Whether Gametophytes Are Reduced or Unreduced in Angiosperms Might Be Determined Metabolically
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Microarray Analyses
2.3. Expression Profiling
2.4. Gene Ontology Analyses
2.5. Expression Verification
2.6. Drought and Heat-Stress Experiments
2.7. In Vitro Pistil Culture Experiments
2.8. Cytoembryological Analyses
3. Results
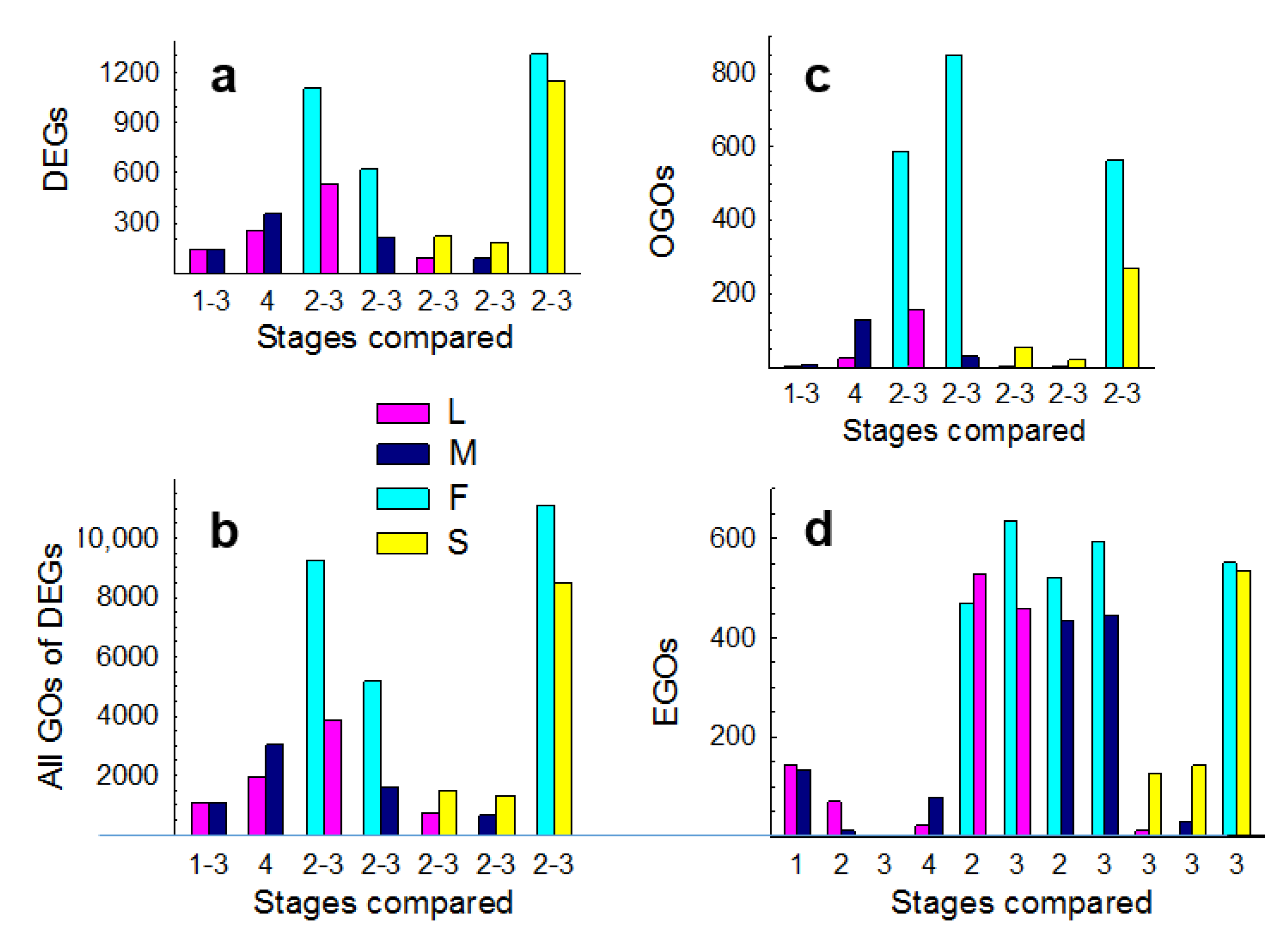
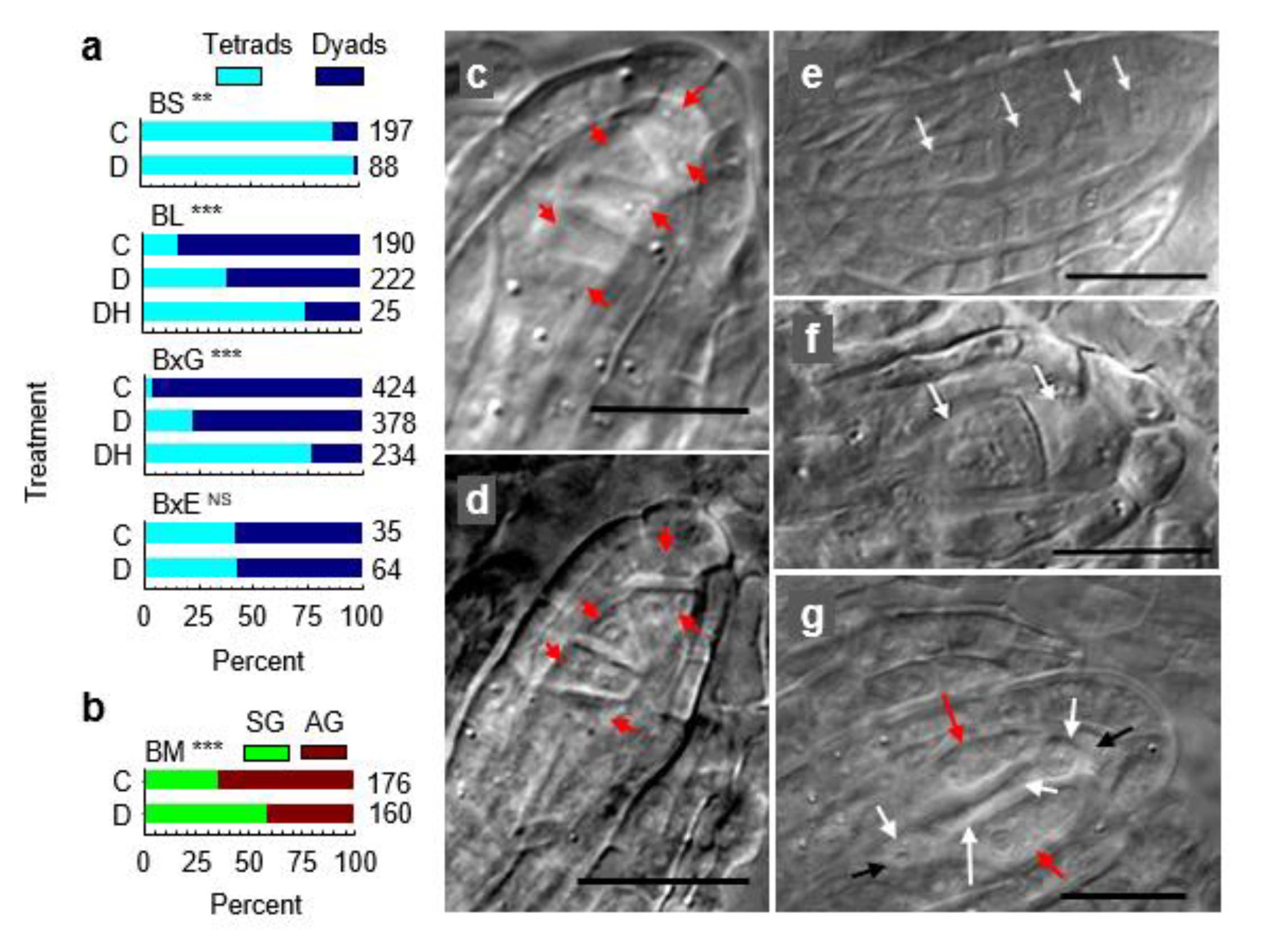
3.1. Expression Profiling
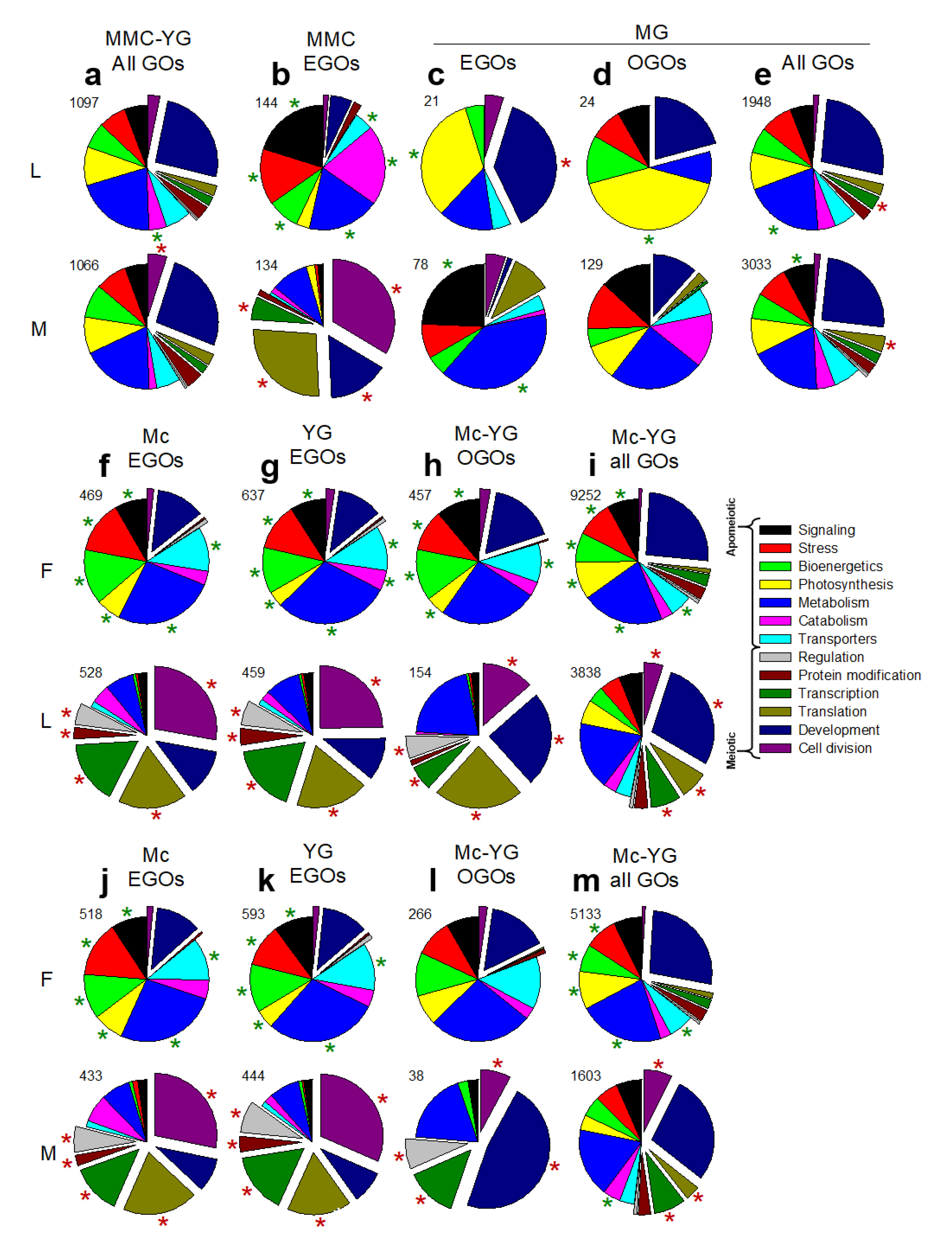
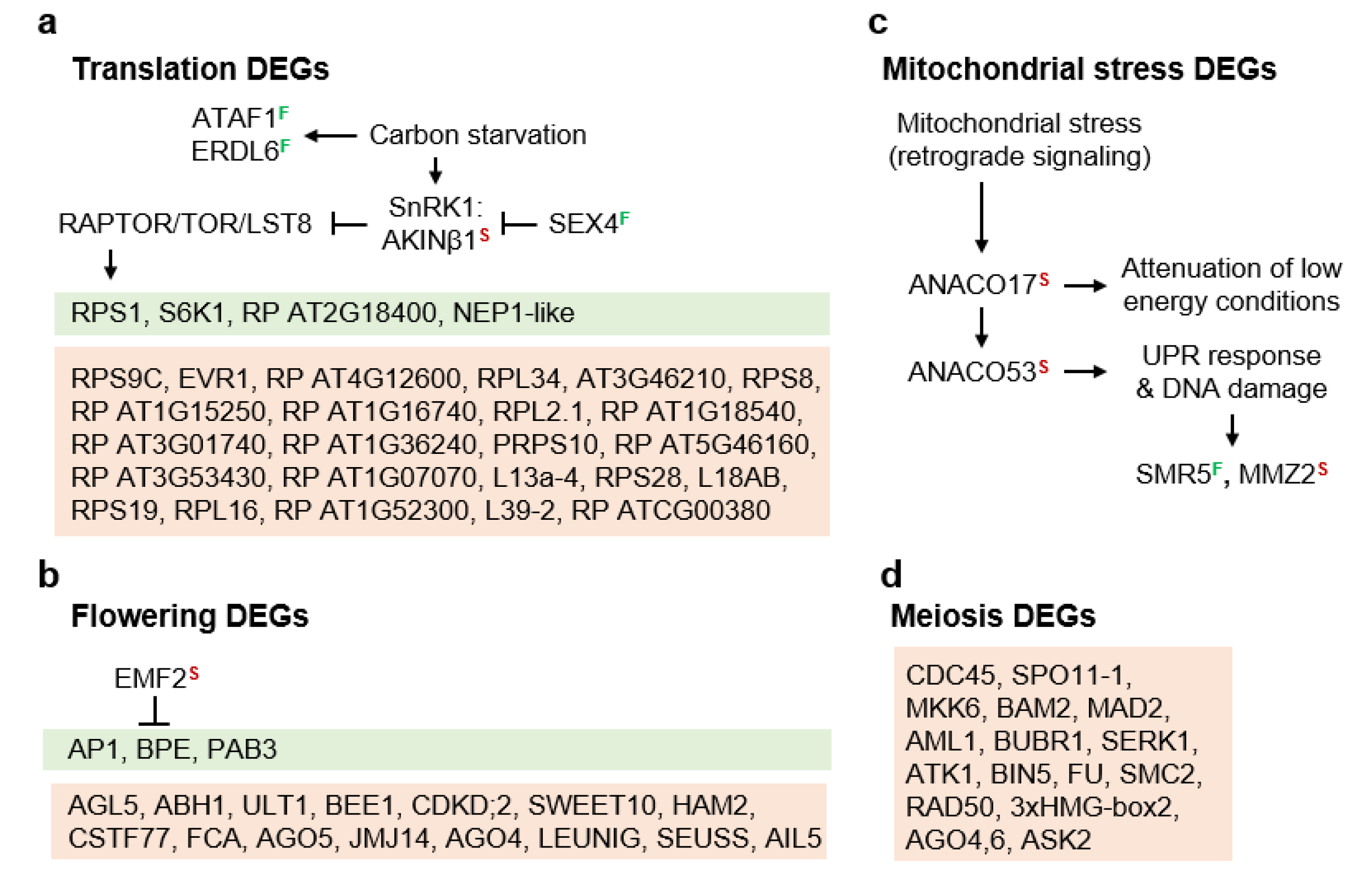
3.2. Molecular Pathways Upregulated in Meiotic Taxa
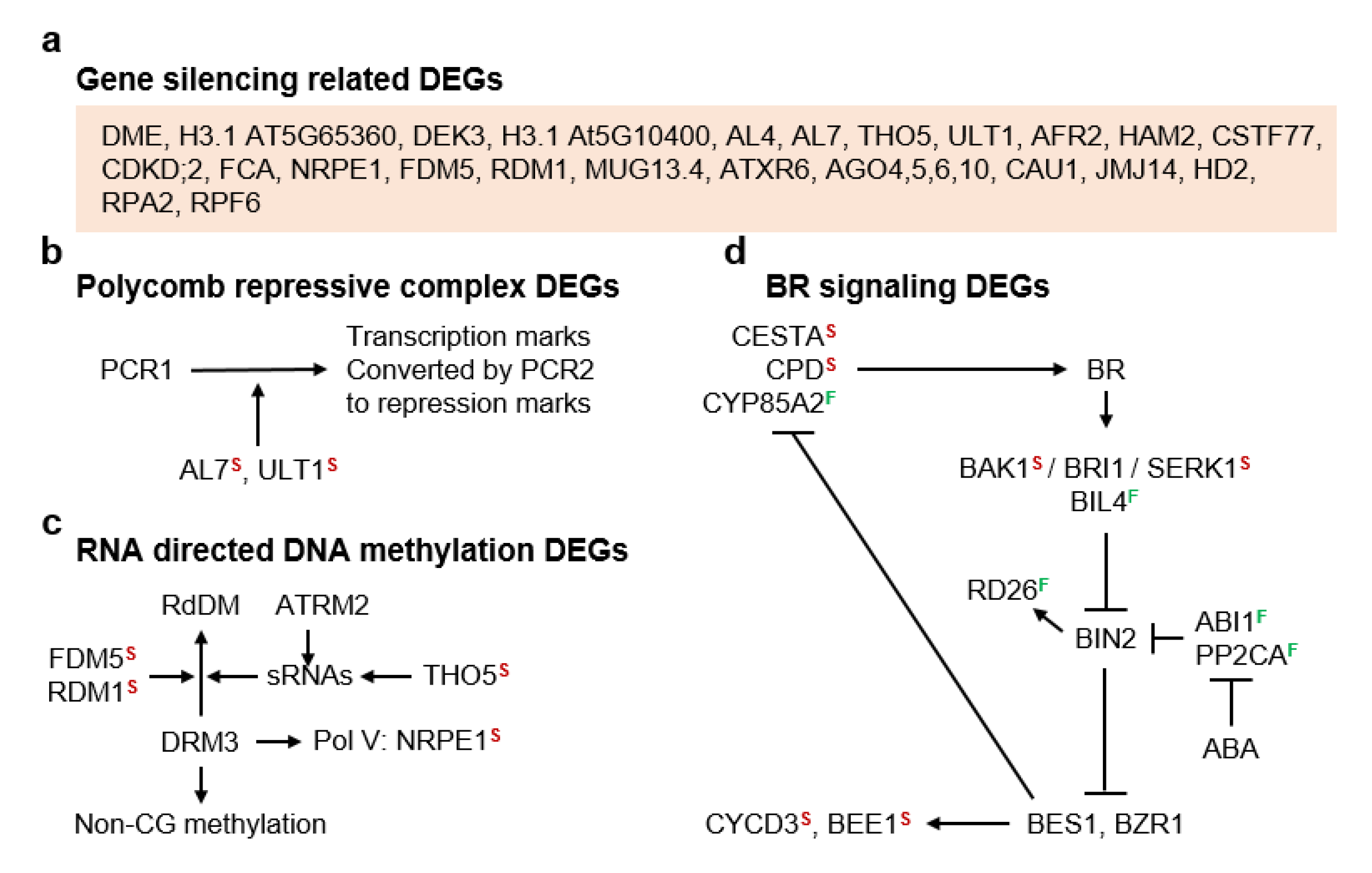
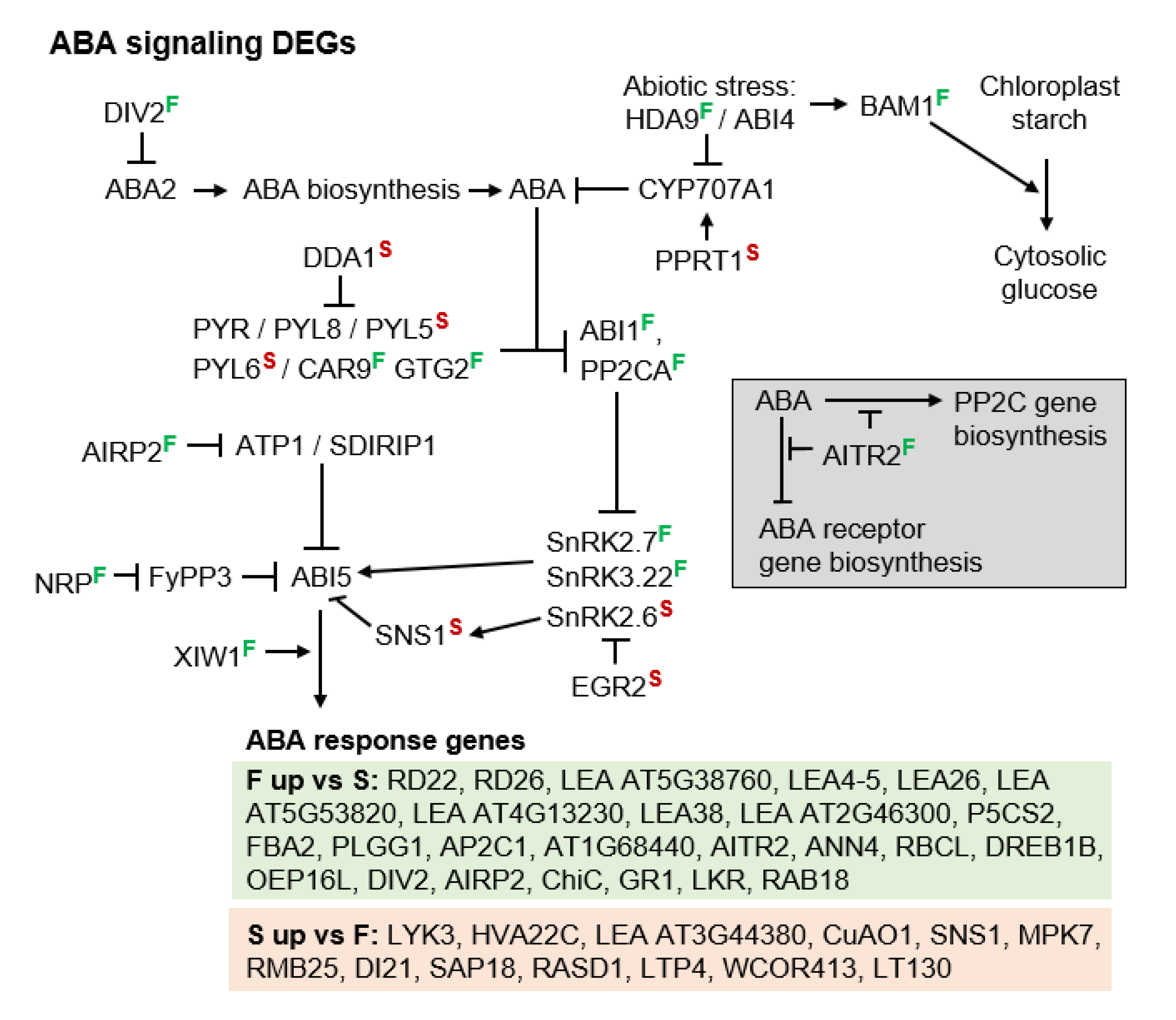
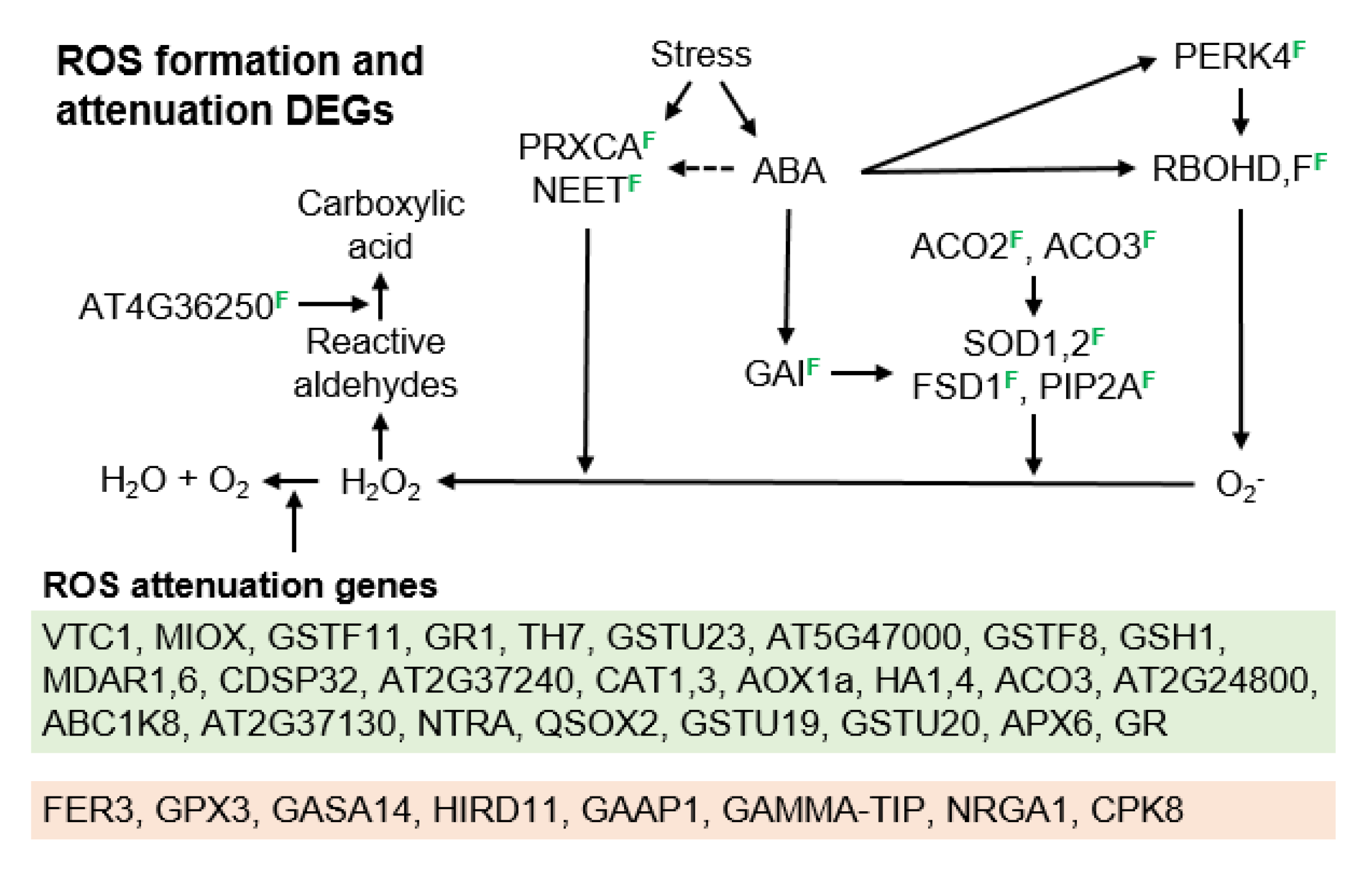
3.3. Molecular Pathways Upregulated in Apomeiotic Taxa
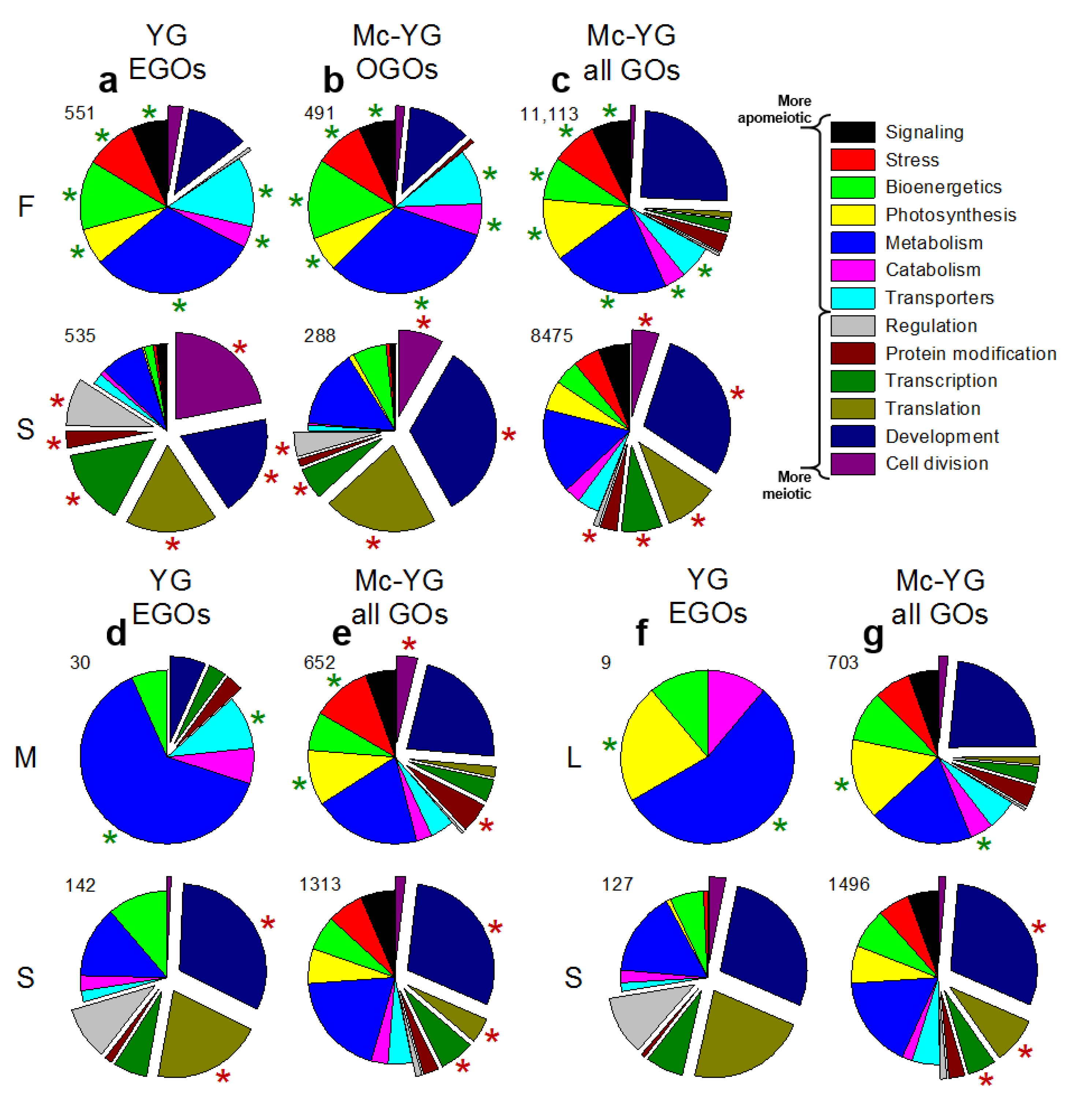
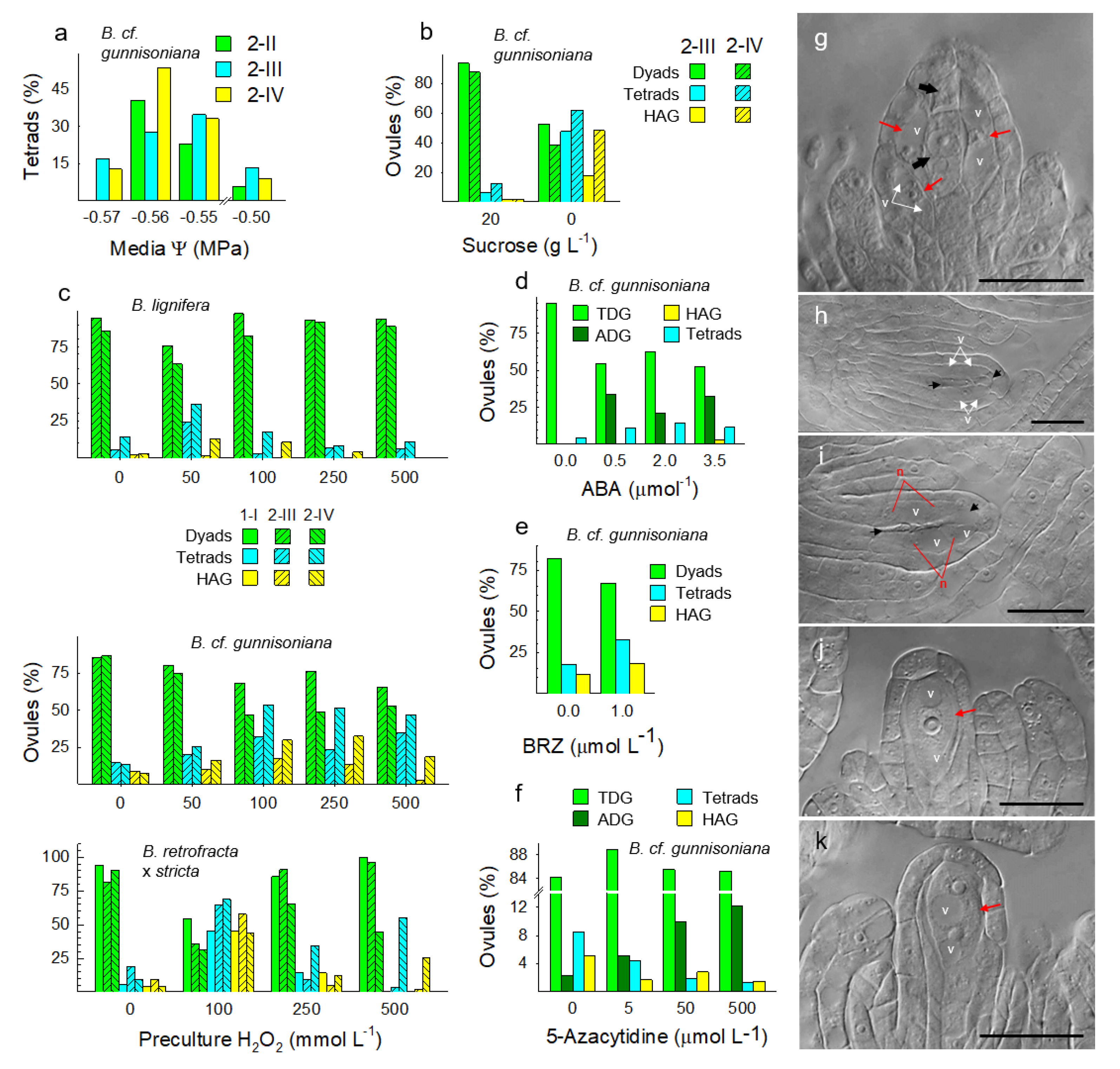
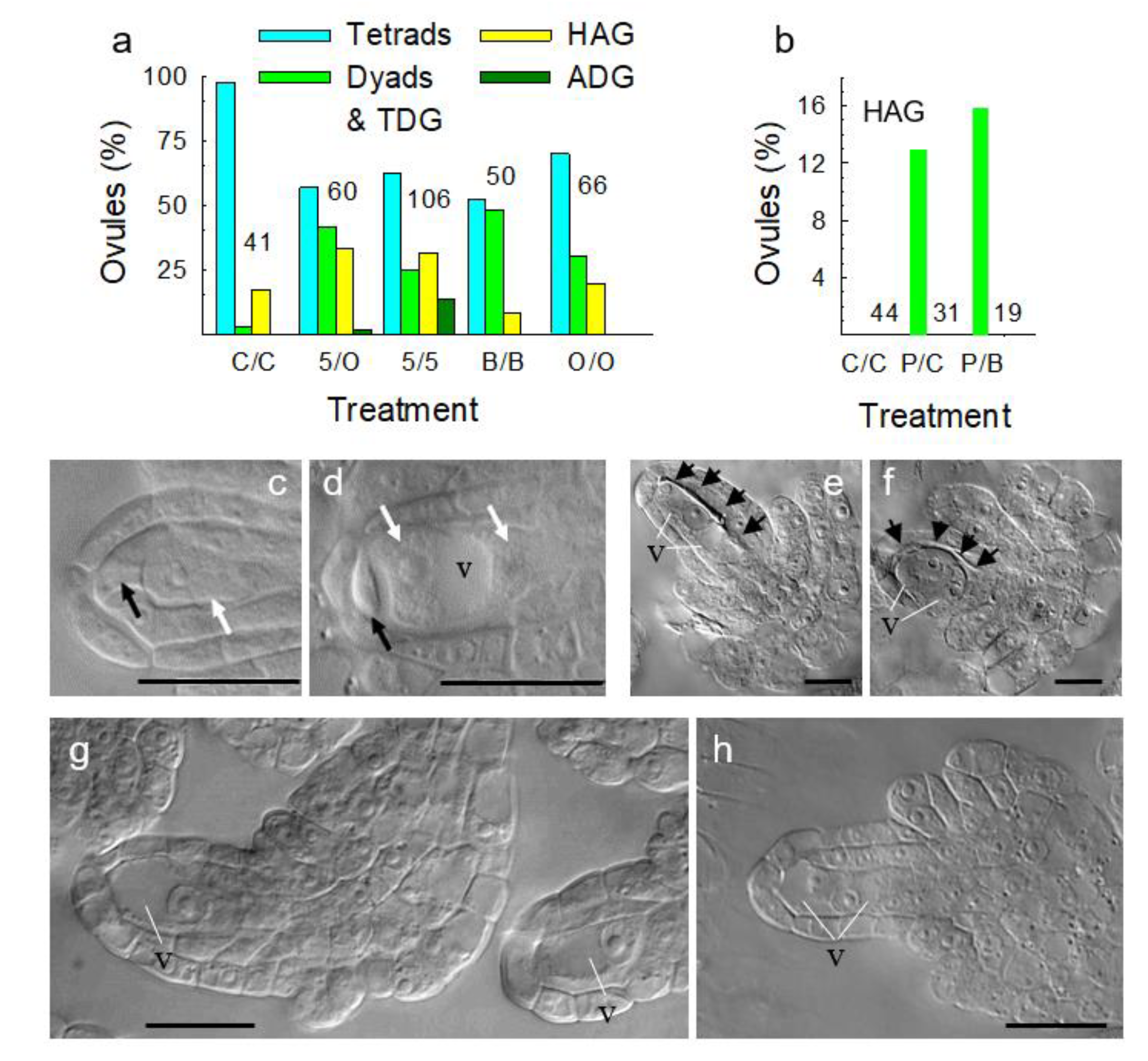
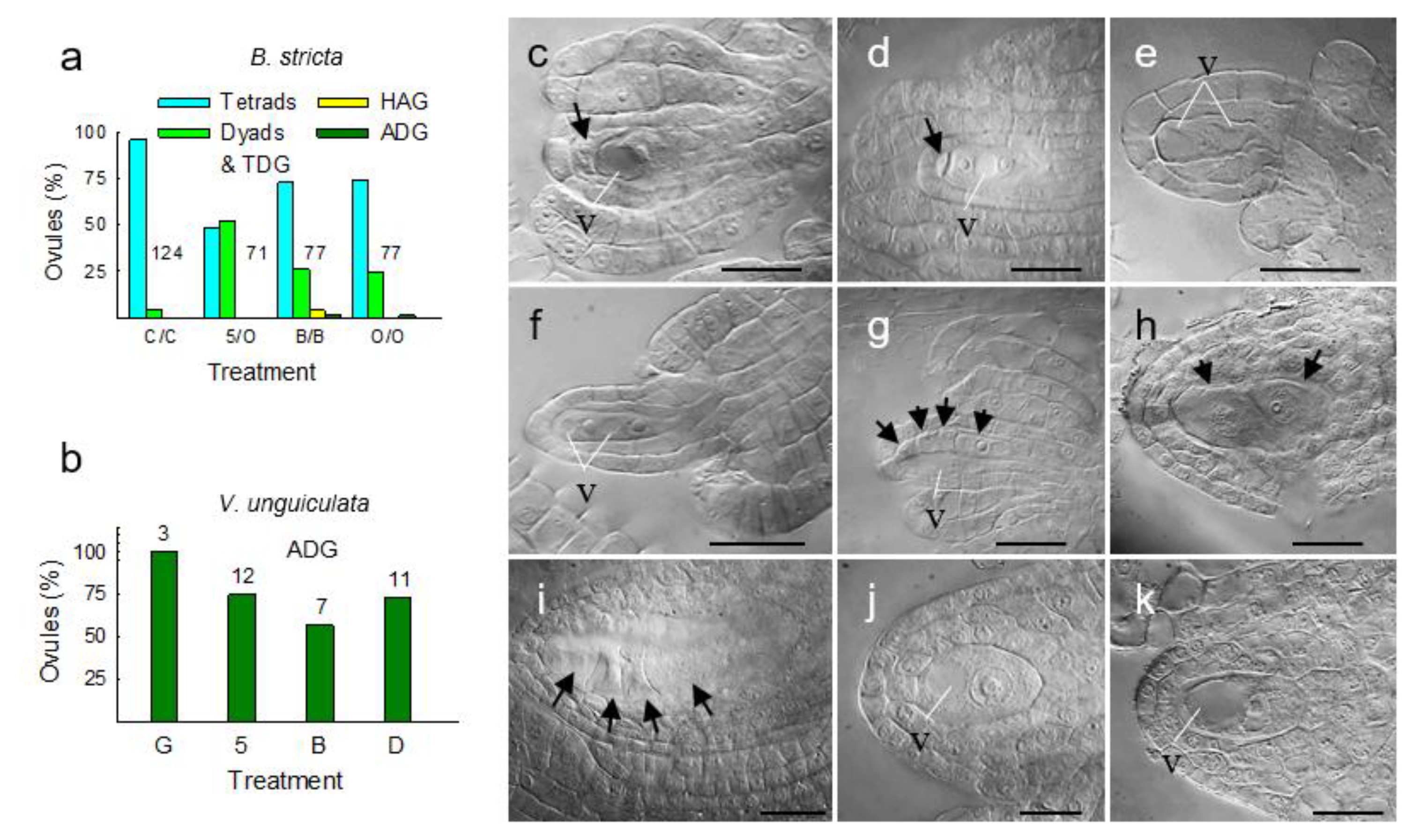
3.4. Metabolically Induced Switching from Apomeiosis to Meiosis
3.5. Metabolically Induced Switching from Meiosis to Apomeiosis
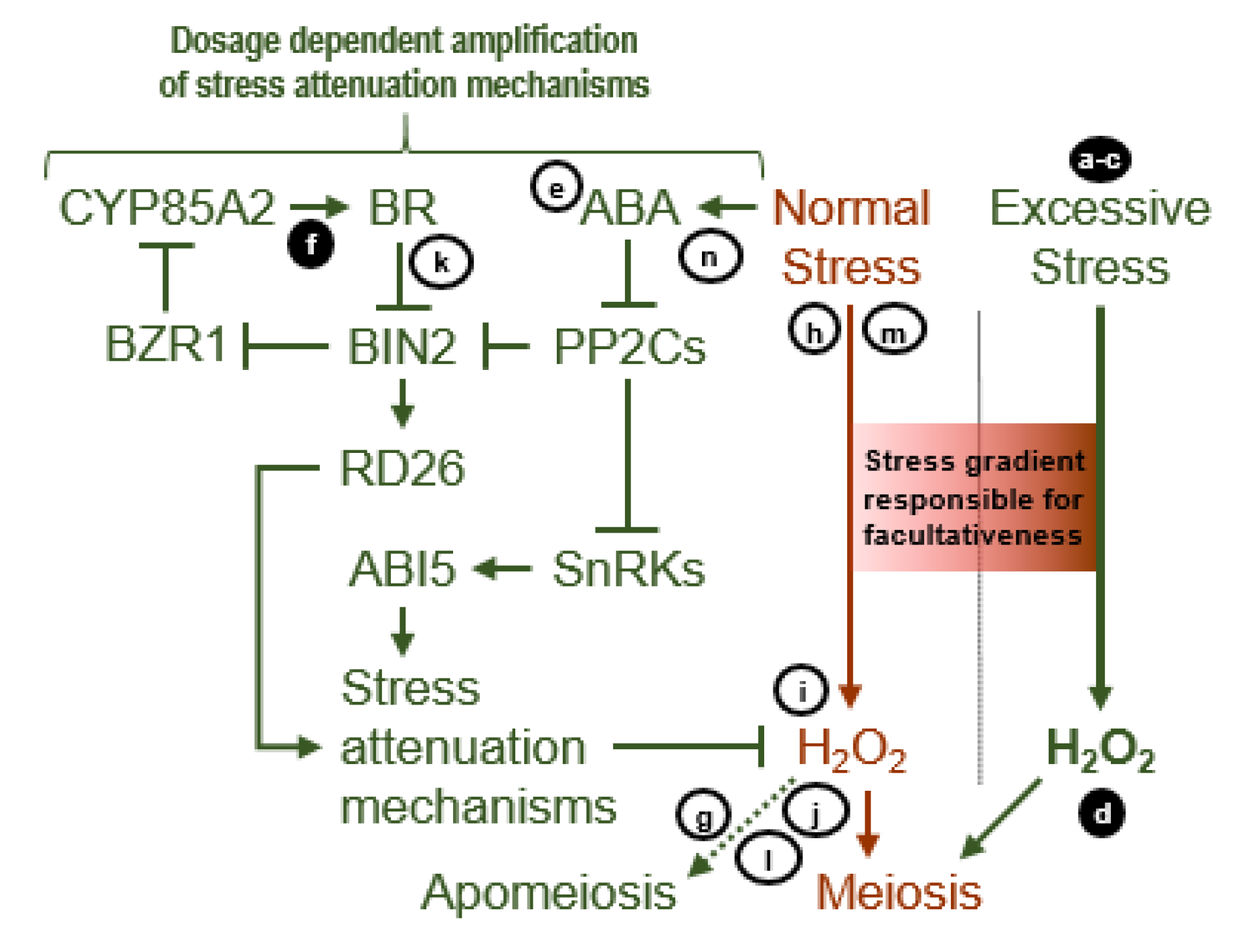
4. Discussion
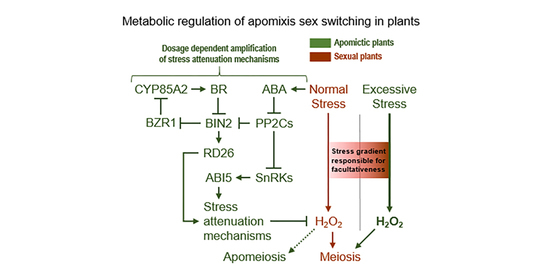
4.1. Sucrose Non-Fermenting-Related Protein Kinases May Be Central to Meiosis Apomeiosis Switching
4.2. Metabolically Inducing Meiosis Apomeiosis Interconversions
4.3. Apomixis Genes: Horses of a Different Color
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Burnett, J.H. Fundamentals of Mycology, 2nd ed.; Edward Arnold: London, UK, 1976. [Google Scholar]
- Suomalainen, E.; Saura, A.; Lokki, J. Cytology and Evolution in Parthenogenesis; CRC Press, Inc.: Boca Raton, FL, USA, 1987; p. 216. [Google Scholar]
- Bilinski, C.A.; Marmiroli, N.; Miller, J.J. Apomixis in Saccharomyces cerevisiae and other eukaryotic micro-organisms. Adv. Microb. Physiol. 1989, 30, 23–52. [Google Scholar]
- Asker, S.E.; Jerling, L. Apomixis in Plants; CRC Press, Inc.: Boca Raton, FL, USA, 1992; p. 298. [Google Scholar]
- Albertini, E.; Barcaccia, G.; Carman, J.G.; Pupilli, F. Did apomixis evolve from sex or was it the other way around? J. Exp. Bot. 2019, 70, 2951–2964. [Google Scholar] [CrossRef]
- Juel, H.O. Vergleichende untersuchungen uber typische und parthenogenetische fortpflanzung bei der gattung Antennaria Kongl Sven. Vetenskars Akad. Handl. 1900, 33, 1–59. [Google Scholar]
- Gustafsson, A. Apomixis in higher plants. I. The mechanisms of apomixis. Lunds Univ. Arsskrift 1946, 42, 1–67. [Google Scholar]
- Hand, M.L.; Koltunow, A.M. The genetic control of apomixis: Asexual seed formation. Genetics 2014, 197, 441–450. [Google Scholar] [CrossRef] [Green Version]
- Nogler, G.A. Gametophytic apomixis. In Embryology of Angiosperms; Johri, B.M., Ed.; Springer-Verlag: New York, NY, USA, 1984; pp. 475–518. [Google Scholar]
- Ozias-Akins, P.; van Dijk, P.J. Mendelian genetics of apomixis in plants. Ann. Rev. Genet. 2007, 41, 509–537. [Google Scholar] [CrossRef]
- Savidan, Y. Apomixis: Genetics and breeding. In Plant Breeding Reviews; Janick, J., Ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2000; Volume 18, pp. 13–86. [Google Scholar]
- Mogie, M. The Evolution of Asexual Reproduction in Plants; Chapman & Hall: London, UK, 1992; p. 276. [Google Scholar]
- Carman, J.G. Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linn. Soc. 1997, 61, 51–94. [Google Scholar] [CrossRef]
- Carman, J.G.; Jamison, M.; Elliott, E.; Dwivedi, K.K.; Naumova, T.N. Apospory appears to accelerate onset of meiosis and sexual embryo sac formation in sorghum ovules. BMC Plant Biol. 2011, 11, 9. [Google Scholar] [CrossRef] [Green Version]
- Horandl, E.; Hadacek, F. The oxidative damage initiation hypothesis for meiosis. Plant Reprod. 2013, 26, 351–367. [Google Scholar] [CrossRef] [Green Version]
- Carman, J.G.; Mateo de Arias, M.; Gao, L.; Zhao, X.; Kowallis, B.M.; Sherwood, D.A.; Srivastava, M.K.; Dwivedi, K.K.; Price, B.J.; Watts, L.; et al. Apospory and diplospory in diploid Boechera (Brassicaceae) may facilitate speciation by recombination-driven apomixis-to-sex reversals. Front. Plant Sci. 2019, 10, 724. [Google Scholar] [CrossRef]
- Li, F.W.; Rushworth, C.A.; Beck, J.B.; Windham, M.D. Boechera microsatellite website: An online portal for species identification and determination of hybrid parentage. Database (Oxford) 2017, 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Windsor, A.J.; Schranz, M.E.; Formanova, N.; Gebauer-Jung, S.; Bishop, J.G.; Schnabelrauch, D.; Kroymann, J.; Mitchell-Olds, T. Partial shotgun sequencing of the Boechera stricta genome reveals extensive microsynteny and promoter conservation with Arabidopsis. Plant Physiol. 2006, 140, 1169–1182. [Google Scholar] [CrossRef] [Green Version]
- Brukhin, V.; Osadtchiy, J.V.; Florez-Rueda, A.M.; Smetanin, D.; Bakin, E.; Nobre, M.S.; Grossniklaus, U. The Boechera genus as a resource for apomixis research. Front. Plant Sci. 2019, 10, 392. [Google Scholar] [CrossRef] [Green Version]
