Recursive Feature Elimination and Random Forest Classification of Natura 2000 Grasslands in Lowland River Valleys of Poland Based on Airborne Hyperspectral and LiDAR Data Fusion
Abstract
1. Introduction
2. Study Areas and Botanical Description
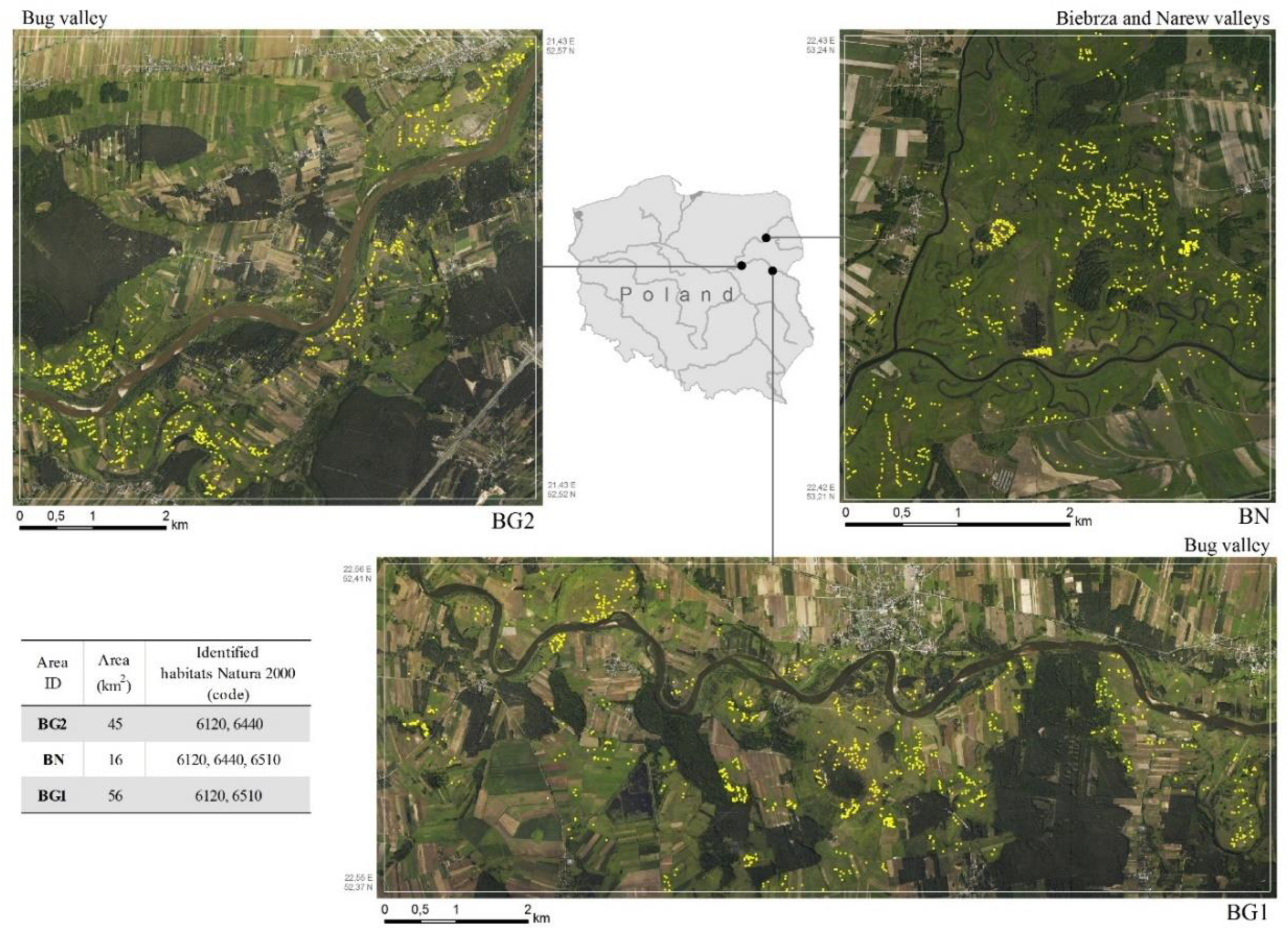
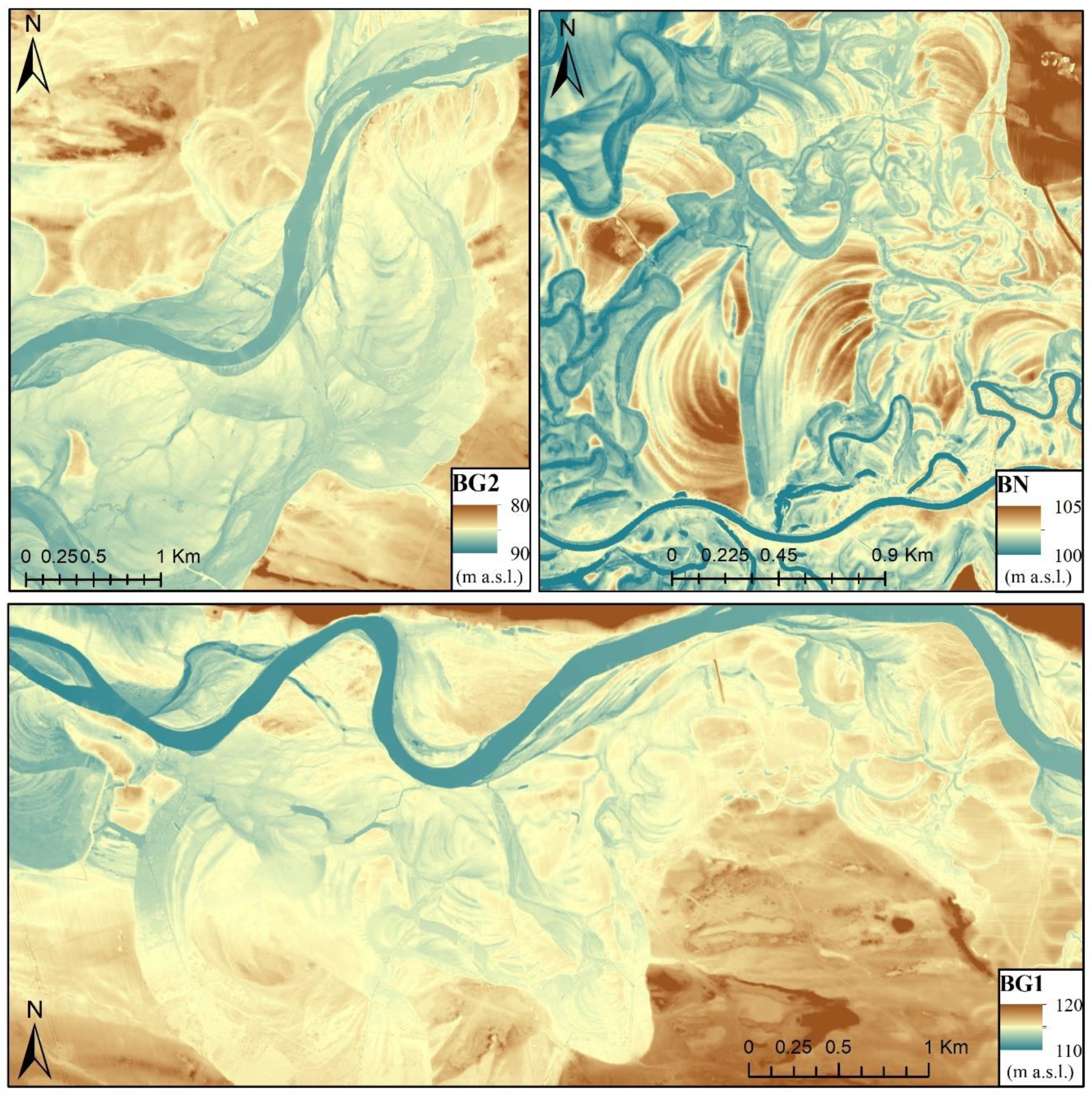
2.1. Study Areas
2.2. Natura 2000 Habitats Descriptions
2.2.1. Habitat 6440: Alluvial Meadows of River Valleys of the Cnidion Dubii
2.2.2. Habitat 6510: Lowland Hay Meadows
2.2.3. Habitat 6120: Xeric and Calcareous Grasslands
3. Methodology
3.1. Airborne Data Acquisition and Botanical Field Measurements
- Hyperspectral scanners (Hyspex: VNIR 0.4–0.9 µm & SWIR 0.9–2.5 µm): 470 spectral bands with 1 m spatial resolution;
- Airborne Laser Scanner (Riegl Lite Mapper LMS-Q680i): point cloud data acquired with 7 points/m2;
- Medium format RGB camera (50Mpix) with 0.1 m spatial resolution.
