Genome Mining and Evaluation of the Biocontrol Potential of Pseudomonas fluorescens BRZ63, a New Endophyte of Oilseed Rape (Brassica napus L.) against Fungal Pathogens
Abstract
:1. Introduction
2. Results
2.1. Antagonistic Activity of P. fluorescens BRZ63 against Fungal Plant Pathogens
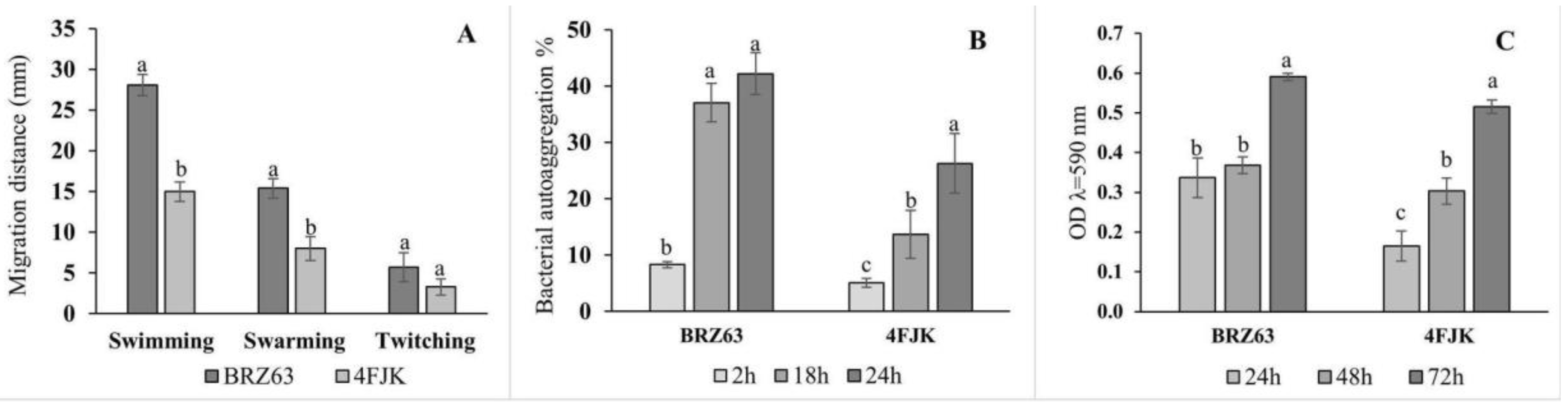
2.2. In Vitro Assessment of Colonization Properties of the BRZ63 Strain
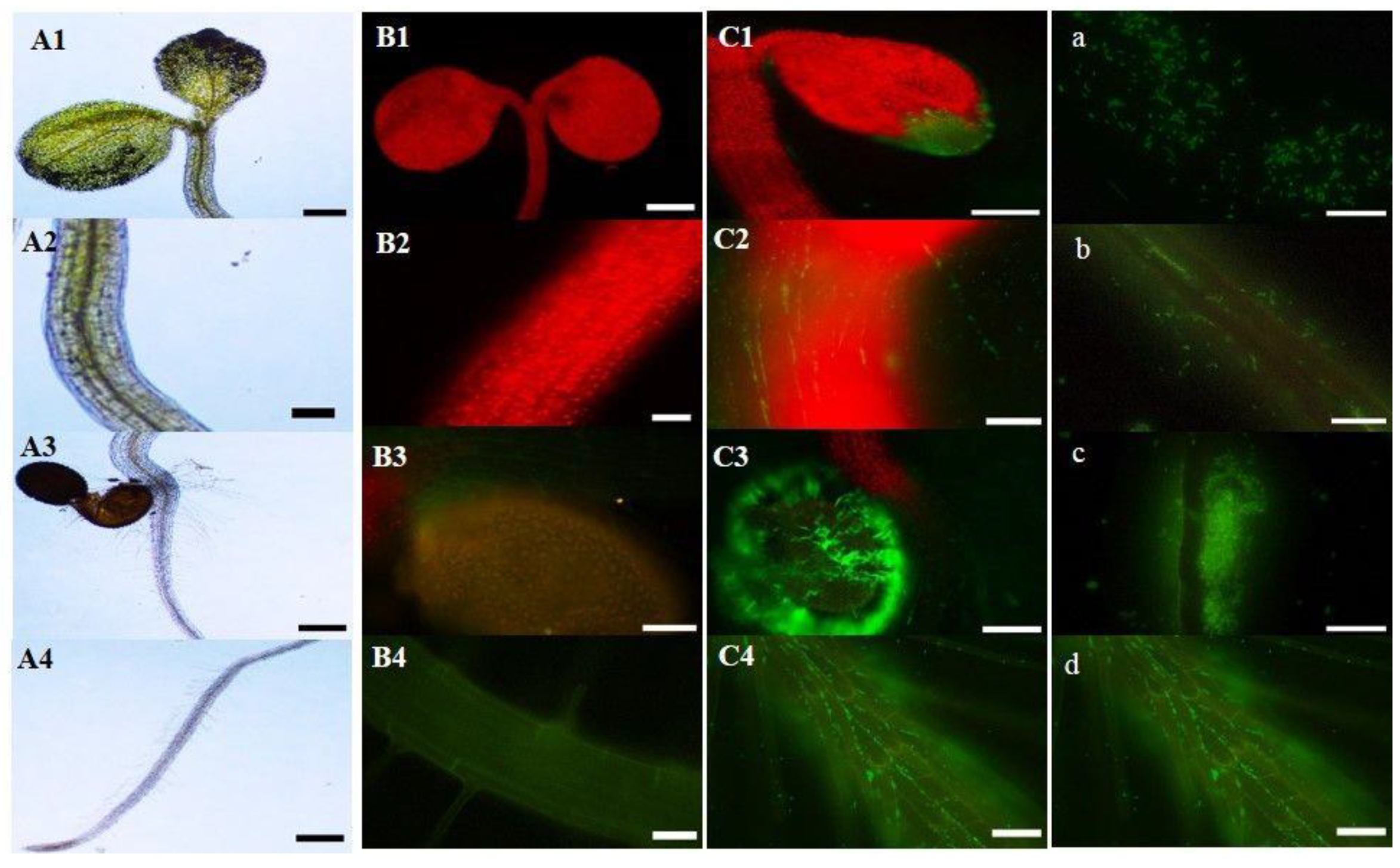
2.3. Inoculation of Arabidopsis Thaliana Col-0 with the EGFP-Labeled Strain
2.4. Effects of the BRZ63 Strain on Plant Development and Disease Protection of Oilseed Rape
2.5. In Vitro Screening for PGP and Biocontrol Traits of the BRZ63 Strain
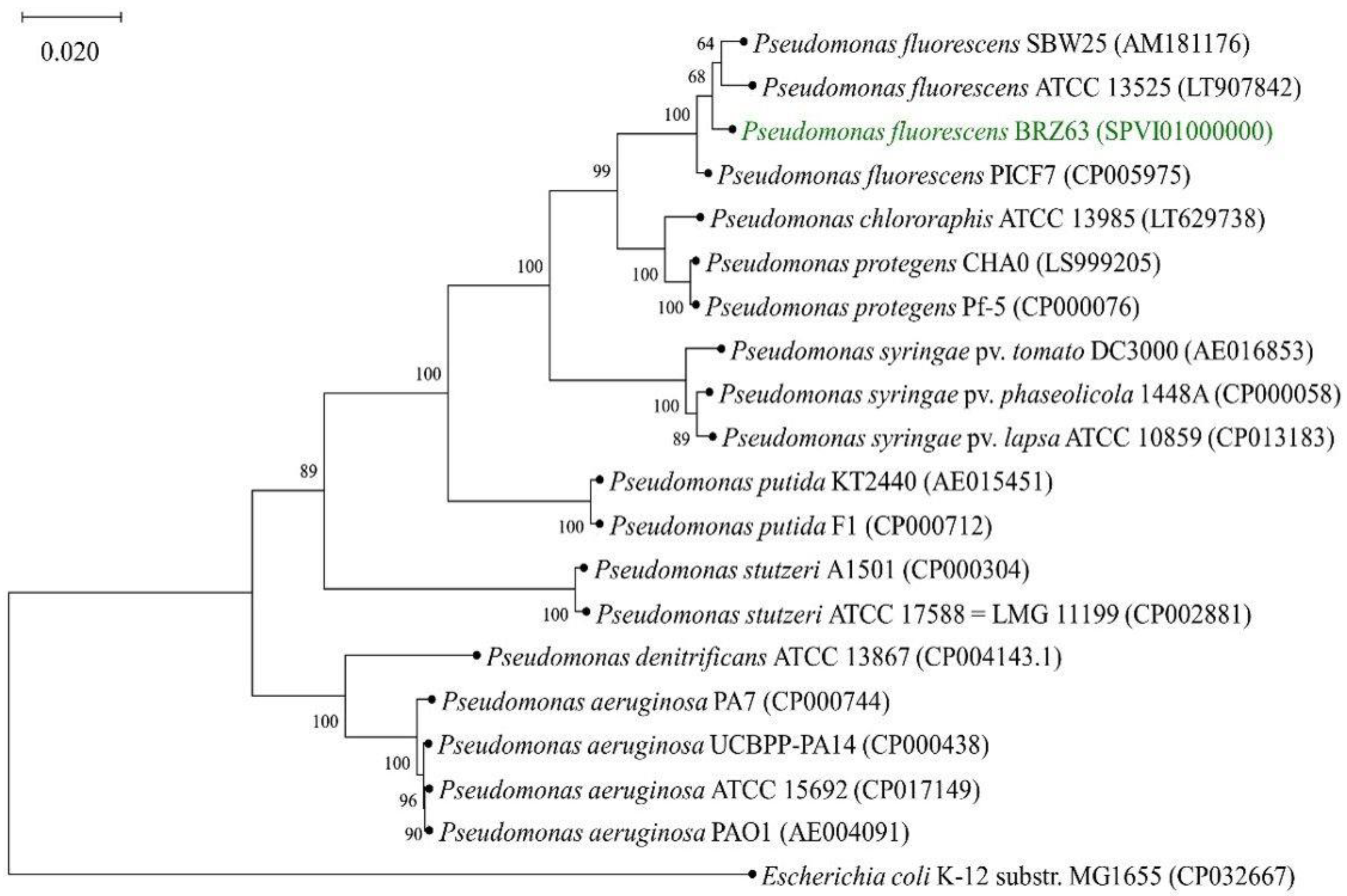
2.6. General Features of P. fluorescens BRZ63 Genome and Phylogenetic Analysis
2.7. The Colonization, Biocontrol, and PGP Features in the BRZ63 Genome
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Isolation and Molecular Identification of the BRZ63 Strain
4.3. Pathogenic Fungi
4.4. Antifungal Activity Assays
4.5. In Vitro Assessment of Colonization, Plant Growth Promotion (PGP), and Biocontrol Traits of the BRZ63 Strain
4.6. Autoaggregation Assay
4.7. Plant Colonization Experiments
4.7.1. Fluorescent Labeling of the BRZ63 Strain
4.7.2. Inoculation of Arabidopsis thaliana Col-0 with the EGFP-Labeled Strain
4.8. Effects of the BRZ63 Strain Inoculation on Oilseed Rape Development and Disease Protection
4.9. Genome Sequencing and Sequence Analysis
4.10. Phylogenetic Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| IAA | Indole-3-acetic acid |
| ACC | 1-aminocyclopropane-1-carboxylate |
| HSD | Honest significant difference |
| CRA | Congo Red Agar |
| EPS | Exopolysaccharide |
| CV | Crystal Violet |
| EGFP | Enhanced Green Fluorescent Protein |
| SA | Salicylic acid |
| ISR | Induced Systemic Resistance |
| DF | Dworkin and Foster |
| PSI | Phosphate Solubilizing Index |
| CAS | Chrome azurol S |
| CTAB | Cetyltrimethylammonium bromide |
| CDS | Protein-encoding sequences |
| COG | Cluster of Orthologous Genes |
| PGP | Plant Growth Promotion |
| LPS | Lipopolysaccharide |
| GH | Glycoside Hydrolases |
| CE | Carbohydrate Esterases |
| TBDRs | TonB-dependent receptors |
| GDH | Glutamate dehydrogenase |
| PQQ | Pyrroloquinoline quinon |
| LB | Luria-Bertani |
| PDA | Potato Dextrose Agar |
| HCN | Hydrogen cyanide |
| CWDEs | Cell Wall Degrading Enzymes |
References
- Abdallah, A.B.R.; Jabnoun-Khiareddine, H.; Nefzi, A.; Mokni-Tlili, S.; Daami-Remadi, M. Endophytic bacteria from Datura stramonium for Fusarium wilt suppression and tomato growth promotion. J. Microb. Biochem. Technol. 2016, 8, 30–41. [Google Scholar] [CrossRef]
- Majid, M.U.; Awan, M.F.; Fatima, K.; Tahir, M.S.; Ali, Q.; Rasid, B.; Rao, A.Q.; Nasit, I.A.; Husnain, T. Phytophthora capsici on chili pepper (Capsicum annuum L.) and its management through genetic and bio-control: A review. Zemdirbyste 2016, 103, 419–430. [Google Scholar] [CrossRef] [Green Version]
- Ajayi-Oyetunde, O.O.; Bradley, C.A. Rhizoctonia solani: Taxonomy, population biology and management of rhizoctonia seedling disease of soybean. Plant. Pathol. 2018, 67, 3–17. [Google Scholar] [CrossRef]
