Alternative Bioremediation Agents against Haloacids, Haloacetates and Chlorpyrifos Using Novel Halogen-Degrading Bacterial Isolates from the Hypersaline Lake Tuz
Abstract
:1. Introduction
2. Results
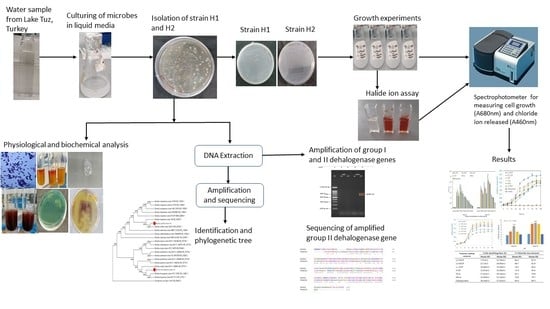
2.1. Enrichment and Isolation of the Bacterial Strains
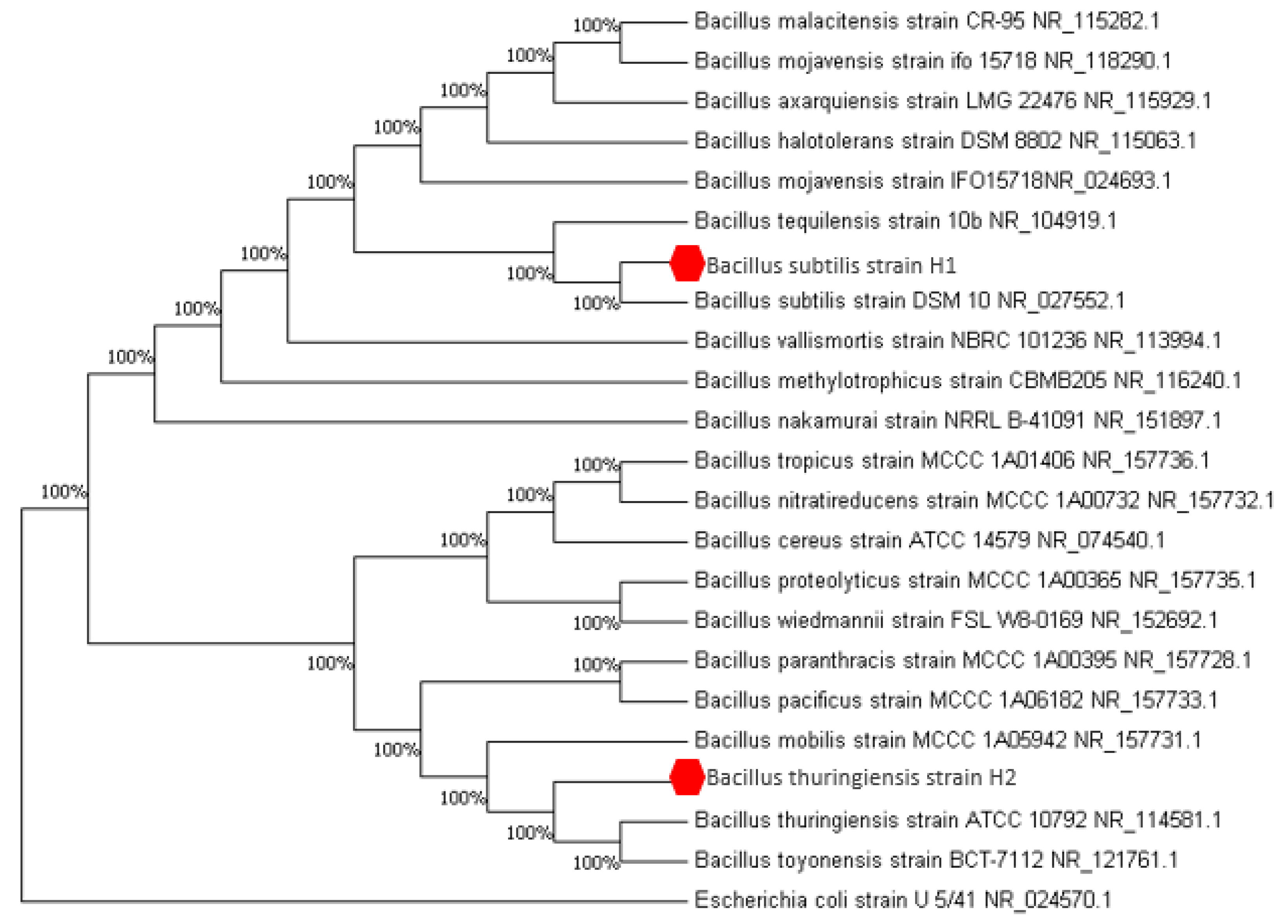
2.2. Bacteria Identification
2.3. Bacterial Growth and Biodegradation Potential of the Isolated Bacteria
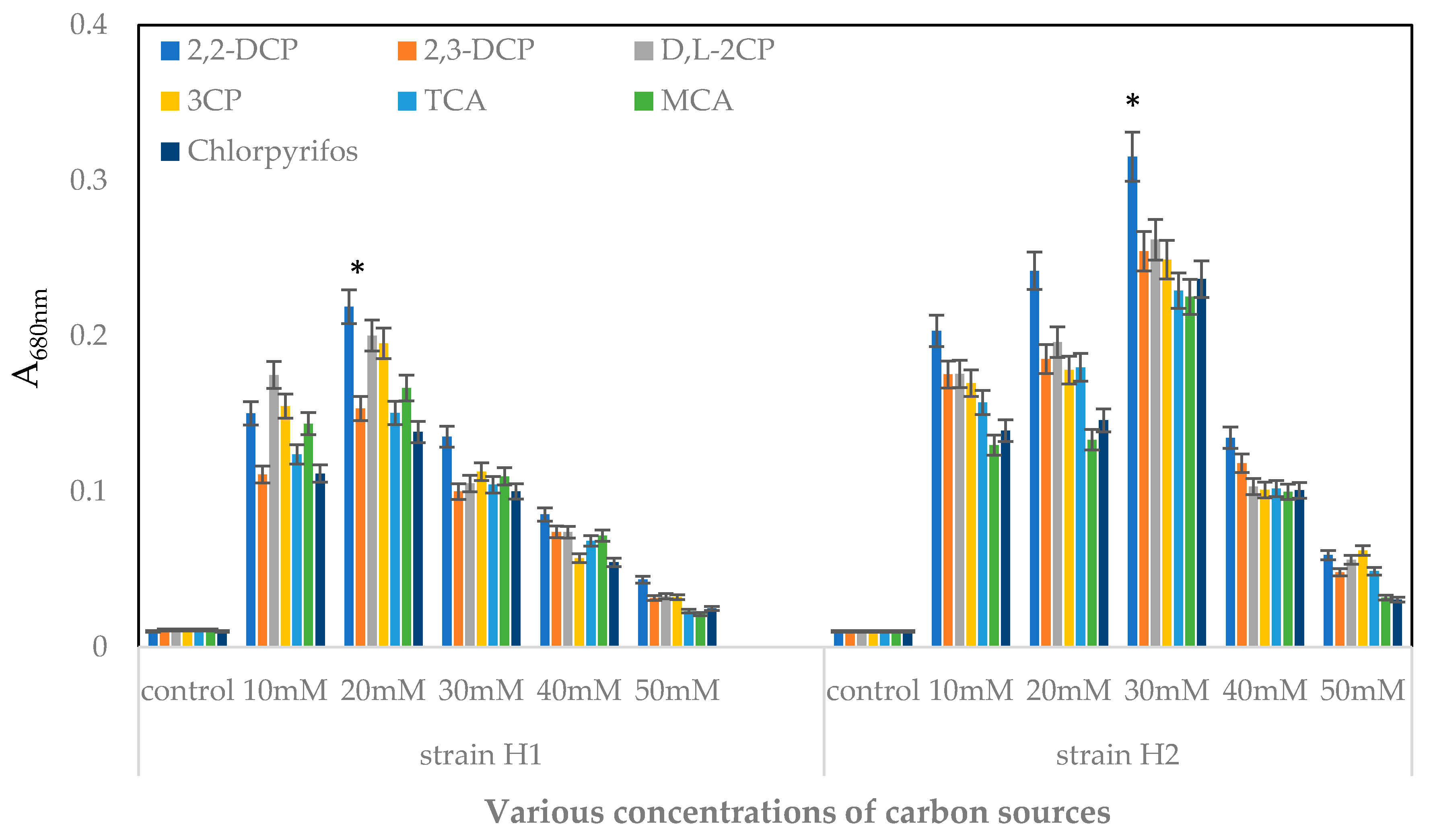
2.3.1. Effects of Various Concentrations of Substrates on Bacterial Growth
2.3.2. Bacterial Growth at Different NaCl Concentrations
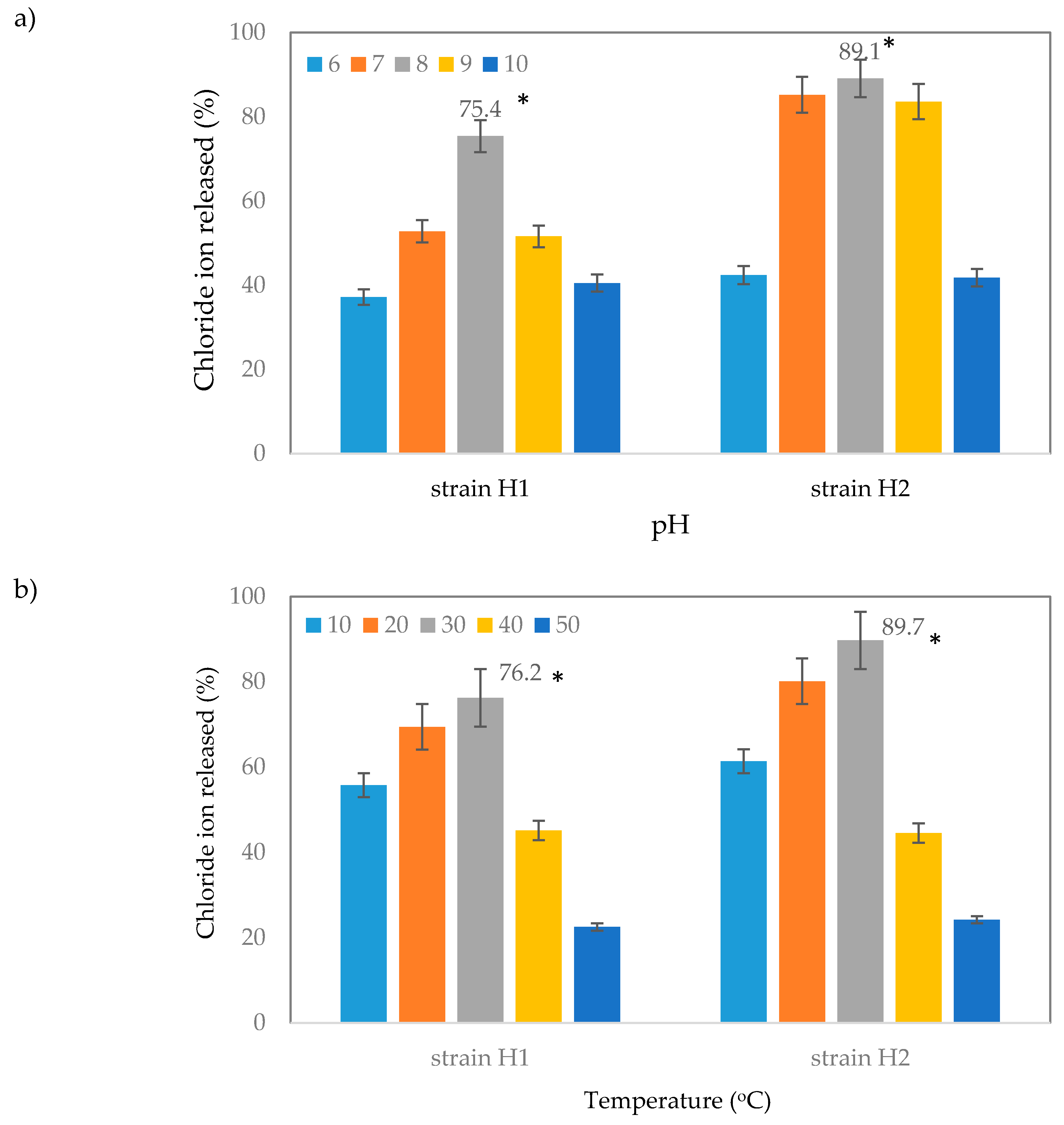
2.3.3. Effect of pH and Temperature on Dehalogenation of 2,2-Dichloropropionic Acid (2,2-DCP) by Strain H1 and H2
2.4. Polymerase Chain Reaction (PCR) Amplification of Putative Dehalogenase Gene
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Chemicals and Media Preparation
4.3. Enrichment and Isolation of Dehalogenase-Producing Bacteria
4.4. Identification of Dehalogenase-Producing Bacteria
4.4.1. Morphological and Biochemical Characterization
4.4.2. Molecular Identification
DNA Extraction, PCR Amplification of the 16S rRNA and Gene Sequencing of Bacteria Strains
Phylogenetic Analysis of 16S rRNA Gene
4.5. Bacteria Growth and Halide Ion Assay
4.5.1. Effects of Various Concentrations of Carbon Source on the Bacterial Growth
4.5.2. Effect of Salinity on Bacterial Growth
4.5.3. Effect of pH and Temperature on the Bacterial Growth and Substrate Degradation
4.6. Amplification of Putative Dehalogenase Gene using Group I and Group II Primers
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Nion, Y.A.; Toyota, K. Recent trends in control methods for bacterial wilt diseases caused by Ralstonia solanacearum. Microbes Environ. 2015, 30. [Google Scholar] [CrossRef] [Green Version]
- Khalili, E.; Javed, M.A.; Huyop, F.; Wahab, R.A. Efficacy and cost study of green fungicide formulated from crude beta-glucosidase. Int. J. Environ. Sci. Technol. 2019, 16, 4503–4518. [Google Scholar] [CrossRef]
