Immune Response to Herpes Simplex Virus Infection and Vaccine Development
Abstract
1. Herpes Simplex Virus and the Immune System
1.1. Introduction
1.2. Overview of the Immune System
1.2.1. The Innate Immune System
1.2.2. The Adaptive Immune System
1.3. The Interplay of Herpes Simplex Infection and the Immune System
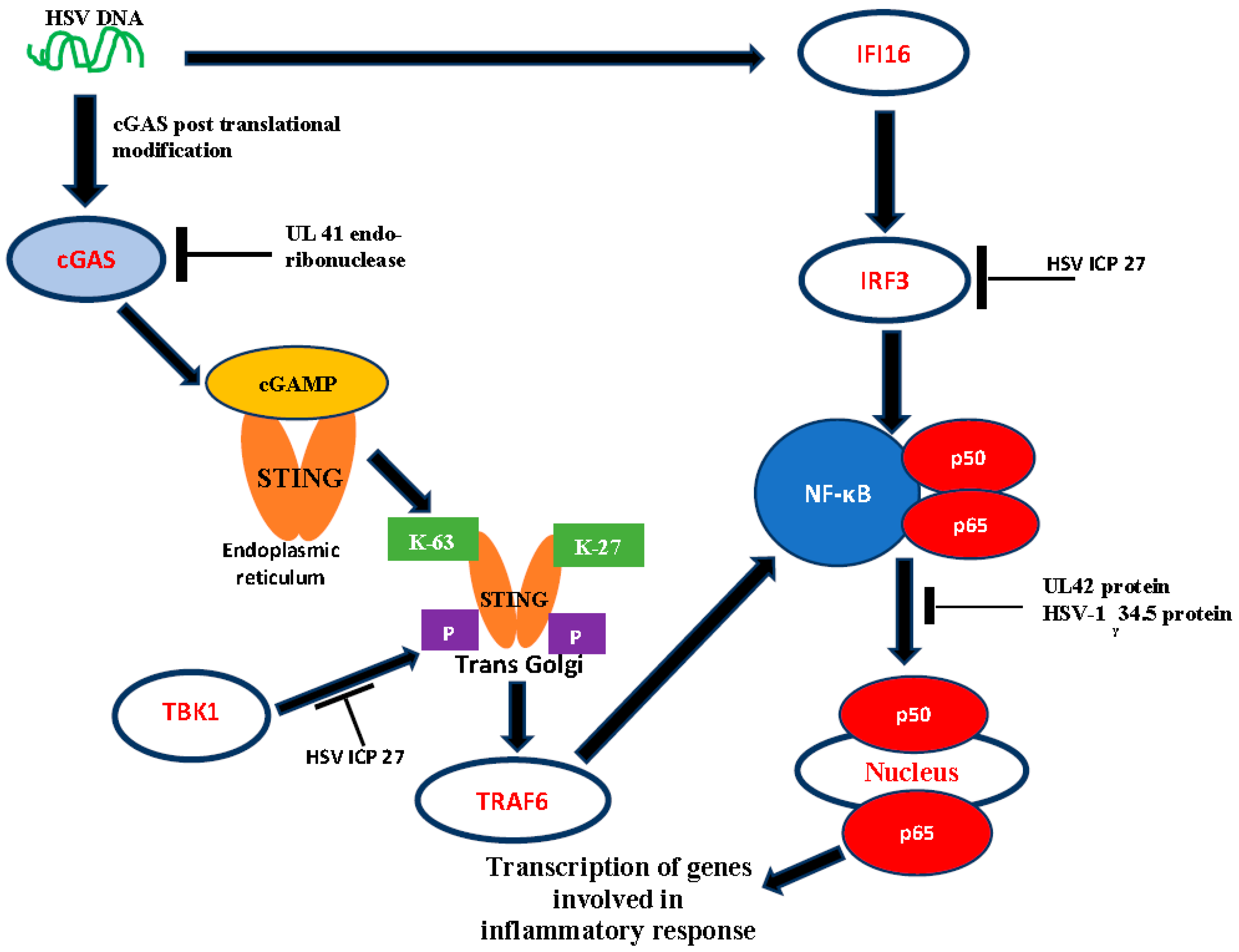
1.4. DNA Sensors as Activators of Host Antiviral Response
2. HSV Immune Evasion Mechanisms
2.1. Modulation of Autophagy
2.2. Interplay of HSV-1 and Host PML Protein
2.3. Modulation of Apoptosis
2.4. Intracellular Cell-to-Cell Propagation
2.5. Inactivation in Expression of Signaling Pathways
2.6. Role of miRNA in HSV Immune Evasion
3. HSV Vaccination and Immunotherapies
3.1. HSV Vaccination
3.2. Potential Vaccine Candidates
3.3. Possible Immunotherapies
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bradley, H.; Markowitz, L.E.; Gibson, T.; McQuillan, G.M. Seroprevalence of herpes simplex virus types 1 and 2-United States, 1999–2010. J. Infect. Dis. 2014, 209, 325–333. [Google Scholar] [CrossRef]
- Okonko, I.O.; Cookey, T.I. Seropositivity and determinants of immunoglobulin-G (IgG) antibodies against herpes simplex virus (HSV) types -1 and -2 in pregnant women in Port Harcourt, Nigeria. Afr. Health Sci. 2015, 15, 737–747. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reward, E.E.; Muo, S.O.; Orabueze, I.N.A.; Ike, A.C. Seroprevalence of herpes simplex virus types 1 and 2 in Nigeria: A systematic review and meta-analyses. Pathog. Glob. Heath 2019, 113, 2229–2237. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, D.I.; Flechtner, J.B.; McNeil, L.K.; Heineman, T.; Oliphant, T.; Tasker, S.; Wald, A.; Hetherington, S.; Genocea Study Group. Therapeutic HSV-2 vaccine decreases recurrent virus shedding and recurrent genital herpes disease. Vaccine 2019, 37, 3443–3450. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, S.; Hook, L.M.; Shaw, C.E.; Friedman, H.M. A trivalent subunit antigen glycoprotein vaccine as immunotherapy for genital herpes in the guinea pig genital infection model. Hum. Vaccines Immunother. 2017, 13, 2785–2793. [Google Scholar] [CrossRef] [PubMed]
- Khodai, T.; Chappell, D.; Christy, C.; Cockle, P.; Eyles, J.; Hammond, D.; Gore, K.; McChuskie, M.J.; Evans, D.M.; Lang, S.; et al. Single and combination herpes simplex virus type 2 glycoprotein vaccines adjuvanted with CpG oligodeoxynucleotides or monophosphoryl lipid A exhibit differential immunity that is not correlated to protection in animal models. Clin. Vaccines Immunol. 2011, 18, 1702–1709. [Google Scholar] [CrossRef]
- Mundle, S.T.; Hernandez, H.; Hamberger, J.; Catalan, J.; Zhou, C.; Stegalkina, S.; Tiffany, A.; Kleanthous, H.; Delagrave, S.; Anderson, S.F. High-purity preparation of HSV-2 vaccine candidate ACAM529 is immunogenic and efficacious in vivo. PLoS ONE 2013, 8, e57224. [Google Scholar] [CrossRef]
- Srivastava, R.; Roy, S.; Coulon, P.G.; Vahed, H.; Prakash, S.; Dhanushkodi, N.; Kim, G.J.; Fouladi, M.A.; Campo, J.; Teng, A.A.; et al. Therapeutic mucosal vaccination of herpes simplex virus 2-infected Guinea pigs with ribonucleotide reductase 2 (RR2) protein boosts antiviral neutralizing antibodies and local tissue-resident CD4+ and CD8+ TRM cells associated with protection against recurrent genital herpes. J. Virol. 2019, 93, e02309-18. [Google Scholar]
- Felsburg, P.J. Overview of the immune system and immunodeficiency diseases. Vet. Clin. N. Am. Small Anim. Pract. 1994, 24, 629–653. [Google Scholar] [CrossRef]
- Matsui, T.; Amagai, M. Dissecting the formation, structure and barrier function of the stratum corneum. Int. Immunol. 2015, 27, 269–280. [Google Scholar] [CrossRef]
- Oliver, W.T.; Wells, J.E. Lysozyme as an alternative to growth promoting antibiotics in swine production. J. Anim. Sci. Biotechnol. 2015, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Chaplin, D.D. Overview of the immune response. J. Allergy Clin. Immunol. 2010, 125 (Suppl. 2), S3–S23. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Zhan, G.; Zheng, C. Evasion of host antiviral innate immunity by HSV-1, an update recruitment of the downstream adaptor TBK1. Virol. J. 2016, 13, 38. [Google Scholar] [CrossRef] [PubMed]
- Peri, P.; Mattila, R.K.; Kantola, H.; Broberg, E.; Karttunen, H.S.; Waris, M.; Vuorinen, T.; Hukkanen, V. Herpes simplex virus type 1 Us3 gene deletion influences toll-like receptor responses in cultured monocytic cells. Virol. J. 2008, 5, 140. [Google Scholar] [CrossRef]
- van Lint, A.L.; Murawski, M.R.; Goodbody, R.E.; Severa, M.; Fitzgerald, K.A.; Finberg, R.W.; Knipe, D.M.; Kurt-Jones, E.A. Herpes simplex virus immediate-early ICP0 protein inhibits Toll-like receptor 2-dependent inflammatory responses and NF-kappaB signaling. J. Virol. 2010, 84, 10802–10811. [Google Scholar] [CrossRef]
- Smith, M.C.; Boutell, C.; Davido, D.J. HSV-1 ICP0: Paving the way for viral replication. Future Virol. 2012, 6, 421–429. [Google Scholar] [CrossRef]
- Gu, H.; Roizman, B. The Two Functions of Herpes Simplex Virus 1 ICP0, Inhibition of Silencing by the CoREST/REST/HDAC Complex and Degradationof PML, Are Executed in Tandem. J. Virol. 2009, 83, 181–187. [Google Scholar] [CrossRef]
- Merkl, P.E.; Orzalli, M.H.; Knipe, D.M. Mechanisms of host IFI16, PML, and Daxxprotein restriction of herpes simplex virus 1replication. J. Virol. 2018, 92, e00057-18. [Google Scholar] [CrossRef]