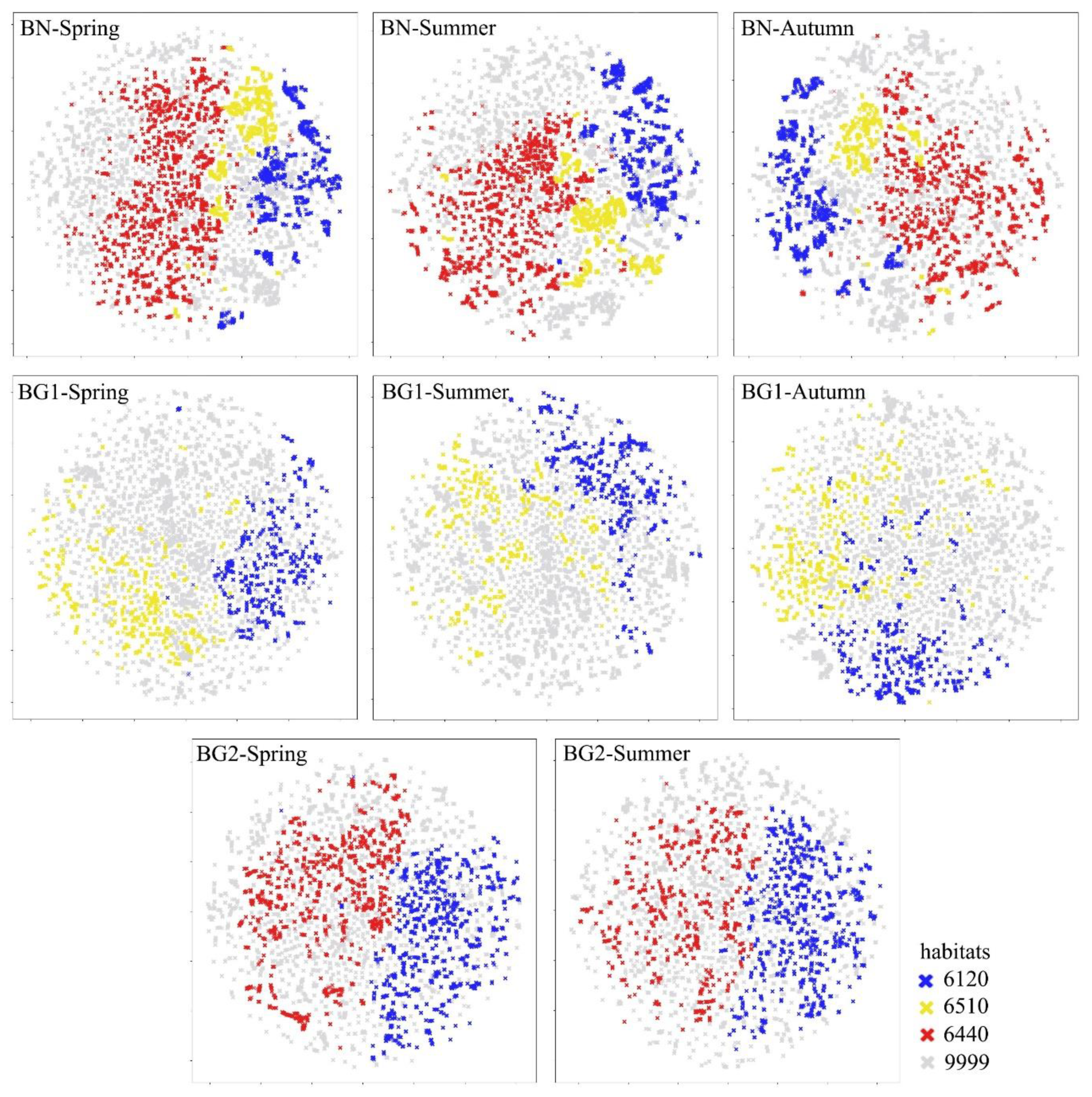
3.2. Reference Botanical Data Quality Assessment
3.3. RS Data Pre-Processing
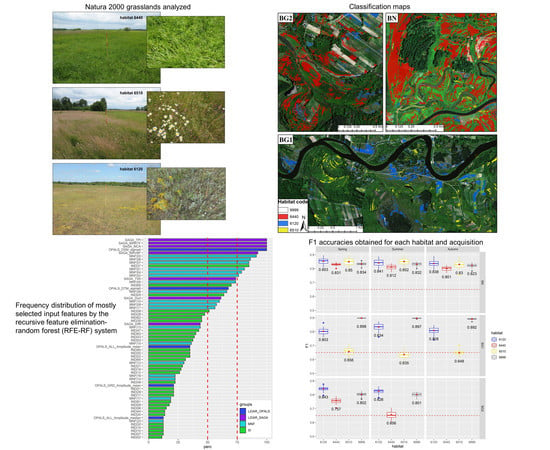
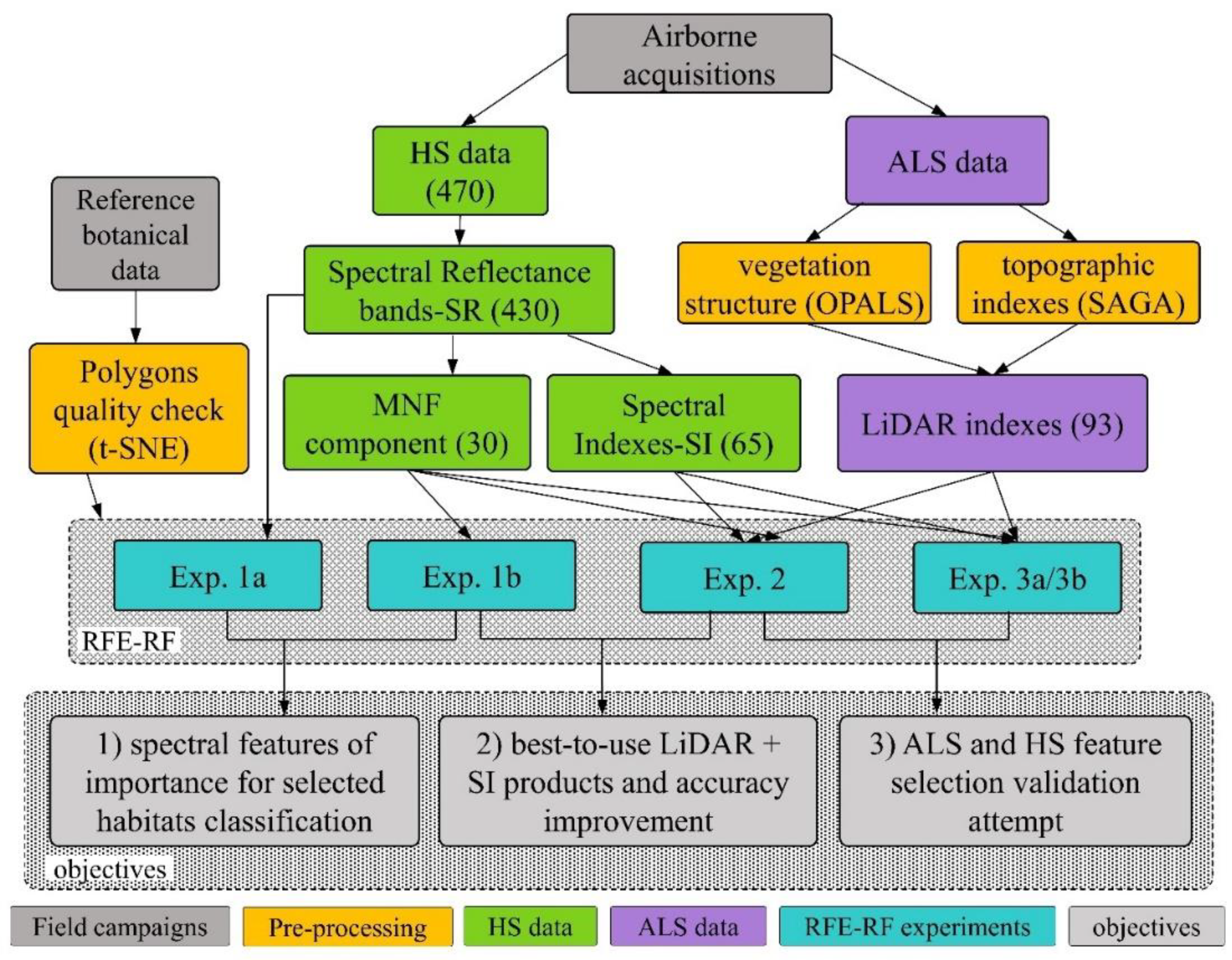
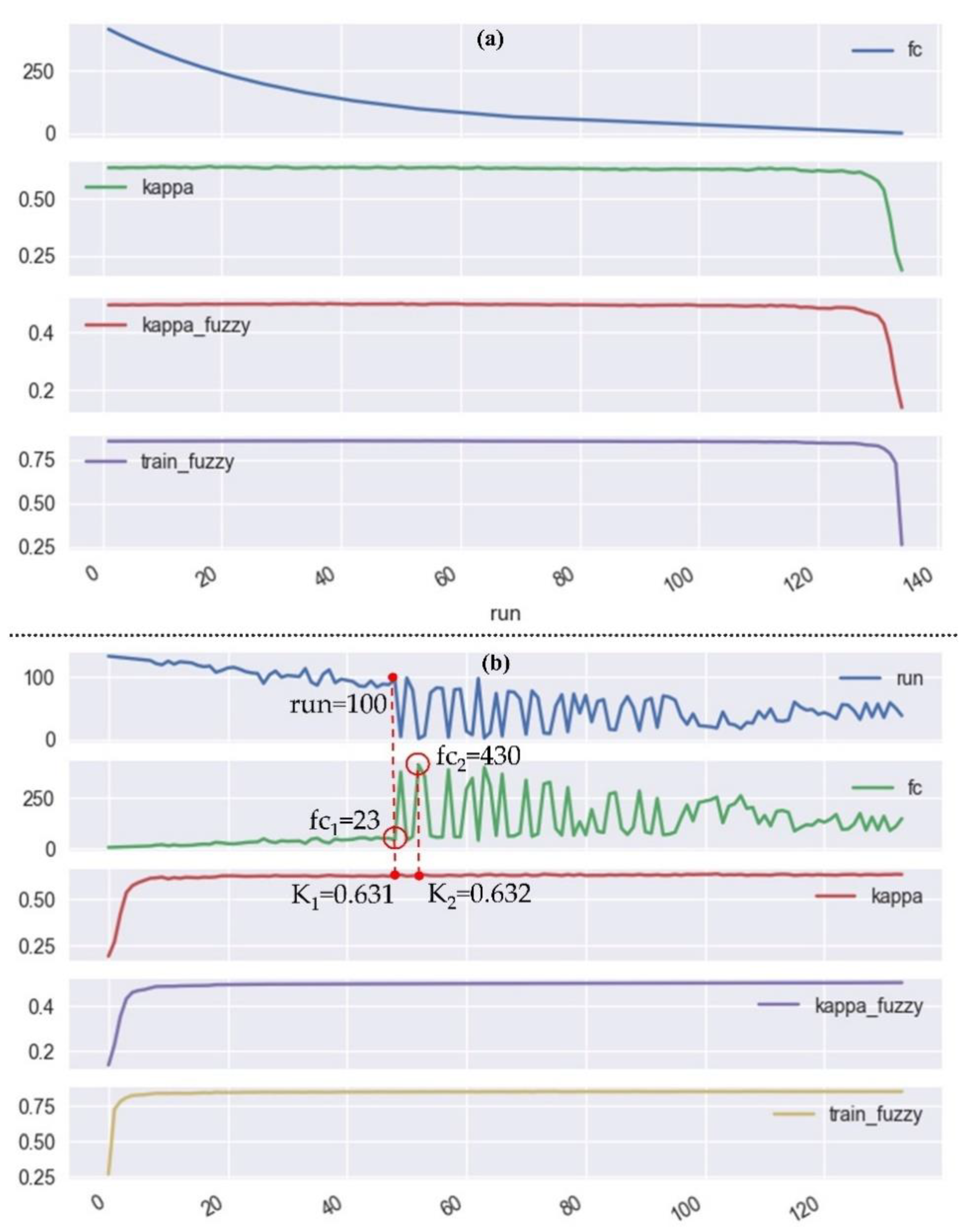
3.4. Recursive Feature Elimination-Random Forest (RFE-RF) Classification System
3.5. Experiments Design
3.5.1. Exp. 1: Spectral Features of Importance and Dimensionality Reduction
3.5.2. Exp. 2: LiDAR Products and Spectral Indices
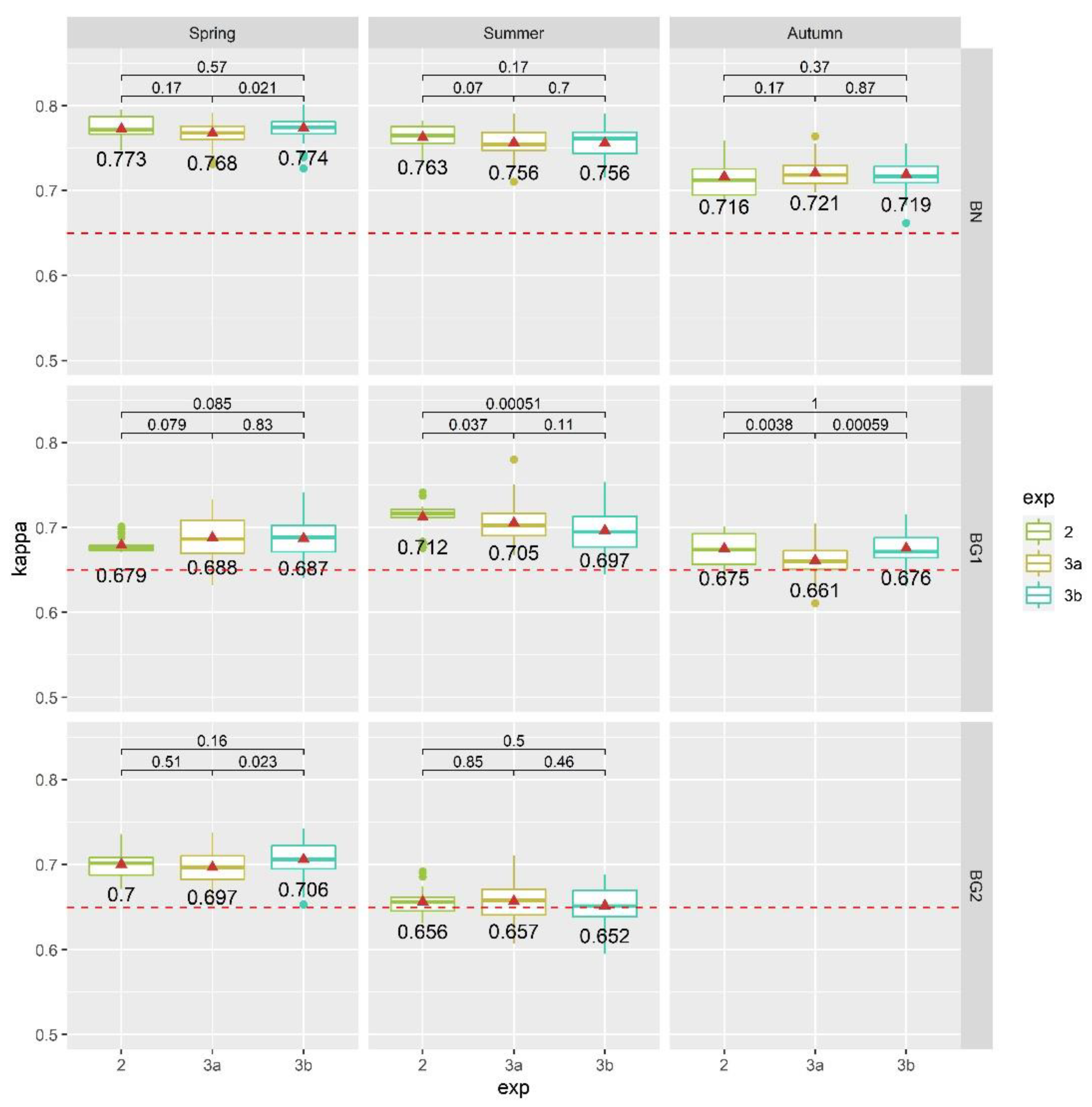
3.5.3. Exp. 3: Feature Selection Validation Attempt