- Douglas, S.H.; Dixon, B.; Griffin, D. Assessing the abilities of intrinsic and specific vulnerability models to indicate groundwater vulnerability to groups of similar pesticides: A comparative study. Phys. Geogr. 2018, 39, 487–505. [Google Scholar] [CrossRef]
- Lu, J.; Yan, X.; Ma, Y.F.; Tian, C.X.; Ding, J.C. Impact of salinity on treatment of saline wastewater by sequencing batch biofilm reactor process. J. Central South Uni. 2014, 21, 1989–1994. [Google Scholar] [CrossRef]
- Lefebvre, O.; Moletta, R. Treatment of organic pollution in industrial saline wastewater: A literature review. Water Res. 2006, 40, 671–3682. [Google Scholar] [CrossRef] [PubMed]
- Aloui, F.; Khoufi, S.; Loukil, S.; Sayadi, S. Performances of an activated sludge process for the treatment of fish processing saline wastewater. Desalination 2009, 246, 389–396. [Google Scholar] [CrossRef]
- Nemati, M.; Abdulghader, M.F.; Gicana, R.G.; Lamis, R.J.S.; Ibrahim, N.; Hamid, A.A.A.; Huyop, F. Identification of putative cof-like hydrolase associated with dehalogenase in enterobacter cloacae mn1 isolated from the contaminated sea-side area of the Philippines. Malays. J. Microbiol. 2013, 9, 253–259. [Google Scholar] [CrossRef] [Green Version]
- Edbeib, M.F.; Wahab, R.A.; Kaya, Y.; Huyop, F. In silico characterization of a novel dehalogenase (DehHX) from the halophile Pseudomonas halophila HX isolated from Tuz Gölü lake, Turkey: Insights into a hypersaline-adapted dehalogenase. Ann. Microbiol. 2017, 1–12. [Google Scholar] [CrossRef]
- Edbeib, M.F.; Wahab, R.A.; Huyop, F. Characterization of an α-haloalkanoic acid–degrading Pseudomonas aeruginosa MX1 isolated from contaminated seawater. Bioremediat. J. 2016, 20, 89–97. [Google Scholar] [CrossRef]
- Abidin, M.H.Z.; Halim, K.B.A.; Huyop, F.; Hamid, T.H.T.A.; Wahab, R.A.; Hamid, A.A.A. The mechanistic role of active site residues in non-stereo haloacid dehalogenase E (DehE). J. Mol. Graph. Model. 2019, 90, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Grattieri, M.; Minteer, S.D. Microbial fuel cells in saline and hypersaline environments: Advancements, challenges and future perspectives. Bioelectrochemistry 2018, 120, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Edbeib, M.F.; Wahab, R.A.; Huyop, F. Halophiles: Biology, adaptation, and their role in decontamination of hypersaline environments. World J. Microbiol. Biotechnol. 