- Na, D.; Son, H.; Gsponer, J. Categorizer: A tool to categorize genes into user-defined biological groups based on semantic similarity. BMC Genom. 2014, 15, 1091. Available online: http://www.biomedcentral.com/1471-2164/15/1091 (accessed on 22 October 2020). [CrossRef] [Green Version]
- Hinderer, E.W., 3rd; Flight, R.M.; Dubey, R.; MacLeod, J.N.; Moseley, H.N.B. Advances in gene ontology utilization improve statistical power of annotation enrichment. PLoS ONE 2019, 14, e0220728. [Google Scholar] [CrossRef] [Green Version]
- Hinderer, E.W., 3rd; Moseley, H.N.B. GOcats: A tool for categorizing Gene Ontology into subgraphs of user-defined concepts. PLoS ONE 2020, 15, e0233311. [Google Scholar] [CrossRef]
- Ostle, B.; Mensing, R.W. Statistics in Research, 3rd ed.; The Iowa State University Press: Ames, UK, 1975; p. 596. [Google Scholar]
- Agresti, A. An Introduction to Categorical Data Analysis, 3rd ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2019. [Google Scholar]
- SYSTAT. SYSTAT for Windows; SYSTAT Software, Inc.: San Jose, CA, USA, 2004. [Google Scholar]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Phsiologia Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Buah, J.N.; Kawamitsu, U.; Sato, S.; Murahama, S. Effects of different types and concentrations of gelling agents on the physical and chemical properties of media and the growth of banana (Musa spp.) in vitro. Plant Prod. Sci. 1999, 2, 138–145. [Google Scholar] [CrossRef]
- Michel, B.E. Evaluation of the water potentials of solutions of polyethylene glycol 8000 both in the absence and presence of other solutes. Plant Physiol. 1983, 72, 66–70. [Google Scholar] [CrossRef] [Green Version]
- Michel, B.E.; Kaufmann, M.R. The osmotic potential of polyethylene glycol 6000. Plant Physiol. 1973, 51, 914–916. [Google Scholar] [CrossRef]
- Schneitz, K.; Hulskamp, M.; Pruitt, R.E. Wild-type ovule development in Arabidopsis thaliana: A light microscope study of cleared whole-mount tissue. Plant J. 1995, 7, 731–749. [Google Scholar] [CrossRef]
- Clark, A.G.; Glanowski, S.; Nielsen, R.; Thomas, P.D.; Kejariwal, A.; Todd, M.A.; Tanenbaum, D.M.; Civello, D.; Lu, F.; Murphy, B.; et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science 2003, 302, 1960–1963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Broeckx, T.; Hulsmans, S.; Rolland, F. The plant energy sensor: Evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J. Exp. Bot 2016, 67, 6215–6252. [Google Scholar] [CrossRef]
- Garapati, P.; Feil, R.; Lunn, J.E.; Van Dijck, P.; Balazadeh, S.; Mueller-Roeber, B. Transcription factor arabidopsis activating factor1 integrates carbon starvation responses with trehalose metabolism. Plant Physiol. 2015, 169, 379–390. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Zhang, P.; Wang, R.; Wan, J.; Ju, Q.; Rothstein, S.J.; Xu, J. The SNAC-A transcription factor ANAC032 reprograms metabolism in Arabidopsis. Plant Cell Physiol. 2019, 60, 999–1010. [Google Scholar] [CrossRef]
- Carvalho, R.F.; Szakonyi, D.; Simpson, C.G.; Barbosa, I.C.; Brown, J.W.; Baena-Gonzalez, E.; Duque, P. The Arabidopsis SR45 splicing factor, a negative regulator of sugar signaling, modulates SNF1-related protein kinase 1 stability. Plant Cell 2016, 28, 1910–1925. [Google Scholar] [CrossRef] [Green Version]
- Poschet, G.; Hannich, B.; Raab, S.; Jungkunz, I.; Klemens, P.A.; Krueger, S.; Wic, S.; Neuhaus, H.E.; Buttner, M. A novel Arabidopsis vacuolar glucose exporter is involved in cellular sugar homeostasis and affects the composition of seed storage compounds. Plant Physiol. 2011, 157, 1664–1676. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Jiang, Q.T.; Wei, L.; Yang, Q.; Zhang, X.W.; Peng, Y.Y.; Chen, G.Y.; Wei, Y.M.; Liu, C.; Zheng, Y.L. Conserved structure and varied expression reveal key roles of phosphoglucan phosphatase gene starch excess 4 in barley. Planta 2014, 240, 1179–1190. [Google Scholar] [CrossRef]
- Crozet, P.; Margalha, L.; Confraria, A.; Rodrigues, A.; Martinho, C.; Adamo, M.; Elias, C.A.; Baena-Gonzalez, E. Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front. Plant Sci. 2014, 5, 190. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Micallef, B.J.; Tetlow, I.J.; Mullen, R.T.; Feil, R.; Lunn, J.E.; Emes, M.J. AKINbeta1, a subunit of SnRK1, regulates organic acid metabolism and acts as a global modulator of genes involved in carbon, lipid, and nitrogen metabolism. J. Exp. Bot. 2020, 71, 1010–1028. [Google Scholar] [CrossRef]
- Liu, M.S.; Chen, L.F.; Lin, C.H.; Lai, Y.M.; Huang, J.Y.; Sung, Z.R. Molecular and functional characterization of broccoli EMBRYONIC FLOWER 2 genes. Plant Cell Physiol. 2012, 53, 1217–1231. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Wang, Q.; Liu, C.; Shang, Y.; Wen, F.; Wang, F.; Liu, W.; Xiao, W.; Li, W. A role for the respiratory chain in regulating meiosis initiation in Saccharomyces cerevisiae. Genetics 2018, 208, 1181–1194. [Google Scholar] [CrossRef] [Green Version]