- Exp. 3a: the selection of optimal features was done separately for each area, meaning that a different best-to-use features selection was used for each case study, as a result of Exp. 2;
- Exp. 3b: the selection of optimal features was the same for all study areas, meaning that a unique best-to-use features selection was extrapolated from all study areas at the same time, as a result of Exp. 2.
4. Results
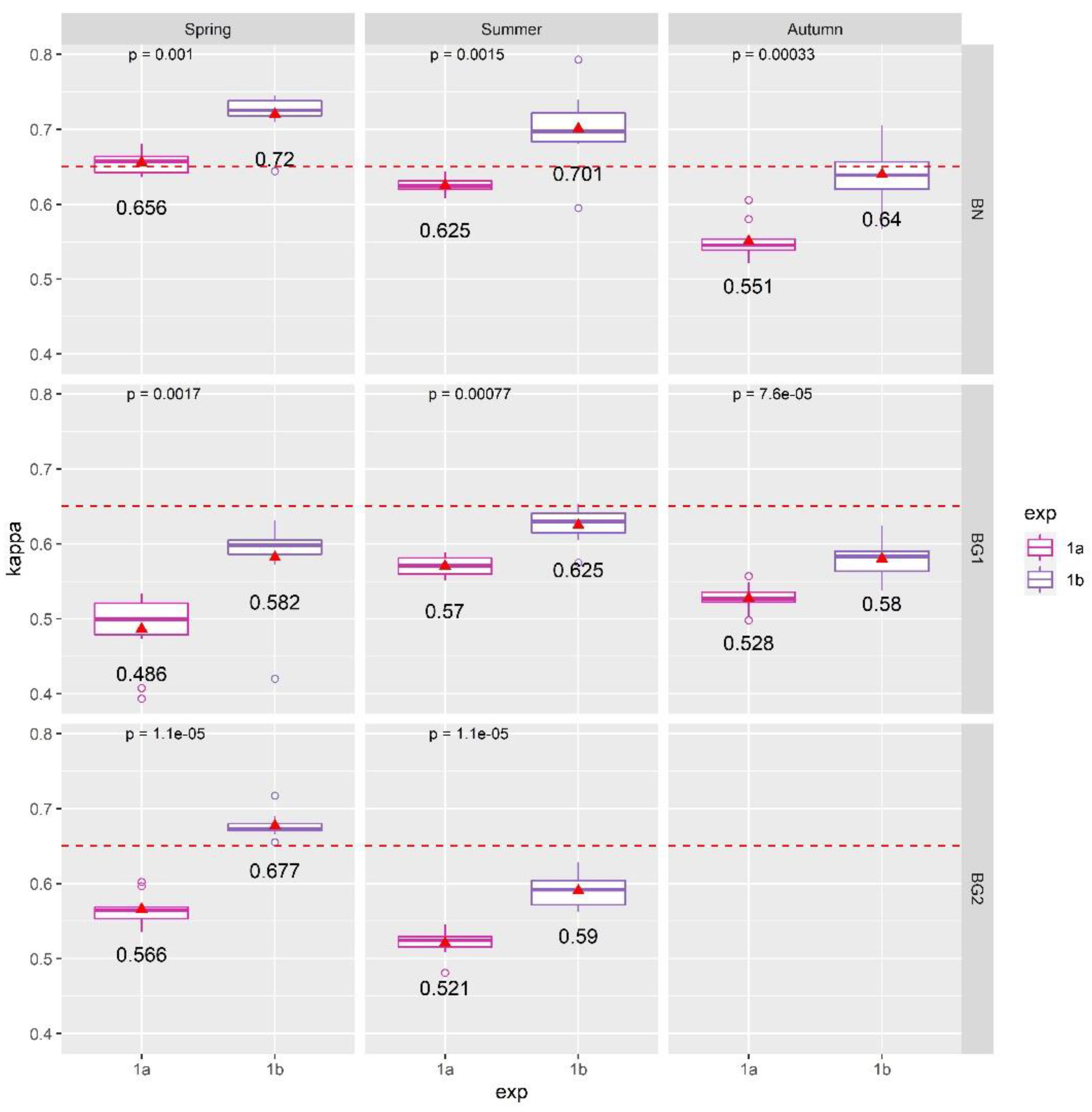
4.1. Results of Exp. 1
- 400–800 nm of the visible spectral range (mainly red and blue);
- 1050–1100 nm of the near-infrared;
- 1250–1400 nm, 1650–1800 nm, 1950–2050 nm, and 2250–2400 nm of the SWIR spectral range.
