Integrated Phytochemical Analysis Based on UPLC-MS and Network Pharmacology Approaches to Explore the Quality Control Markers for the Quality Assessment of Trifolium pratense L.
Abstract
1. Introduction
2. Results and Discussion
2.1. Metabolomics Profiling Analysis of Red Clover by UPLC-ESI-Orbitrap MS/MS
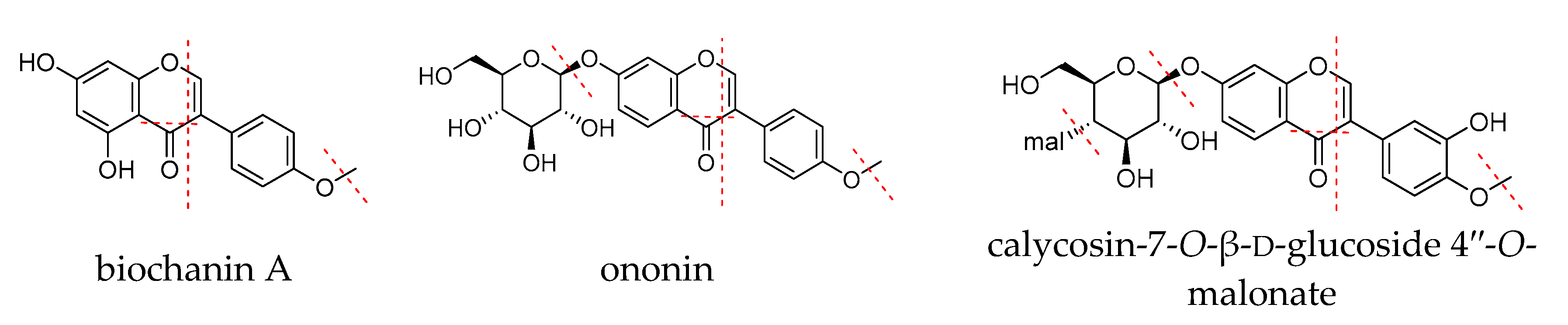
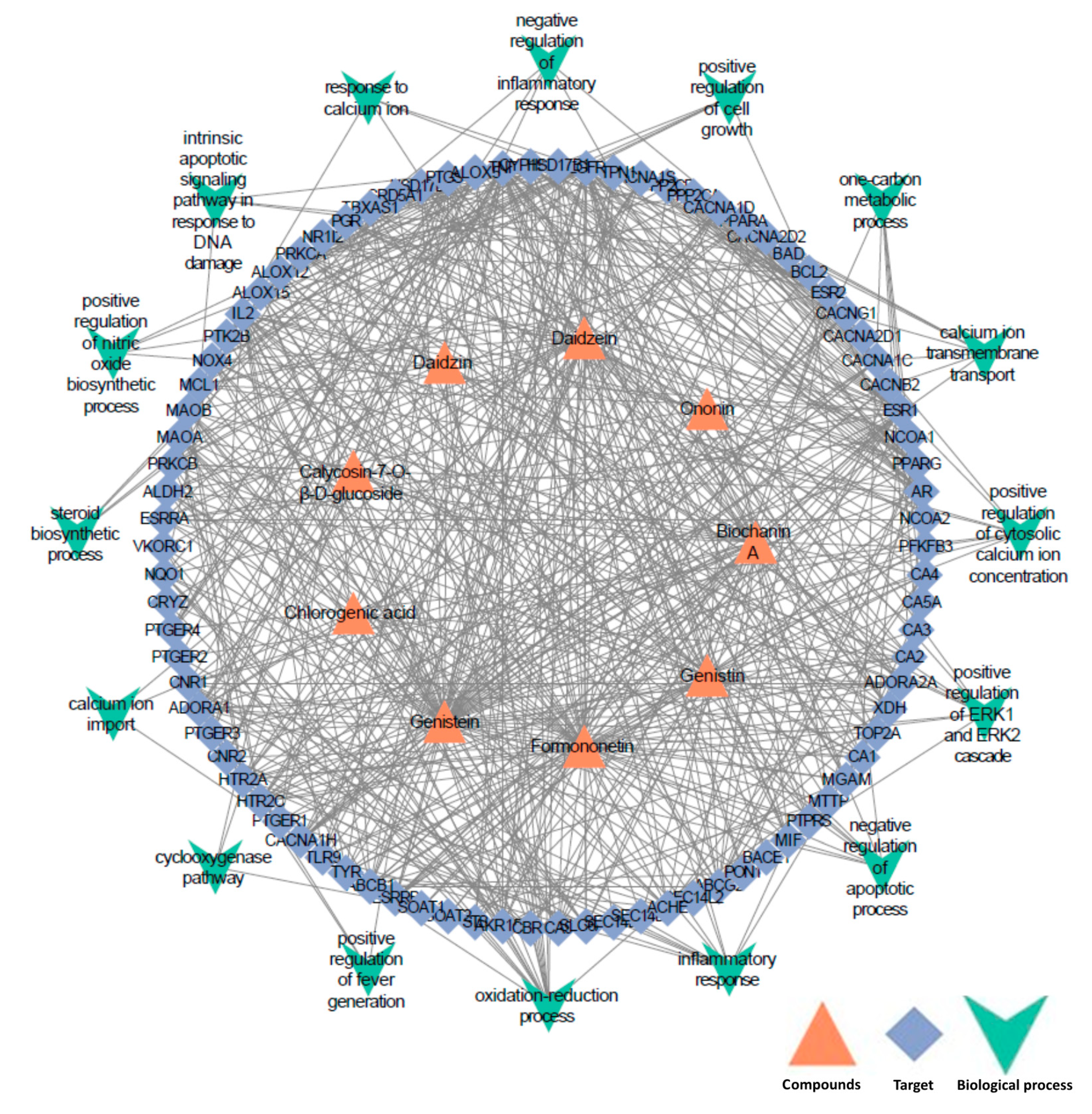
2.2. Quality Control Markers Screening and Validation by Network Pharmacology and Blood-Absorbed Components Analysis
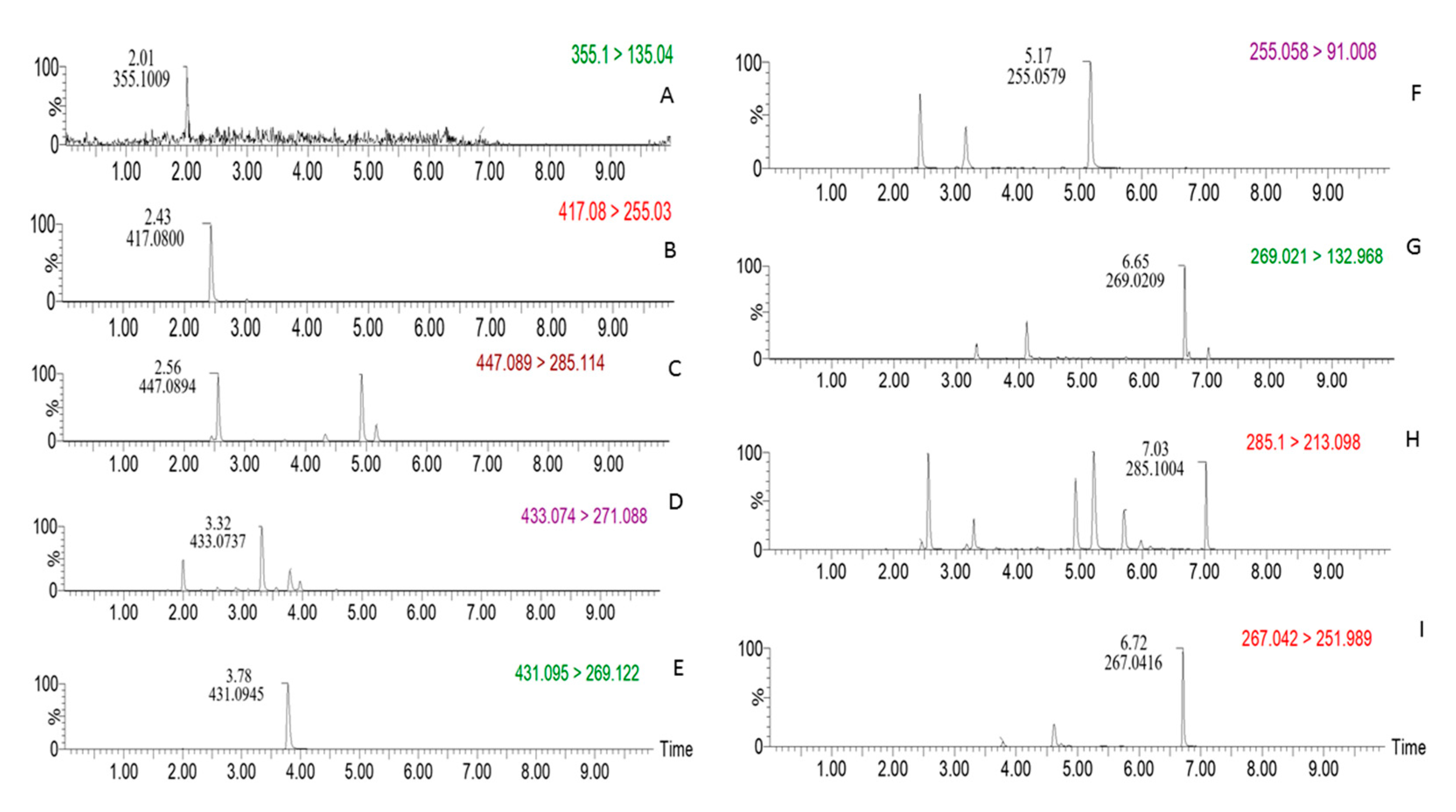
2.3. UPLC-MS Method Validation
2.3.1. Specificity
2.3.2. Linearity and Limit of Quantitation
2.3.3. Linearity and Limit of Quantitation Precision, Repeatability and Stability
2.3.4. Accuracy
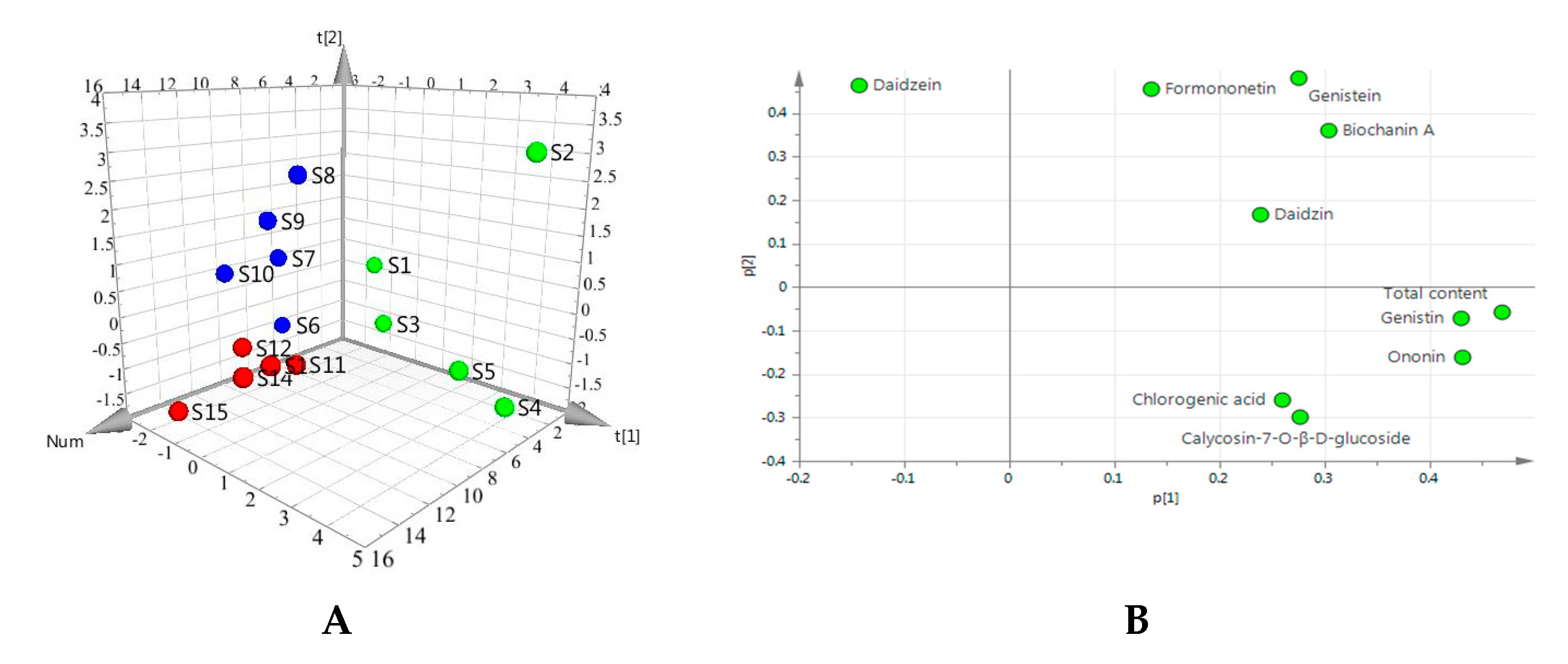
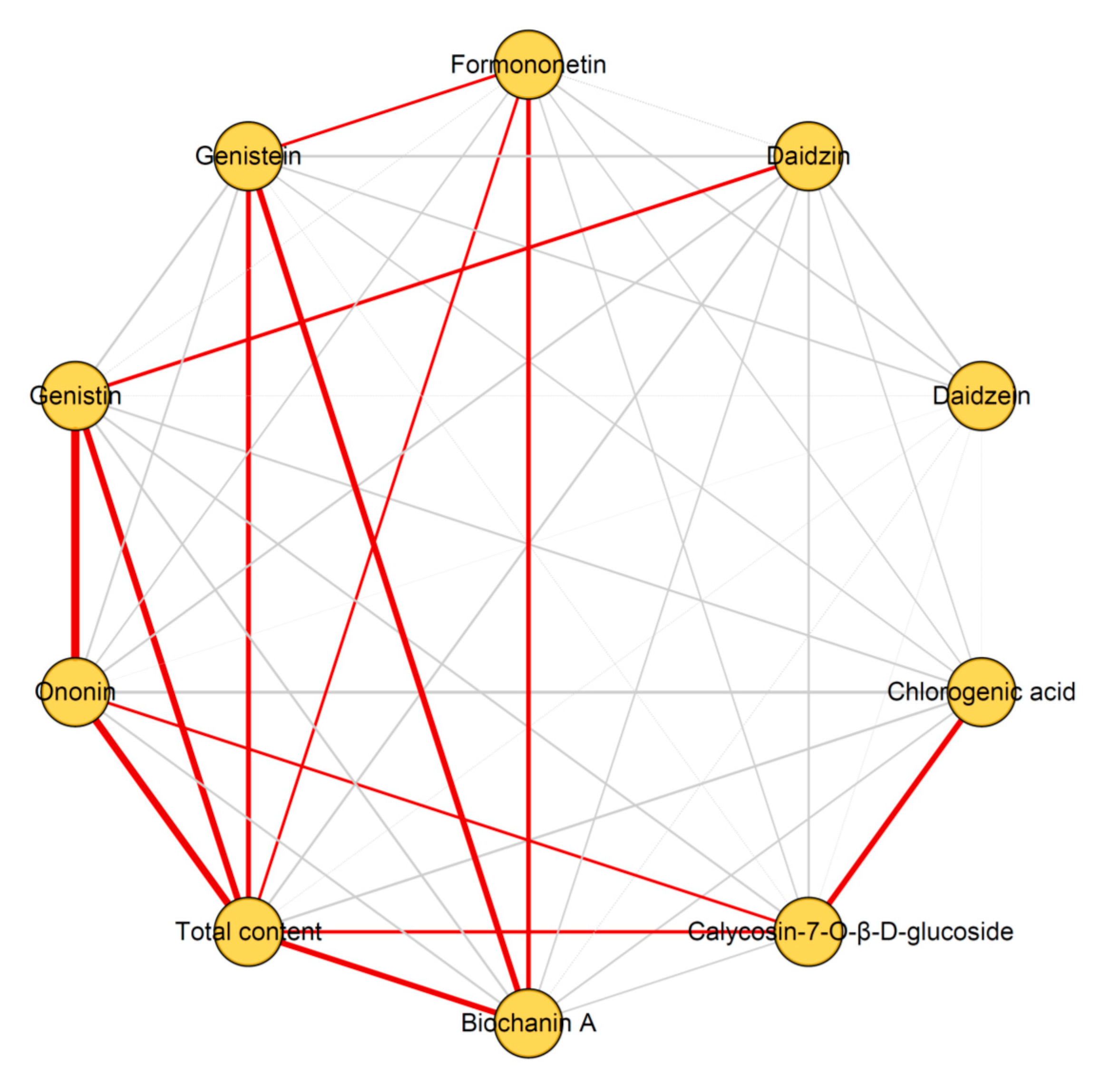
2.4. Determination of the Nine Quality Control Markers in 15 Batches of Red Clover
3. Materials and Methods
3.1. Instruments and Chemicals
3.2. Red Clover Sample Collection
3.3. Preparation of Reference Solutions
3.4. Metabolomics Profiling Analysis by UPLC-ESI-Orbitrap MS/MS
3.4.1. Sample Preparation for Compounds Identification
3.4.2. Chromatographic and Mass Spectrometric Conditions
3.5. Quality Control Markers Screening and Validation by Network Pharmacology and Blood-Absorbed Components Analysis
3.5.1. Quality Control Markers Screening by Network Pharmacology
3.5.2. Blood-Absorbed Components Analysis for the Validation of Quality Control Markers
3.6. Development and Validation of UPLC-MS/MS Method for Quality Control of Red Clover
3.6.1. Sample Preparation for Quality Evaluation
3.6.2. Chromatographic and Mass Spectrometric Conditions
3.6.3. Methodology Validation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Liu, Y.; Liu, S.H.; Wang, P. Research progress of Trifolium pratense. Chin. Herbal Med. 2007, 801–804. [Google Scholar]
