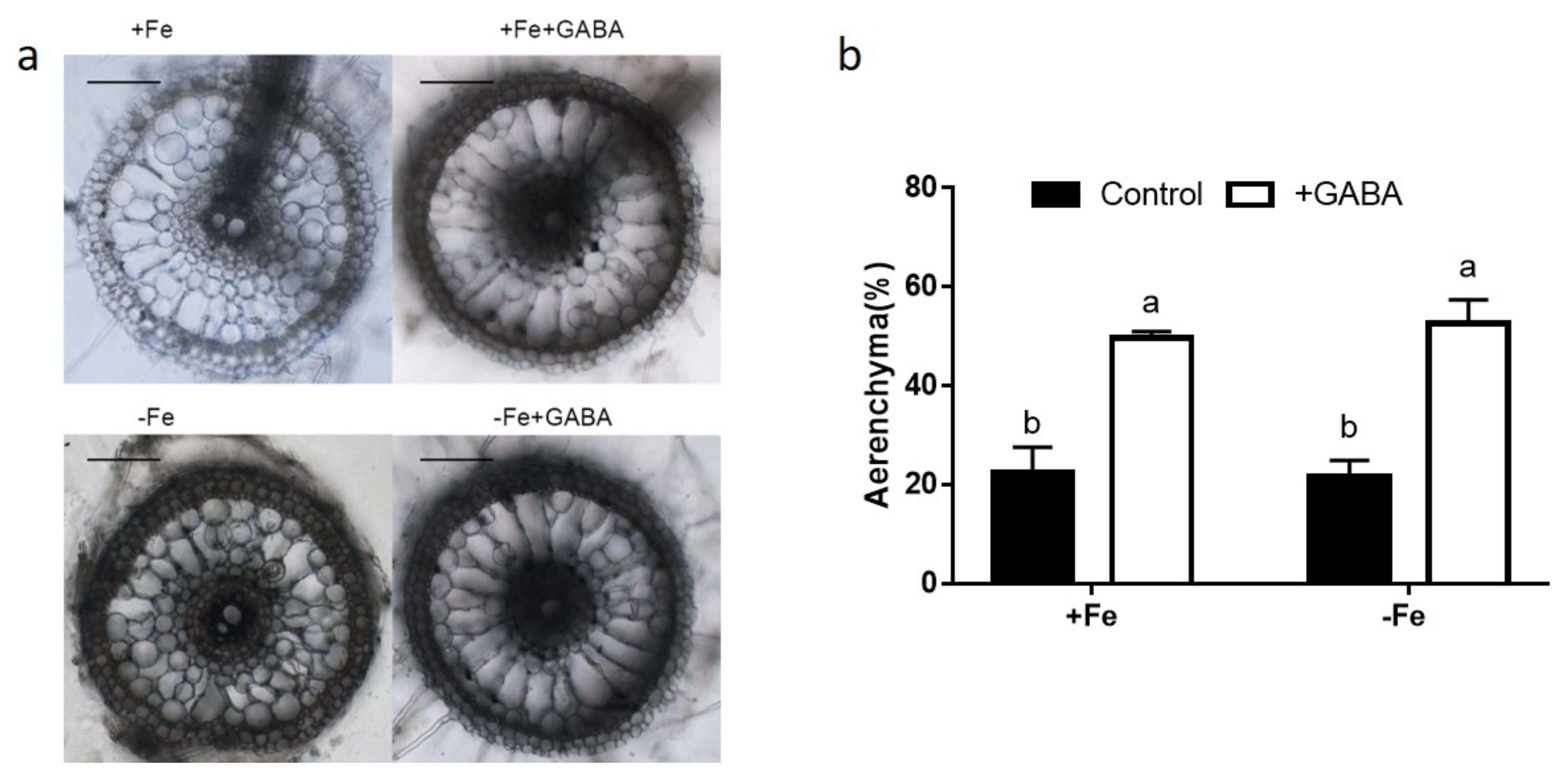
2.4. GABA Induced Aerenchyma Formation
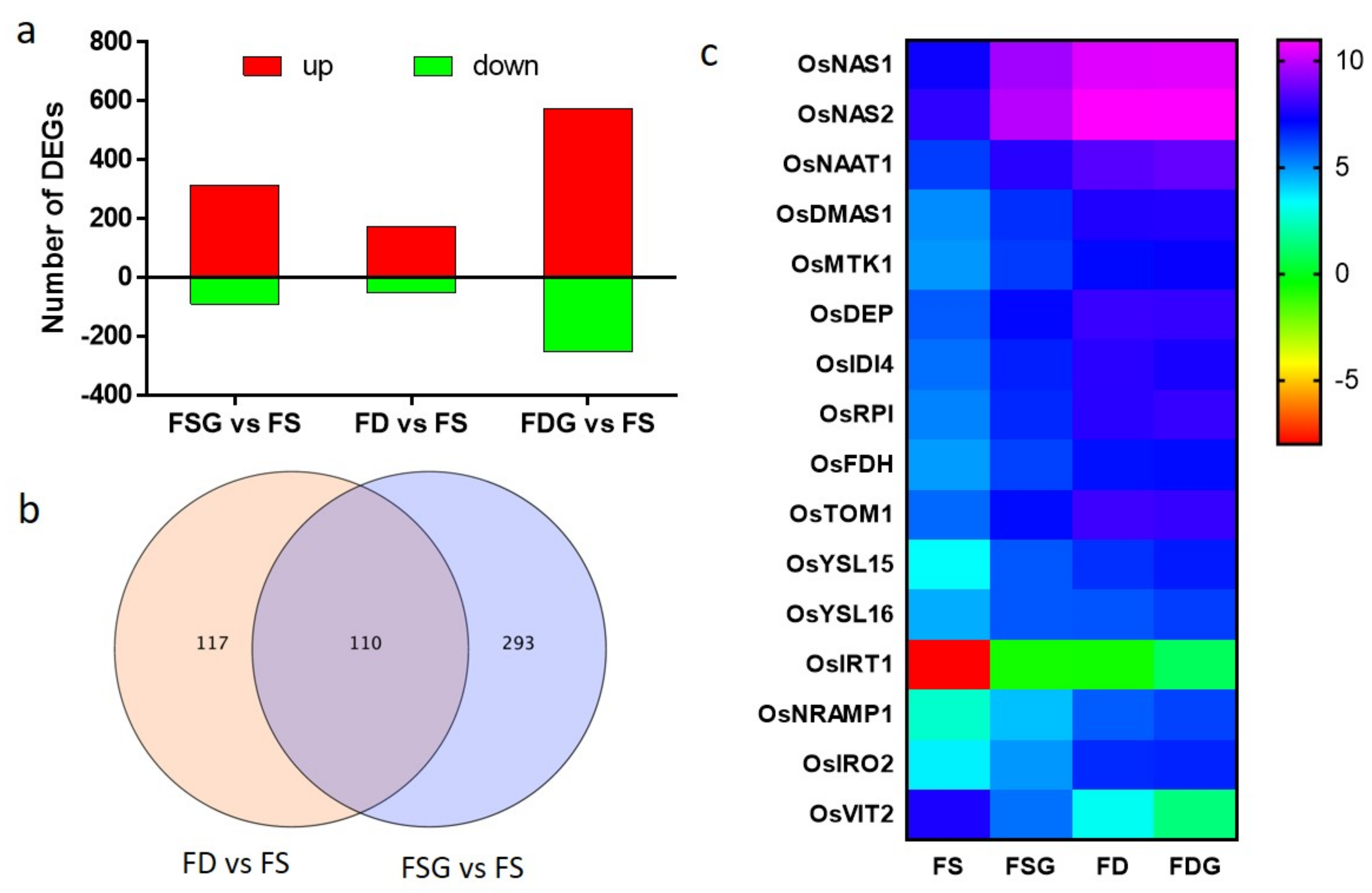
2.5. Transcriptomic Analysis of Roots Treated with GABA Under conditions of FD and FS
2.6. FD-Related Genes Were Induced in GABA-Treated Roots Under Conditions of FS
3. Discussion
4. Materials and Methods
4.1. Plant Materials and GABA Treatment
4.2. Rice Transformation
4.3. Determination of GABA
4.4. Analysis of Xylem Sap
4.5. Analysis of Chlorophyll Content
4.6. Detedmination of Fe Concentration
4.7. Perls’ Blue Staining
4.8. Analysis of Aerenchyma in Roots
4.9. RNA-seq and Analysis
4.10. Quantitative Real-Time RT-PCR
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GABA | γ-Aminobutyric acid |
| Fe | Iron |
| GAD | Glutamate decarboxylase |
| IRT | Iron-regulated transporters |
| MA | Mugineic acid |
| YSL | Yellow stripe/yellow stripe 1-like |
| NAS | Nicotianamine synthase |
| NAAT | Nicotianamine aminotransferase |
| DMAS | Deoxymugineic acid synthase |
| DMA | Deoxymugineic acid |
| RNA-seq | RNA sequencing |
| DEGs | Differentially expressed genes |
| FS | Fe-sufficiency |
| FSG | with GABA under Fe-sufficient conditions |
| FD | Fe-deficiency |
| FDG | with GABA under Fe-deficient conditions |
References
- Kobayashi, T.; Nishizawa, N.K. Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 2012, 63, 131–152. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, L.; Yang, A. The molecular mechanisms underlying iron deficiency responses in rice. Int. J. Mol. Sci. 2020, 21, 43. [Google Scholar] [CrossRef] [PubMed]
- Onaga, G.; Drame, K.; Ismail, A. Understanding the regulation of iron nutrition: Can it contribute to improving iron toxicity tolerance in rice? Funct. Plant Biol. 2016, 43, 709–726. [Google Scholar] [CrossRef] [PubMed]
- Bashir, K.; Hanada, K.; Shimizu, M.; Seki, M.; Nakanishi, H.; Nishizawa, N. Transcriptomic analysis of rice in response to iron deficiency and excess. Rice 2014, 7, 18. [Google Scholar] [CrossRef]
- Nozoye, T.; Nagasaka, S.; Kobayashi, T.; Takahashi, M.; Sato, Y.; Uozumi, N.; Nakanishi, H.; Nishizawa, N. Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 2011, 286, 5446–5454. [Google Scholar] [CrossRef]
