State-of-the-Art Technology of Model Organisms for Current Human Medicine
Abstract
1. Introduction
2. Regenerative Medicine
3. Biological Detection and Diagnosis
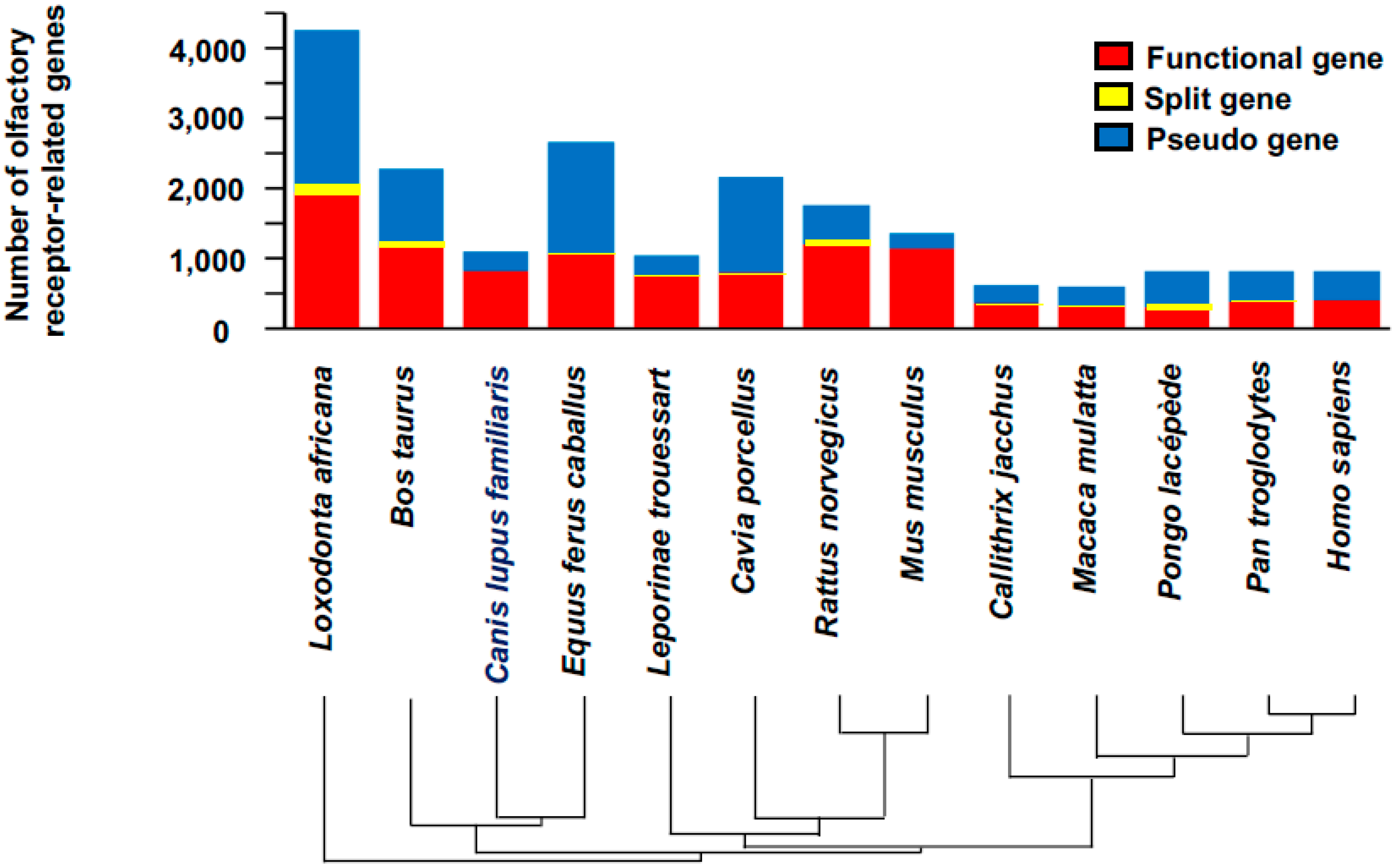
Dogs
4. Caenorhabditis elegans (C. elegans)
5. Discovery of Drug Targets Using Animal Models
5.1. Mus musculus (Mice)
5.2. Rattus norvegicus (Rat)
5.3. Drosophila melanogaster (Fruit Flies)
5.4. Bombyx mori (Silk Moth)
5.5. Schmidtea mediterranea (Flatworms)
5.6. Ciona intestinalis (Sea Vase)
5.7. Strongylocentrotus purpuratus (Sea Urchin)
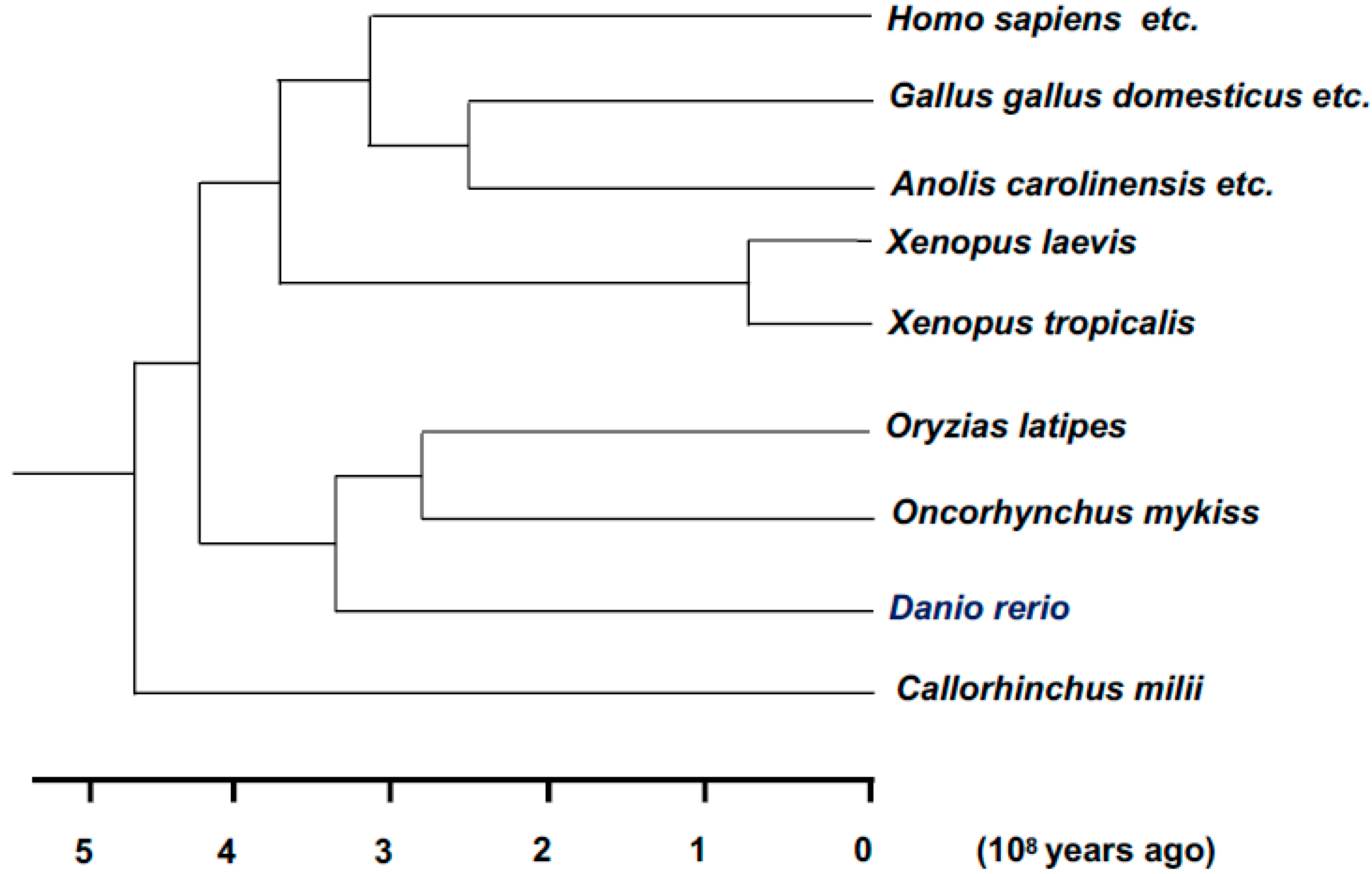
5.8. Xenopus laevis (African Clawed Frog)
5.9. Xenopus tropicalis (Western Clawed Frog)
5.10. Danio rerio (Zebrafish)
5.11. C. elegans
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Yanai, I.; Lercher, M. Renaissance minds in 21st century science. Genome Biol. 2020, 21, 67. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, H. Encyclopædia Britannica, 11th ed.; Schleiden, M.J., Ed.; Cambridge University Press: Cambridge, UK, 1911. [Google Scholar]
- Brotman, D.J.; Deitcher, S.R.; Lip, G.Y.; Matzdorff, A.C. Virchow’s triad revisited. South Med. J. 2004, 97, 213–214. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D.; Crick, F.H.C. Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Kankala, R.K.; Wang, S.B.; Chen, A.Z. Microengineered organ-on-a-chip platforms towards personalized medicine. Curr. Pharm. Des. 2018, 24, 5354–5366. [Google Scholar] [CrossRef] [PubMed]
- Perestrelo, A.R.; Águas, A.C.; Rainer, A.; Forte, G. Microfluidic organ/body-on-a-chip devices at the convergence of biology and microengineering. Sensors 2015, 15, 31142–31170. [Google Scholar] [CrossRef]
- Liu, Y.; Jia, Y.; Yang, K.; Li, R.; Xiao, X.; Wang, Z. Anti-HIV agent azidothymidine decreases Tet(X)-mediated bacterial resistance to tigecycline in Escherichia coli. Commun. Biol. 2020, 3, 1–10. [Google Scholar] [CrossRef]
- Ibhazehiebo, K.; Rho, J.M.; Kurrasch, D.M. Metabolism-based drug discovery in zebrafish: An emerging strategy to uncover new anti-seizure therapies. Neuropharmacology 2020, 167, 107988. [Google Scholar] [CrossRef]
- Wasala, N.B.; Chen, S.J.; Duan, D. Duchenne muscular dystrophy animal models for high-throughput drug discovery and precision medicine. Expert Opin. Drug Discov. 2020, 15, 443–456. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, J.; An, X.; Duan, H. Recent progress on the organic and metal complex-based fluorescent probes for monitoring nitric oxide in living biological systems. Org. Biomol. Chem. 2020, 18, 1522–1549. [Google Scholar] [CrossRef]