- del Rıo, L.E.; Bradley, C.A.; Henson, R.A.; Endres, G.J.; Hanson, B.K.; Mckay, K.; Halvorson, M.; Porter, P.M.; Le Gare, D.G.; Lamey, H.A. Impact of Sclerotinia stem rot on yield of canola. Plant Dis. 2007, 91, 191–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lally, R.D.; Galbally, P.; Moreira, A.S.; Spink, J.; Ryan, D.; Germaine, K.J.; Dowling, D.N. Application of endophytic Pseudomonas fluorescens and a bacterial consortium to Brassica napus can increase plant height and biomass under greenhouse and field conditions. Front. Plant. Sci. 2017, 8, 2193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aktar, M.W.; Sengupta, D.; Chowdhury, A. Impact of pesticides use in agriculture: Their benefits and hazards. Interdiscip. Toxicol. 2009, 2, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bardin, M.; Ajouz, S.; Comby, M.; Lopez-Ferber, M.; Graillot, B.; Siegwart, M.; Nicot, P.C. Is the efficacy of biological control against plant diseases likely to be more durable than that of chemical pesticides? Front. Plant. Sci. 2015, 6, 566. [Google Scholar] [CrossRef]
- Eljounaidi, K.; Kyu, S.; Bae, H. Bacterial endophytes as potential biocontrol agents of vascular wilt diseases—Review and future prospects. Biol. Control 2016, 103, 62–68. [Google Scholar] [CrossRef]
- Kandel, S.L.; Firrincieli, A.; Joubert, P.M.; Okubara, P.; Natalie, D.; McGeorge, K.M.; Mugnozza, G.S.; Harfouche, A.; Kim, S.H.; Doty, S.L. An in vitro study of bio-control and plant growth promotion potential of salicaceae endophytes. Front. Microbiol. 2017, 8, 386. [Google Scholar] [CrossRef] [Green Version]
- Yadav, A.; Yadav, K. Exploring the potential of endophytes in agriculture: A minireview. Adv. Plants Agric. Res. 2017, 6, 102–106. [Google Scholar] [CrossRef] [Green Version]
- Bakker, P.A.; Berendsen, R.L.; Doornbos, R.F.; Wintermans, P.C.; Pieterse, C.M. The rhizosphere revisited: Root microbiomics. Front. Plant. Sci. 2013, 4, 165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.M.J.; Schenke, P.M. Inner Plant Values: Diversity, Colonization and Benefits from Endophytic Bacteria. Front. Microbiol. 2017, 8, 2552. [Google Scholar] [CrossRef]
- Verma, S.K.; Kingsley, K.L.; Bergen, M.S.; Kowalski, K.P.; White, J.F. Fungal disease prevention in seedlings of rice (Oryza sativa) and other grasses by growth-promoting seed-associated endophytic bacteria from invasive Phragmites australis. Microorganisms 2018, 6, 21. [Google Scholar] [CrossRef] [Green Version]
- Benhamou, N.; Gagné, S.; Quéré, D.L.; Dehbi, L. Bacterial mediated induced resistance in cucumber: Beneficial effect of the endophytic bacterium Serratia plymuthica on the protection against infection by Pythium ultimum. Biochem. Cell Biol. 2000, 90, 45–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinski, A.; Betekhtin, A.; Hupert-Kocurek, K.; Mur, L.A.J.; Hasterok, R. Defining the Genetic Basis of Plant–Endophytic Bacteria Interactions. Int. J. Mol. Sci. 2019, 20, 1947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Jia, J.; Atkinson, S.; Cámara, M.; Gao, K.; Li, H.; Cao, J. Biocontrol potential of an endophytic Serratia sp. G3 and its mode of action. World J. Microbiol. Biotechnol. 2010, 26, 1465–1471. [Google Scholar] [CrossRef]
- Vinayarani, G.; Prakash, H.S. Growth promoting rhizospheric and endophytic bacteria from Curcuma longa L. as biocontrol agents against rhizome rot and leaf blight diseases. Plant. Pathol. J. 2018, 3, 218–235. [Google Scholar]