- Curie, C.; Panaviene, Z.; Loulergue, C.; Dellaporta, S.L.; Briat, J.F.; Walker, E.L. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 2001, 409, 346–349. [Google Scholar] [CrossRef]
- Inoue, H.; Kobayashi, T.; Nozoye, T.; Takahashi, M.; Kakei, Y.; Suzuki, K.; Nakazono, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J. Biol. Chem. 2009, 284, 3470–3479. [Google Scholar] [CrossRef]
- Masuda, H.; Shimochi, E.; Hamada, T.; Senoura, T.; Kobayashi, T.; Aung, M.S.; Ishimaru, Y.; Ogo, Y.; Nakanishi, H.; Nishizawa, N.K. A new transgenic rice line exhibiting enhanced ferric iron reduction and phytosiderophore production confers tolerance to low iron availability in calcareous soil. PLoS ONE 2017, 12, e0173441. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nozoye, T.; Nishizawa, N.K. Iron transport and its regulation in plants. Free Radic. Biol. Med. 2019, 133, 11–20. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Masuda, H.; Bashir, K.; Inoue, H.; Tsukamoto, T.; Takahashi, M.; Nakanishi, H.; Aoki, N.; Hirose, T.; Ohsugi, R.; et al. Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 2010, 62, 379–390. [Google Scholar] [CrossRef]
- Bughio, N.; Yamaguchi, H.; Nishizawa, N.; Nakanishi, H.; Mori, S. Cloning an iron-regulated metal transporter from rice. J. Exp. Bot. 2002, 53, 1677–1682. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Suzuki, M.; Tsukamoto, T.; Suzuki, K.; Nakazono, M.; Kobayashi, T.; Wada, Y.; Watanabe, S.; Matsuhashi, S.; Takahashi, M.; et al. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 2006, 45, 335–346. [Google Scholar] [CrossRef]
- Takahashi, R.; Ishimaru, Y.; Senoura, T.; Shimo, H.; Ishikawa, S.; Arao, T.; Nakanishi, H.; Nishizawa, N. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J. Exp. Bot. 2011, 62, 4843–4850. [Google Scholar] [CrossRef] [PubMed]
- Kinnersley, A.; Turano, F. Gamma aminobutyric acid (GABA) and plant responses to stress. Crit. Rev. Plant Sci. 2000, 19, 479–509. [Google Scholar] [CrossRef]