- Chen, Y. Recent developments of fluorescent probes for detection and bioimaging of nitric oxide. Nitric Oxide 2020, 98, 1–19. [Google Scholar] [CrossRef]
- Amilan Jose, D.; Sharma, N.; Sakla, R.; Kaushik, R.; Gadiyaram, S. Fluorescent nanoprobes for the sensing of gasotransmitters hydrogen sulfide (H2S), nitric oxide (NO) and carbon monoxide (CO). Methods 2019, 168, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Okita, K.; Ichisaka, T.; Yamanaka, S. Generation of germline-competent induced pluripotent stem cells. Nature 2007, 448, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Hatou, S.; Shimmura, S. Corneal endothelial cell derivation methods from ES/iPS cells. Inflamm. Regen. 2019, 39, 19. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, S.; Sawa, Y. Building a new strategy for treating heart failure using induced pluripotent stem cells. J. Cardiol. 2018, 72, 445–448. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.; Mizuguchi, H. Generation of human pluripotent stem cell-derived hepatocyte-like cells for drug toxicity screening. Drug Metab. Pharmacokinet. 2017, 32, 12–20. [Google Scholar] [CrossRef]
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced pluripotent stem cell technology: A decade of progress. Nat. Rev. Drug Discov. 2017, 16, 115–130. [Google Scholar] [CrossRef]
- Zhou, Q.; Melton, D.A. Pancreas regeneration. Nature 2018, 557, 351–358. [Google Scholar] [CrossRef]
- Kawamoto, K.; Ohashi, T.; Konno, M.; Nishida, N.; Koseki, J.; Matsui, H.; Sakai, D.; Kudo, T.; Eguchi, H.; Satoh, T.; et al. Cell-free culture conditioned medium elicits pancreatic β cell lineage-specific epigenetic reprogramming in mice. Oncol. Lett. 2018, 16, 3255–3259. [Google Scholar] [CrossRef]
- McKimpson, W.M.; Accili, D. Reprogramming cells to make insulin. J. Endocr. Soc. 2019, 3, 1214–1226. [Google Scholar] [CrossRef] [PubMed]
- Vegas, A.J.; Veiseh, O.; Gürtler, M.; Millman, J.R.; Pagliuca, F.W.; Bader, A.R.; Doloff, J.C.; Li, J.; Chen, M.; Olejnik, K.; et al. Long-term glycemic control using polymer-encapsulated human stem cell-derived beta cells in immune-competent mice. Nat. Med. 2016, 22, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Yamaguchi, T.; Hamanaka, S.; Kato-Itoh, M.; Yamazaki, Y.; Ibata, M.; Sato, H.; Lee, Y.S.; Usui, J.; Knisely, A.S.; et al. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell 2010, 142, 787–799. [Google Scholar] [CrossRef]