- Oza, M.J.; Kulkarni, Y.A. Trifolium pratense (Red Clover) Improve SIRT1 Expression and Glycogen Content in High Fat Diet-Streptozotocin Induced Type 2 Diabetes in Rats. Chem. Biodivers. 2020, 17, e2000019. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.B.; Yang, Y.R. Trifolium pratense isoflavones improve pulmonary vascular remodelling in broiler chickens. J. Anim. Physiol. Anim. Nutr. 2016, 100, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Circosta, C.; De Pasquale, R.; Palumbo, D.R.; Samperi, S.; Occhiuto, F. Effects of isoflavones from red clover (Trifolium pratense) on skin changes induced by ovariectomy in rats. Phytother. Res. 2006, 20, 1096–1099. [Google Scholar] [CrossRef]
- Booth, N.L.; Piersen, C.E.; Banuvar, S.; Geller, S.E.; Shulman, L.P.; Farnsworth, N.R. Clinical studies of red clover (Trifolium pratense) dietary supplements in menopause: A literature review. Menopause 2006, 13, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Cecotti, R.; Carpana, E.; Bergomi, P.; Tava, A. Volatile constituents of Trifolium pratense spp. nivale quantified at different growth stages, and evaluation of their antimicrobial activity. Nat. Prod. Commun. 2013, 8, 1625–1628. [Google Scholar] [CrossRef]
- Khazaei, M.; Pazhouhi, M. Protective effect of hydroalcoholic extracts of Trifolium pratense L. on pancreatic beta cell line (RIN-5F) against cytotoxicty of streptozotocin. Res. Pharm. Sci. 2018, 13, 324–331. [Google Scholar]
- Jarred, R.A.; Keikha, M.; Dowling, C.; McPherson, S.J.; Clare, A.M.; Husband, A.J.; Pedersen, J.S.; Frydenberg, M.; Risbridger, G.P. Induction of apoptosis in low to moderate-grade human prostate carcinoma by red clover-derived dietary isoflavones. Cancer Epidemiol. Biomark. Prev. 2002, 11, 1689–1696. [Google Scholar]
- Katz, A.E. Flavonoid and botanical approaches to prostate health. J. Altern. Complement Med. 2002, 8, 813–821. [Google Scholar] [CrossRef]
- Atkinson, C.; Warren, R.M.; Sala, E.; Dowsett, M.; Dunning, A.M.; Healey, C.S.; Runswick, S.; Day, N.E.; Bingham, S.A. Red-clover-derived isoflavones and mammographic breast density: A double-blind, randomized, placebo-controlled trial [ISRCTN42940165]. Breast Cancer Res. BCR 2004, 6, R170–R179. [Google Scholar] [CrossRef]