- Gu, H. What role does cytoplasmic ICP0 play in HSV-1 infection? Future Virol. 2018, 13, 6. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, S.; Wang, K.; Zheng, C. Herpes simplex virus 1 DNA polymerase processivity factor UL42 inhibits TNF-a-induced NF-jB activation by interacting with p65/RelA and p50/NF-jB1. Med. Microbiol. Immunol. 2013, 202, 313–325. [Google Scholar] [CrossRef]
- Chen, Z.J. Ubiquitin signaling in the NF-kappaB pathway. Nat. Cell Biol. 2005, 7, 758–765. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Ma, Y.; Yan, Z.; Prabhakar, B.S.; He, B. Activation of NF-kappaB in CD8+ dendritic cells ex vivo by the gamma134.5 null mutant correlates with immunity against herpes simplex virus 1. J. Virol. 2012, 86, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Cotter, C.R.; Kim, W.K.; Nguyen, M.L.; Yount, J.S.; Lopez, C.B.; Blaho, J.A.; Moran, T.M. The virion host shutoff protein of herpes simplex virus 1 blocks the replication-independent activation of NF-kappaB in dendritic cells in the absence of type I interferon signaling. J. Virol. 2011, 85, 12662–12672. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Wang, S.; Lin, R.; Mossman, K.L.; Zheng, C. Herpes simplex virus 1 tegument protein US11 downmodulates the RLR signaling pathway via direct interaction with RIG-I and MDA-5. J. Virol. 2012, 86, 3528–3540. [Google Scholar] [CrossRef] [PubMed]
- Horng, T.; Barton, G.M.; Flavell, R.A.; Medzhitov, R. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature 2002, 420, 329–333. [Google Scholar] [CrossRef]
- Finberg, R.W.; Knipe, D.M.; Kurt-Jones, E.A. Herpes simplex virus and Toll-like receptors. Viral Immunol. 2005, 18, 457–465. [Google Scholar] [CrossRef]
- Schenkel, J.M.; Masopust, D. Tissue-resident memory T cells. Immunity 2014, 41, 886–897. [Google Scholar] [CrossRef]
- Schenkel, J.M.; Fraser, K.A.; Beura, L.K.; Kristen, E.P.; Vaiva, V.; Masopust, D. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science 2014, 346, 98–101. [Google Scholar] [CrossRef]
- Zheng, S.G. Regulatory T cells vs Th17: Differentiation of Th17 versus Treg, are the mutually exclusive? Am. J. Clin. Exp. Immunol. 2013, 3, 94–106. [Google Scholar]
- Suvas, S.; Kumaraguru, U.; Pack, C.D.; Lee, S.; Rouse, B.T. CD4+CD25+ T cells regulate virus-specific primary and memory CD8+ T cell responses. J. Exp. Med. 2003, 198, 889–901. [Google Scholar] [CrossRef]
- Manickan, E.; Rouse, R.J.; Yu, Z.; Wire, W.S.; Rouse, B.T. Genetic immunization against herpes simplex virus. Protection is mediated by CD4+ T lymphocytes. J. Immunol. 1995, 155, 259–265. [Google Scholar] [PubMed]
- Yu, W.; Geng, S.; Suo, Y.; Wei, X.; Cai, Q.; Wu, B.; Zhou, X.; Shi, Y.; Wang, B. Critical role of regulatory T Cells in the latency and stress-induced reactivation of HSV-1. Cell Rep. 2018, 25, 2379–2389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, H.; Wei, B. Immune response of T cells during herpes simplex virus type 1 (HSV-1) infection. J. Zhejiang Univ. Sci. B Biomed. Biotech. 2017, 18, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.M.; Yang, Q.; Xie, X.Q.; Liao, C.Y.; Lin, H.; Liu, T.T.; Yin, L.; Shu, H.B. Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity 2016, 45, 555–569. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Ye, B.; Wang, S.; Zhu, X.; Du, Y.; Xiong, Z.; Tian, Y.; Fan, Z. Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat. Immunol. 2016, 17, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Zhang, X.W.; Jin, J.; Du, X.X.; Lian, T.; Yang, J.; Zhou, X.; Jiang, Z.; Su, X.D. Nonspecific DNA binding of cGAS N terminus promotes cGAS activation. J. Immunol. 2017, 198, 3627–3636. [Google Scholar] [CrossRef]
- Du, M.; Chen, Z.J. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 2018, 361, 704–709. [Google Scholar] [CrossRef]
- Zhang, C.; Shang, G.; Gui, X.; Zhang, X.; Bai, X.C.; Chen, Z.J. Structural basis of STING binding with and phosphorylation by TBK1. Nature 2019, 567, 394–398. [Google Scholar] [CrossRef]
- Ishikawa, H.; Barber, G.N. The STING pathway and regulation of innate immune signaling in response to DNA pathogens. Cell. Mol. Life Sci. 2011, 68, 1157–1165. [Google Scholar] [CrossRef]
- Abe, T.; Barber, G.N. Cytosolic-DNA-Mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-B activation through TBK1. J. Virol. 2014, 88, 5328–5341. [Google Scholar] [CrossRef]
- Zhang, Z.; Yuan, B.; Bao, M.; Lu, N.; Kim, T.; Liu, Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011, 12, 959–965. [Google Scholar] [CrossRef]
- Horan, K.A.; Hansen, K.; Jakobsen, M.R.; Holm, C.K.; Søby, S.; Unterholzner, L.; Thompson, M.; West, J.A.; Iversen, M.B.; Rasmussen, S.B.; et al. Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J. Immunol. 2013, 190, 2311–2319. [Google Scholar] [CrossRef] [PubMed]
- Unterholzner, L.; Keating, S.E.; Baran, M.; Horan, K.A.; Jensen, S.B.; Sharma, S.; Siros, C.M.; Jin, I.; Latz, E.; Xiao, T.S.; et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010, 11, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Karaba, A.H.; Figueroa, A.; Massaccesi, G.; Botto, S.; DeFilippis, V.R.; Cox, A.L. Herpes simplex virus type 1 inflammasome activation in proinflammatory human macrophages is dependent on NLRP3, ASC, and Caspase-1. PLoS ONE 2020, 15, e0229570. [Google Scholar] [CrossRef] [PubMed]
- Koelle, D.M.; Corey, L. Recent progress in herpes simplex virus immunobiology and vaccine research. Clin. Microbiol. Rev. 2003, 16, 96–113. [Google Scholar] [CrossRef]
- Ravanan, P.; Srikumar, F.I.; Talwar, P. Autophagy: The spotlight for cellular stress responses. Life Sci. 2017, 188, 53–67. [Google Scholar] [CrossRef]