2016, 32, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Le Borgne, S.; Paniagua, D.; Vazquez-Duhalt, R. Biodegradation of organic pollutants by halophilic bacteria and archaea. J. Mol. Microbiol. Biotechnol. 2008, 15, 74–92. [Google Scholar] [CrossRef] [PubMed]
- Vymazal, J.; Březinová, T. Long term treatment performance of constructed wetlands for wastewater treatment in mountain areas: Four case studies from the Czech Republic. Ecol. Eng. 2014, 71, 578–583. [Google Scholar] [CrossRef]
- Erin, A.G.; Fenical, W.; Jensen, P.R. Phylogenetic Diversity of Gram-Positive Bacteria Cultured from Marine Sediments. Appl. Environ. Microbiol. 2007, 73, 3272–3282. [Google Scholar]
- Hashem, A.; Tabassum, B.; Abd_Allah, E.F. Bacillus subtilis: A plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi J. Biol. Sci. 2019, 26, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Arai, R.; Kukimoto-Niino, M.; Kuroishi, C.; Bessho, Y.; Shirouzu, M.; Yokoyama, S. Crystal structure of the probable haloacid dehalogenase PH0459 from Pyrococcus horikoshii OT3. Protein Sci. 2006, 15, 373–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thasif, S.; Hamdan, S.; Huyop, F. Degradation of d,l-2-chloropropionic acid by bacterial dehalogenases that shows stereospecificity and its partial enzymatic characteristics. Biotechnology 2009, 8, 264–269. [Google Scholar]
- Zulkifly, A.H.; Roslan, D.D.; Hamid, A.A.A.; Hamdan, S.; Huyop, F. Biodegradation of low concentration of monochloroacetic acid-degrading Bacillus sp. TW1 isolated from Terengganu water treatment and distribution plant. J. Appl. Sci. 2010, 10, 2940–2944. [Google Scholar] [CrossRef]
- Muslem, W.H.; Edbeib, M.F.; Aksoy, H.M.; Kaya, Y.; Hamid, A.A.A.; Hood, M.H.M.; Wahab, R.A.; Huyop, F. Biodegradation of 3-chloropropionic acid (3-CP) by Bacillus cereus WH2 and its in silico enzyme-substrate docking analysis. J. Biomol. Struct. Dyn. 2019, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bollag, J.M.; Alexander, M. Bacterial dekalogenation of chlorinated aliphatic acids. Soil. Biol. Biochem. 1971, 3, 91–96. [Google Scholar] [CrossRef]