- Flythe, M.; Kagan, I. Antimicrobial effect of red clover (Trifolium pratense) phenolic extract on the ruminal hyper ammonia-producing bacterium, Clostridium sticklandii. Curr. Microbiol. 2010, 61, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Dobrucka, R.; Dlugaszewska, J. Biosynthesis and antibacterial activity of ZnO nanoparticles using Trifolium pratense flower extract. Saudi J. Biol. Sci. 2016, 23, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Krenn, L.; Paper, D.H. Inhibition of angiogenesis and inflammation by an extract of red clover (Trifolium pratense L.). Phytomedicine 2009, 16, 1083–1088. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Rimbach, G.; Moini, H.; Weber, S.; Packer, L. ESR and cell culture studies on free radical-scavenging and antioxidant activities of isoflavonoids. Toxicology 2002, 179, 171–180. [Google Scholar] [CrossRef]
- Liu, B.J.; Guo, Y.S.; Qu, Y.P.; Diao, P.F. Study on the antioxidant effect of total flavonoids of Trifolium pratense in vitro. Chin. J. Pharm. 2009, 44, 1234–1237. [Google Scholar]
- Kaurinovic, B.; Popovic, M.; Vlaisavljevic, S.; Schwartsova, H.; Vojinovic-Miloradov, M. Antioxidant profile of Trifolium pratense L. Molecules 2012, 17, 11156–11172. [Google Scholar] [CrossRef]
- Arash, K.E.; Rosna, M.T.; Sadegh, M.; Behrooz, B. Antioxidant Activity and Total Phenolic and Flavonoid Content of Various Solvent Extracts from In Vivo and In Vitro Grown Trifolium pratense L. (Red Clover). Biomed Res. Int. 2015. [Google Scholar] [CrossRef]
- Occhiuto, F.; Zangla, G.; Samperi, S.; Palumbo, D.R.; Pino, A.; De Pasquale, R.; Circosta, C. The phytoestrogenic isoflavones from Trifolium pratense L. (Red clover) protects human cortical neurons from glutamate toxicity. Phytomedicine 2008, 15, 676–682. [Google Scholar] [CrossRef]
- Chen, H.Q.; Wang, X.J.; Jin, Z.Y.; Xu, X.M.; Zhao, J.W.; Xie, Z.J. Protective effect of isoflavones from Trifolium pratense on dopaminergic neurons. Neurosci. Res. 2008, 62, 123–130. [Google Scholar] [CrossRef]
- Kami, T. Qualitative and quantitative analyses of the essential oils of red and Ladino white clovers. J. Agric. Food Chem. 1978, 26, 1194–1197. [Google Scholar] [CrossRef]