- Turan, A.; Grosche, L.; Krawczyk, A.; Muhl-Zurbes, P.; Drassner, C.; Duthorn, A.; Kummer, M.; Hasenberg, M.; Voortamann, S.; Jastrow, H.; et al. Autophagic degradation of lamins facilitate the nuclear egress of herpes simplex virus type 1. J. Cell Biol. 2019, 218, 508–523. [Google Scholar] [CrossRef]
- DuRaine, G.; Wisner, T.W.; Howard, P.; Johnson, C.D. Kinesin-1 proteins KIF5A, -5B, and -5C promote anterograde transport of herpes simplex virus enveloped virions in proteins. J. Virol. 2018, 92, e01269-18. [Google Scholar] [CrossRef]
- Rubio, R.M.; Mohr, I. Inhibition of ULK1 and Beclin1 by an α-herpesvirus Akt-like Ser/Thr kinase limits autophagy to stimulate virus replication. Proc. Natl. Acad. Sci. USA 2019, 116, 26941–26950. [Google Scholar] [CrossRef]
- Xu, X.; He, Y.; Fan, S.; Feng, M.; Jiang, G.; Wang, L.; Zhang, L.; Liao, Y.; Qihan, L. Reducing viral inhibition of host cellular apoptosis strengthens the immunogenicity and protective efficacy of an attenuated HSV-1 strain. Virol. Sin. 2019, 34, 673–687. [Google Scholar] [CrossRef]
- Zhang, L.; Qin, Y.; Chen, M. Viral strategies for triggering and manipulating mitophagy. Autophagy 2018, 14, 1665–1673. [Google Scholar] [CrossRef] [PubMed]
- Waisner, H.; Kalamvoki, M. The ICP0 protein of herpes simplex virus 1 (HSV-1) downregulates major autophagy adaptor proteins sequestosome 1 and optineurin during the early stages of HSV-1 infection. J. Virol. 2019, 93, e01258-19. [Google Scholar] [CrossRef]
- Gu, H. Infected cell protein O functional domains and their coordination in herpes simplex virus replication. World J. Virol. 2016, 5, 1–13. [Google Scholar] [CrossRef]
- Lussignol, M.; Queval, C.; Bernet-Camard, M.; Cotte-Laffitte, J.; Beau, I.; Codogno, P.; Esclatine, A. The herpes simplex virus 1 Us11 protein inhibits autophagy through its interaction with the protein kinase PKR. J. Virol. 2013, 87, 859–871. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Matrenec, R.; Gack, M.U.; He, B. Disassembly of the TRIM23-TBK1 complex by the Us11 protein of herpes simplex virus 1 impairs autophagy. J. Virol. 2019, 93, e00497-19. [Google Scholar] [CrossRef]
- Liu, X.; Main, D.; Ma, Y.; He, B. Herpes simplex virus 1 inhibits TANK-Binding kinase 1 through formation of the Us11-Hsp90 complex. J. Virol. 2018, 92, e00402-18. [Google Scholar] [CrossRef]
- O’Connell, D.; Liang, C. Autophagy interaction with herpes simplex virus type -1 infection. Autophagy 2016, 12, 451–459. [Google Scholar] [CrossRef]
- Lussignol, M.; Esclatine, A. Herpes virus and autophagy: “All right, everybody be cool, this is a robbery!”. Viruses 2017, 9, 372. [Google Scholar] [CrossRef] [PubMed]
- Yakoub, A.M.; Shukla, D. Herpes simplex virus-1 fine-tunes host’s autophagic response to infection: A comprehensive analysis in productive infection models. PLoS ONE 2015, 10, e0124646. [Google Scholar] [CrossRef]
- Xu, P.; Roizman, B. The SP100 component of ND10 enhances accumulation of PML and suppresses replication and the assembly of HSV replication compartments. Proc. Natl. Acad. Sci. USA 2017, 114, E3823–E3829. [Google Scholar] [CrossRef]
- Rodriguez, M.C.; Dybas, J.M.; Hughes, J.; Weitzman, M.D.; Boutell, C. The HSV-1 ubiquitin ligase ICP0: Modifying the cellular proteome to promote infection. Virus Res. 2020, 285, 198015. [Google Scholar] [CrossRef] [PubMed]
- Stamminger, T.; Tavalai, N. Interplay between herpesvirus infection and host defense by PML Nuclear Bodies. Viruses 2009, 1, 1240–1264. [Google Scholar]
- Wang, S.; Long, J.; Zheng, C. The potential link between PML NBs and ICP0 in regulating lytic and latent infection of HSV-1. Protein Cell 2012, 3, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Stamminger, T.; Scherer, M. Emerging role of PML Nuclear Bodies in innate immune signaling. J. Virol. 2016, 90, 5850–5854. [Google Scholar]
- Cohen, C.; Corpet, A.; Roubille, S.; Maroui, M.A.; Poccardi, N.; Rousseau, A.; Kleijwegt, C.; Binda, O.; Texier, P.; Sawtell, N.; et al. Promyelocytic leukemia (PML) nuclear bodies (NBs) induce latent/quiescent HSV-1 genomes chromatinization through a PML NB/Histone H3.3/H3.3 chaperone axis. PLoS Pathog. 2018, 14, e1007313. [Google Scholar] [CrossRef]
- Rai, T.S.; Glass, M.; Cole, J.J.; Rather, M.I.; Marsden, M.; Neilson, M.; Brock, C.; Humphreys, I.R.; Everett, R.D.; Adams, P.D. Histone chaperone HIRA deposits histone H3.3 onto foreign viral DNA and contributes to anti-viral intrinsic immunity. Nucleic Acids Res. 2017, 45, 11673–11683. [Google Scholar] [CrossRef]
- Cabral, J.M.; Oh, H.S.; Knipe, D.M. ATRX promotes maintenance of herpes simplex virus heterochromatin during chromatin stress. eLife 2018, 7, e40228. [Google Scholar] [CrossRef]
- McFarlane, S.; Orr, A.; Roberts, A.P.E.; Conn, K.L.; Llier, V.; Loney, C.; Filipe, A.S.; Smollett, K.; Gu, Q.; Robertson, N.; et al. The histone chaperon HIRA promotes the induction of host innate immune defenses in response to HSV-1 infection. PLoS Pathog. 2019, 15, e1007667. [Google Scholar] [CrossRef]
- Sahin, U.; Lallemand-Breitenbach, V.; de The, H. PML nuclear bodies: Regulation, function and therapeutic perspectives. J. Pathol. 2014, 234, 289–291. [Google Scholar] [CrossRef]
- Cuchet-Lourenco, D.; Boutell, C.; Lukashchuk, V.; Grant, K.; Sykes, A.; Murray, J.; Orr, A.; Everett, R.D. SUMO pathway dependent recruitment of cellular repressors to herpes simplex virus type 1 genomes. PLoS Pathog. 2011, 7, e1002123. [Google Scholar] [CrossRef]