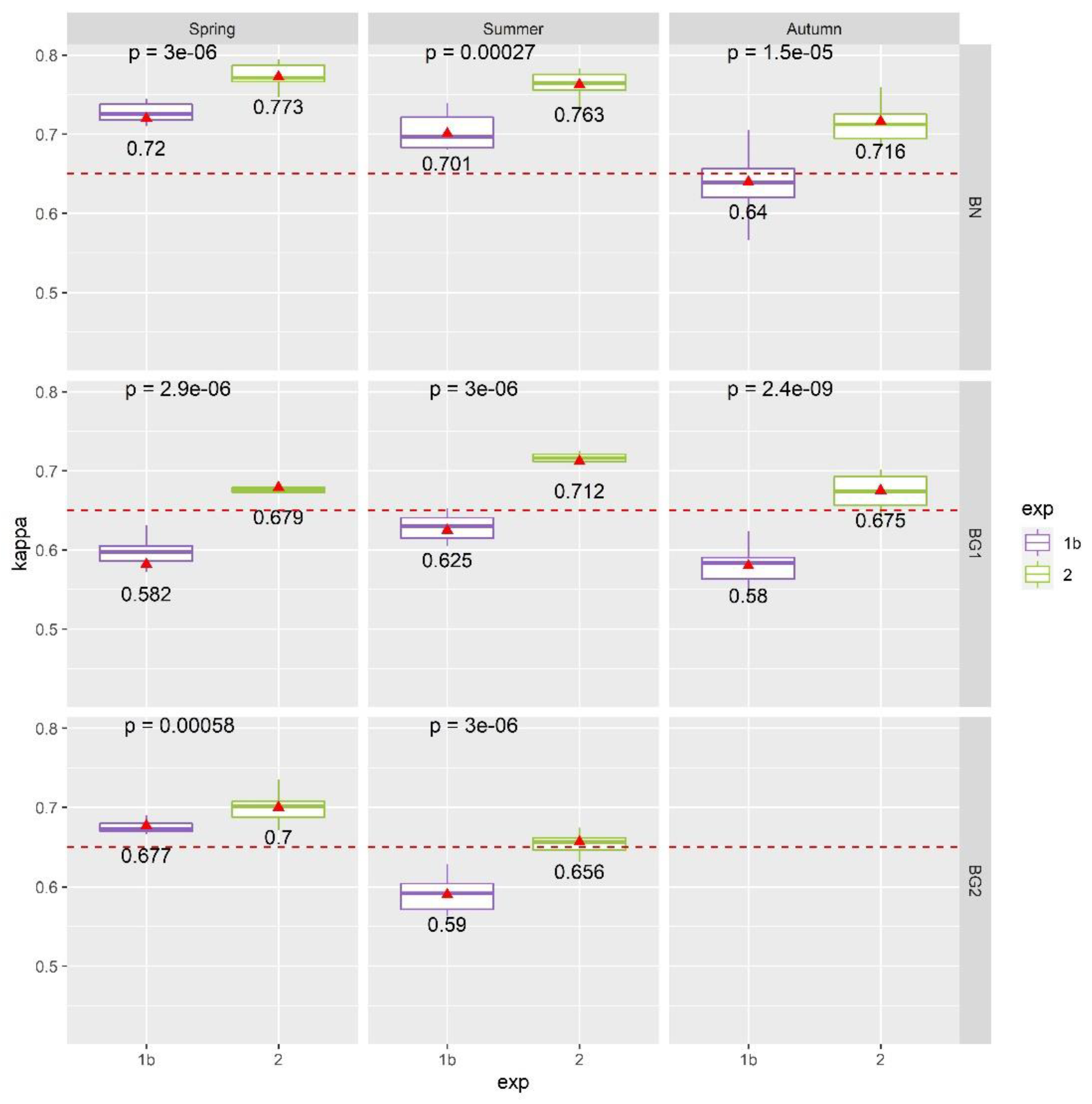
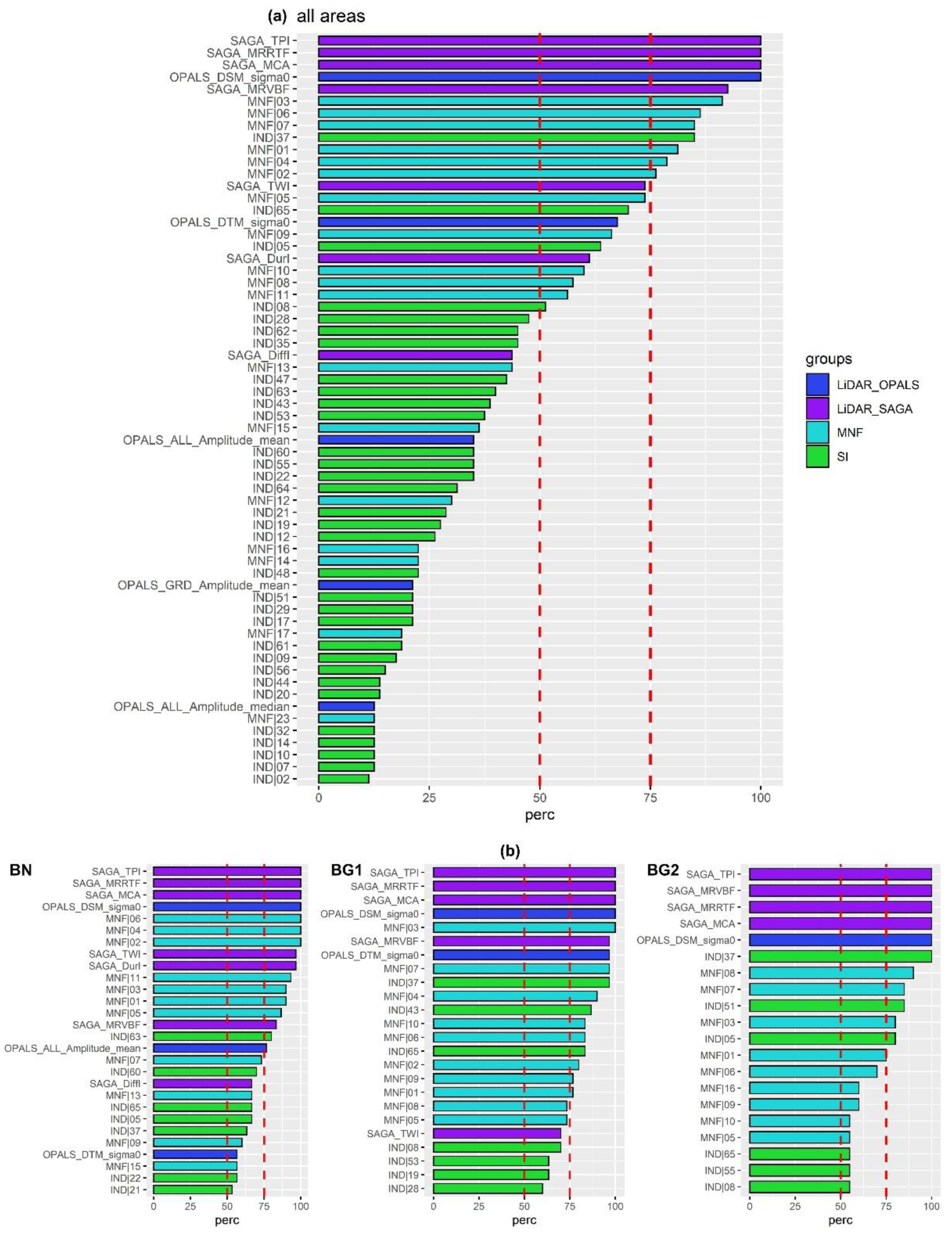
4.2. Results of Exp. 2
- SAGA_TPI (topographic position index);
- SAGA_MRRTF (multiresolution index of the ridge top flatness);
- SAGA_MCA (modified catchment area);
- OPALS_DSM_Sigma0: DSM standard deviation of the unit weight.