- Beermann, F.; Hansmann, I. Follicular maturation, luteinization and first meiotic division in oocytes after inhibiting mitochondrial function in mice with chloramphenicol. Mutat. Res. 1986, 160, 47–54. [Google Scholar] [CrossRef]
- Meng, X.; Li, L.; De Clercq, I.; Narsai, R.; Xu, Y.; Hartmann, A.; Claros, D.L.; Custovic, E.; Lewsey, M.G.; Whelan, J.; et al. ANAC017 coordinates organellar functions and stress responses by reprogramming retrograde signaling. Plant Physiol. 2019, 180, 634–653. [Google Scholar] [CrossRef] [Green Version]
- Gladman, N.P.; Marshall, R.S.; Lee, K.H.; Vierstra, R.D. The proteasome stress regulon is controlled by a pair of NAC transcription factors in Arabidopsis. Plant Cell 2016, 28, 1279–1296. [Google Scholar] [CrossRef] [Green Version]
- Cavalier-Smith, T. Origin of the cell nucleus, mitosis and sex: Roles of intracellular coevolution. Biol. Direct. 2010, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- Bernstein, H.; Bernstein, C. Evolutionary origin and adaptive function of meiosis. In Meiosis; Bernstein, H., Bernstein, C., Eds.; InTech: Rijeka, Croatia, 2013; p. 123. [Google Scholar]
- Mirzaghaderi, G.; Horandl, E. The evolution of meiotic sex and its alternatives. Proc. Biol. Sci. 2016, 283. [Google Scholar] [CrossRef]
- Walker, J.; Gao, H.; Zhang, J.; Aldridge, B.; Vickers, M.; Higgins, J.D.; Feng, X. Sexual-lineage-specific DNA methylation regulates meiosis in Arabidopsis. Nat. Genet. 2018, 50, 130–137. [Google Scholar] [CrossRef]
- Peng, L.; Wang, L.; Zhang, Y.; Dong, A.; Shen, W.H.; Huang, Y. Structural analysis of the Arabidopsis AL2-PAL and PRC1 complex provides mechanistic insight into active-to-repressive chromatin state switch. J. Mol. Biol. 2018, 430, 4245–4259. [Google Scholar] [CrossRef]
- Tyler, L.; Miller, M.J.; Fletcher, J.C. The trithorax group factor ULTRAPETALA1 regulates developmental as well as biotic and abiotic stress response genes in Arabidopsis. G3 (Bethesda) 2019, 9, 4029–4043. [Google Scholar] [CrossRef] [Green Version]
- Wils, C.R.; Kaufmann, K. Gene-regulatory networks controlling inflorescence and flower development in Arabidopsis thaliana. Biochim. Biophys. Acta Gene. Regul. Mech. 2017, 1860, 95–105. [Google Scholar] [CrossRef]
- Planas-Riverola, A.; Gupta, A.; Betegon-Putze, I.; Bosch, N.; Ibanes, M.; Cano-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146. [Google Scholar] [CrossRef] [Green Version]
- Lv, M.; Li, J. Molecular mechanisms of brassinosteroid-mediated responses to changing environments in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 2737. [Google Scholar] [CrossRef] [Green Version]
- Oh, E.; Zhu, J.Y.; Wang, Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802–809. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Lee, Y.J.; Liu, B. Establishment of a mitotic model system by transient expression of the D-type cyclin in differentiated leaf cells of tobacco (Nicotiana benthamiana). New Phytol. 2020, 226, 1213–1220. [Google Scholar] [CrossRef]
- Wang, F.; Gao, Y.; Liu, Y.; Zhang, X.; Gu, X.; Ma, D.; Zhao, Z.; Yuan, Z.; Xue, H.; Liu, H. BES1-regulated BEE1 controls photoperiodic flowering downstream of blue light signaling pathway in Arabidopsis. New Phytol. 2019, 223, 1407–1419. [Google Scholar] [CrossRef]
- Poppenberger, B.; Rozhon, W.; Khan, M.; Husar, S.; Adam, G.; Luschnig, C.; Fujioka, S.; Sieberer, T. CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J. 2011, 30, 1149–1161. [Google Scholar] [CrossRef] [Green Version]
- Zhiponova, M.K.; Vanhoutte, I.; Boudolf, V.; Betti, C.; Dhondt, S.; Coppens, F.; Mylle, E.; Maes, S.; Gonzalez-Garcia, M.P.; Cano-Delgado, A.I.; et al. Brassinosteroid production and signaling differentially control cell division and expansion in the leaf. New Phytol. 2013, 197, 490–502. [Google Scholar] [CrossRef]
- Santiago, J.; Henzler, C.; Hothorn, M. Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 2013, 341, 889–892. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Huang, J.; Ahsan, N.; Biener, G.; Paprocki, J.; Thelen, J.J.; Raicu, V.; Zhao, D. Two SERK receptor-like kinases interact with EMS1 to control anther cell fate determination. Plant Physiol. 2017, 173, 326–337. [Google Scholar] [CrossRef] [Green Version]
- Northey, J.G.; Liang, S.; Jamshed, M.; Deb, S.; Foo, E.; Reid, J.B.; McCourt, P.; Samuel, M.A. Farnesylation mediates brassinosteroid biosynthesis to regulate abscisic acid responses. Nat. Plants 2016, 2, 16114. [Google Scholar] [CrossRef]
- Yamagami, A.; Saito, C.; Nakazawa, M.; Fujioka, S.; Uemura, T.; Matsui, M.; Sakuta, M.; Shinozaki, K.; Osada, H.; Nakano, A.; et al. Evolutionarily conserved BIL4 suppresses the degradation of brassinosteroid receptor BRI1 and regulates cell elongation. Sci. Rep. 2017, 7, 5739. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Tang, B.; Xie, Z.; Nolan, T.; Ye, H.; Song, G.Y.; Walley, J.; Yin, Y. GSK3-like kinase BIN2 phosphorylates RD26 to potentiate drought signaling in Arabidopsis. Plant J. 2019, 100, 923–937. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Nishimura, N.; Kitahata, N.; Kuromori, T.; Ito, T.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006, 140, 115–126. [Google Scholar] [CrossRef] [Green Version]