- Vlaisavljevic, S.; Kaurinovic, B.; Popovic, M.; Djurendic-Brenesel, M.; Vasiljevic, B.; Cvetkovic, D.; Vasiljevic, S. Trifolium pratense L. as a potential natural antioxidant. Molecules 2014, 19, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Howes, J.B.; Howes, L.G. Content of isoflavone-containing preparations. Med. J. Aust. 2002, 176, 135–136. [Google Scholar] [CrossRef] [PubMed]
- Clifton-Bligh, P.B.; Baber, R.J.; Fulcher, G.R.; Nery, M.L.; Moreton, T. The effect of isoflavones extracted from red clover (Rimostil) on lipid and bone metabolism. Menopause 2001, 8, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, C.; Compston, J.E.; Day, N.E.; Dowsett, M.; Bingham, S.A. The effects of phytoestrogen isoflavones on bone density in women: A double-blind, randomized, placebo-controlled trial. Am. J. Clin. Nutr. 2004, 79, 326–333. [Google Scholar] [CrossRef]
- Coon, J.T.; Pittler, M.H.; Ernst, E. Trifolium pratense isoflavones in the treatment of menopausal hot flushes: A systematic review and meta-analysis. Phytomedicine 2007, 14, 153–159. [Google Scholar] [CrossRef]
- Luís, Â.; Domingues, F.; Pereira, L.J.C. Effects of Red clover on perimenopausal and postmenopausal women′s blood lipid profile: A meta-analysis. Climacteric 2018, 21, 446–453. [Google Scholar] [CrossRef]
- Renda, G.; Yalçın, F.N.; Nemutlu, E.; Akkol, E.K.; Süntar, I.; Keleş, H.; Ina, H.; Çalış, I.; Ersöz, T. Comparative assessment of dermal wound healing potentials of various Trifolium L. extracts and determination of their isoflavone contents as potential active ingredients. J. Ethnopharmacol. 2013, 148, 423–432. [Google Scholar] [CrossRef]
- Li, T.X.; Hu, Y.T.; Chu, C.S.; Gou, Q.X.; Chang, Y. Determination of six isoflavones in Trifolium pratense by one test and multiple evaluation method. Chin. Herbal Med. 2018, 41, 2619–2622. [Google Scholar]
- Ma, Q.; Zhou, Y.X.; Lei, H.M.; Wang, C.H. Determination of daidzein, genistein, formononetin and biochanin A in Trifolium pratense L. by HPLC. Chin. Tradit. Pat. Med. 2005, 1193–1195. [Google Scholar]
- He, L.; Zhou, P.; Wen, Z.H.; Zhang, G.P. Analysis of volatile components of Trifolium pratense from different habitats by HS-SPME-GC-MS. Chin. Herbal Med. 2018, 1122–1128. [Google Scholar]