- Bouche, N.; Fromm, H. GABA in plants: Just a metabolite? Trends Plant Sci. 2004, 9, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.A.; Tyerman, S.D.; Gilliham, M.; Xu, B. γ-Aminobutyric acid (GABA) signalling in plants. Cell Mol. Life Sci. 2017, 74, 1577–1603. [Google Scholar] [CrossRef]
- Bown, A.; Shelp, B. Plant GABA: Not just a metabolite. Trends Plant Sci. 2016, 21, 811–813. [Google Scholar] [CrossRef]
- Bown, A.W.; Shelp, B.J. Does the GABA shunt regulate cytosolic GABA? Trends Plant Sci. 2020, 25, 422–424. [Google Scholar] [CrossRef]
- Hijaz, F.; Killiny, N. The use of deuterium-labeled gamma-aminobutyric (D6-GABA) to study uptake, translocation, and metabolism of exogenous GABA in plants. Plant Methods. 2020, 16, 1. [Google Scholar] [CrossRef]
- Sasaki, T.; Yamamoto, Y.; Ezaki, B.; Katsuhara, M.; Ahn, S.J.; Ryan, P.R.; Delhaize, E.; Matsumoto, H. A wheat gene encoding an aluminum-activated malate transporter. Plant J. 2004, 37, 645–653. [Google Scholar] [CrossRef]
- Seifikalhor, M.; Aliniaeifard, S.; Bernard, F.; Seif, M.; Latifi, M.; Hassani, B.; Didaran, F.; Bosacchi, M.; Rezadoost, H.; Li, T. γ-Aminobutyric acid confers cadmium tolerance in maize plants by concerted regulation of polyamine metabolism and antioxidant defense systems. Sci. Rep. 2020, 10, 3356. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Gautam, A.; Dubey, A.; Ranjan, R.; Pandey, A.; Kumari, B.; Singh, G.; Mandotra, S.; Chauhan, P.; Srikrishna, S.; et al. GABA mediated reduction of arsenite toxicity in rice seedling through modulation of fatty acids, stress responsive amino acids and polyamines biosynthesis. Ecotoxico. Environ. Saf. 2019, 173, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.; Kamran, M.; Sullivan, W.; Chirkova, L.; Okamoto, M.; Degryse, F.; McLaughlin, M.; Gilliham, M.; Tyerman, S. Aluminum-activated malate transporters can facilitate GABA transport. Plant Cell. 2018, 30, 1147–1164. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhu, C.; Yang, N.; Gan, L.; Xia, K. gamma-Aminobutyric acid addition alleviates ammonium toxicity by limiting ammonium accumulation in rice (Oryza sativa) seedlings. Physiol. Plantarum. 2016, 158, 389–401. [Google Scholar] [CrossRef]