- Martínez-García, P.M.; Ruano-Rosa, D.; Schilirò, E.; Prieto, P.; Ramos, C.; Rodríguez-Palenzuela, P. Complete genome sequence of Pseudomonas fluorescens strain PICF7, an indigenous root endophyte from olive (Olea europaea L.) and effective biocontrol agent against Verticillium dahliae. Stand. Genomic Sci. 2015, 10, 10. [Google Scholar] [CrossRef] [Green Version]
- Preston, G.M. Plant perceptions of plant growth-promoting Pseudomonas. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2004, 359, 907–918. [Google Scholar] [CrossRef] [Green Version]
- Chlebek, D.; Hupert-Kocurek, K. Endophytic bacteria in the phytodegradation of persistent organic pollutions. Post. Microbiol. 2019, 1, 70–79. [Google Scholar]
- Bajpai, A.; Johri, B.N. Endophytic Pseudomonads and Their Metabolites. In Endophytes and Secondary Metabolites; Jha, S., Ed.; Springer: Cham, Switzerland, 2019; pp. 33–59. [Google Scholar]
- Stepanovic, S.; Vukovic, D.; Hola, V.; Di Bonaventura, G.; Djukic, S.; Cirkovic, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Bloemberg, G.V.; Wijfjes, A.H.M.; Lamers, G.E.M.; Stuurman, N.; Lugtenberg, B.J.J. Simultaneous imaging of Pseudomonas fluorescens WCS365 populations expressing three different autofluorescent proteins in the rhizosphere: New perspectives for studying microbial communities. Mol. Plant. Microbe Interact. 2000, 13, 1170–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimbrel, J.A.; Givan, S.A.; Halgren, A.B.; Creason, A.L.; Mills, D.I.; Banowetz, G.M.; Armstrong, D.J.; Chang, J.H. An improved, high-quality draft genome sequence of the Germination-Arrest Factor-producing Pseudomonas fluorescens WH6. BMC Genom. 2010, 11, 522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelkner, J.; Tejerizo, G.T.; Hassa, J.; Lin, T.W.; Witte, J.; Verwaaijen, B.; Winkler, A.; Bunk, B.; Spröer, C.; Overmann, J.; et al. Genetic potential of the biocontrol agent Pseudomonas brassicacearum (formerly P. trivialis) 3Re2-7 unraveled by genome sequencing and mining, comparative genomics and transcriptomics. Genes 2019, 10, 601. [Google Scholar] [CrossRef] [Green Version]
- Sheibani-Tezerji, R.; Rattei, T.; Sessitsch, A.; Trognitz, F.; Mitter, B. Transcriptome profiling of the endophyte Burkholderia phytofirmans PsJN indicates sensing of the plant environment and drought stress. mBio 2016, 6, e00621-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Li, H.B.; Bai, Y.; Wang, J.; Nie, M.; Li, B.; Xiao, M. The use of Pseudomonas fluorescens P13 to control sclerotinia stem rot (Sclerotinia sclerotiorum) of oilseed rape. J. Microbiol. 2011, 49, 884–889. [Google Scholar] [CrossRef] [PubMed]
- Hammoudi, O.; Salman, M.; Abuamsha, R.; Ehlers, R. Effectiveness of bacterial and fungal isolates to control Phoma lingam on oilseed rape Brassica napus. Am. J. Plant. Sci. 2012, 3, 773–779. [Google Scholar] [CrossRef] [Green Version]
- Redondo-Nieto, M.; Barret, M.; Morrissey, J.; Germaine, K.; Martínez-Granero, F.; Barahona, E.; Dowling, D. Genome sequence reveals that Pseudomonas fluorescens F113 possesses a large and diverse array of systems for rhizosphere function and host interaction. BMC Genom. 2013, 14, 54. [Google Scholar] [CrossRef] [Green Version]
- Nesemann, K.; Braus-Stromeyer, S.A.; Harting, R.; Höfer, A.; Kusch, H.; Ambrosio, A.B.; Braus, G.H. Fluorescent pseudomonads pursue media-dependent strategies to inhibit growth of pathogenic Verticillium fungi. Appl. Microbiol. Biotechnol. 2018, 102, 817–831. [Google Scholar] [CrossRef]