- SAGA_ MRVBF (multiresolution index of valley bottom flatness);
- SAGA_TWI (topographic wetness index);
- Spectral MNF components [1:7];
- Spectral Index nr.37 (NDNI: normalized difference nitrogen index).
- LiDAR products: SAGA_TWI, SAGA Duration of Insolation (SAGA_DurI), OPALS mean amplitude (ALL_Amplitude_mean), SAGA_MRVBF;
- SI: nr.63 (WV-NHFD: WorldView non-homogeneous feature difference);
- Spectral MNF components: [01,03,05,07,11].
- LiDAR products: SAGA_ MRVBF and OPALS_DTM_sigma0 (DTM standard deviation of the unit weight);
- SI: nr.37 (NDNI), nr.43 (PRI: photochemical reflectance index), nr.65 (WVWI: WorldView Water Index);
- MNF components: [01;02;04-10].
- SI: nr.51 (SIPI: structure insensitive pigment index) and nr.05 (CRI1: carotenoid reflectance index 1);
- MNF components: [01;03;07,08].
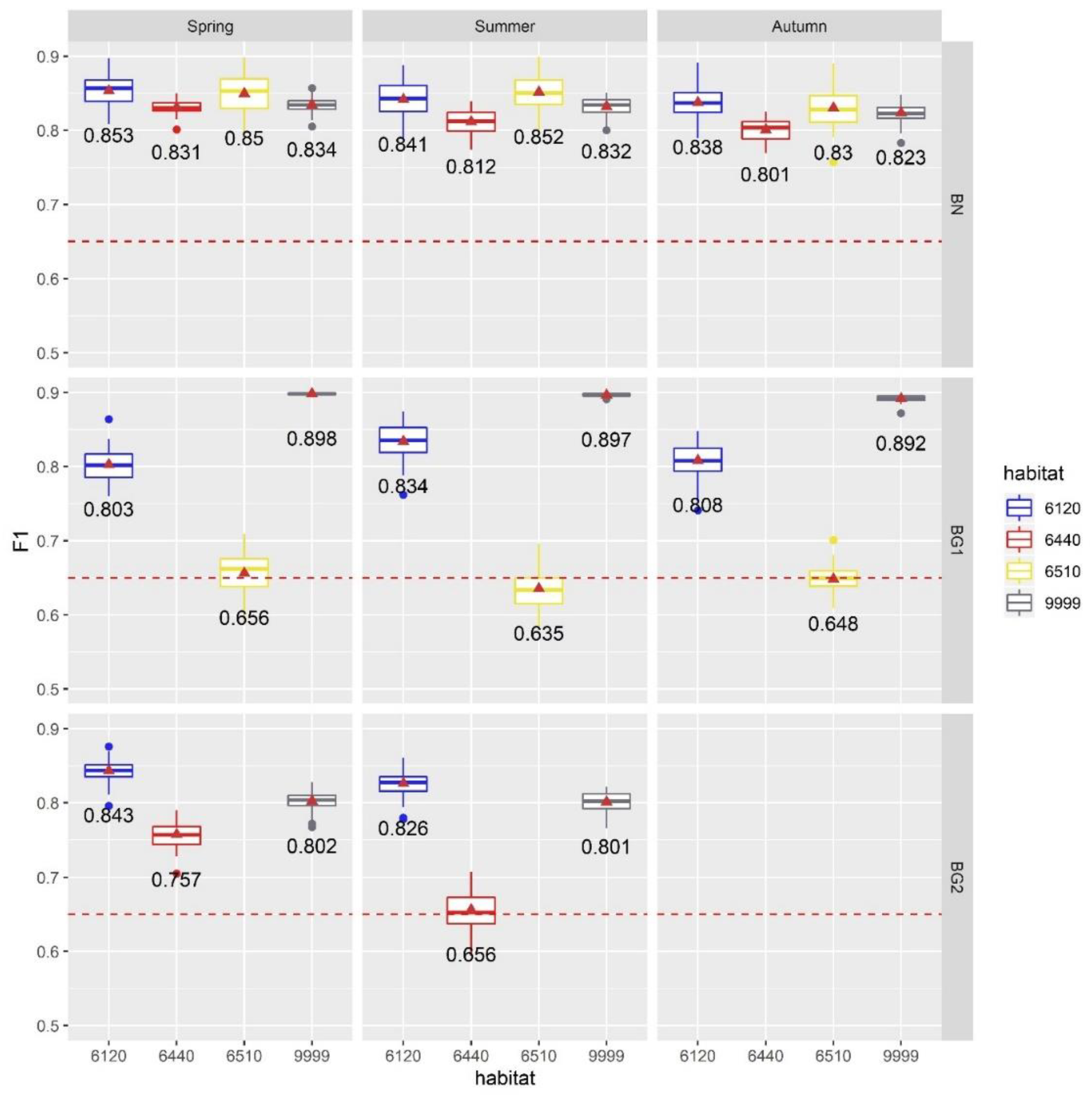
4.3. Results of Exp. 3
5. Discussion
5.1. The Importance of Different Hyperspectral Channels and Dimensionality Reduction Techniques for Mapping Meadows and Dry Grasslands Habitats in the Selected River Valleys
5.2. LiDAR Products and Spectral Indices Selection to Enhance Habitats Classification Performances
5.2.1. LiDAR-Based Features of Importance
5.2.2. Spectral Indices of Importance
5.2.3. Best-to-Use HS+ALS Products for Mapping Meadows and Dry Grasslands Habitats in the Selected River Valleys
- LiDAR (SAGA) products: DurI, MRRTF, MRVBF, TPI, MCA, TWI;
- LiDAR (OPALS) products: DSM_Sigma0, DTM_Sigma0;
- SI: CR1, CM, NDNI, WVWI;
- MNF: 1-11.
5.3. Considerations on the Computational Efficiency of the RFE-RF System
6. Conclusions
- The MNF dimensionality reduction method outperformed RFE: part of the inherent spectral information was lost during RFE feature selection and therefore the classification accuracy significantly reduced. On the other hand, with the RFE technique, it was possible to highlight the important HS channels that are necessary for mapping the investigated habitats: VIS, NIR, and SWIR are all required spectral channels;
- By selecting and using only the original input bands, without any kind of data-transformation, the RFE-RF system proved to be a very efficient and useful setup for automated hyperspectral and LiDAR data processing, highlighting the added value of the fusion between these complementary and diverse datasets. It is therefore possible to use a common selection of 24 features (instead of 188) to distinguish the investigated habitats of this study and still obtain very similar classification accuracies among all considered study areas;
- LiDAR-based products, depicting the variable topographic micro-reliefs of the investigated river valleys, proved to be the most selected features producing also a significant enhancement in the classification accuracy. In particular: topographic position index (TPI), multiresolution index of the ridge top flatness (MRRTF), multiresolution index of valley bottom flatness (MRVBF), modified catchment area (MCA), topographic wetness index (TWI), DSM_Sigma0 and DTM_Signma0 proved to be necessary adjuncts for mapping Natura 2000 habitats 6120, 6440 and 6510;
- The meaningfulness of the selected products is strongly linked to the habitats’ characteristics. It was remarkable noticing that since habitat 6510 is mostly found in higher terraces, while habitat 6440 is usually found in low depressions of the floodplain, the MRRTF and MRVBF products were retained as the most relevant classification features. In fact, MRRTF and MRVBF have been conceived for the identification of high-flat areas and flat valley bottoms respectively [99]. Likewise, since habitats 6120, 6510, and 6440 are all connected to periodic floods in different ways, the TWI and MCA products, reported in literature to be good indicators of floods [101], have also been selected as best-to-use features;
- The common feature selection also showed the importance of using HS data with high spectral resolution covering a broad part of the spectral range, so to compute specific spectral indices (SI) which significantly contributed to enhancing the final classification accuracies. The high heterogeneity of the habitats is well pictured by the selection of different SI in different study areas. However, it was proved that using only CRI1, CM, NDNI, and WVWI, together with the other topographical products and MNF components, it was possible to obtain satisfiable classification accuracies over all investigated areas. All of them are strictly linked to the highly variable characteristics and conditions of the diverse habitats analyzed: vegetation stress and health (CRI1), presence of different soil types and minerals (CM), nitrogen concentrations (NDNI), and moisture content (WVWI). The selection of these specific indicators highlight also the importance of the SWIR, NIR and visible channels for mapping Natura 2000 habitats 6440, 6120, 6510 and confirms the selected HS channels in the first part of our analysis;
- A great time-effort needs to be envisaged to collect a high number of field-based samples (between 1000 and 1500 polygons) in order to achieve similar classification results. This is a very important step in order to embark on a classification problem of this kind.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sikorska, D.; Sikorski, P.; Archiciński, P.; Chormański, J.; Hopkins, R.J. You Can’t See the Woods for the Trees: Invasive Acer negundo L. in Urban Riparian Forests Harms Biodiversity and Limits Recreation Activity. Sustainability 2019, 11, 5838. [Google Scholar] [CrossRef]
- European Parliament-Council of the European Union. EC Council Directive 1992/43/EEC on the Conservation of Natural Habitats and of Wild Fauna and Flora; European Parliament-Council of the European Union: Brussel, Belgium, 1992. [Google Scholar]
- Habel, J.C.; Dengler, J.; Janišová, M.; Török, P.; Wellstein, C.; Wiezik, M. European grassland ecosystems: Threatened hotspots of biodiversity. Biodivers. Conserv. 2013, 22, 2131–2138. [Google Scholar] [CrossRef]
- Hopkins, A.; Holz, B. Grassland for agriculture and nature conservation: Production, quality and multi-functionality. Agron. Res. 2006, 4, 3–20. [Google Scholar]
- Leuschner, C.; Ellenberg, H. Ecology of Central European Non-Forest Vegetation: Coastal to Alpine, Natural to Man-Made Habitats; Springer International Publishing: Cham, Switzerland, 2017; Volume 2. [Google Scholar]
- Rychnovska, M. Structure and Functioning of Seminatural Meadows, Developments in Agricultural and Managed-Forest Ecology; Palacky University: Olomouc, Czech Republic, 1993; Volume 27, p. 385. [Google Scholar]
- Czech, H.A.; Parsons, K.C. Agricultural wetlands and waterbirds: A review. Waterbirds 2002, 25, 56–65. [Google Scholar]
- Ma, Z.; Cai, Y.; Li, B.; Chen, J. Managing Wetland Habitats for Waterbirds: An International Perspective. Wetlands 2009, 30, 15–27. [Google Scholar] [CrossRef]
- Czochański, J.T.; Wiśniewski, P. River valleys as ecological corridors – structure, function and importance in the conservation of natural resources. Ecol. Quest. 2018, 29, 77–87. [Google Scholar] [CrossRef]