- Carman, J.G. The evolution of gametophytic apomixis. In Embryology of Flowering Plants, Vol. 3: The Systems of Reproduction; Batygina, T.B., Ed.; Russian Academy of Sciences: St. Petersburg, Russia, 2000; pp. 218–245. [Google Scholar]
- Bar-Zvi, D.; Lupo, O.; Levy, A.A.; Barkai, N. Hybrid vigor: The best of both parents, or a genomic clash? Curr. Opin. Syst. Biol. 2017, 6, 22–27. [Google Scholar] [CrossRef]
- Kirk, H.; Choi, Y.H.; Kim, H.K.; Verpoorte, R.; van der Meijden, E. Comparing metabolomes: The chemical consequences of hybridization in plants. New Phytol. 2005, 167, 613–622. [Google Scholar] [CrossRef]
- Hauser, F.; Li, Z.; Waadt, R.; Schroeder, J.I. SnapShot: Abscisic acid signaling. Cell 2017, 171, 1708–1708 e1700. [Google Scholar] [CrossRef]
- Fang, Q.; Wang, Q.; Mao, H.; Xu, J.; Wang, Y.; Hu, H.; He, S.; Tu, J.; Cheng, C.; Tian, G.; et al. AtDIV2, an R-R-type MYB transcription factor of Arabidopsis, negatively regulates salt stress by modulating ABA signaling. Plant Cell Rep. 2018, 37, 1499–1511. [Google Scholar] [CrossRef]
- Baek, D.; Shin, G.; Kim, M.C.; Shen, M.; Lee, S.Y.; Yun, D.J. Histone deacetylase HDA9 with ABI4 contributes to abscisic acid homeostasis in drought stress response. Front. Plant Sci. 2020, 11, 143. [Google Scholar] [CrossRef]
- Liu, Y.; Pei, L.; Xiao, S.; Peng, L.; Liu, Z.; Li, X.; Yang, Y.; Wang, J. AtPPRT1 negatively regulates salt stress response in Arabidopsis seedlings. Plant Signal. Behav. 2020, 15, 1732103. [Google Scholar] [CrossRef]
- Pei, L.; Peng, L.; Wan, X.; Xiong, J.; Liu, Z.; Li, X.; Yang, Y.; Wang, J. Expression pattern and function analysis of AtPPRT1, a novel negative regulator in ABA and drought stress responses in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 394. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Huang, S.C.; Wise, A.; Castanon, R.; Nery, J.R.; Chen, H.; Watanabe, M.; Thomas, J.; Bar-Joseph, Z.; Ecker, J.R. A transcription factor hierarchy defines an environmental stress response network. Science 2016, 354. [Google Scholar] [CrossRef] [Green Version]
- Tian, H.; Chen, S.; Yang, W.; Wang, T.; Zheng, K.; Wang, Y.; Cheng, Y.; Zhang, N.; Liu, S.; Li, D.; et al. A novel family of transcription factors conserved in angiosperms is required for ABA signalling. Plant Cell Environ. 2017, 40, 2958–2971. [Google Scholar] [CrossRef]
- Rodriguez, L.; Gonzalez-Guzman, M.; Diaz, M.; Rodrigues, A.; Izquierdo-Garcia, A.C.; Peirats-Llobet, M.; Fernandez, M.A.; Antoni, R.; Fernandez, D.; Marquez, J.A.; et al. C2-domain abscisic acid-related proteins mediate the interaction of PYR/PYL/RCAR abscisic acid receptors with the plasma membrane and regulate abscisic acid sensitivity in Arabidopsis. Plant Cell 2014, 26, 4802–4820. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, S.; Roy Choudhury, S.; Hicks, L.M.; Pandey, S. Quantitative proteomics-based analysis supports a significant role of GTG proteins in regulation of ABA response in Arabidopsis roots. J. Proteome Res. 2013, 12, 1487–1501. [Google Scholar] [CrossRef]
- Irigoyen, M.L.; Iniesto, E.; Rodriguez, L.; Puga, M.I.; Yanagawa, Y.; Pick, E.; Strickland, E.; Paz-Ares, J.; Wei, N.; De Jaeger, G.; et al. Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell 2014, 26, 712–728. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Lv, J.; Shi, Y.; Gao, J.; Hua, J.; Song, C.; Gong, Z.; Yang, S. EGR2 phosphatase regulates OST1 kinase activity and freezing tolerance in Arabidopsis. EMBO J. 2019, 38. [Google Scholar] [CrossRef]
- Umezawa, T.; Sugiyama, N.; Takahashi, F.; Anderson, J.C.; Ishihama, Y.; Peck, S.C.; Shinozaki, K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 2013, 6, rs8. [Google Scholar] [CrossRef]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 1717–1722. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Hao, H.; Zhang, Y.; Bai, Y.; Zhu, W.; Qin, Y.; Yuan, F.; Zhao, F.; Wang, M.; Hu, J.; et al. SOS2-LIKE PROTEIN KINASE5, an SNF1-RELATED PROTEIN KINASE3-type protein kinase, is iImportant for abscisic acid responses in Arabidopsis through phosphorylation of ABSCISIC ACID-INSENSITIVE5. Plant Physiol. 2015, 168, 659–676. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Wang, C.; Xue, Y.; Liu, X.; Chen, S.; Song, C.; Yang, Y.; Guo, Y. Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 2019, 10, 1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Wan, W.; Jiang, G.; Xi, Y.; Huang, H.; Cai, J.; Chang, Y.; Duan, C.G.; Mangrauthia, S.K.; Peng, X.; et al. Nucleocytoplasmic trafficking of the Arabidopsis WD40 repeat protein XIW1 regulates ABI5 stability and abscisic acid responses. Mol. Plant 2019, 12, 1598–1611. [Google Scholar] [CrossRef]