- Offield, M.F.; Jetton, T.L.; Labosky, P.A.; Ray, M.; Stein, R.W.; Magnuson, M.A.; Hogan, B.L.; Wright, C.V. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 1996, 122, 983–989. [Google Scholar]
- Mori, M.; Furuhashi, K.; Danielsson, J.A.; Hirata, Y.; Kakiuchi, M.; Lin, C.S.; Ohta, M.; Riccio, P.; Takahashi, Y.; Xu, X.; et al. Generation of functional lungs via conditional blastocyst complementation using pluripotent stem cells. Nat. Med. 2019, 25, 1691–1698. [Google Scholar] [CrossRef]
- Matsunari, H.; Nagashima, H.; Watanabe, M.; Umeyama, K.; Nakano, K.; Nagaya, M.; Kobayashi, T.; Yamaguchi, T.; Sumazaki, R.; Herzenberg, L.A.; et al. Blastocyst complementation generates exogenic pancreas in vivo in apancreatic cloned pigs. Proc. Natl. Acad. Sci. USA 2013, 19, 4557–4562. [Google Scholar] [CrossRef] [PubMed]
- Asai, A.; Konno, M.; Kawamoto, K.; Isotani, A.; Mori, M.; Eguchi, H.; Doki, Y.; Arai, T.; Ishii, H. Hereditary pancreatitis model by blastocyst complementation in mouse. Oncotarget 2020, in press. [Google Scholar] [CrossRef]
- Lo, G.K.; Macpherson, K.; MacDonald, H.; Roberts, W.A. A comparative study of memory for olfactory discriminations: Dogs (Canis familiaris), rats (Rattus norvegicus) and humans (Homo sapiens). J. Comp. Psychol. 2019. [Google Scholar] [CrossRef]
- Guerrero-Flores, H.; Apresa-García, T.; Garay-Villar, Ó.; Sánchez-Pérez, A.; Flores-Villegas, D.; Bandera-Calderón, A.; García-Palacios, R.; Rojas-Sánchez, T.; Romero-Morelos, P.; Sánchez-Albor, V.; et al. A non-invasive tool for detecting cervical cancer odor by trained scent dogs. BMC Cancer 2017, 17, 79. [Google Scholar] [CrossRef]
- Seo, I.S.; Lee, H.G.; Koo, B.; Koh, C.S.; Park, H.Y.; Im, C.; Shin, H.C. Cross detection for odor of metabolic waste between breast and colorectal cancer using canine olfaction. PLoS ONE 2018, 13, e0192629. [Google Scholar] [CrossRef]
- Niimura, Y.; Matsui, A.; Touhara, K. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 2014, 24, 1485–1496. [Google Scholar] [CrossRef] [PubMed]
- Hirotsu, T.; Sonoda, H.; Uozumi, T.; Shinden, Y.; Mimori, K.; Maehara, Y.; Ueda, N.; Hamakawa, M. A highly accurate inclusive cancer screening test using Caenorhabditis elegans scent detection. PLoS ONE 2015, 10, e0118699. [Google Scholar] [CrossRef] [PubMed]