- Jiménez, J.A.; Novinscak, A.; Filion, M. Pseudomonas fluorescens LBUM677 differentially increases plant biomass, total oil content and lipid composition in three oilseed crops. J. Appl. Microbiol. 2020, 128, 1119–1127. [Google Scholar] [CrossRef]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial endophyte colonization and distribution within plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandhya, V.; Shrivastava, M.; Ali, S.Z.; Prasad, V.S.S.K. Endophytes from maize with plant growth promotion and biocontrol activity under drought stress. Russ. Agric. Sci. 2017, 43, 22–34. [Google Scholar] [CrossRef]
- Li, H.; Parmar, S.; Sharma, V.K.; White, J.F. Seed endophytes and their potential applications. In Seed Endophytes; Verma, S.K., White, J.F., Jr., Eds.; Springer: Cham, Switzerland, 2019; pp. 35–54. [Google Scholar]
- Sturrock, C.J.; Woodhall, J.; Brown, M.; Walker, C.; Mooney, S.J.; Ray, R.V. Effects of damping-off caused by Rhizoctonia solani anastomosis group 2-1 on roots of wheat and oil seed rape quantified using X-ray Computed Tomography and real-time PCR. Front. Plant. Sci. 2015, 6, 461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, D.; Kalita, P. Reducing Postharvest Losses during Storage of Grain Crops to Strengthen Food Security in Developing Countries. Foods 2017, 6, 8. [Google Scholar] [CrossRef] [Green Version]
- Drizou, F.; Graham, N.S.; Bruce, T.J.A.; Ray, R.V. Development of high-throughput methods to screen disease caused by Rhizoctonia solani AG 2-1 in oilseed rape. Plant. Methods 2017, 13, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Zhou, Q.; Strelkov, S.E.; Hwang, S.F. Genetic Diversity and Aggressiveness of Fusarium spp. Isolated from Canola in Alberta, Canada. Plant. Dis. 2015, 98, 727–738. [Google Scholar] [CrossRef] [Green Version]
- Machowicz-Stefaniak, Z. Occurence and chracterization of Colletotrichum dematium (Fr.) Grove. Pol. J. Microbiol. 2010, 59, 191–200. [Google Scholar] [CrossRef]
- Loper, J.E.; Kobayashi, D.Y.; Paulsen, I.T. The genomic sequence of Pseudomonas fluorescens Pf-5: Insights into biological control. Phytopathology 2007, 97, 233–238. [Google Scholar] [CrossRef] [Green Version]
- Trippe, K.; McPhail, K.; Armstrong, D.; Azevedo, M.; Banowetz, G. Pseudomonas fluorescens SBW25 produces furanomycin, a non-proteinogenic amino acid with selective antimicrobial properties. BMC Microbiol. 2013, 13, 111. [Google Scholar] [CrossRef] [Green Version]
- Hardoim, P.R.; van Overbeek, L.S.; Elsas, J.D. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008, 16, 463–471. [Google Scholar] [CrossRef]
- Pankievicz, V.C.; Camilios-Neto, D.; Bonato, P.; Balsanelli, E.; Tadra-Sfeir, M.Z.; Faoro, H.; Monteiro, R.A. RNA-seq transcriptional profiling of Herbaspirillum seropedicae colonizing wheat (Triticum aestivum) roots. Plant. Mol. Biol. 2016, 90, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Monchy, S.; Taghavi, S.; Zhu, W.; Ramos, J.; van der Lelie, D. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbiol. Rev. 2011, 35, 299–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weert, S.; Vermeiren, H.; Mulders, I.H.; Kuiper, I.; Hendrickx, N.; Bloemberg, G.V.; Vanderleyden, J.; de Mot, R.; Lugtenberg, B.J. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol. Plant. Microbe. Interact. 2002, 15, 1173–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seneviratne, G.; Weerasekara, M.L.M.A.W.; Seneviratne, K.A.C.N.; Zavahir, J.S.; Kecskes, M.L.; Kennedy, I.R. Importance of Biofilm Formation in Plant Growth Promoting Rhizobacterial Action. Plant Growth and Health Promoting Bacteria. Microbiology Monographs; Springer: Berlin/Heidelberg, Germany, 2011; Volume 18, pp. 81–95. [Google Scholar]