- He, C.L.; Zhang, G.P.; Wang, R.Y.; Zhou, P. HS-SPME-GC-MS and principal component analysis of volatile oil components in different parts of Trifolium pratense. Chin. J. Exp. Pharmacol. 2018, 24, 71–81. [Google Scholar]
- Wu, Q.; Wang, M.; Simon, J.E. Determination of isoflavones in red clover and related species by high-performance liquid chromatography combined with ultraviolet and mass spectrometric detection. J. Chromatogr. A 2003, 1016, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Zhang, J.L.; Zhou, Y.X.; Lei, H.M.; Wang, C.H. Determination of isoflavones in Trifolium pratense by with high performance liquid chromatography electrospray ionization mass spectrometry. Anal. Chem. 2006, 34, 247–250. [Google Scholar]
- Occhiuto, F.; Pasquale, R.D.; Guglielmo, G.; Palumbo, D.R.; Zangla, G.; Samperi, S.; Renzo, A.; Circosta, C. Effects of phytoestrogenic isoflavones from red clover (Trifolium pratense L.) on experimental osteoporosis. Phytother. Res. 2007, 21, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.A.; Moon, S.M.; Han, S.H.; Kim, J.S.; Kim, D.K.; Kim, C.S. The Effect of the Prethanol Extract of Trifolium pratense Leaves on Interleukin-1beta-Induced Cartilage Matrix Degradation in Primary Rat Chondrocytes. Cells Tissues Organs 2018, 206, 95–105. [Google Scholar] [CrossRef]
- Ling, S.; Dai, A.; Williams, M.R.; Husband, A.J.; Nestel, P.J.; Komesaroff, P.A.; Sudhir, K. The isoflavone metabolite cis-tetrahydrodaidzein inhibits ERK-1 activation and proliferation in human vascular smooth muscle cells. J. Cardiovasc. Pharmacol. 2004, 43, 622–628. [Google Scholar] [CrossRef]
- Sun, J.; Geng, T.; Pan, Q.; Xiao, F. Research progress of isoflavones in Trifolium pratense. Foreign Med. 2007, 22, 150–155. [Google Scholar]
- Duan, H.; Chai, J.; Sheng, Z.; Yao, Y.; Yin, H.; Liang, L.; Shen, C.; Lin, J. Effect of burn injury on apoptosis and expression of apoptosis-related genes/proteins in skeletal muscles of rats. Apoptosis 2009, 14, 52–65. [Google Scholar] [CrossRef]
- Teede, H.J.; McGrath, B.P.; DeSilva, L.; Cehun, M.; Fassoulakis, A.; Nestel, P.J. Isoflavones reduce arterial stiffness: A placebo-controlled study in men and postmenopausal women. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 1066–1071. [Google Scholar] [CrossRef]