- Mesri, S.; Wahab, R.A.; Huyop, F. Degradation of 3-chloropropionic acid (3CP) by Pseudomonas sp. B6p isolated from a rice paddy field. Ann. Microbiol. 2009, 59, 447–451. [Google Scholar] [CrossRef]
- Jing, N.H.; Wahab, R.A.; Taha, A.M.; Rashid, N.A.A.; Huyop, F. A further characterization of 3-chloropropionic acid dehalogenase from Rhodococcus sp. HJ1. Res. J. Microbiol. 2008, 3, 482–488. [Google Scholar]
- Lin, C.; Yang, L.; Xu, G.; Wu, J. Biodegradation and metabolic pathway of β-chlorinated aliphatic acid in Bacillus sp. CGMCC no. 4196. Appl. Microbiol. Biotechnol. 2011, 90, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Sufian, N.H.M.; Naim, M.A.; Hamid, T.H.T.A.; Huyop, F.; Hamid, A.A.A. Isolation and identification of 3-chloropropionic acid degrading bacterium from marine sponge. J. Teknologi. 2015, 77. [Google Scholar] [CrossRef] [Green Version]
- Muslem, W.H.; Huyop, F.; Zakaria, I.I.; Wahab, R.A. Isolation and characterization of a biodegrading 3-chloropropionic acid Burkholderia cepacia WH1 isolated from abandoned agricultural land. Asia Pac. J. Mol. Biol. Biotechnol. 2015, 23, 268–279. [Google Scholar]
- Bagherbaigi, S.; Gicana, R.G.; Lamis, R.J.; Nemati, M.; Huyop, F. Characterisation of Arthrobacter sp. S1 that can degrade α and β-haloalkanoic acids isolated from contaminated soil. Ann. Microbiol. 2013, 63, 1363–1369. [Google Scholar] [CrossRef]
- Jabeen, H.; Iqbal, S.; Anwar, S. Biodegradation of chlorpyrifos and 3, 5, 6-trichloro-2-pyridinol by a novel rhizobial strain Mesorhizobium sp. HN3. Water Environ. J. 2015, 29, 151–160. [Google Scholar] [CrossRef]
- Abraham, J.; Silambarasan, S. Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol using a novel bacterium Ochrobactrum sp. JAS2: A proposal of its metabolic pathway. Pestic. Biochem. Physiol. 2016, 126, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Mnif, I.; Ghribi, D. Potential of bacterial derived biopesticides in pest management. Crop Prot. 2015, 77, 52–64. [Google Scholar] [CrossRef]