- Everett, R.D.; Boutell, C.; Pheasant, K.; Cuchet-Lourenco, D.; Orr, A. Sequences related to SUMO interaction motifs in herpes simplex virus 1 protein ICP0 act cooperatively to stimulate virus infection. J. Virol. 2013, 88, 2763–2774. [Google Scholar] [CrossRef]
- Zheng, Y.; Gu, H. Identification of three redundant segments responsible for herpes simplex virus ICP0 to fuse with ND10 nuclear bodies. J. Virol. 2015, 89, 4214–4226. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zheng, Y.; Sumrat, S.K.; Gu, H. A tale of two PMLs: Elements regulating a differential substrate recognition by the ICP0 E3 ubiquitin ligase of herpes simplex virus 1. J. Virol. 2016, 90, 10875–10885. [Google Scholar] [CrossRef] [PubMed]
- Hembram, D.S.S.; Negi, H.; Biswas, P.; Tripathi, V.; Bhushan, L.; Shet, D.; Kumar, V.; Das, R. The viral SUMO-targeted ubiquitin ligase ICP0 Is phosphorylated and activated by host kinase Chk2. J. Mol. Biol. 2020, 432, 1952–1977. [Google Scholar] [CrossRef] [PubMed]
- Fada, J.B.; Kaadi, E.; Samrat, S.; Zheng, Y.; Gu, H. Regulations of SUMO-SIM Interaction on the ICP0-mediated degradation of PML Isoform II and its associated proteins in HSV-1 infection. J. Virol. 2020. [Google Scholar] [CrossRef]
- Xu, P.; Mallon, S.; Roizman, B. PML plays both inimical and beneficial roles in HSV-1 replication. Proc. Natl. Acad. Sci. USA 2016, 113, E3022–E3028. [Google Scholar] [CrossRef]
- Full, F.; Ensser, A. Early nuclear events after herpesviral infection. J. Clin. Med. 2019, 8, 1408. [Google Scholar] [CrossRef]
- He, S.; Han, J. Manipulation of host cell death pathways by herpes simplex virus. Curr. Top. Microbiol. Immunol. 2020. [Google Scholar] [CrossRef]
- He, Q.; Liu, H.; Haung, C.; Wang, R.; Luo, M.; Lu, W. Herpes simplex virus 1-induced blood-brain barrier damage involves apoptosis associated with GM130-mediated golgi stress. Front. Mol. Neurosci. 2020, 13, 2. [Google Scholar] [CrossRef]
- Li, H.; Gao, Q.; Shao, Y.; Sun, B.; Wang, F.; Qiao, Y.; Wang, N.; Liu, S. Gallid herpesvirus 1 initiates apoptosis is uninfected cells through paracrine repression of p53. J. Virol. 2018, 92, e00529-18. [Google Scholar] [CrossRef]
- Jaggi, U.; Matundan, H.H.; Tormanen, K.; Wang, S.; Yu, J.; Mott, K.R.; Ghiasi, H. Expression of murine CD80 by herpes simplex virus 1 in place of latency-associated transcript (LAT) can compensate for latency reactivation and anti-apoptotic functions of LAT. J. Virol. 2020, 94, e01798-19. [Google Scholar] [CrossRef] [PubMed]
- Adlakha, M.; Livingston, C.M.; Bezsonva, I.; Weller, S.K. The herpes simplex virus 1 immediate early protein ICP22 is a functional mimic of a cellular J protein. J. Virol. 2020, 94, e01564-19. [Google Scholar] [CrossRef] [PubMed]
- Aubert, M.; Krantz, E.M.; Jerome, K.R. Herpes simplex virus genes, Us3, Us5, and Us12 differentially regulate cytotoxic T lymphocyte -induced cytotoxicity. Viral Immunol. 2006, 19, 391–408. [Google Scholar] [CrossRef] [PubMed]
- Smith, G. Herpesvirus transport to the nervous system and back again. Annu. Rev. Microbiol. 2012, 66, 153–176. [Google Scholar] [CrossRef] [PubMed]
- Lahmidi, S.; Strunk, U.; Smiley, J.R.; Pearson, A.; Duplay, P. Herpes simplex virus 1 infection of T cells causes VP11/12-dependent phosphorylation and degradation of the cellular protein Dok-2. Virology 2017, 511, 66–73. [Google Scholar] [CrossRef]
- Huang, J.; You, H.; Su, C.; Li, Y.; Chen, S.; Zheng, C. Herpes simplex virus 1 tegument protein VP22 abrogates cGAS/STING-mediated antiviral innateimmunity. J. Virol. 2018, 92, e00841-18. [Google Scholar] [CrossRef]
- Pan, S.; Liu, X.; Ma, Y.; Cao, Y.; He, B. Herpes simplex virus 1 γ134.5 protein inhibits STING activation that restricts viral replication. J. Virol. 2018, 9, e01015-18. [Google Scholar] [CrossRef]
- Su, C.; Zheng, C. Herpes simplex virus 1 abrogates the cGAS/STING-mediated cytosolic DNA-sensing pathway via its virion host shutoff protein, UL41. J. Virol. 2017, 91, e02414-16. [Google Scholar] [CrossRef]
- Martin, C.; Leyton, L.; Hott, M.; Arancibia, Y.; Spichiger, C.; McNiven, M.A.; Court, F.A.; Concha, M.I.; Burgos, P.V.; Otth, C. Herpes simplex virus type 1 neuronal infection perturbs golgi apparatus integrity through activation of Src tyrosine kinase and Dyn-2 GTPase. Front. Cell. Infect. Microbiol. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Koyanagi, N.; Imai, T.; Shindo, K.; Sato, A.; Fujii, W.; Ichinohe, T.; Takemura, N.; Kakuta, S.; Uematsu, S.; Kiyono, H. Herpes simplex virus 1 evasion of CD8+ T cell accumulation contributes to viral encephalitis. J. Clin. Investig. 2017, 127, 3784–3795. [Google Scholar] [CrossRef]
- Zhang, D.; Su, C.; Zheng, C. Herpes simplex virus 1 serine protease VP24 blocks the DNA-sensing signal pathway by abrogating activation of interferon regulatory factor 3. J. Virol. 2016, 90, 5824–5829. [Google Scholar] [CrossRef] [PubMed]
- Xiong, R.; Rao, P.; Kim, S.; Li, M.; Wen, X.; Yuan, W. Herpes simplex virus 1 US3 phosphorylates cellular KIF3A to downregulate CD1d expression. J. Virol. 2015, 89, 6646–6655. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Su, C.; Zheng, C. Herpes simplex virus 1 tegument protein UL41 counteracts IFIT3 antiviral innate immunity. J. Virol. 2016, 90, 11056–11061. [Google Scholar] [CrossRef] [PubMed]
- Rao, P.; Wen, X.; Lo, J.H.; Kim, S.; Li, X.; Chen, S.; Feng, X.; Akbari, O.; Yuan, W. Herpes simplex virus 1 specifically targets human CD1d antigen presentation to enhance its pathogenicity. J. Virol. 2018, 92, e01490-18. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Su, C. Herpes simplex virus 1 infection dampens the immediate early antiviral innate immunity signaling from peroxisomes by tegument protein VP16. Virol. J. 2017, 14, 35. [Google Scholar] [CrossRef]