- Tovi, N.; Frenk, S.; Hadar, Y.; Minz, D. Host Specificity and Spatial Distribution Preference of Three Pseudomonas Isolates. Front. Microbiol. 2019, 9, 3263. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Salmerón, J.E.; Moreno-Hagelsieb, G.; Santoyo, G. Genome comparison of Pseudomonas fluorescens UM270 with related fluorescent strains unveils genes involved in rhizosphere competence and colonization. J. Genom. 2017, 5, 91–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meneses, C.H.S.G.; Rouws, L.F.M.; Simoes-Araujo, J.L.; Vidal, M.S.; Baldani, J.I. Exopolysaccharide production is required for biofilm formation and plant colonization by the nitrogen-fixing endophyte Gluconacetobacter diazotrophicus. Mol. Plant. Microbe Interact. 2011, 24, 1448–1458. [Google Scholar] [CrossRef] [Green Version]
- Costa, O.Y.A.; Raaijmakers, J.M.; Kuramae, E.E. Microbial Extracellular Polymeric Substances: Ecological Function and Impact on Soil Aggregation. Front. Microbiol. 2018, 9, 1636. [Google Scholar] [CrossRef] [Green Version]
- Chang, W.S.; van de Mortel, M.; Nielsen, L.; de Guzman, G.N.; Li, X.; Halverson, L.J. Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J. Bacteriol. 2007, 189, 8290–8299. [Google Scholar] [CrossRef] [Green Version]
- Maleki, S.; Almaas, E.; Zotchev, S.; Valla, S.; Ertesvåg, H. Alginate biosynthesis factories in Pseudomonas fluorescens: Localization and correlation with alginate production level. Appl. Environ. Microbiol. 2016, 82, 41227–41236. [Google Scholar] [CrossRef] [Green Version]
- Bogino, P.C.; de las Mercedes, O.M.; Sorroche, F.G.; Giordano, W. The role of bacterial biofilms and surface components in plant-bacterial associations. Int. J. Mol. Sci. 2013, 14, 15838–15859. [Google Scholar] [CrossRef] [Green Version]
- Vanitha, S.; Ramjegathesh, R. Bio Control Potential of Pseudomonas fluorescens against Coleus Root Rot Disease. J. Plant. Pathol. Microb. 2014, 5, 216. [Google Scholar]
- Zhao, L.F.; Xua, Y.J.; Lai, X.H. Antagonistic endophytic bacteria associated with nodules of soybean (Glycine max L.) and plant growth-promoting properties. Braz. J. Microbiol. 2018, 49, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Nagarajkumar, M.; Bhaskaran, R.; Velazhahan, R. Involvement of secondary metabolites and extracellular lytic enzymes produced by Pseudomonas fluorescens in inhibition of Rhizoctonia solani, the rice sheath blight pathogen. Microbiol. Res. 2004, 159, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Solanki, M.K.; Robert, A.S.; Singh, R.K.; Kumar, S.; Pandey, A.K.; Srivastava, A.K.; Arora, D.K. Characterization of mycolytic enzymes of Bacillus strains and their bio-protection role against Rhizoctonia solani in tomato. Curr. Microbiol. 2012, 65, 330–336. [Google Scholar] [CrossRef]