- Fira, D.; Dimkić, I.; Berić, T.; Lozo, J.; Stanković, S. Biological control of plant pathogens by bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Akcay, K.; Kaya, Y. Isolation, characterization and molecular identification of a halotolerant Bacillus megaterium CTBmeg1 able to grow on halogenated compounds. Biotechnol. Biotechnol. Equip. 2019, 33, 945–953. [Google Scholar] [CrossRef] [Green Version]
- Sorokin, D.Y.; Tourova, T.P.; Galinski, E.A.; Belloch, C.; Tindall, B.J. Extremely halophilic denitrifying bacteria from hypersaline inland lakes, Halovibrio denitrificans sp. nov. and Halospina denitrificans gen. nov., sp. nov., and evidence that the genus name Halovibrio Fendrich 1989 with the type species Halovibrio variabilis should be associated with DSM 3050. Int. J. Syst. Evol. Microbiol. 2006, 56, 379–388. [Google Scholar] [PubMed]
- Lu, H.; Xing, P.; Zhai, L.; Li, H.; Wu, Q. Halomonas montanilacus sp. nov., isolated from hypersaline Lake Pengyanco on the Tibetan Plateau. Int. J. Syst. Evol. Microbiol. 2020, 70, 2859–2866. [Google Scholar] [CrossRef] [PubMed]
- Kajale, S.; Deshpande, N.; Pali, S.; Shouche, Y.; Sharma, A. Natrialba swarupiae sp. nov., a halophilic archaeon isolated from a hypersaline lake in India. Int. J. Syst Evol. Microbiol. 2020, 70, 1876–1881. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liang, P.; Zhang, X.; Huang, X. Bioelectrochemical systems-driven directional ion transport enables low-energy water desalination, pollutant removal, and resource recovery. Bioresour. Technol. 2016, 215, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Castro, D.J.; Gomez-Altuve, A.; Reina, J.C.; Rodríguez, M.; Sampedro, I.; Llamas, I.; Martínez-Checa, F. Roseovarius bejariae sp. nov., a moderately halophilic bacterium isolated from a hypersaline steep-sided riverbed. Int. J. Syst Evol. Microbiol. 2020, ijsem004154. [Google Scholar] [CrossRef]
- Ibrahim, I.M.; Konnova, S.A.; Sigida, E.N.; Lyubun, E.V.; Muratova, A.Y.; Fedonenko, Y.P.; Elbanna, K. Bioremediation potential of a halophilic Halobacillus sp. strain, EG1HP4QL: Exopolysaccharide production, crude oil degradation, and heavy metal tolerance. Extremophiles 2020, 24, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Ben Abdallah, M.; Karray, F.; Sayadi, S. Production of Polyhydroxyalkanoates by Two Halophilic Archaeal Isolates from Chott El Jerid Using Inexpensive Carbon Sources. Biomolecules 2020, 10, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeki Camur, M.; Mutlu, H. Major-ion geochemistry and mineralogy of the salt lake (Tuz Gölü) basin, Turkey. Chem. Geol. 1996, 127, 313–329. [Google Scholar] [CrossRef]