- Liu, Z.; Qin, Q.; Wu, C.; Li, H.; Shou, J.; Yang, Y.; Gu, M.; Ma, C.; Lin, W.; Zou, Y.; et al. Downregulated NDRI protein kinase inhibits innate immune response by initiating an miRNA146a-STAT1 feedback loop. Nat. Commun. 2018, 9, 1–16. [Google Scholar]
- Liu, Y.; Yang, H.L.; Zhong, F.F.; Fan, J.Y. Anti-apoptotic function of herpes simplex virus-2 latency-associated transcript RL1 sequence and screening of its encoded microRNAs. Clin. Exp. Dermatol. 2016, 41, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Kuang, L.; Deng, Y.; Liu, X.; Zou, Z.; Mi, L. Differential expression of mRNA and miRNA in guinea pigs following infection with HSV2v. Exp. Ther. Med. 2017, 14, 2577–2583. [Google Scholar] [CrossRef]
- Zhang, Y.; Dai, J.; Tang, T.; Zhou, L.; Zhou, M. MicroRNA-649 promotes HSV-1 replication by directly targeting MALT1. J. Med. Virol. 2017, 89, 1069–1079. [Google Scholar] [CrossRef]
- Bhela, S.; Mulik, S.; Gimenez, F.; Reddy, P.B.J.; Richardson, R.L.; Varanasi, S.K.; Jaggi, U.; Xu, J.; Lu, P.Y.; Rouse, B.T. Role of miRNA-155 in the pathogenesis of herpetic stromal keratitis. Am. J. Pathol. 2015, 185, 1073–1084. [Google Scholar] [CrossRef]
- Enk, J.; Levi, A.; Weisblum, Y.; Yamin, R.; Charpak-Amikam, Y.; Wolf, D.G.; Mandelboim, O. HSV1 microRNA modulation of GPI anchoring and downstream immune evasion. Cell Rep. 2016, 17, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Ru, J.; Sun, H.; Fan, H.; Wang, C.; Li, Y.; Liu, M.; Tang, H. MiR-23α facilitates the replication of HSV-1 through the suppression of the interferon regulatory factor 1. PLoS ONE 2014, 9, e114021. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Cui, S.; Li, Y.; Yang, G.; Wang, P.; Fikrig, E.; You, F. MiR-221 negatively regulates innate antiviral response. PLoS ONE 2018, 13, e0200385. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, J.; Liu, Y.; Fan, J.; Yang, H. HSV-2-encoded miRNA-H4 regulates cell cycle progression and Act-D-induced apoptosis in HeLa cells by targeting CDKL2 and CDKN2A. Virol. Sin. 2019, 34, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Shabani, M.; Esfahani, B.N.; Ehdaei, B.S.; Moghim, S.; Mirzaei, A.; Sharifi, M.; Mouhebat, L. Inhibition of herpes simplex virus type 1 replication by novel has-miR-7704 in vitro. Res. Pharm. Sci. 2019, 14, 167–174. [Google Scholar]
- Wang, L.; Chen, X.; Zhou, X.; Roizman, B.; Zhou, G.G. miRNAs targeting ICP4 and delivered to susceptible cells in exosomes block HSV-1 replication in a dose-dependent manner. Mol. Ther. 2018, 26, 1032–1039. [Google Scholar] [CrossRef]
- Wang, X.; Diao, C.; Yang, X.; Yang, Z.; Liu, M.; Li, X.; Tang, H. ICP4-induced miR-101 attenuates HSV- replication. Sci. Rep. 2016, 6, 23205. [Google Scholar] [CrossRef]
- Bernstein, D.I.; Wald, A.; Warren, T.; Fife, K.; Tyring, S.; Lee, P.; Wagoner, N.V.; Margaret, A.; Flechtner, J.B.; Tasker, S.; et al. Therapeutic vaccine for genital herpes simplex virus-2 infection: Findings from a randomized trial. J. Infect. Dis. 2017, 215, 856–864. [Google Scholar] [CrossRef]
- van Wagoner, N.; Fife, K.; Leone, P.A.; Bernstein, D.I.; Warren, T.; Panther, L.; Novak, R.M.; Beigi, R.; Kriesel, J.; Tyring, S.; et al. Effect of different doses of GEN-003, a therapeutic vaccine for genital herpes simplex virus-2, on viral shedding and lesions: Results of a randomized placebo-controlled trial. J. Infect. Dis. 2018, 218, 1890–1899. [Google Scholar] [CrossRef]
- Bernard, M.C.; Barban, V.; Pradezynski, F.; deMontfort, A.; Ryall, R.; Caillet, C.; Londono-Hayes, P. Immunogenicity, protective efficacy, and non-replicative status of HSV-2 vaccine candidate HSV529 in mice and guinea pigs. PLoS ONE 2015, 10, e0121518. [Google Scholar] [CrossRef]
- Dropulic, L.K.; Oestreich, M.C.; Pietz, H.L.; Laing, K.J.; Hunsberger, S.; Lumbard, K.; Garabedian, D.; Turk, S.P.; Chen, A.; Hornung, R.L.; et al. A randomized, double-blinded, placebo-controlled, phase I study of a replication-defective herpes simplex virus (HSV) type 2 vaccine, HSV529, in adults with or without HSV infection. J. Infect. Dis. 2019, 220, 990–1000. [Google Scholar] [CrossRef]
- Delagrave, S.; Hernandez, H.; Zhou, C.; Hamberger, J.F.; Mundle, S.T.; Catalan, J.; Baloglu, S.; Anderson, S.F.; DiNapoli, J.M.; Londono-Hayes, P.; et al. Immunogenicity and efficacy of intramuscular replication-defective and subunit vaccines against herpes simplex virus type 2 in the mouse genital model. PLoS ONE 2012, 7, e46714. [Google Scholar] [CrossRef]
- Diaz, F.M.; Knipe, D.M. Protection from genital herpes disease, seroconversion, and latent infection in a non-lethal murine genital infection model by immunization with an HSV-2 replication-defective mutant virus. Virology 2016, 488, 61–67. [Google Scholar] [CrossRef]
- Diaz, F.; Gregory, S.; Nakashima, H.; Viapiano, M.S.; Knipe, D.M. Intramuscular delivery of replication-defective herpes simplex virus gives antigen expression in muscle syncitia and improved protection against pathogenic HSV-2 strains. Virology 2018, 513, 129–135. [Google Scholar] [CrossRef]
- Chandra, J.; Woo, W.P.; Dutton, J.L.; Xu, Y.; Li, B.; Kinrade, S.; Druce, J.; Finlayson, N.; Griffin, P.; Laing, K.J.; et al. Immune response to a HSV-2 polynucleotide immunotherapy COR-1 in HSV-2 positive subjects: A randomized double blinded phase I/IIa trial. PLoS ONE 2019, 14, e0226320. [Google Scholar] [CrossRef]