- Ahmad, Z.; Wu, J.; Chen, L.; Dong, W. Isolated Bacillus subtilis strain 330-2 and its antagonistic genes identified by the removing PCR. Sci. Rep. 2017, 7, 1777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goswami, D.; Borah, S.N.; Lahkar, J.; Handique, P.J.; Deka, S. Antifungal properties of rhamnolipid produced by Pseudomonas aeruginosa DS9 against Colletotrichum falcatum. J. Basic Microbiol. 2015, 55, 1265–1274. [Google Scholar] [CrossRef]
- Das, P.; Yang, X.P.; Ma, L.Z. Analysis of biosurfactants from industrially viable Pseudomonas strain isolated from crude oil suggests how rhamnolipids congeners affect emulsification property and antimicrobial activity. Front. Microbiol. 2014, 5, 696. [Google Scholar] [CrossRef]
- Abalos, A.; Pinazo, A.; Infante, M.R.; Casals, M.; Garcia, F.; Manresa, A. Physicochemical and antimicrobial properties of new rhamnolipids produced by Pseudomonas aeruginosa AT10 from soybean oil refinery wastes. Langmuir 2001, 17, 1367–1371. [Google Scholar] [CrossRef]
- Djavaheri, M.; Mercado-Blanco, J.; Versluis, C.; Meyer, J.M.; Van Loon, L.C.; Bakker, P.A.H.M. Iron-regulated metabolites produced by Pseudomonas fluorescens WCS374r are not required for eliciting induced systemic resistance against Pseudomonas syringae pv. tomato in Arabidopsis. Microbiol. Open 2012, 1, 311–325. [Google Scholar] [CrossRef] [Green Version]
- Mercado-Blanco, J.; Rodríguez-Jurado, D.; Hervás, A.; Jiménez-Díaz, R.M. Suppression of Verticillium wilt in olive planting stocks by root-associated fluorescent Pseudomonas spp. Biol. Control 2004, 30, 474–486. [Google Scholar] [CrossRef]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Etesami, H.; Alikhani, H.A.; Hosseini, H.M. Indole-3-acetic acid (IAA) production trait, a useful screening to select endophytic and rhizosphere competent bacteria for rice growth promoting agents. MethodsX 2015, 2, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Leon, R.; Rojas-Solis, D.; Contreras-Perez, M.; Orozco-Mosqueda, M.D.; Macias-Rodriguez, L.I.; Reyes-de la Cruz, H.; Valencia-Cantero, E.; Santoyo, G. Characterization of the antifungal and plant growth-promoting effects of diffusible and volatile organic compounds produced by Pseudomonas fluorescens strains. Biol. Control 2015, 81, 83–92. [Google Scholar] [CrossRef]
- Kaur, C.; Selvakumar, G.; Ganeshamurthy, A.N. Draft genome sequence of phosphate-solubilizing bacterium Paraburkholderia tropica strain P-31 isolated from pomegranate (Punica granatum) rhizosphere. Genome Announc. 2016, 4, e00844-16. [Google Scholar] [CrossRef] [Green Version]
- Otieno, N.; Lally, R.D.; Kiwanuka, S.; Lloyd, A.; Ryan, D.; Germaine, K.J.; Dowling, D.N. Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 2015, 6, 745. [Google Scholar] [CrossRef] [Green Version]
- Pawlik, M.; Piotrowska-Seget, Z. Endophytic bacteria associated with Hieracium piloselloides: Their potential for hydrocarbon-utilizing and plant growth-promotion. J. Toxicol. Environ. Health Part A 2015, 78, 860–870. [Google Scholar] [CrossRef]
- Pacwa-Płociniczak, M.; Płaza, G.A.; Poliwoda, A.; Piotrowska-Seget, Z. Characterization of hydrocarbon-degrading and biosurfactant-producing Pseudomonas sp. P-1 strain as a potential tool for bioremediation of petroleum contaminated soil. Environ. Sci. Pollut. Res. 2014, 21, 9385–9395. [Google Scholar] [CrossRef] [Green Version]