- Litchfield, C.D.; Gillevet, P.M. Microbial diversity and complexity in hypersaline environments: A preliminary assessment. J. Ind. Microbiol. Biotechnol. 2002, 28, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Oren, A. Microbial life at high salt concentrations: Phylogenetic and metabolic diversity. Saline Syst. 2008, 4, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garg, S.K.; Tripathi, M. Microbial strategies for discoloration and detoxification of azo dyes from textile effluents. Res. J. Microbiol. 2017, 12, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Samimi, M.; Shahriari Moghadam, M. Phenol biodegradation by bacterial strain O-CH1 isolated from seashore. Glob. J. Environ. Sci. Manag. 2020, 6, 109–118. [Google Scholar]
- Brandt, K.K.; Ingvorsen, K. Desulfobacter halotolerans sp. nov. a halotolerant acetate-oxidizing sulfate-reducing bacterium isolated from sediments of Great Salt Lake, Utah. Syst. Appl. Microbiol. 1997, 20, 366–373. [Google Scholar] [CrossRef]
- Garcia, N.F.L.; Santos, F.R.D.S.; Gonçalves, F.A.; Paz, M.F.D.; Fonseca, G.G.; Leite, R.S.R. Production of β-glucosidase on solid-state fermentation by Lichtheimia ramosa in agroindustrial residues: Characterization and catalytic properties of the enzymatic extract. Electron. J. Biotechnol. 2015, 18, 314–319. [Google Scholar] [CrossRef] [Green Version]
- Altin, T.B.; Barak, B.; Altın, B.N. Change in precipitation and temperature amounts over three decades in central anatolia, turkey. Atmos. Clim. Sci. 2012, 2, 107. [Google Scholar] [CrossRef] [Green Version]
- Ezeilo, U.R.; Wahab, R.A.; Tin, L.C.; Zakaria, I.I.; Huyop, F.; Mahat, N.A. Fungal-assisted valorization of raw oil palm leaves for production of cellulase and xylanase in solid state fermentation media. Waste Biomass. Valorization 2020, 11, 3133–3149. [Google Scholar] [CrossRef]