- Bischoff, A.; Warthemann, G.; Klotz, S. Succession of floodplain grasslands following reduction in land use intensity: The importance of environmental conditions, management and dispersal. J. Appl. Ecol. 2009, 46, 241–249. [Google Scholar] [CrossRef]
- Galvánek, D.; Ripka, J. Vegetation development after a large scale restoration of species-rich grasslands in a Central European floodplain. Wetl. Ecol. Manag. 2017, 26, 373–381. [Google Scholar] [CrossRef]
- Bakker, J.P.; Berendse, F. Constraints in the restoration of ecological diversity in grassland and heathland communities. Trends Ecol. Evol. 1999, 14, 63–68. [Google Scholar] [CrossRef]
- Pykälä, J. Mitigating Human Effects on European Biodiversity through Traditional Animal Husbandry. Conserv. Boil. 2000, 14, 705–712. [Google Scholar] [CrossRef]
- Reidsma, P.; Tekelenburg, T.; Berg, M.V.D.; Alkemade, R. Impacts of land-use change on biodiversity: An assessment of agricultural biodiversity in the European Union. Agric. Ecosyst. Environ. 2006, 114, 86–102. [Google Scholar] [CrossRef]
- Kotecky, P.; Prach, K. Recovery of alluvial meadows after an extreme summer flood: A case study. Ecohydrol. Hydrobiol. 2005, 5, 32–38. [Google Scholar]
- Gerard, M.; El Kahloun, M.; Mertens, W.; Verhagen, B.; Meire, P. Impact of flooding on potential and realised grassland species richness. Vegetatio 2007, 194, 85–98. [Google Scholar] [CrossRef]
- Kącki, Z. Comprehensive syntaxonomy of Molinion meadows in southwestern Poland. Acta Bot. Silesiaca. Monogr. 2007, 2, 134. [Google Scholar]
- Ellwanger, G.; Runge, S.; Wagner, M.; Ackermann, W.; Neukirchen, M.; Frederking, W.; Müller, C.; Ssymank, A.; Sukopp, U. Current status of habitat monitoring in the European Union according to Article 17 of the Habitats Directive, with an emphasis on habitat structure and functions and on Germany. Nat. Conserv. 2018, 29, 57–78. [Google Scholar] [CrossRef]
- Feilhauer, H.; Dahlke, C.; Doktor, D.; Lausch, A.; Schmidtlein, S.; Schulz, G.; Stenzel, S. Mapping the local variability of Natura 2000 habitats with remote sensing. Appl. Veg. Sci. 2014, 17, 765–779. [Google Scholar] [CrossRef]
- Šmihula, D. Waves of technological innovations and the end of the information revolution. J. Econ. Int. Financ. 2010, 2, 58–67. [Google Scholar]
- Rose, R.; Byler, D.; Eastman, J.R.; Fleishman, E.; Geller, G.; Goetz, S.J.; Guild, L.; Hamilton, H.; Hansen, M.; Headley, R.; et al. Ten ways remote sensing can contribute to conservation. Conserv. Boil. 2014, 29, 350–359. [Google Scholar] [CrossRef]
- Zimmermann, N.E.; Washington-Allen, R.A.; Ramsey, R.D.; Schaepman, M.E.; Mathys, L.; Kötz, B.; Kneubühlerx, M.; Edwards, T.C. Modern Remote Sensing for Environmental Monitoring of Landscape States and Trajectories. In Landscape Series; Springer: Dordrecht, The Netherlands, 2007; Volume 8, pp. 65–91. [Google Scholar]
- Vaz, A.S.; Alcaraz-Segura, D.; Vicente, J.R.; Honrado, J.P. The Many Roles of Remote Sensing in Invasion Science. Front. Ecol. Evol. 2019, 7, 1–5. [Google Scholar] [CrossRef]
- Chi, M.; Plaza, J.; Benediktsson, J.A.; Sun, Z.; Shen, J.; Zhu, Y. Big Data for Remote Sensing: Challenges and Opportunities. Proc. IEEE 2016, 104, 2207–2219. [Google Scholar] [CrossRef]
- Du, J.; Watts, J.D.; Jiang, L.; Lu, H.; Cheng, X.; Duguay, C.; Farina, M.; Qiu, Y.; Kim, Y.; Kimball, J.S.; et al. Remote Sensing of Environmental Changes in Cold Regions: Methods, Achievements and Challenges. Remote Sens. 2019, 11, 1952. [Google Scholar] [CrossRef]
- Demarchi, L.; Bizzi, S.; Piégay, H. Regional hydromorphological characterization with continuous and automated remote sensing analysis based on VHR imagery and low-resolution LiDAR data. Earth Surf. Process. Landf. 2017, 42, 531–551. [Google Scholar] [CrossRef]
- Corbane, C.; Lang, S.; Pipkins, K.; Alleaume, S.; Deshayes, M.; Millán, V.E.G.; Strasser, T.; Borre, J.V.; Toon, S.; Michael, F. Remote sensing for mapping natural habitats and their conservation status – New opportunities and challenges. Int. J. Appl. Earth Obs. Geoinf. 2015, 37, 7–16. [Google Scholar] [CrossRef]
- Ichter, J.; Evans, D.; Richard, D. Terrestrial Habitat Mapping in Europe: An Overview; European Environment Agency: Copenhagen, Denmark, 2014. [Google Scholar]
- Franke, J.; Keuck, V.; Siegert, F. Assessment of grassland use intensity by remote sensing to support conservation schemes. J. Nat. Conserv. 2012, 20, 125–134. [Google Scholar] [CrossRef]
- Schuster, C.; Schmidt, T.; Conrad, C.; Kleinschmit, B.; Förster, M. Grassland habitat mapping by intra-annual time series analysis – Comparison of RapidEye and TerraSAR-X satellite data. Int. J. Appl. Earth Obs. Geoinf. 2015, 34, 25–34. [Google Scholar] [CrossRef]
- Strasser, T.; Lang, S. Object-based class modelling for multi-scale riparian forest habitat mapping. Int. J. Appl. Earth Obs. Geoinf. 2015, 37, 29–37. [Google Scholar] [CrossRef]
- Stuart, M.; Mcgonigle, A.; Willmott, J. Hyperspectral Imaging in Environmental Monitoring: A Review of Recent Developments and Technological Advances in Compact Field Deployable Systems. Sensors 2019, 19, 3071. [Google Scholar] [CrossRef]
- Adão, T.; Hruška, J.; Pádua, L.; Bessa, J.; Peres, E.; Morais, R.; Sousa, J.J. Hyperspectral Imaging: A Review on UAV-Based Sensors, Data Processing and Applications for Agriculture and Forestry. Remote Sens. 2017, 9, 1110. [Google Scholar] [CrossRef]
- Khan, M.J.; Khan, H.S.; Yousaf, A.; Khurshid, K.; Abbas, A. Modern Trends in Hyperspectral Image Analysis: A Review. IEEE Access 2018, 6, 14118–14129. [Google Scholar] [CrossRef]
- Ghamisi, P.; Yokoya, N.; Li, J.; Liao, W.; Liu, S.; Plaza, J.; Rasti, B.; Plaza, J. Advances in Hyperspectral Image and Signal Processing: A Comprehensive Overview of the State of the Art. IEEE Geosci. Remote Sens. Mag. 2017, 5, 37–78. [Google Scholar] [CrossRef]
- Chan, J.C.-W.; Paelinckx, D. Evaluation of Random Forest and Adaboost tree-based ensemble classification and spectral band selection for ecotope mapping using airborne hyperspectral imagery. Remote Sens. Environ. 2008, 112, 2999–3011. [Google Scholar] [CrossRef]
- Delalieux, S.; Somers, B.; Haest, B.; Kooistra, L.; Mucher, S.; Borre, J.V. Monitoring heathland habitat status using hyperspectral image classification and unmixing. In Proceedings of the 2010 2nd Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing, Reykjavik, Iceland, 14–16 June 2010; pp. 1–4. [Google Scholar] [CrossRef]
- Delalieux, S.; Somers, B.; Haest, B.; Spanhove, T.; Borre, J.V.; Mucher, S. Heathland conservation status mapping through integration of hyperspectral mixture analysis and decision tree classifiers. Remote Sens. Environ. 2012, 126, 222–231. [Google Scholar] [CrossRef]
- Mucher, S.; Kooistra, L.; Vermeulen, M.; Borre, J.V.; Haest, B.; Haveman, R. Quantifying structure of Natura 2000 heathland habitats using spectral mixture analysis and segmentation techniques on hyperspectral imagery. Ecol. Indic. 2013, 33, 71–81. [Google Scholar] [CrossRef]