- Kusumoto, H.; Tashiro, K.; Shimaoka, S.; Tsukasa, K.; Baba, Y.; Furukawa, S.; Furukawa, J.; Suenaga, T.; Kitazono, M.; Tanaka, S.; et al. Behavioural response alteration in Caenorhabditis elegans to urine after surgical removal of cancer: Nematode-NOSE (N-NOSE) for postoperative evaluation. Biomark. Cancer 2019, 11, 1179299X19896551. [Google Scholar] [CrossRef] [PubMed]
- Kusumoto, H.; Tashiro, K.; Shimaoka, S.; Tsukasa, K.; Baba, Y.; Furukawa, S.; Furukawa, J.; Niihara, T.; Hirotsu, T.; Uozumi, T. Efficiency of gastrointestinal cancer detection by nematode-NOSE (N-NOSE). In Vivo 2020, 34, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Ueda, Y.; Kawamoto, K.; Konno, M.; Noguchi, K.; Kaifuchi, S.; Satoh, T.; Eguchi, H.; Doki, Y.; Hirotsu, T.; Mori, M.; et al. Application of C. elegans cancer screening test for the detection of pancreatic tumor in genetically engineered mice. Oncotarget 2019, 10, 5412–5418. [Google Scholar] [CrossRef] [PubMed]
- Miklos, G.L.G.; Rubin, M.G. The role of the genome project in determining gene function: Insights from model organisms. Cell 1996, 86, 521–529. [Google Scholar] [CrossRef]
- Reinke, V.; White, P.K. Developmental genomic approaches in model organisms. Ann. Rev. Genom. Hum. Gen. 2002, 3, 153–178. [Google Scholar] [CrossRef] [PubMed]
- Aspesi, D.; Pinna, G. Animal models of post-traumatic stress disorder and novel treatment targets. Behav. Pharmacol. 2019, 30, 130–150. [Google Scholar] [CrossRef]
- Montalbano, A.; Canver, M.C.; Sanjana, N.E. High-throughput approaches to pinpoint function within the noncoding genome. Mol. Cell 2017, 68, 44–59. [Google Scholar] [CrossRef]
- Ung, Y.T.; Ong, C.E.; Pan, Y. Current high-throughput approaches of screening modulatory effects of Xenobiotics on cytochrome P450 (CYP) enzymes. High Throughput 2018, 7, 29. [Google Scholar] [CrossRef]
- Markert, L.C. Fertilization of mammalian eggs by sperm injection. J. Exp. Zool. 1983, 228, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Capecchi, M.R. Gene targeting in mice: Functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 2005, 6, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Eshraghi, M.; Ramírez-Jarquín, U.N.; Shahani, N.; Nuzzo, T.; De Rosa, A.; Swarnkar, S.; Galli, N.; Rivera, O.; Tsaprailis, G.; Scharager-Tapia, C.; et al. RasGRP1 is a causal factor in the development of l-DOPA-induced dyskinesia in Parkinson’s disease. Sci. Adv. 2020, 6, eaaz7001. [Google Scholar] [CrossRef] [PubMed]