- Zhu, T.; Wu, Y.; Yang, X.; Chen, W.; Gong, Q.; Liu, X. The asparagine-rich protein NRP facilitates the degradation of the PP6-type phosphatase FyPP3 to promote ABA response in Arabidopsis. Mol. Plant 2018, 11, 257–268. [Google Scholar] [CrossRef] [Green Version]
- Oh, T.R.; Kim, J.H.; Cho, S.K.; Ryu, M.Y.; Yang, S.W.; Kim, W.T. AtAIRP2 E3 ligase affects ABA and high-salinity responses by stimulating Its ATP1/SDIRIP1 substrate turnover. Plant Physiol. 2017, 174, 2515–2531. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Kang, K.; Gan, L.; Ning, S.; Xiong, J.; Song, S.; Xi, L.; Lai, S.; Yin, Y.; Gu, J.; et al. Drought-responsive genes, late embryogenesis abundant group3 (LEA3) and vicinal oxygen chelate, function in lipid accumulation in Brassica napus and Arabidopsis mainly via enhancing photosynthetic efficiency and reducing ROS. Plant Biotechnol. J. 2019, 17, 2123–2142. [Google Scholar] [CrossRef] [Green Version]
- Lu, W.; Tang, X.; Huo, Y.; Xu, R.; Qi, S.; Huang, J.; Zheng, C.; Wu, C.A. Identification and characterization of fructose 1,6-bisphosphate aldolase genes in Arabidopsis reveal a gene family with diverse responses to abiotic stresses. Gene 2012, 503, 65–74. [Google Scholar] [CrossRef]
- Hernandez-Sanchez, I.E.; Maruri-Lopez, I.; Molphe-Balch, E.P.; Becerra-Flora, A.; Jaimes-Miranda, F.; Jimenez-Bremont, J.F. Evidence for in vivo interactions between dehydrins and the aquaporin AtPIP2B. Biochem. Biophys. Res. Commun. 2019, 510, 545–550. [Google Scholar] [CrossRef]
- Seok, H.Y.; Nguyen, L.V.; Park, H.Y.; Tarte, V.N.; Ha, J.; Lee, S.Y.; Moon, Y.H. Arabidopsis non-TZF gene AtC3H17 functions as a positive regulator in salt stress response. Biochem. Biophys. Res. Commun. 2018, 498, 954–959. [Google Scholar] [CrossRef]
- Ebeed, H.T.; Stevenson, S.R.; Cuming, A.C.; Baker, A. Conserved and differential transcriptional responses of peroxisome associated pathways to drought, dehydration and ABA. J. Exp. Bot. 2018, 69, 4971–4985. [Google Scholar] [CrossRef]
- Singh, A.; Yadav, A.K.; Kaur, K.; Sanyal, S.K.; Jha, S.K.; Fernandes, J.L.; Sharma, P.; Tokas, I.; Pandey, A.; Luan, S.; et al. A protein phosphatase 2C, AP2C1, interacts with and negatively regulates the function of CIPK9 under potassium-deficient conditions in Arabidopsis. J. Exp. Bot. 2018, 69, 4003–4015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shubchynskyy, V.; Boniecka, J.; Schweighofer, A.; Simulis, J.; Kvederaviciute, K.; Stumpe, M.; Mauch, F.; Balazadeh, S.; Mueller-Roeber, B.; Boutrot, F.; et al. Protein phosphatase AP2C1 negatively regulates basal resistance and defense responses to Pseudomonas syringae. J. Exp. Bot. 2017, 68, 1169–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman, J.M.; Muhlemann, J.K.; Gayomba, S.R.; Muday, G.K. RBOH-dependent ROS synthesis and ROS scavenging by plant specialized metabolites to modulate plant development and stress responses. Chem. Res. Toxicol. 2019, 32, 370–396. [Google Scholar] [CrossRef] [PubMed]
- Kaman-Toth, E.; Danko, T.; Gullner, G.; Bozso, Z.; Palkovics, L.; Pogany, M. Contribution of cell wall peroxidase- and NADPH oxidase-derived reactive oxygen species to Alternaria brassicicola-induced oxidative burst in Arabidopsis. Mol. Plant Pathol. 2019, 20, 485–499. [Google Scholar] [CrossRef] [Green Version]
- Zandalinas, S.I.; Song, L.; Sengupta, S.; McInturf, S.A.; Grant, D.G.; Marjault, H.B.; Castro-Guerrero, N.A.; Burks, D.; Azad, R.K.; Mendoza-Cozatl, D.G.; et al. Expression of a dominant-negative AtNEET-H89C protein disrupts iron-sulfur metabolism and iron homeostasis in Arabidopsis. Plant J. 2020, 101, 1152–1169. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, X.; Yang, L.; Tang, M.; Wang, K.; Wang, L.; Bai, L.; Song, C. Hydrogen peroxide plays an important role in PERK4-mediated abscisic acid-regulated root growth in Arabidopsis. Funct. Plant Biol. 2019, 46, 165–174. [Google Scholar] [CrossRef]
- Angelos, E.; Brandizzi, F. NADPH oxidase activity is required for ER stress survival in plants. Plant J. 2018, 96, 1106–1120. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Yi, H.; Xue, M.; Yi, M. miR398 and miR395 are involved in response to SO2 stress in Arabidopsis thaliana. Ecotoxicology 2017, 26, 1181–1187. [Google Scholar] [CrossRef]
- Panda, S.K.; Sunkar, R. Nutrient- and other stress-responsive microRNAs in plants: Role for thiol-based redox signaling. Plant Signal. Behav. 2015, 10, e1010916. [Google Scholar] [CrossRef]
- Dvorak, P.; Krasylenko, Y.; Ovecka, M.; Basheer, J.; Zapletalova, V.; Samaj, J.; Takac, T. FSD1: Developmentally-regulated plastidial, nuclear and cytoplasmic enzyme with anti-oxidative and osmoprotective role. Plant Cell Environ. 2020. [Google Scholar] [CrossRef]