- Siala, R.; Chobba, I.B.; Vallaeys, T.; Triki, M.A.; Jrad, M.; Cheffi, M.; Ayedi, I.; Elleuch, A.; Nemsi, A.; Cerqueira, F.; et al. Analysis of the Cultivable Endophytic Bacterial Diversity in the Date Palm (Phoenix dactylifera L.) and Evaluation of Its Antagonistic Potential against Pathogenic Fusarium Species that Cause Date Palm Bayound Disease. J. Appl. Environ. Microbiol. 2016, 4, 93–104. [Google Scholar]
- Vijayalakshmi, R.; Kairunnisa, K.; Sivvaswamy, S.N.; Dharan, S.; Nataranjan, S. Enzyme production and antimicrobial activity of endophytic bacteria isolated from medicinal plants. Indian J. Sci. Technol. 2016, 9, 1–8. [Google Scholar] [CrossRef]
- Kuddus, S.M.; Ahmad, R.I.Z. Isolation of novel chitinolytic bacteria and production optimization of extracellular chitinase. J. Genet. Eng. Biotechnol. 2013, 11, 39–46. [Google Scholar]
- Naveed, M.; Mitter, B.; Yousaf, S.; Pastar, M.; Afzal, M.; Sessitsch, A. The endophyte Enterobacter sp. FD17: A maize growth enhancer selected based on rigorous testing of plant beneficial traits and colonization characteristics. Biol. Fertil. Soils 2014, 50, 249–262. [Google Scholar] [CrossRef]
- Freeman, D.J.; Falkiner, F.R.; Keane, C.T. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 1989, 42, 872–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szilagyi-Zecchin, V.J.; Ikeda, A.C.; Hungria, M.; Adamoski, D.; Kava-Cordeiro, V.; Glienke, C.; Galli-Terasawa, L.V. Identification and characterization of endophytic bacteria from corn (Zea mays L.) roots with biotechnological potential in agriculture. AMB Express 2014, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Cappuccino, J.G.; Sherman, N. Biochemical activities of microorganisms. Microbiology, A Laboratory Manual; The Benjamin/Cummings Publishing Co.: San Francisco, CA, USA, 1992; pp. 188–247. [Google Scholar]
- Pikovskaya, R.I. Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Microbiology 1948, 17, 362–370. [Google Scholar]
- Pande, A.; Pandey, P.; Mehra, S.; Singh, M.; Kaushik, S. Phenotypic and genotypic characterization of phosphate solubilizing bacteria and their efficiency on the growth of maize. J. Genet. Eng. Biotechnol. 2017, 15, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Johnston-Monje, D.; Raizada, M.N. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 2013, 6, e20396. [Google Scholar] [CrossRef] [Green Version]
- Syamala, M.; Sivaji, M. Functional characterization of various plant growth promoting activity of Pseudomonas fluorescens and Bacillus subtilis from Aloe vera rhizosphere. J. Pharmacogn. Phytochem. 2017, 6, 120–122. [Google Scholar]
- Ahmad, F.; Ahmad, I.; Khan, M.S. Screening of free-living rhizospheric bacteria for their multiple growth promoting activities. Microbiol. Res. 2008, 163, 173–181. [Google Scholar] [CrossRef]
- Shoeb, E.; Ahmed, N.; Akhter, J.; Badar, U.; Siddiqui, K.; Ansari, F.; Baig, R. Screening and characterization of biosurfactant-producing bacteria isolated from the Arabian Sea coast of Karachi. Turkish J. Biol. 2015, 39, 210–216. [Google Scholar] [CrossRef]
- Nigris, S.; Baldan, E.; Tondello, A.; Zanella, F.; Vitulo, N.; Favaro, G.; Marcato, S. Biocontrol traits of Bacillus licheniformis GL174, a culturable endophyte of Vitis vinifera cv. Glera. BMC Microbiol. 2018, 18, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prieto, P.; Mercado-Blanco, J. Endophytic colonisation of olive roots by the biocontrol strain Pseudomonas fluorescens PICF7. FEMS Microbiol. Ecol. 2008, 64, 297–306. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acid Res. 2019, 47, 309–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.; Zhu, Z.; Fu, L.; Niu, B.; Li, W. WebMGA: A customizable web server for fast metagenomic sequence analysis. BMC Genom. 2011, 12, 444. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Mao, X.; Yang, J.; Chen, X.; Mao, F.; Xu, Y. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012, 40, W445–W451. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [Green Version]