- Dutton, J.L.; Woo, W.P.; Chandra, J.; Xu, Y.; Li, B.; Finlayson, N.; Griffin, P.; Frazer, I.H. An escalating dose study to assess the safety, tolerability and immunogenicity of a herpes simplex virus DNA vaccine COR-1. Hum. Vaccines Immunother. 2016, 12, 3079–3088. [Google Scholar] [CrossRef]
- Awasthi, S.; Hook, L.M.; Shaw, C.E.; Pahar, B.; Stagray, J.A.; Liu, D.; Veazey, R.S.; Friedman, H.M. An HSV-2 trivalent vaccine is immunogenic in Rhesus Macaques and highly efficacious in guinea pigs. PLoS Pathog. 2017, 13, e1006141. [Google Scholar] [CrossRef]
- Hook, L.M.; Awasthi, S.; Dubin, J.; Flechtner, J.; Long, D.; Friedman, H.M. A trivalent gC2/gD2/gE2 vaccine for herpes simplex virus generates antibody responses that block immune evasion domains on gC2 better than natural infection. Vaccine 2019, 37, 664–669. [Google Scholar] [CrossRef]
- Bourne, N.; Bravo, F.J.; Francotte, M.; Bernstein, D.I.; Myers, M.G.; Slaoui, M.; Stanberry, L.R. Herpes simplex virus (HSV) type 2 glycoprotein D subunit vaccine and protection against genital HSV-1 or HSV-2 disease in guinea pigs. J. Infect. Dis. 2003, 187, 542–549. [Google Scholar] [CrossRef]
- Leroux-Roels, G.; Clement, F.; Vande-Papeliere, P.; Fourneau, M.; Heineman, T.C.; Dublin, G. Immunogenicity and safety of different formulations of an adjuvanted glycoprotein D genital herpes vaccine in healthy adults: A double-blind randomized trial. Hum. Vaccines Immunother. 2013, 9, 1254–1262. [Google Scholar] [CrossRef]
- Hook, L.M.; Cairns, T.M.; Awasthi, S.; Brooks, B.D.; Ditto, N.T.; Eisenberg, R.J.; Cohen, G.H.; Friedman, H.M. Vaccine-induced antibodies to herpes simplex virus glycoprotein D epitopes involved in virus entry and cell-to-cell spread correlate with protection against genital disease in guinea pigs. PLoS Pathog. 2018, 14, e1007095. [Google Scholar] [CrossRef]
- Group, H.S.V.S.; Abu-Elyazeed, R.R.; Heineman, T.; Dublin, G.; Fourneau, M.; Leroux-Roels, I.; Leroux-Roels, G.; Richardus, J.H.; Ostergaard, L.; Diez-Domingo, J.; et al. Safety and immunogenicity of a glycoprotein D genital herpes vaccine in healthy girls 10–17 years of age: Results from a randomized, controlled, double-blind trial. Vaccine 2013, 31, 6136–6143. [Google Scholar]
- Awasthi, S.; Berthe, R.B.; Friedman, M. Better neutralization of herpes simplex virus type 1 (HSV-1) than HSV-2 by antibody from recipients of GlaxoSmithKline HSV-2 glycoprotein D2 subunit vaccine. J. Infect. Dis. 2014, 210, 571–575. [Google Scholar] [CrossRef]
- Davido, D.J.; Tu, E.M.; Wang, H.; Korom, M.; Casals, A.G.; Reddy, P.J.; Mostafa, H.H.; Combs, B.; Haenchen, S.D.; Morrison, L.A. Attenuated herpes simplex virus 1 (HSV-1) expressing a mutant form of ICP6 stimulates a strong immune response that protects mice against HSV-1 induced corneal disease. J. Virol. 2018, 92, e01036-18. [Google Scholar] [CrossRef]
- Mostafa, H.H.; Thompson, T.W.; Konen, A.J.; Haenchen, S.D.; Hillard, J.G.; MacDonald, S.J.; Morrison, L.A.; Davido, D.J. Herpes simplex virus 1 mutant with point mutations in UL39 is impaired for acute viral replication in mice, establishment of latency, and explant-induced reactivation. J. Virol. 2018, 92, e01654-17. [Google Scholar] [CrossRef]
- Akhrameyeva, N.V.; Zhang, P.; Sugiyama, N.; Behar, S.M.; Yao, F. Development of a glycoprotein D-expressing dominant-negative and replication-defective herpes simplex virus 2 (HSV-2) recombinant viral vaccine against HSV-2 infection in mice. J. Virol. 2011, 85, 5036–5047. [Google Scholar] [CrossRef]
- Zhang, P.; Xie, L.; Balliet, J.W.; Casimiro, D.R.; Yao, F. A herpes simplex virus 2 (HSV-2) glycoprotein D-expressing non-replicating dominant-negative HSV-2 virus vaccine is superior to a gD2 subunit vaccine against HSV-2 genital infection in guinea pigs. PLoS ONE 2014, 9, e101373. [Google Scholar]
- Mo, A.; Musselli, C.; Chen, H.; Pappas, J.; Leclair, K.; Liu, A.; Chicz, R.M.; Truneh, A.; Monks, S.; Levey, D.L.; et al. A heat shock protein based polyvalent vaccine targeting HSV-2: CD4(+) and CD8(+) cellular immunity and protective efficacy. Vaccine 2011, 29, 8530–8541. [Google Scholar] [CrossRef]
- Wald, A.; Koelle, D.M.; Fife, K.; Warren, T.; Leclair, K.; Chicz, R.M.; Monks, S.; Levey, D.L.; Musselli, C.; Srivastava, P.K. Safety and immunogenicity of long HSV-2 peptides complexed with rhHsc70 in HSV-2 seropositive persons. Vaccine 2011, 39, 8520–8529. [Google Scholar] [CrossRef]
- Odegard, J.M.; Flynn, P.A.; Campbell, D.J.; Robbins, S.H.; Dong, L.; Wang, K.; Ter Meulen, J.; Cohen, J.I.; Koelle, D.M. A novel HSV-2 subunit vaccine induces GLA-dependent CD4 and CD8 T cell responses and protective immunity in mice and guinea pigs. Vaccine 2016, 34, 101–109. [Google Scholar] [CrossRef]
- Stanfield, B.A.; Stahl, J.; Chouljenko, V.N.; Subramanian, R.; Charles, A.-S.; Saied, A.A.; Walker, J.D.; Kousoulas, K.G. A single intramuscular vaccination of mice with the HSV-1 VC2 virus with mutations in the glycoprotein k and the membrane protein UL20 confers full protection against lethal intravaginal challenge with virulent HSV-1 and HSV-2 strains. PLoS ONE 2014, 9, e109890. [Google Scholar] [CrossRef] [PubMed]
- Stanfield, B.A.; Pahar, B.; Chouljenko, V.N.; Veazey, R.; Kousoulas, K.G. Vaccination of rhesus macaques with the live-attenuated HSV-1 vaccine VC2 stimulates the proliferation of mucosal T cells and germinal center responses resulting in sustained production of highly neutralizing antibodies. Vaccine 2017, 35, 536–543. [Google Scholar] [CrossRef]