- Martiniere, A.; Fiche, J.B.; Smokvarska, M.; Mari, S.; Alcon, C.; Dumont, X.; Hematy, K.; Jaillais, Y.; Nollmann, M.; Maurel, C. Osmotic stress activates two reactive oxygen species pathways with distinct effects on protein nanodomains and diffusion. Plant Physiol. 2019, 179, 1581–1593. [Google Scholar] [CrossRef] [Green Version]
- Achard, P.; Renou, J.P.; Berthome, R.; Harberd, N.P.; Genschik, P. Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 2008, 18, 656–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujiwara, A.; Togawa, S.; Hikawa, T.; Matsuura, H.; Masuta, C.; Inukai, T. Ascorbic acid accumulates as a defense response to Turnip mosaic virus in resistant Brassica rapa cultivars. J. Exp. Bot. 2016, 67, 4391–4402. [Google Scholar] [CrossRef] [Green Version]
- Nepal, N.; Yactayo-Chang, J.P.; Medina-Jimenez, K.; Acosta-Gamboa, L.M.; Gonzalez-Romero, M.E.; Arteaga-Vazquez, M.A.; Lorence, A. Mechanisms underlying the enhanced biomass and abiotic stress tolerance phenotype of an Arabidopsis MIOX over-expresser. Plant Direct. 2019, 3, e00165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selinski, J.; Scheibe, R.; Day, D.A.; Whelan, J. Alternative oxidase is positive for plant performance. Trends Plant Sci. 2018, 23, 588–597. [Google Scholar] [CrossRef]
- Chen, C.; Letnik, I.; Hacham, Y.; Dobrev, P.; Ben-Daniel, B.H.; Vankova, R.; Amir, R.; Miller, G. ASCORBATE PEROXIDASE6 protects Arabidopsis desiccating and germinating seeds from stress and mediates cross talk between reactive oxygen species, abscisic acid, and auxin. Plant Physiol. 2014, 166, 370–383. [Google Scholar] [CrossRef] [Green Version]
- Muller-Schussele, S.J.; Wang, R.; Gutle, D.D.; Romer, J.; Rodriguez-Franco, M.; Scholz, M.; Buchert, F.; Luth, V.M.; Kopriva, S.; Dormann, P.; et al. Chloroplasts require glutathione reductase to balance reactive oxygen species and maintain efficient photosynthesis. Plant J. 2020. [Google Scholar] [CrossRef]
- Tossounian, M.A.; Van Molle, I.; Wahni, K.; Jacques, S.; Gevaert, K.; Van Breusegem, F.; Vertommen, D.; Young, D.; Rosado, L.A.; Messens, J. Disulfide bond formation protects Arabidopsis thaliana glutathione transferase tau 23 from oxidative damage. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 775–789. [Google Scholar] [CrossRef]
- Horvath, E.; Bela, K.; Galle, A.; Riyazuddin, R.; Csomor, G.; Csenki, D.; Csiszar, J. Compensation of mutation in Arabidopsis glutathione transferase (AtGSTU) genes under control or salt stress conditions. Int. J. Mol. Sci. 2020, 21, 2349. [Google Scholar] [CrossRef] [Green Version]
- Dixon, D.P.; Edwards, R. Glutathione transferases. Arab. Book 2010, 8, e0131. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Wu, R.; Feng, J.; Feng, T.; Wang, C.; Hu, J.; Zhan, N.; Li, Y.; Ma, X.; Ren, B.; et al. Transnitrosylation mediated by the non-canonical catalase ROG1 regulates nitric oxide signaling in plants. Dev. Cell 2020, 53, 444–457.e445. [Google Scholar] [CrossRef]
- da Fonseca-Pereira, P.; Daloso, D.M.; Gago, J.; de Oliveira Silva, F.M.; Condori-Apfata, J.A.; Florez-Sarasa, I.; Tohge, T.; Reichheld, J.P.; Nunes-Nesi, A.; Fernie, A.R.; et al. The mitochondrial thioredoxin system contributes to the metabolic responses under drought episodes in Arabidopsis. Plant Cell Physiol. 2019, 60, 213–229. [Google Scholar] [CrossRef] [Green Version]
- Baune, M.C.; Lansing, H.; Fischer, K.; Meyer, T.; Charton, L.; Linka, N.; von Schaewen, A. The Arabidopsis plastidial glucose-6-phosphate transporter GPT1 is dually targeted to peroxisomes via the endoplasmic reticulum. Plant Cell 2020, 32, 1703–1726. [Google Scholar] [CrossRef] [Green Version]
- Bissoli, G.; Munoz-Bertomeu, J.; Bueso, E.; Sayas, E.; Vilcara, E.A.; Felipo, A.; Ninoles, R.; Rubio, L.; Fernandez, J.A.; Serrano, R. An Arabidopsis mutant over-expressing subtilase SBT4.13 uncovers the role of oxidative stress in the inhibition of growth by intracellular acidification. Int. J. Mol. Sci. 2020, 21, 1173. [Google Scholar] [CrossRef] [Green Version]
- Manara, A.; DalCorso, G.; Leister, D.; Jahns, P.; Baldan, B.; Furini, A. AtSIA1 AND AtOSA1: Two Abc1 proteins involved in oxidative stress responses and iron distribution within chloroplasts. New Phytol. 2014, 201, 452–465. [Google Scholar] [CrossRef]
- Bocher, T.W. Cytological and embryological studies in the amphi-apomictic Arabis holboellii complex. KongelDanske Vidensk-SelskabBiolSkr 1951, 6, 1–59. [Google Scholar]
- Contento, A.L.; Kim, S.J.; Bassham, D.C. Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol. 2004, 135, 2330–2347. [Google Scholar] [CrossRef] [Green Version]
- Griffin, P.T.; Niederhuth, C.E.; Schmitz, R.J. A comparative analysis of 5-azacytidine- and zebularine-induced DNA demethylation. G3 (Bethesda) 2016, 6, 2773–2780. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Fan, M.; Qin, Z.; Lv, H.; Wang, M.; Zhang, Z.; Zhou, W.; Zhao, N.; Li, X.; Han, C.; et al. Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat. Commun. 2018, 9, 1063. [Google Scholar] [CrossRef]