- Odongo, V.O.; Hamm, N.A.; Milton, E.J. Spatio-temporal assessment of Tuz Gölü, Turkey as a potential radiometric vicarious calibration site. Remote Sens. 2014, 6, 2494–2513. [Google Scholar] [CrossRef] [Green Version]
- Hareland, W.A.; Crawford, R.L.; Chapman, P.J.; Dagley, S.T. Metabolic function and properties of 4-hydroxyphenylacetic acid 1-hydroxylase from Pseudomonas acidovorans. J. Bacteriol. 1975, 121, 272–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergey, D.; Holt, J.; Krieg, P. Bergey’s Manual of Determinative Bacteriology; Williams and Wilkins: Baltimore, MD, USA, 1994. [Google Scholar]
- Fulton, C.K.; Cooper, R.A. Catabolism of sulfamate by Mycobacterium sp. CF1. Environ. Microbiol. 2005, 7, 378–381. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergmann, J.; Sanik, J.J. Determination of trace amounts of chlorine in naphtha. Anal. Chem. 1957, 29, 241–243. [Google Scholar] [CrossRef]
- Joghee, N.N.; Jayaraman, G. Biochemical changes induced by salt stress in halotolerant bacterial isolates are media dependent as well as species specific. Prep. Biochem. Biotechnol. 2016, 46, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Hill, K.E.; Marchesi, J.R.; Weightman, A.J. Investigation of two evolutionarily unrelated halocarboxylic acid dehalogenase gene families. J. Bacteriol. 1999, 181, 2535–2547. [Google Scholar] [CrossRef] [PubMed] [Green Version]


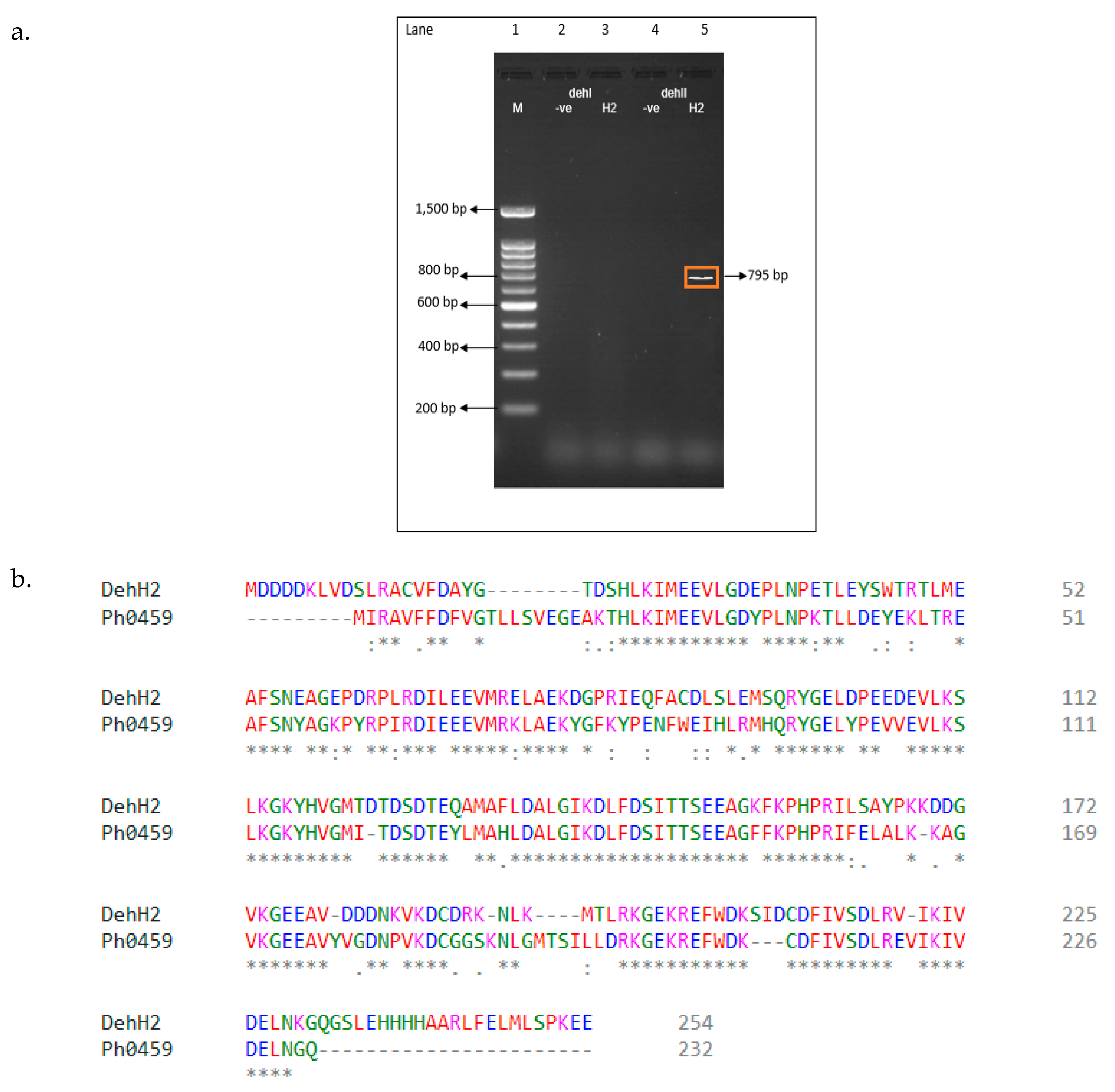
| Features | Strain H1 | Strain H2 |
|---|---|---|
| Size | Medium | Tiny |
| Shape | Circular | Circular |
| Pigmentation | Light cream | Cream |
| Texture | Smooth/slime | Smooth/damp |
| Elevation | Convex | Raised |
| Margin | Entire | Entire |
| Cell morphology | Rod-shape | Cocci |
| Biochemical Characteristics | ||
| Motility | + | + |
| Gram staining | + | + |
| Catalase | + | + |
| Amylase | + | + |
| Oxidase | + | + |
| Methyl red | + | + |
| VP | + | + |
| Indole | − | − |
| Urease | + | + |
| Gelatine liquefaction | − | − |
| Simon citrate | − | + |
| Nitrate | + | + |
| Casein | + | − |
| H2S production | − | − |
| Glucose | + | + |
| Lactose | + | − |
| Maltose | + | + |
| Sucrose | + | + |
| Various Carbon Sources | Cells Doubling Time (h) | % Chloride Ion Released | ||