- Bernstein, D.I.; Pullum, D.A.; Cardin, R.D.; Bravo, F.J.; Dixon, D.A.; Kousoulas, K.G. The HSV-1 live attenuated VC2 vaccine provides protection against HSV-2 genital infection in the guinea pig model of genital herpes. Vaccine 2019, 37, 61–68. [Google Scholar] [CrossRef]
- Shlapobersky, M.; Marshak, J.O.; Dong, L.; Huang, M.L.; Wei, Q.; Chu, A.; Rolland, A.; Sullivan, S.; Koelle, D.M. Vaxfectin-adjuvinated plasmid DNA vaccine improves protection and immunogenicity in a murine model of genital herpes infection. J. Gen. Virol. 2012, 93, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Vaselenak, R.L.; Shlapobersky, M.; Pyles, R.B.; Wei, Q.; Sullivan, S.M.; Bourne, N. A vaxfectin (®)-adjuvanted HSV-2 plasmid DNA vaccine is effective for prophylactic and therapeutic use in the guinea pig model of genital herpes. Vaccine 2012, 30, 7046–7051. [Google Scholar] [CrossRef] [PubMed]
- Geltz, J.J.; Gershburg, E.; Halford, W.P. Herpes simplex virus 2 (HSV-2) infected cells proteins are among the most dominant antigens of a live-attenuated HSV-2 vaccine. PLoS ONE 2015, 10, e0116091. [Google Scholar] [CrossRef] [PubMed]
- Halford, W.P.; Geltz, J.; Messer, R.J.; Hasenkrug, K.J. Antibodies are required for complete vaccine-induced protection against herpes simplex virus-2. PLoS ONE 2015, 10, e0145228. [Google Scholar] [CrossRef]
- Govander, S.; Harandi, A.M.; Lindqvist, M.; Bergstrom, T.; Liljeqvist, J.A. Glycoprotein G of herpes simplex virus 2 as a novel vaccine antigen for immunity to genital and neurological diseases. J. Virol. 2012, 86, 7544–7553. [Google Scholar] [CrossRef]
- Onnheim, K.; Ekblad, M.; Gorander, S.; Bergstrom, T.; Liljeqvist, J.A. Vaccination with the secreted glycoprotein G of herpes simplex virus 2 induces protective immunity after genital infection. Viruses 2016, 8, 110. [Google Scholar] [CrossRef]
- Awasthi, S.; Zumbrun, E.E.; Si, H.; Wang, F.; Shaw, C.E.; Cai, M.; Lubinski, J.M.; Barrett, S.M.; Balliet, J.W.; Flynn, J.A. Live attenuated herpes simplex virus 2 glycoprotein E deletion mutant as a vaccine candidate defective in neuronal spread. J. Virol. 2012, 86, 4586–4598. [Google Scholar] [CrossRef]
- Cortesi, R.; Ravani, L.; Rinaldi, F.; Marconi, P.; Drechsler, M.; Manservigi, M.; Argnani, R.; Menegatti, E.; Esposito, E.; Manservigi, R. Intranasal immunization in mice with non-ionic surfactants vesicles containing HSV immunogens: A preliminary study as possible vaccine against genital herpes. Int. J. Pharm. 2013, 440, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Dutton, J.L.; Li, B.; Woo, W.P.; Marshak, J.O.; Xu, Y.; Huang, M.L.; Dong, L.; Fraser, I.H.; Koelle, D.M. A novel DNA vaccine technology conveying protection against a lethal herpes simplex viral challenge in mice. PLoS ONE 2013, 8, e76407. [Google Scholar] [CrossRef] [PubMed]
- Koshizuka, T.; Ishioka, K.; Kobayashi, T.; Ikuta, K.; Suzutani, T. Protection from lethal herpes simplex virus type 1 infection by vaccination with a UL41-deficient recombinant strain. Fukushima J. Med. Sci. 2016, 62, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Truong, N.R.; Smith, J.B.; Sandgren, K.J.; Cunningham, A.L. Mechanisms of immune control of mucosal HSV infection: A guide to rational vaccine design. Front. Immunol. 2019, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, Y.; Qihan, L. Characteristics of herpes simplex virus infection and pathogenesis suggest a strategy for vaccine development. Rev. Med. Virol. 2019, 29, e2054. [Google Scholar] [CrossRef]
- Sandgren, K.J.; Truong, N.R.; Smith, J.B.; Bertram, K.; Cunningam, A.L. Vaccines for herpes simplex: Recent progress driven by viral and adjuvant immunology. Methods Mol. Biol. 2020, 2060, 31–56. [Google Scholar]
- Kim, C.H.; Oh, S.D.; Park, H.J.; Kim, H.; Seo, Y.B.; Yoo, J.H.; Jang, S.H.; Shin, J.; Kim, W.C.; Kwon, S.M.; et al. Multivalent DNA vaccine protects against genital herpes by T-cell immune induction in vaginal mucosa. Antivir. Res. 2020, 177, 104755. [Google Scholar] [CrossRef]
- Petro, C.D.; Weinrick, B.; Khajoueinejad, N.; Burn, C.; Sellers, R.; Jacobs, W.R., Jr.; Herold, B.C. HSV-2 ∆gD elicits FcγR-effector antibodies that protect against clinical isolates. JCI Insight 2016, 1, e88529. [Google Scholar] [CrossRef]
- Ratamal-Diaz, A.; Weiss, K.A.; Tognarelli, E.I.; Freire, M.; Bueno, S.M.; Herold, B.C.; Jacobs, W.R., Jr.; Gonzalez, P.A. US6 gene deletion in herpes simplex virus type 2 enhances dendritic cell function and T cell activation. Front. Immunol. 2017, 8, 1523. [Google Scholar] [CrossRef]
- Burn, C.; Ramsey, N.; Garforth, S.J.; Almo, S.; Jacobs, W.R., Jr.; Herold, B.C. A herpes simplex virus (HSV)-2 single-cycle candidate vaccine deleted in glycoprotein D protects male mice from lethal skin challenge with clinical isolates of HSV-1 and HSV-2. J. Infect. Dis. 2018, 217, 754–758. [Google Scholar]
- Kao, C.M.; Goymer, J.; Loh, L.N.; Mahant, A.; Aschner, B.C.; Herold, B.C. Murine model of maternal immunization demonstrates protective role for antibodies that mediate antibody-dependent cellular cytotoxicity in protecting neonates from herpes simplex virus type 1 and type 2. J. Infect. Dis. 2020, 221, 729–738. [Google Scholar] [CrossRef]
- Skoberne, M.; Cardin, R.; Lee, A.; Kazimirova, A.; Zielinski, V.; Garvie, D.; Lundberg, A.; Larson, S.; Bravo, F.J.; Bernstein, D.I.; et al. An adjuvanted herpes simplex virus 2 subunit vaccine elicits a T cell response in mice and is an effective therapeutic vaccine in guinea pigs. J. Virol. 2013, 87, 3930–3940. [Google Scholar] [CrossRef]