- Haest, B.; Thoonen, G.; Borre, J.V.; Spanhove, T.; Delalieux, S.; Bertels, L.; Kooistra, L.; Mücher, C.A.; Scheunders, P. An object-based approach to quantity and quality assessment of heathland habitats in the framework of natura 2000 using hyperspectral airborne ahs images. In Proceedings of the GEOBIA 2010 Conference, Ghent, Belgium, 29 June 2010. [Google Scholar]
- Zlinszky, A.; Schroiff, A.; Kania, A.; Deák, B.; Mücke, W.; Vári, Á.; Székely, B.; Pfeifer, N. Categorizing Grassland Vegetation with Full-Waveform Airborne Laser Scanning: A Feasibility Study for Detecting Natura 2000 Habitat Types. Remote Sens. 2014, 6, 8056–8087. [Google Scholar] [CrossRef]
- Vierling, K.T.; Vierling, L.A.; Gould, W.; Martinuzzi, S.; Clawges, R. Lidar: Shedding new light on habitat characterization and modeling. Front. Ecol. Environ. 2008, 6, 90–98. [Google Scholar] [CrossRef]
- Johansen, K.; Tiede, D.; Blaschke, T.; Arroyo, L.A.; Phinn, S. Automatic Geographic Object Based Mapping of Streambed and Riparian Zone Extent from LiDAR Data in a Temperate Rural Urban Environment, Australia. Remote Sens. 2011, 3, 1139–1156. [Google Scholar] [CrossRef]
- Onojeghuo, A.O.; Blackburn, G.A. Characterising Reedbeds Using LiDAR Data: Potential and Limitations. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 6, 935–941. [Google Scholar] [CrossRef]
- Crespo-Peremarch, P.; Tompalski, P.; Coops, N.C.; Ruiz, L.A. Characterizing understory vegetation in Mediterranean forests using full-waveform airborne laser scanning data. Remote Sens. Environ. 2018, 217, 400–413. [Google Scholar] [CrossRef]
- Onojeghuo, A.O.; Onojeghuo, A.R. Object-based habitat mapping using very high spatial resolution multispectral and hyperspectral imagery with LiDAR data. Int. J. Appl. Earth Obs. Geoinf. 2017, 59, 79–91. [Google Scholar] [CrossRef]
- Onojeghuo, A.O.; Blackburn, G.A. Optimising the use of hyperspectral and LiDAR data for mapping reedbed habitats. Remote Sens. Environ. 2011, 115, 2025–2034. [Google Scholar] [CrossRef]
- Ramdani, F. Urban Vegetation Mapping from Fused Hyperspectral Image and LiDAR Data with Application to Monitor Urban Tree Heights. J. Geogr. Inf. Syst. 2013, 5, 404–408. [Google Scholar] [CrossRef]
- Hladik, C.; Schalles, J.; Alber, M. Salt marsh elevation and habitat mapping using hyperspectral and LIDAR data. Remote Sens. Environ. 2013, 139, 318–330. [Google Scholar] [CrossRef]
- Marcinkowska-ochtyra, A.; Gryguc, K.; Ochtyra, A.; Kope, D.; Jaroci, A. Multitemporal Hyperspectral Data Fusion with Topographic Indices — Improving Classification of Natura 2000 Grassland Habitats. Remote Sens. 2019, 11, 2264. [Google Scholar] [CrossRef]
- Sankey, T.; McVay, J.; Swetnam, T.; McClaran, M.P.; Heilman, P.; Nichols, M. UAV hyperspectral and lidar data and their fusion for arid and semi-arid land vegetation monitoring. Remote Sens. Ecol. Conserv. 2017, 4, 20–33. [Google Scholar] [CrossRef]
- Dashti, H.; Poley, A.; Glenn, N.F.; Ilangakoon, N.T.; Spaete, L.; Roberts, D.; Enterkine, J.; Flores, A.N.; Ustin, S.L.; Mitchell, J.J. Regional Scale Dryland Vegetation Classification with an Integrated Lidar-Hyperspectral Approach. Remote Sens. 2019, 11, 2141. [Google Scholar] [CrossRef]
- Bellman, R. Adaptive Control Processes; Princeton Univ. Press: Princeton, NJ, USA, 1961. [Google Scholar]
- Bruce, L.; Koger, C.; Li, J. Dimensionality reduction of hyperspectral data using discrete wavelet transform feature extraction. IEEE Trans. Geosci. Remote Sens. 2002, 40, 2331–2338. [Google Scholar] [CrossRef]
- Kiala, Z.; Mutanga, O.; Odindi, J.; Peerbhay, K. Feature Selection on Sentinel-2 Multispectral Imagery for Mapping a Landscape Infested by Parthenium Weed. Remote Sens. 2019, 11, 1892. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, R.; Wang, S.; Wang, F. Feature Selection Method Based on High-Resolution Remote Sensing Images and the Effect of Sensitive Features on Classification Accuracy. Sensors 2018, 18, 2013. [Google Scholar] [CrossRef]
- Demarchi, L.; Canters, F.; Cariou, C.; Licciardi, G.A.; Chan, J.C.-W. Assessing the performance of two unsupervised dimensionality reduction techniques on hyperspectral APEX data for high resolution urban land-cover mapping. Isprs J. Photogramm. Remote Sens. 2014, 87, 166–179. [Google Scholar] [CrossRef]
- Hasani, H.; Samadzadegan, F.; Reinartz, P. A metaheuristic feature-level fusion strategy in classification of urban area using hyperspectral imagery and LiDAR data. Eur. J. Remote Sens. 2017, 50, 222–236. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Fusion of Hyperspectral and LIDAR Remote Sensing Data for Classification of Complex Forest Areas. IEEE Trans. Geosci. Remote Sens. 2008, 46, 1416–1427. [Google Scholar] [CrossRef]
- Ghamisi, P.; Mura, M.D.; Benediktsson, J.A. A Survey on Spectral–Spatial Classification Techniques Based on Attribute Profiles. IEEE Trans. Geosci. Remote Sens. 2014, 53, 2335–2353. [Google Scholar] [CrossRef]
- Dian, Y.; Pang, Y.; Dong, Y.; Li, Z. Urban Tree Species Mapping Using Airborne LiDAR and Hyperspectral Data. J. Indian Soc. Remote Sens. 2016, 44, 595–603. [Google Scholar] [CrossRef]
- Khodadadzadeh, M.; Li, J.; Prasad, S.; Plaza, J. Fusion of Hyperspectral and LiDAR Remote Sensing Data Using Multiple Feature Learning. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 2971–2983. [Google Scholar] [CrossRef]
- Liu, X.; Bo, Y. Object-Based Crop Species Classification Based on the Combination of Airborne Hyperspectral Images and LiDAR Data. Remote Sens. 2015, 7, 922–950. [Google Scholar] [CrossRef]
- Pan, Z.; Glennie, C.; Fernandez-Diaz, J.C.; Shrestha, R.; Carter, B.; Hauser, D.; Singhania, A.; Sartori, M. Fusion of bathymetric LiDAR and hyperspectral imagery for shallow water bathymetry. 2016 IEEE Int. Geosci. Remote Sens. Symp. (Igarss) 2016, 2016, 3792–3795. [Google Scholar] [CrossRef]
- Pullanagari, R.R.; Kereszturi, G.; Yule, I. Integrating Airborne Hyperspectral, Topographic, and Soil Data for Estimating Pasture Quality Using Recursive Feature Elimination with Random Forest Regression. Remote Sens. 2018, 10, 1117. [Google Scholar] [CrossRef]
- Darst, B.F.; Malecki, K.; Engelman, C.D. Using recursive feature elimination in random forest to account for correlated variables in high dimensional data. Bmc Genet. 2018, 19, 65. [Google Scholar] [CrossRef]
- Bahl, A.; Hellack, B.; Balas, M.; Dinischiotu, A.; Wiemann, M.; Brinkmann, J.; Luch, A.; Renard, B.Y.; Haase, A. Recursive feature elimination in random forest classification supports nanomaterial grouping. NanoImpact 2019, 15, 100179. [Google Scholar] [CrossRef]
- Granitto, P.M.; Furlanello, C.; Biasioli, F.; Gasperi, F. Recursive feature elimination with random forest for PTR-MS analysis of agroindustrial products. Chemom. Intell. Lab. Syst. 2006, 83, 83–90. [Google Scholar] [CrossRef]