- Reeve, J.L.; Duffy, A.M.; O′Brien, T.; Samali, A. Don′t lose heart—therapeutic value of apoptosis prevention in the treatment of cardiovascular disease. J. Cell. Mol. Med. 2005, 9, 609–622. [Google Scholar] [CrossRef]
- Locasale, J.W. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 2013, 13, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.; Hobiger, S.; Jungbauer, A. Red clover extract: A source for substances that activate peroxisome proliferator-activated receptor alpha and ameliorate the cytokine secretion profile of lipopolysaccharide-stimulated macrophages. Menopause 2010, 17, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Lam, A.N.; Demasi, M.; James, M.J.; Husband, A.J.; Walker, C. Effect of red clover isoflavones on cox-2 activity in murine and human monocyte/macrophage cells. Nutr. Cancer 2004, 49, 89–93. [Google Scholar] [CrossRef]
- Liu, Z.Y.; Guo, F.F.; Wang, Y.; Li, C.; Zhang, X.L.; Li, H.L.; Diao, L.H.; Gu, J.Y.; Wang, W.; Li, D.; et al. BATMAN-TCM: A Bioinformatics Analysis Tool for Molecular mechANism of Traditional Chinese Medicine. Sci. Rep. 2016, 6, 21146. [Google Scholar] [CrossRef] [PubMed]
- Brohée, S.; Faust, K.; Lima-Mendez, G.; Vanderstocken, G.; van Helden, J. Network Analysis Tools: From biological networks to clusters and pathways. Nat. Protoc. 2008, 3, 1616–1629. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, 1–11. [Google Scholar] [CrossRef]
- da Huang, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |
| Compound Name | Linear Range (ng/mL) | R2 | Limit of Quantitation (ng/mL) | Reproducibility RSD (%) (n = 6) | Average Recovery Rate (%) | ||
|---|---|---|---|---|---|---|---|
| Low a (n = 3) | Middle (n = 3) | High (n = 3) | |||||
| Chlorogenic acid | 20.97~419.42 | 0.9991 | 20.97 | 1.8 | 103.4 | 104.2 | 105.2 |
| Daidzin | 1.45~289.14 | 0.9995 | 0.014 | 2.5 | 96.5 | 105.2 | 97.8 |
| Calycosin-7-O-β-d-glucoside | 1.28~255.43 | 0.9999 | 0.013 | 2.8 | 96.6 | 103.5 | 103.5 |
| Genistin | 1.20~239.08 | 0.9997 | 0.24 | 3.8 | 97.8 | 101.4 | 97.3 |
| Ononin | 1.66~165.58 | 0.9998 | 0.017 | 1.5 | 100.4 | 104.1 | 97.7 |