- Avram, O.; Rapoport, D.; Portugez, S.; Pupko, T. M1CR0B1AL1Z3R-a user-friendly web server for the analysis of large-scale microbial genomics data. Nucleic Acids Res. 2019, 47, W88–W92. [Google Scholar] [CrossRef] [Green Version]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]



| Features | Strain BRZ63 | Strain 4FJK |
|---|---|---|
| Exopolysaccharides production | + | − |
| Siderophore production | + | + |
| Endoglucanase production Chitinase production Protease production | + | − |
| + | − | |
| − | − | |
| Acetoin and 2,3-butanediol production | − | + |
| IAA production (μg/mL) | 59.62 ± 1.11 a | 34.53 ± 0.97 b |
| SA production (μg/mL) | 17.83 ± 0.95 a | 55.22 ± 1.36 b |
| ACC deaminase production | + | + |
| Ammonia production | + | + |
| HCN production | − | + |
| Catalase production | + | − |
| Oxidase production | + | − |
| Bacterial Strain | Colony Diameter (mm) | Zone of Solubilization (mm) | Phosphate Solubilization Index (PSI) |
|---|---|---|---|
| BRZ63 | 7.84 ± 0.2 | 52.3 ± 0.12 a | 7.67 ± 0.33 a |
| 4FJK | 10.1 ± 0.3 | 7.7 ± 0.2 b | 1.76 ± 0.19 b |
| Test | Strain BRZ63 | Strain P-1 | |
|---|---|---|---|
| Methylene blue agar test | + | + | |
| Oil-spreading (mm) | 40.30 ± 3.50 a | 44.01 ± 1.52 a | |
| Emulsification index (%) | diesel oil | 32.51 ± 2.53 a | 58.33 ± 4.81 b |
| cyclohexane | 78.79 ± 2.62 a | 77.37 ± 2.63 a | |
| Attribute | Value |
|---|---|
| Genome size (bp) | 6,335,040 bp |
| Contigs | 363 |
| G+C content (%) | 64 |
| Genes (total) | 6120 |
| CDSs (total) | 6043 |
| Genes (coding) | 5915 |
| Protein genes | 5915 |
| RNA genes | 77 |
| rRNAs | 7, 3, 1 (5S, 16S, 23S) |
| Complete rRNAs | 7, 1, 1 (5S, 16S, 23S) |
| Partial rRNAs | 2 (16S) |
| tRNAs | 62 |
| ncRNAs | 4 |
| Pseudogenes | 128 |
| BioProject ID | PRJNA529642 |
| GenBank accession number | SPVI00000000.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chlebek, D.; Pinski, A.; Żur, J.; Michalska, J.; Hupert-Kocurek, K. Genome Mining and Evaluation of the Biocontrol Potential of Pseudomonas fluorescens BRZ63, a New Endophyte of Oilseed Rape (Brassica napus L.) against Fungal Pathogens. Int. J. Mol. Sci. 2020, 21, 8740. https://doi.org/10.3390/ijms21228740
Chlebek D, Pinski A, Żur J, Michalska J, Hupert-Kocurek K. Genome Mining and Evaluation of the Biocontrol Potential of Pseudomonas fluorescens BRZ63, a New Endophyte of Oilseed Rape (Brassica napus L.) against Fungal Pathogens. International Journal of Molecular Sciences. 2020; 21(22):8740. https://doi.org/10.3390/ijms21228740
Chicago/Turabian StyleChlebek, Daria, Artur Pinski, Joanna Żur, Justyna Michalska, and Katarzyna Hupert-Kocurek. 2020. "Genome Mining and Evaluation of the Biocontrol Potential of Pseudomonas fluorescens BRZ63, a New Endophyte of Oilseed Rape (Brassica napus L.) against Fungal Pathogens" International Journal of Molecular Sciences 21, no. 22: 8740. https://doi.org/10.3390/ijms21228740