- Harun-Or-Rashid, M.; Pappenhagen, N.; Zubricky, R.; Coughlin, L.; Jassim, A.H.; Inman, D.M. MCT2 overexpression rescues metabolic vulnerability and protects retinal ganglion cells in two models of glaucoma. Neurobiol. Dis. 2020, 141, 104944. [Google Scholar] [CrossRef] [PubMed]
- Bogomolovas, J.; Feng, W.; Yu, M.D.; Huang, S.; Zhang, L.; Trexler, C.L.; Gu, Y.; Spinozzi, S.; Chen, J. Atypical ALPK2 kinase is not essential for cardiac development and function. Am. J. Physiol. Heart Circ. Physiol. 2020. Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Zan, Y.; Haag, J.D.; Chen, K.S.; Shepel, L.A.; Wigington, D.; Wang, Y.R.; Hu, R.; Lopez-Guajardo, C.C.; Brose, H.L.; Porter, K.I.; et al. Production of knockout rats using ENU mutagenesis and a yeast-based screening assay. Nat. Biotechnol. 2003, 21, 645–651. [Google Scholar] [CrossRef]
- Bainbridge, S.P.; Bownes, M. Staging the metamorphosis of Drosophila melanogaster. J. Embryol. Exp. Morphol. 1981, 66, 57–80. [Google Scholar]
- Thurmond, J.; Goodman, J.L.; Strelets, V.B.; Attrill, H.; Gramates, L.S.; Marygold, S.J.; Matthews, B.B.; Millburn, G.; Antonazzo, G.; Trovisco, V.; et al. The FlyBase Consortium: FlyBase 2.0: The next generation. Nucleic Acid. Res. 2019, 47, D759–D765. [Google Scholar] [CrossRef]
- Ohyama, T.; Schneider-Mizell, C.M.; Fetter, R.D.; Aleman, J.V.; Franconville, R.; Rivera-Alba, M.; Mensh, B.D.; Branson, K.M.; Simpson, J.H.; Truman, J.W.; et al. A multilevel multimodal circuit enhances action selection in Drosophila. Nature 2015, 520, 633–639. [Google Scholar] [CrossRef]
- Takemura, S.Y.; Bharioke, A.; Lu, Z.; Nern, A.; Vitaladevuni, S.; Rivlin, P.K.; Katz, W.T.; Olbris, D.J.; Plaza, S.M.; Winston, P.; et al. A visual motion detection circuit suggested by Drosophila connectomics. Nature 2013, 500, 175–181. [Google Scholar] [CrossRef]
- Venken, K.J.; Simpson, J.H.; Bellen, H.J. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron 2011, 72, 202–230. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Wei, Z.G.; Zhang, Y.Q. Dissolution and regeneration of silk from silkworm Bombyx mori in ionic liquids and its application to medical biomaterials. Int. J. Biol. Macromol. 2020, 143, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, A.A. Planarian regeneration: Its end is its beginning. Cell 2006, 124, 241–245. [Google Scholar] [CrossRef]