- Yokosho, K.; Yamaji, N.; Ueno, D.; Mitani, N.; Ma, J.F. OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol. 2009, 149, 297–305. [Google Scholar] [CrossRef]
- Green, L.S.; Rogers, E.E. FRD3 controls iron localization in Arabidopsis. Plant Physiol. 2004, 136, 2523–2531. [Google Scholar] [CrossRef]
- Ogo, Y.; Kakei, Y.; Itai, R.N.; Kobayashi, T.; Nakanishi, H.; Takahashi, H.; Nakazono, M.; Nishizawa, N.K. Spatial transcriptomes of iron-deficient and cadmium-stressed rice. New Phytol. 2014, 201, 781–794. [Google Scholar] [CrossRef]
- Ogo, Y.; Itai, R.N.; Inoue, H.; Kobayashi, T.; Suzuki, M.; Takahashi, M.; Mori, S.; Nishizawa, N. Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J. Exp. Bot. 2006, 57, 2867–2878. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Y.; Yi, H.; Gong, J. Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 2012, 72, 400–410. [Google Scholar] [CrossRef]
- Ramesh, S.; Tyerman, S.; Xu, B.; Bose, J.; Kaur, S.; Conn, V.; Domingos, P.; Ullah, S.; Wege, S.; Shabala, S.; et al. GABA signalling modulates plant growth by directly regulating the activity of plant-specific anion transporters. Nat. Commun. 2015, 6, 7879. [Google Scholar] [CrossRef]
- Sheteiwy, M.S.; Shao, H.B.; Qi, W.C.; Hamoud, Y.A.; Shaghaleh, H.; Khan, N.U.; Yang, R.P.; Tang, B.P. GABA-alleviated oxidative injury induced by salinity, osmotic Stress and their combination by regulating cellular and molecular signals in Rice. Int. J. Mol. Sci. 2019, 20, 22. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Du, N.; Li, Y.; Zheng, S.; Shen, S.; Piao, F. Gamma-aminobutyric acid enhances tolerance to iron deficiency by stimulating auxin signaling in cucumber (Cucumis sativus L.). Ecotoxicol. Environ. Saf. 2020, 192, 110285. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Itai, R.; Nishizawa, N. Iron deficiency responses in rice roots. Rice 2014, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Nishizawa, N. Iron sensors and signals in response to iron deficiency. Plant Sci. 2014, 224, 36–43. [Google Scholar] [CrossRef]