| Daidzein | 1.51~151.16 | 0.9992 | 0.30 | 2.5 | 103.7 | 104.3 | 100.9 |
| Genistein | 12.56~627.98 | 0.9993 | 12.56 | 2.1 | 108.9 | 101.3 | 97.0 |
| Formononetin | 1.17~233.16 | 0.9997 | 0.50 | 1.2 | 102.5 | 95.4 | 97.5 |
| Biochanin A | 1.77~884.35 | 0.9997 | 0.71 | 2.2 | 104.7 | 103.4 | 97.5 |
| Sample Number | Content of Compounds in Red Clover Extract (ng/mL) | Location | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chlorogenic Acid | Daidzin | Calycosin-7-O-β-d-glucoside | Genistin | Ononin | Daidzein | Genistein | Formononetin | Biochanin A | Total Content | ||
| S1 | 85 | 7361 | 3450 | 5175 | 33647 | 5662 | 3933 | 35193 | 11729 | 106235 | Gansu |
| S2 | 1717 | 18395 | 571 | 28649 | 72213 | 4678 | 9323 | 48149 | 40433 | 224128 | |
| S3 | 5710 | 2880 | 2812 | 7970 | 35019 | 3146 | 5696 | 34597 | 19793 | 117623 | |
| S4 | 7695 | 11353 | 29628 | 18027 | 72187 | 3562 | 4823 | 50069 | 28870 | 226214 | |
| S5 | 1291 | 7952 | 8447 | 24379 | 80374 | 2342 | 4072 | 36096 | 26756 | 191709 | |
| Average (S1–S5) | 3300 | 9588 | 8982 | 16840 | 58688 | 3878 | 5569 | 40821 | 25516 | 173182 | |
| S6 | 397 | 5767 | 2202 | 3790 | 14945 | 4058 | 2233 | 22354 | 4359 | 60105 | Hubei |
| S7 | 739 | 12847 | 3094 | 3065 | 16570 | 6484 | 3280 | 44849 | 5792 | 96720 | |
| S8 | 872 | 8382 | 2674 | 6294 | 18462 | 6727 | 7860 | 57704 | 35219 | 144194 | |
| S9 | 217 | 2834 | 1430 | 2326 | 16204 | 5783 | 6203 | 75028 | 45784 | 155809 | |
| S10 | 209 | 7003 | 1650 | 3152 | 11080 | 5944 | 4183 | 36855 | 8209 | 78285 | |
| Average (S6–S10) | 487 | 7367 | 2210 | 3725 | 15452 | 5799 | 4752 | 47358 | 19873 | 107023 | |
| S11 | 4023 | 3955 | 5301 | 9726 | 39039 | 2501 | 4873 | 46977 | 34518 | 150913 | Shanxi |
| S12 | - | 5457 | 3254 | 6584 | 33063 | 3033 | 3734 | 43176 | 23020 | 121321 | |
| S13 | - | 4155 | 5628 | 8145 | 60609 | 3268 | 5434 | 74243 | 20612 | 182094 | |
| S14 | - | 4701 | 5162 | 12056 | 47835 | 2475 | 4997 | 41490 | 19992 | 138708 | |
| S15 | 1345 | 2142 | 2778 | 6023 | 24024 | 1854 | 3152 | 33998 | 15880 | 91196 | |
| Average (S11–S15) | 2684 | 4082 | 4425 | 8507 | 40914 | 2626 | 4438 | 47977 | 22804 | 136846 | |