- Harada, Y.; Takagaki, Y.; Sunagawa, M.; Saito, T.; Yamada, L.; Taniguchi, H.; Shoguchi, E.; Sawada, H. Mechanism of self-sterility in a hermaphroditic chordate. Science 2008, 3208, 548–550. [Google Scholar] [CrossRef][Green Version]
- Consortium, S.U.G.S. The genome of the sea urchin Strongylocentrotus purpuratus. Science 2006, 314, 941–952. [Google Scholar]
- Blum, M.; Ott, T. Xenopus: An undervalued model organism to study and model human genetic disease. Cells Tissues Organs 2018, 205, 303–313. [Google Scholar] [CrossRef]
- Session, A.M.; Uno, Y.; Kwon, T.; Chapman, J.A.; Toyoda, A.; Takahashi, S.; Fukui, A.; Hikosaka, A.; Suzuki, A.; Kondo, M.; et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 2016, 538, 336–343. [Google Scholar] [CrossRef]
- Shi, Z.; Tian, D.; Xin, H.; Lian, J.; Guo, X.; Chen, Y. Targeted integration of genes in Xenopus tropicalis. Genesis 2017, 55. [Google Scholar] [CrossRef]
- Dahm, R.; Geisler, R. Learning from small fry: The zebrafish as a genetic model organism for aquaculture fish species. Mar. Biotechnol. N. Y. 2006, 8, 329–345. [Google Scholar] [CrossRef]
- Muniandy, Y. The use of larval Zebrafish (Danio rerio) model for Identifying new anxiolytic drugs from herbal medicine. Zebrafish 2018, 15, 321–339. [Google Scholar] [CrossRef]
- Hisaoka, K.K.; Firlit, F.C. Further studies on the embryonic development of the zebrafish, Brachydanio rerio (Hamilton-Buchanan). J. Molphol. 1960, 107, 205–212. [Google Scholar] [CrossRef]
- George, S.; Xia, T.; Rallo, R.; Zhao, Y.; Ji, Z.; Lin, S.; Wang, X.; Zhang, H.; France, B.; Schoenfeld, D.; et al. Use of a high-throughput screening approach coupled with in vivo zebrafish embryo screening to develop hazard ranking for engineered nanomaterials. ACS Nano 2011, 5, 1805–1817. [Google Scholar] [CrossRef]
- Lage, O.M.; Ramos, M.C.; Calisto, R.; Almeida, E.; Vasconcelos, V.; Vicente, F. Current Screening Methodologies in drug discovery for selected human diseases. Mar. Drugs 2018, 16, 279. [Google Scholar] [CrossRef] [PubMed]
- Howe, D.G.; Bradford, Y.M.; Eagle, A.; Fashena, D.; Frazer, K.; Kalita, P.; Mani, P.; Martin, R.; Moxon, S.T.; Paddock, H.; et al. The Zebrafish model organism database: New support for human disease models, mutation details, gene expression phenotypes and searching. Nucleic Acid. Res. 2017, 45, D758–D768. [Google Scholar] [CrossRef]