- Bernstein, D.I.; Cardin, R.D.; Bravo, F.J.; Hamouda, T.; Pullum, D.A.; Cohen, G.; Bitko, V.; Fattom, A. Intranasal nanoemulsion-adjuvanted HSV-2 subunit vaccine is effective as a prophylactic and therapeutic vaccine using the guinea pig model of genital herpes. Vaccine 2019, 37, 6470–6477. [Google Scholar] [CrossRef]
- Bernstein, D.I.; Morello, C.S.; Cardin, R.D.; Bravo, F.J.; Kraynyak, K.A.; Spector, D.H. A vaccine containing highly purified virus particles in adjuvant provides high level protection against genital infection and disease in guinea pigs challenged intravaginally with homologous and heterogenous strains of herpes simplex virus type 2. Vaccine 2020, 38, 79–89. [Google Scholar] [CrossRef]
- Egan, K.; Hook, L.M.; Latourette, P.; Desmond, A.; Awasthi, S.; Friedman, H.M. Vaccines to prevent genital herpes. Transl. Res. 2020. [Google Scholar] [CrossRef]
| S/N | Name of Vaccine | Type of Vaccine | Antigens | Adjuvants | Mode of Action | Phase of Trial | Company/Institute |
|---|---|---|---|---|---|---|---|
| 1 | GEN-003 [4,108,109] | Therapeutic | gD2, ICP4 | Matrix-M2 | Stimulates both humoral and cellular immune response | Phase II | Genocea Biosciences |
| 2 | HSV529(ACAM 529)/d15-29 [7,110,111,112,113,114] | Prophylactic | Replication-deficient derived from dl5-29 | Not Applicable | Stimulates production of neutralizing antibody and mild CD4+ T-cells | Phase I | Sanofi |
| 3 | COR-1 [115,116] | Therapeutic | HSV-2 DNA | Vaxfectin | Cell-mediated immune response | Phase I/IIa | VGXI Inc. (Texas, USA) under license from Admedus |
| 4 | Trivalent Vaccine [5,117,118] | Therapeutic | gC2, gD2, gE2 | CpG and alum | Blocks virus entry by gD2 and immune evasion by gC2 and gE2. Induces plasma- and mucosa-neutralizing antibodies, stimulates CD4 T cell response | Clinical phase | Harvey M. Friedman Penn Institute of Immunology, University Pennsylvania |
| 5 | gD2 subunit vaccine [6,119,120,121,122,123] | Prophylactic | gD2 | AS04, MPL and alum, | Produces neutralizing antibodies to gD2 | Phase III | Glaxosmithkline |
| 6 | KOS-NA [124,125] | Prophylactic | Mutation in UL39 encoding ICP6 (Live attenuated) | Not applicable | Anti-apoptosis effect as a result of diminished ICP6 protein levels | Pre-clinical | David J. David (University of Kansas, USA) and Lynda Annemarison (St. Louis University, USA) |
| 7 | HSV-2 CJ2-gD2 [126,127] | Prophylactic | Replication defective, expressing gD2 | Not applicable | Elicits neutralizing antibody | Pre-clinical | Department of Surgery and the Department of Medicine Brigham Hospital and Women Hospital and Harvard Medical School, Boston |
| 8 | HerpV [128,129] | Therapeutic | 32 HSV-2 peptides | QS-21 | Elicits CD4+ and CD8+ T cell responses | Completed Phase II | Agenus |
| 9 | G103 [130] | Prophylactic | gD, UL19, UL25 | GLA | Elicits antigen-specific binding and neutralizing antibody responses, including CD4 and CD8 effector and memory T cells | Pre-clinical | Immune-Design (Sanofi) |
| 10 | VC2 [131,132,133] | Prophylactic | Mutations in gK and UL20 (life attenuated) | Not Applicable | Induces humoral and cellular immunity | Pre-clinical | Lousiana State University |
| 11 | Vaxfectin®- gD2/UL46/UL47 [134,135] | Prophylactic | gD, VP11/12, VP13/14 | Vaxfectin | Induces neutralizing antibody and stimulates CD8+ T cells | Phase II | Vical |
| 12 | RR2 [8] | Therapeutic | RR2 protein | CPG and alum | Boosts high neutralizing antibodies, enhance number of functioning IFN-γ | Pre-clinical | - |
| 13 | HSV-2 0∆NLS [136,137] | Prophylactic | Live HSV-2 ICP0- (Live attenuated) | Not applicable | Stimulates the humoral and cellular immune response | Phase I | Rational Vaccines Inc. (RVs) |
| 14 | sgG-2 [138,139] | Prophylactic vaccine candidate | gD | CpG and alum | Stimulates IgG antibody response | Pre-clinical | - |
| 15 | gE2 [140] | Prophylactic | gE | Live attenuated gE deletion mutant | CPG and alum | Stimulates neutralizing antibody | - |
| 16 | gB1s-NISV [141] | Therapeutic | gB | CpG | Generates gB-specific IgG antibody and lymphoproliferative responses | Pre-clinical | - |
| 17 | Codon optimized polynucleotide vaccine [142] | Therapeutic | gD | Plasmid encoded | Induces both B and T cell responses | Phase II | Admedus |
| 18 | VR∆41 [143] | Prophylactic | Live attenuated | Not Applicable | Spreads to the CNS from the site of inoculation, evoke potent immune reaction within the CNS without the induction of lethal encephalitis | Pre-clinical | Fukushima Medical University School of Medicine, Japan |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ike, A.C.; Onu, C.J.; Ononugbo, C.M.; Reward, E.E.; Muo, S.O. Immune Response to Herpes Simplex Virus Infection and Vaccine Development. Vaccines 2020, 8, 302. https://doi.org/10.3390/vaccines8020302
Ike AC, Onu CJ, Ononugbo CM, Reward EE, Muo SO. Immune Response to Herpes Simplex Virus Infection and Vaccine Development. Vaccines. 2020; 8(2):302. https://doi.org/10.3390/vaccines8020302
Chicago/Turabian StyleIke, Anthony C., Chisom J. Onu, Chukwuebuka M. Ononugbo, Eleazar E. Reward, and Sophia O. Muo. 2020. "Immune Response to Herpes Simplex Virus Infection and Vaccine Development" Vaccines 8, no. 2: 302. https://doi.org/10.3390/vaccines8020302
APA StyleIke, A. C., Onu, C. J., Ononugbo, C. M., Reward, E. E., & Muo, S. O. (2020). Immune Response to Herpes Simplex Virus Infection and Vaccine Development. Vaccines, 8(2), 302. https://doi.org/10.3390/vaccines8020302