- Joshi, R.; Kumar, P. Lysigenous aerenchyma formation involves non-apoptotic programmed cell death in rice (Oryza sativa L.) roots. Physiol. Mol. Biol. Plants. 2012, 18, 1–9. [Google Scholar] [CrossRef]
- Aung, M.; Masuda, H. How does rice defend against excess iron? Physiological and molecular mechanisms. Front. Plant Sci. 2020, 11, 1102. [Google Scholar] [CrossRef]
- Yamauchi, T.; Yoshioka, M.; Fukazawa, A.; Mori, H.; Nishizawa, N.K.; Tsutsumi, N.; Yoshioka, H.; Nakazono, M. An NADPH oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen-deficient conditions. Plant Cell. 2017, 29, 775–790. [Google Scholar] [CrossRef]
- Kathiresan, A.; Tung, P.; Chinnappa, C.C.; Reid, D.M. γ-Aminobutyric acid stimulates ethylene biosynthesis in sunflower. Plant Physiol. 1997, 115, 129–135. [Google Scholar] [CrossRef]
- Bao, H.; Chen, X.; Lv, S.; Jiang, P.; Feng, J.; Fan, P.; Nie, L.; Li, Y. Virus-induced gene silencing reveals control of reactive oxygen species accumulation and salt tolerance in tomato by gamma-aminobutyric acid metabolic pathway. Plant Cell Environ. 2015, 38, 600–613. [Google Scholar] [CrossRef]
- Aurisano, N.; Bertani, A.; Reggiani, R. Anaerobic accumulation of 4-aminobutyrate in rice seedlings-causes and significance. Phytochemistry 1995, 38, 1147–1150. [Google Scholar] [CrossRef]
- Zhang, X.; Shabala, S.; Koutoulis, A.; Shabala, L.; Johnson, P.; Hayes, D.; Nichols, D.; Zhou, M. Waterlogging tolerance in barley is associated with faster aerenchyma formation in adventitious roots. Plant Soil. 2015, 394, 355–372. [Google Scholar] [CrossRef]
- Rajhi, I.; Yamauchi, T.; Takahashi, H.; Nishiuchi, S.; Shiono, K.; Watanabe, R.; Mliki, A.; Nagamura, Y.; Tsutsumi, N.; Nishizawa, N.K.; et al. Identification of genes expressed in maize root cortical cells during lysigenous aerenchyma formation using laser microdissection and microarray analyses. New Phytol. 2011, 190, 351–368. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.; Asch, F. Iron toxicity in rice-conditions and management concepts. J. Plant J. Soil Sci. Plant Nutr. 2005, 168, 558–573. [Google Scholar] [CrossRef]