- MacRae, C.A.; Peterson, R.T. Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 2015, 14, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Swinney, D.C.; Anthony, J. How were new medicines discovered? Nat. Rev. Drug Discov. 2011, 10, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Carretero, M.; Solis, G.M.; Petrascheck, M. C. elegans as model for drug discovery. Curr. Top. Med. Chem. 2017, 17, 2067–2076. [Google Scholar] [CrossRef]
- Shaye, D.D.; Greenwald, I. OrthoList: A compendium of C. elegans genes with human orthologs. PLoS ONE 2011, 6, e20085. [Google Scholar] [CrossRef]
- Leinwand, S.G.; Chalasani, S.H. Neuropeptide signaling remodels chemosensory circuit composition in Caenorhabditis elegans. Nat. Neurosci. 2013, 16, 1461–1467. [Google Scholar] [CrossRef]
- Kauffman, A.L.; Ashraf, J.M.; Corces-Zimmerman, M.R.; Landis, J.N.; Murphy, C.T. Insulin signaling and dietary restriction differentially influence the decline of learning and memory with age. PLoS Biol. 2010, 8, e1000372. [Google Scholar] [CrossRef]
- Ardiel, E.L.; Rankin, C.H. An elegant mind: Learning and memory in Caenorhabditis elegans. Learn. Mem. 2010, 17, 191–201. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konno, M.; Asai, A.; Kitagawa, T.; Yabumoto, M.; Ofusa, K.; Arai, T.; Hirotsu, T.; Doki, Y.; Eguchi, H.; Ishii, H. State-of-the-Art Technology of Model Organisms for Current Human Medicine. Diagnostics 2020, 10, 392. https://doi.org/10.3390/diagnostics10060392
Konno M, Asai A, Kitagawa T, Yabumoto M, Ofusa K, Arai T, Hirotsu T, Doki Y, Eguchi H, Ishii H. State-of-the-Art Technology of Model Organisms for Current Human Medicine. Diagnostics. 2020; 10(6):392. https://doi.org/10.3390/diagnostics10060392
Chicago/Turabian StyleKonno, Masamitsu, Ayumu Asai, Toru Kitagawa, Masami Yabumoto, Ken Ofusa, Takahiro Arai, Takaaki Hirotsu, Yuichiro Doki, Hidetoshi Eguchi, and Hideshi Ishii. 2020. "State-of-the-Art Technology of Model Organisms for Current Human Medicine" Diagnostics 10, no. 6: 392. https://doi.org/10.3390/diagnostics10060392
APA StyleKonno, M., Asai, A., Kitagawa, T., Yabumoto, M., Ofusa, K., Arai, T., Hirotsu, T., Doki, Y., Eguchi, H., & Ishii, H. (2020). State-of-the-Art Technology of Model Organisms for Current Human Medicine. Diagnostics, 10(6), 392. https://doi.org/10.3390/diagnostics10060392