| Average (S1–S15) | 2025 | 7012 | 5205 | 9691 | 38351 | 4101 | 4920 | 45385 | 22731 | 139017 | |
| Compound Name | ESI | Molecular Formula | Parent Ion (m/z) | Daughter Ion (m/z) | Cone Voltage (V) | Collision Energy (eV) |
|---|---|---|---|---|---|---|
| chlorogenic acid | + | C16H18O9 | 355.10 | 135.04 | 30 | 40 |
| daidzin | + | C21H20O9 | 417.08 | 255.03 | 48 | 12 |
| calycosin-7-O-β-d-glucoside | + | C22H22O10 | 447.09 | 285.11 | 52 | 16 |
| genistin | + | C21H20O10 | 433.07 | 271.09 | 24 | 16 |
| ononin | + | C22H22O9 | 431.09 | 269.12 | 50 | 14 |
| daidzein | + | C15H10O4 | 255.06 | 91.01 | 82 | 18 |
| genistein | − | C15H10O5 | 269.02 | 132.97 | 74 | 28 |
| formononetin | − | C16H12O4 | 267.04 | 251.99 | 58 | 16 |
| biochanin A | + | C16H12O5 | 285.10 | 213.00 | 74 | 34 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, L.; Gao, W.; Zhang, Y.; Liu, C.; Wang, G.; Wu, H.; Gao, W. Integrated Phytochemical Analysis Based on UPLC-MS and Network Pharmacology Approaches to Explore the Quality Control Markers for the Quality Assessment of Trifolium pratense L. Molecules 2020, 25, 3787. https://doi.org/10.3390/molecules25173787
Luo L, Gao W, Zhang Y, Liu C, Wang G, Wu H, Gao W. Integrated Phytochemical Analysis Based on UPLC-MS and Network Pharmacology Approaches to Explore the Quality Control Markers for the Quality Assessment of Trifolium pratense L. Molecules. 2020; 25(17):3787. https://doi.org/10.3390/molecules25173787
Chicago/Turabian StyleLuo, Liyu, Wenya Gao, Yan Zhang, Chang Liu, Guopeng Wang, Hongwei Wu, and Wenyuan Gao. 2020. "Integrated Phytochemical Analysis Based on UPLC-MS and Network Pharmacology Approaches to Explore the Quality Control Markers for the Quality Assessment of Trifolium pratense L." Molecules 25, no. 17: 3787. https://doi.org/10.3390/molecules25173787
APA StyleLuo, L., Gao, W., Zhang, Y., Liu, C., Wang, G., Wu, H., & Gao, W. (2020). Integrated Phytochemical Analysis Based on UPLC-MS and Network Pharmacology Approaches to Explore the Quality Control Markers for the Quality Assessment of Trifolium pratense L. Molecules, 25(17), 3787. https://doi.org/10.3390/molecules25173787