- Rasmussen, R.D.; Hole, D.; Hess, J.R.; Carman, J.G. Wheat kernel dormancy and +abscisic acid level following exposure to fluridone. J. Plant Physiol. 1997, 150, 440–445. [Google Scholar] [CrossRef]
- Bartels, P.G.; Watson, C.W. Inhibition of carotenoid synthesis by fluridone and norflurazon. Weed Sci. 1978, 26, 198–203. [Google Scholar] [CrossRef]
- Bowman, J.L.; Kohchi, T.; Yamato, K.T.; Jenkins, J.; Shu, S.; Ishizaki, K.; Yamaoka, S.; Nishihama, R.; Nakamura, Y.; Berger, F.; et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 2017, 171, 287–304e215. [Google Scholar] [CrossRef] [PubMed]
- Jamsheer, K.M.; Jindal, S.; Laxmi, A. Evolution of TOR-SnRK dynamics in green plants and its integration with phytohormone signaling networks. J. Exp. Bot. 2019, 70, 2239–2259. [Google Scholar] [CrossRef]
- Szymanska, K.P.; Polkowska-Kowalczyk, L.; Lichocka, M.; Maszkowska, J.; Dobrowolska, G. SNF1-related protein kinases SnRK2.4 and SnRK2.10 modulate ROS homeostasis in plant response to salt stress. Int. J. Mol. Sci. 2019, 20, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilroy, S.; Bialasek, M.; Suzuki, N.; Gorecka, M.; Devireddy, A.R.; Karpinski, S.; Mittler, R. ROS, calcium, and electric signals: Key mediators of rapid systemic signaling in plants. Plant Physiol. 2016, 171, 1606–1615. [Google Scholar] [CrossRef]
- Bheri, M.; Pandey, G.K. Protein phosphatases meet reactive oxygen species in plant signaling networks. Environ. Exp. Bot. 2019, 161, 26–40. [Google Scholar] [CrossRef]
- Mhamdi, A.; Van Breusegem, F. Reactive oxygen species in plant development. Development 2018, 145. [Google Scholar] [CrossRef] [Green Version]
- Uzilday, B.; Ozgur, R.; Sekmen, A.H.; Turkan, I. Endoplasmic reticulum stress regulates glutathione metabolism and activities of glutathione related enzymes in Arabidopsis. Funct. Plant Biol. 2018, 45, 284–296. [Google Scholar] [CrossRef]
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Nukarinen, E.; Nagele, T.; Pedrotti, L.; Wurzinger, B.; Mair, A.; Landgraf, R.; Bornke, F.; Hanson, J.; Teige, M.; Baena-Gonzalez, E.; et al. Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 2016, 6, 31697. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.C.; Liu, X.; Fu, L.; Hou, Y.J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell 2018, 69, 100–112e106. [Google Scholar] [CrossRef] [Green Version]
- Jamsheer, K.M.; Sharma, M.; Singh, D.; Mannully, C.T.; Jindal, S.; Shukla, B.N.; Laxmi, A. FCS-like zinc finger 6 and 10 repress SnRK1 signalling in Arabidopsis. Plant J. 2018, 94, 232–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soto-Burgos, J.; Bassham, D.C. SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PLoS ONE 2017, 12, e0182591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberger, C.L.; Chen, J. To grow or not to grow: TOR and SnRK2 coordinate growth and stress response in Arabidopsis. Mol. Cell 2018, 69, 3–4. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Yuan, M.; Wang, R.; Yang, Y.; Wang, C.; Oses-Prieto, J.A.; Kim, T.W.; Zhou, H.W.; Deng, Z.; Gampala, S.S.; et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 2011, 13, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhu, J.Y.; Roh, J.; Marchive, C.; Kim, S.K.; Meyer, C.; Sun, Y.; Wang, W.; Wang, Z.Y. TOR Signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Curr. Biol. 2016, 26, 1854–1860. [Google Scholar] [CrossRef]
- Caldana, C.; Martins, M.C.M.; Mubeen, U.; Urrea-Castellanos, R. The magic ’hammer’ of TOR: The multiple faces of a single pathway in the metabolic regulation of plant growth and development. J. Exp. Bot. 2019, 70, 2217–2225. [Google Scholar] [CrossRef]
- Rodriguez, M.; Parola, R.; Andreola, S.; Pereyra, C.; Martinez-Noel, G. TOR and SnRK1 signaling pathways in plant response to abiotic stresses: Do they always act according to the "yin-yang" model? Plant Sci. 2019, 288, 110220. [Google Scholar] [CrossRef]
- Pellino, M.; Hojsgaard, D.; Horandl, E.; Sharbel, T.F. Chasing the apomictic factors in the Ranunculus auricomus complex: Exploring gene expression patterns in microdissected sexual and apomictic ovules. Genes (Basel) 2020, 11, 728. [Google Scholar] [CrossRef]
- Ulum, F.B.; Costa Castro, C.; Horandl, E. Ploidy-dependent effects of light stress on the mode of reproduction in the Ranunculus auricomus complex (Ranunculaceae). Front. Plant Sci. 2020, 11, 104. [Google Scholar] [CrossRef] [Green Version]
- Shah, J.N.; Kirioukhova, O.; Pawar, P.; Tayyab, M.; Mateo, J.L.; Johnston, A.J. Depletion of key meiotic genes and transcriptome-wide abiotic stress reprogramming mark early preparatory events ahead of apomeiotic transition. Front. Plant Sci. 2016, 7, 1539. [Google Scholar] [CrossRef] [Green Version]
- Khanday, I.; Skinner, D.; Yang, B.; Mercier, R.; Sundaresan, V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 2019, 565, 91–95. [Google Scholar] [CrossRef]
- Muller, M.; Munne-Bosch, S. Ethylene response factors: A key regulatory hub in hormone and stress signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef] [Green Version]