|---|---|---|---|---|
| Strain H1 | Strain H2 | Strain H1 | Strain H2 | |
| 2,2-DCP | 7.57 ± 0.1 | 12.79 ± 0.2 | 84.5 | 87.9 |
| 2,3-DCP | 22.7 ± 0.1 | 18.00 ± 0.2 | 68.7 | 82.9 |
| D, L-2CP | 16.96 ± 0.3 | 13.59 ± 0.1 | 74.5 | 85.6 |
| 3-CP | 12.01 ± 0.2 | 13.28 ± 0.2 | 71.9 | 81.6 |
| TCA | 17.41 ± 0.1 | 18.16 ± 0.3 | 81.1 | 78.8 |
| MCA | 14.30 ± 0.2 | 15.14 ± 0.1 | 76.8 | 79.7 |
| Chlorpyrifos | 18.16 ± 0.3 | 17.02 ± 0.3 | 69.4 | 80.3 |
| Basal Salts (10× Concentration) | Trace Metals (10× Concentration) | ||
|---|---|---|---|
| Components | Amount (g/L) | Components | Amount (g/L) |
| K2HPO4.3H2O NaH2PO4.2H2O (NH4)2SO4 | 42.5 10.0 25.0 | Nitriloacetic acid MgSO4 FeSO4.7H2O MnSO4.4H2O ZnSO4.H2O CoCl2 | 1.0 2.0 0.12 0.03 0.03 0.01 |
| Preparation of minimal/enrichment media in 100 mL volume | |||
| Liquid minimal medium | Solid minimal medium | ||
| Medium components | Amount (mL) | Flask A (mL) | Flask B (mL) |
| Distilled water | To final volume of 100 mL | To final volume of 50 mL | 50 |
| Basal salts (10×) | 10 | 10 | __ |
| Trace metals (10×) | 10 | 10 | __ |
| 1M Carbon source(s) | Varies (1 or above) | Varies (1 or above) | __ |
| Agar No1 | __ | __ | 1.5 g |
| Total | 100 | 50 | 50 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oyewusi, H.A.; Wahab, R.A.; Kaya, Y.; Edbeib, M.F.; Huyop, F. Alternative Bioremediation Agents against Haloacids, Haloacetates and Chlorpyrifos Using Novel Halogen-Degrading Bacterial Isolates from the Hypersaline Lake Tuz. Catalysts 2020, 10, 651. https://doi.org/10.3390/catal10060651
Oyewusi HA, Wahab RA, Kaya Y, Edbeib MF, Huyop F. Alternative Bioremediation Agents against Haloacids, Haloacetates and Chlorpyrifos Using Novel Halogen-Degrading Bacterial Isolates from the Hypersaline Lake Tuz. Catalysts. 2020; 10(6):651. https://doi.org/10.3390/catal10060651
Chicago/Turabian StyleOyewusi, Habeebat Adekilekun, Roswanira Abdul Wahab, Yilmaz Kaya, Mohamed Faraj Edbeib, and Fahrul Huyop. 2020. "Alternative Bioremediation Agents against Haloacids, Haloacetates and Chlorpyrifos Using Novel Halogen-Degrading Bacterial Isolates from the Hypersaline Lake Tuz" Catalysts 10, no. 6: 651. https://doi.org/10.3390/catal10060651