- Nishimura, A.; Aichi, I.; Matsuoka, M. A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 2006, 1, 2796–2802. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Shibato, J.; Agrawal, G.K.; Fujihara, S.; Iwahashi, H.; Kim, d.H.; Shim, I.S.; Rakwal, R. Gene transcription in the leaves of rice undergoing salt-induced morphological changes (Oryza sativa L.). Mol. Cells. 2007, 24, 45–59. [Google Scholar]
- Wang, B.; Li, G.; Zhang, W.H. Brassinosteroids are involved in Fe homeostasis in rice (Oryza sativa L.). J. Exp. Bot. 2015, 66, 2749–2761. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. AgriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
- Li, H.; Wu, H.; Qi, Q.; Li, Z.; Chen, S.; Ding, Q.; Wang, Q.; Yan, Z.; Gai, Y.; Jiang, X.; et al. Gibberellins play a role in regulating tomato fruit ripening. Plant Cell Physiol. 2019, 60, 1619–1629. [Google Scholar] [CrossRef]
- Yang, A.; Li, Y.; Xu, Y.; Zhang, W. A receptor-like protein RMC is involved in regulation of iron acquisition in rice. J. Exp. Bot. 2013, 64, 5009–5020. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, C.; Qi, Q.; Niu, H.; Wu, J.; Yang, N.; Gan, L. γ-Aminobutyric Acid Suppresses Iron Transportation from Roots to Shoots in Rice Seedlings by Inducing Aerenchyma Formation. Int. J. Mol. Sci. 2021, 22, 220. https://doi.org/10.3390/ijms22010220
Zhu C, Qi Q, Niu H, Wu J, Yang N, Gan L. γ-Aminobutyric Acid Suppresses Iron Transportation from Roots to Shoots in Rice Seedlings by Inducing Aerenchyma Formation. International Journal of Molecular Sciences. 2021; 22(1):220. https://doi.org/10.3390/ijms22010220
Chicago/Turabian StyleZhu, Changhua, Qi Qi, Huijiao Niu, Jiaqi Wu, Na Yang, and Lijun Gan. 2021. "γ-Aminobutyric Acid Suppresses Iron Transportation from Roots to Shoots in Rice Seedlings by Inducing Aerenchyma Formation" International Journal of Molecular Sciences 22, no. 1: 220. https://doi.org/10.3390/ijms22010220
APA StyleZhu, C., Qi, Q., Niu, H., Wu, J., Yang, N., & Gan, L. (2021). γ-Aminobutyric Acid Suppresses Iron Transportation from Roots to Shoots in Rice Seedlings by Inducing Aerenchyma Formation. International Journal of Molecular Sciences, 22(1), 220. https://doi.org/10.3390/ijms22010220