- Oliver, C.; Santos, J.L.; Pradillo, M. Accurate chromosome segregation at first meiotic division requires AGO4, a protein involved in RNA-dependent DNA methylation in Arabidopsis thaliana. Genetics 2016, 204, 543–553. [Google Scholar] [CrossRef] [Green Version]
- Shashkova, S.; Welkenhuysen, N.; Hohmann, S. Molecular communication: Crosstalk between the Snf1 and other signaling pathways. FEMS Yeast Res. 2015, 15, fov026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honigberg, S.M.; Lee, R.H. Snf1 kinase connects nutritional pathways controlling meiosis in Saccharomyces cerevisiae. Mol. Cell Biol. 1998, 18, 4548–4555. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.S.; Chao, Y.C.; Tseng, T.W.; Huang, C.K.; Lo, P.C.; Lu, C.A. Two MYB-related transcription factors play opposite roles in sugar signaling in Arabidopsis. Plant Mol. Biol. 2017, 93, 299–311. [Google Scholar] [CrossRef]
- Haufler, C.H.; Pryer, K.M.; Schuettpelz, E.; Sessa, E.B.; Farrar, D.R.; Moran, R.; Schneller, J.J.; Watkins, J.E.; Windham, M.D. Sex and the single gametophyte: Revising the homosporous vascular plant life cycle in light of contemporary research. BioScience 2016, 66, 928–937. [Google Scholar] [CrossRef] [Green Version]
- Herr, J.M.J. The origin of the ovule. Am. J. Bot. 1995, 82, 547–564. [Google Scholar] [CrossRef]
- Stebbins, G.L. Apomixis in the angiosperms. Bot. Rev. 1941, 7, 507–542. [Google Scholar] [CrossRef]
- Ernst, A. Bastardierung als Ursache der Apogamie im Pflanzenreich; Fischer: Jena, Germany, 1918. [Google Scholar]
- Krisko, A.; Leroy, M.; Radman, M.; Meselson, M. Extreme anti-oxidant protection against ionizing radiation in bdelloid rotifers. Proc. Natl. Acad. Sci. USA 2012, 109, 2354–2357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flot, J.F.; Hespeels, B.; Li, X.; Noel, B.; Arkhipova, I.; Danchin, E.G.; Hejnol, A.; Henrissat, B.; Koszul, R.; Aury, J.M.; et al. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 2013, 500, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Pattiwael, M.R.; Mangindaan, R.E.P.; Prabowo, R.; Rumengan, I.F.M. Acute and chronic effect of cyaniade on survival rate, behavior, and reproduction of Daphnia sp. Aquat. Sci. Manag. 2013, 1, 52. [Google Scholar] [CrossRef]
- Jedlicka, P.; Jedlickova, V.; Lee, H.J. Expression of stress-related genes in the parthenogenetic forms of the pea aphid, Acyrthosiphon pisum. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2015, 180, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Sabbe, K.; Chepurnov, V.A.; Vyverman, W.; Mann, D.G. Apomixis in Achnanthes (Bacillariophyceae); development of a model system for diatom reproductive biology. Eur. J. Phycol. 2004, 39, 327–341. [Google Scholar] [CrossRef] [Green Version]
- Nedelcu, A.M. Sex as a response to oxidative stress: Stress genes co-opted for sex. P Roy. Soc. B-Biol. Sci. 2005, 272, 1935–1940. [Google Scholar] [CrossRef] [Green Version]
- Nedelcu, A.M.; Marcu, O.; Michod, R.E. Sex as a response to oxidative stress: A twofold increase in cellular reactive oxygen species activates sex genes. P Roy. Soc. B-Biol. Sci. 2004, 271, 1591–1596. [Google Scholar] [CrossRef] [Green Version]
- Nedelcu, A.M.; Michod, R.E. Sex as a response to oxidative stress: The effect of antioxidants on sexual induction in a facultatively sexual lineage. P Roy. Soc. B-Biol. Sci. 2003, 270, S136–S139. [Google Scholar] [CrossRef] [Green Version]
- Fiore-Donno, A.M.; Novozhilov, Y.K.; Meyer, M.; Schnittler, M. Genetic structure of two protist species (Myxogastria, Amoebozoa) suggests asexual reproduction in sexual Amoebae. PLoS ONE 2011, 6, e22872. [Google Scholar] [CrossRef] [Green Version]
- Collins, O.R. Myxomycete biosystematics: Some recent developments and future research opportunities. Bot. Rev. 1979, 45, 145–201. [Google Scholar] [CrossRef]
- Collins, O.N.R.; Gong, T. Genetical relatedness of a former apomict and a heterothallic isolate in Didymium Iridis (Myxomycetes). Mycologia 1985, 77, 300–307. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mateo de Arias, M.; Gao, L.; Sherwood, D.A.; Dwivedi, K.K.; Price, B.J.; Jamison, M.; Kowallis, B.M.; Carman, J.G. Whether Gametophytes Are Reduced or Unreduced in Angiosperms Might Be Determined Metabolically. Genes 2020, 11, 1449. https://doi.org/10.3390/genes11121449
Mateo de Arias M, Gao L, Sherwood DA, Dwivedi KK, Price BJ, Jamison M, Kowallis BM, Carman JG. Whether Gametophytes Are Reduced or Unreduced in Angiosperms Might Be Determined Metabolically. Genes. 2020; 11(12):1449. https://doi.org/10.3390/genes11121449
Chicago/Turabian StyleMateo de Arias, Mayelyn, Lei Gao, David A. Sherwood, Krishna K. Dwivedi, Bo J. Price, Michelle Jamison, Becky M. Kowallis, and John G. Carman. 2020. "Whether Gametophytes Are Reduced or Unreduced in Angiosperms Might Be Determined Metabolically" Genes 11, no. 12: 1449. https://doi.org/10.3390/genes11121449