- Frassy, F.; Via, G.D.; Maianti, P.; Marchesi, A.; Nodari, F.R.; Gianinetto, M. Minimum noise fraction transform for improving the classification of airborne hyperspectral data: Two case studies. In Proceedings of the 2013 5th Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Gainesville, FL, USA, 26–28 June 2013; pp. 1–4. [Google Scholar] [CrossRef]
- Qian, S.-E. Dimensionality Reduction of Hyperspectral Imagery. In Optical Satellite Signal Processing and Enhancement; Society of Photo-Optical Instrumentation Engineers, SPIE eBooks: Bellingham, WA, USA, 2013. [Google Scholar] [CrossRef]
- Priyadarshini, K.N.; Sivashankari, V.; Shekhar, S.; Balasubramani, K. Comparison and Evaluation of Dimensionality Reduction Techniques for Hyperspectral Data Analysis. Proceedings 2019, 24, 6. [Google Scholar] [CrossRef]
- Luo, G.; Chen, G.; Tian, L.; Qin, K.; Qian, S.-E. Minimum Noise Fraction versus Principal Component Analysis as a Preprocessing Step for Hyperspectral Imagery Denoising. Can. J. Remote Sens. 2016, 42, 106–116. [Google Scholar] [CrossRef]
- Banaszuk, H.; Micun, K. Formation and evolution of river valleys in large melt-out depressions in the North Podlasie Lowland. Pr. I Stud. Geogr. 2009, 41, 25–36. [Google Scholar]
- Wierzbicki, G.; Ostrowski, P.; Falkowski, T.; Mazgajski, M. Geological setting control of flood dynamics in lowland rivers (Poland). Sci. Total. Environ. 2018, 636, 367–382. [Google Scholar] [CrossRef] [PubMed]
- Kazuń, A. Alluvial meadows of Cnidion dubii Bal.-Tul. 1966 in the Middle Oder River Valley (Natura 2000 site “Łęgi Odrzańskie”, SW Poland). Steciana 2015, 18, 49–55. [Google Scholar] [CrossRef]
- Kącki, Z.; Czarniecka, M.; Swacha, G. Statistical determination of diagnostic, constant and dominant species of the higher vegetation units of Poland. Monogr. Bot. 2014, 103, 1–267. [Google Scholar] [CrossRef]
- Załuski, T. 6440 Łąki selernicowe (Cnidion dubii). In Sprawozdanie z prac monitoringowych w roku 2010; Cierlik, G., Makomaska-Juchiewicz, M., Mróz, W., Perzanowska, J., Król, W., Baran, P.A.Z., Eds.; Instytut Ochrony Przyrody PAN: Kraków, Poland, 2010; Volume 1, pp. 182–200. [Google Scholar]
- Jermaczek-Sitak, M. Charakter i stan zachowania łąk selernicowych Cnidion w zachodniej Polsce a warunki wodne. Przegląd Przyr. 2011, 22, 83–90. [Google Scholar]
- Rodríguez-Rojo, M.P.; Jiménez-Alfaro, B.; Jandt, U.; Bruelheide, H.; Rodwell, J.S.; Schamineée, J.; Perrin, P.; Kącki, Z.; Willner, W.; Fernández-González, F.; et al. Diversity of lowland hay meadows and pastures in Western and Central Europe. Appl. Veg. Sci. 2017, 20, 702–719. [Google Scholar] [CrossRef]
- Kucharski, L. Vegetation of oat-grass meadows in Central Poland. Steciana 2015, 18, 119–125. [Google Scholar] [CrossRef]
- Faust, C.; Süss, K.; Storm, C.; Schwabe, A. Threatened inland sand vegetation in the temperate zone under different types of abiotic and biotic disturbances during a ten-year period. Flora Morphol. Distrib. Funct. Ecol. Plants 2011, 206, 611–621. [Google Scholar] [CrossRef]
- Willner, W.; Roleček, J.; Korolyuk, A.; Dengler, J.; Chytrý, M.; Janišová, M.; Lengyel, A.; Aćić, S.; Becker, T.; Ćuk, M.; et al. Formalized classification of semi-dry grasslands in central and eastern Europe. Preslia 2019, 91, 25–49. [Google Scholar] [CrossRef]
- Marcinkowska-Ochtyra, A.; Jarocińska, A.; Bzdęga, K.; Tokarska-Guzik, B. Classification of Expansive Grassland Species in Different Growth Stages Based on Hyperspectral and LiDAR Data. Remote Sens. 2018, 10, 2019. [Google Scholar] [CrossRef]
- Sławik, Ł.; Niedzielko, J.; Kania, A.; Piórkowski, H.; Kopeć, D. Multiple Flights or Single Flight Instrument Fusion of Hyperspectral and ALS Data? A Comparison of their Performance for Vegetation Mapping. Remote Sens. 2019, 11, 970. [Google Scholar] [CrossRef]
- Halladin-Dąbrowska, A.; Kania, A.; Kopeć, D. The t-SNE Algorithm as a Tool to Improve the Quality of Reference Data Used in Accurate Mapping of Heterogeneous Non-Forest Vegetation. Remote Sens. 2019, 12, 39. [Google Scholar] [CrossRef]
- Pelletier, C.; Valero, S.; Inglada, J.; Champion, N.; Sicre, C.M.; Dedieu, G. Effect of Training Class Label Noise on Classification Performances for Land Cover Mapping with Satellite Image Time Series. Remote Sens. 2017, 9, 173. [Google Scholar] [CrossRef]
- Ge, Y.; Bai, H.; Wang, J.; Cao, F. Assessing the quality of training data in the supervised classification of remotely sensed imagery: A correlation analysis. J. Spat. Sci. 2012, 57, 135–152. [Google Scholar] [CrossRef]
- van der Maaten, L.; Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Schläpfer, D.; Richter, R. Geo-atmospheric processing of airborne imaging spectrometry data. Part 1: Parametric orthorectification. Int. J. Remote Sens. 2002, 23, 2609–2630. [Google Scholar] [CrossRef]
- Richter, R.; Schläpfer, D. Geo-atmospheric processing of wide-FOV airborne imaging spectrometry data. Int. Symp. Remote Sens. 2002, 4545, 264–273. [Google Scholar] [CrossRef]
- ITT Visual Information Solutions. ENVI User’s Guide. Available online: http://www.harrisgeospatial.com/portals/0/pdfs/envi/ENVI_User_Guide.pdf (accessed on 1 June 2020).
- Mandlburger, G.; Otepka, J.; Karel, W.; Wagner, W.; Pfeifer, N. Orientation and processing of Airborne Laser Scanning data (OPALS)-Concept and first results of a comprehensive ALS software. In Proceedings of the Laser Scanning 2009, IAPRS, Paris, France, 1–2 September 2009. [Google Scholar]
- Conrad, O.; Bechtel, B.; Bock, M.; Dietrich, H.; Fischer, E.; Gerlitz, L.; Wehberg, J.; Wichmann, V.; Böhner, J. System for Automated Geoscientific Analyses (SAGA) v. 2.1.4. Geosci. Model Dev. 2015, 8, 1991–2007. [Google Scholar] [CrossRef]
- Kania, A.; Kopeć, D.; Niedzielko, J.; Sławik, Ł. Automated and efficient workflow for large airborne remote sensing vegetation mapping and research of Natura 2000 habitats. In Proceedings of the ICEI 2018: 10th International Conference on Ecological Informatics, Jena, Germany, 24–28 September 2018. [Google Scholar]
- Guthrie, N.; Kotz, S.; Johnson, N.L. Breakthrough in Statistics. J. Am. Stat. Assoc. 1993, 88, 388. [Google Scholar] [CrossRef]
- Andrade, C. The P Value and Statistical Significance: Misunderstandings, Explanations, Challenges, and Alternatives. Indianj. Psychol. Med. 2019, 41, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Boehner, J.; Antonic, O. Chapter 8: Land Surface Parameters Specific to Topo-Climatology. Dev. Soil Sci. 2009, 33, 195–226. [Google Scholar] [CrossRef]
- Wilson, J.P.; Gallant, J.C. Terrain Analysis - Principles and Applications; John Wiley & Sons, Inc.: New York, NY, USA, 2000. [Google Scholar]
- Gallant, J.C.; Dowling, T.I. A multiresolution index of valley bottom flatness for mapping depositional areas. Water Resour. Res. 2003, 39, 39. [Google Scholar] [CrossRef]
- Guisan, A.; Weiss, S.B. GLM versus CCA spatial modeling of plant species distribution. Plant Ecol. 1999, 143, 107–122. [Google Scholar] [CrossRef]
- Boehner, J.; Selige, T. Spatial prediction of soil attributes using terrain analysis and climate regionalisation. In SAGA—Analysis and Modelling Applications; Boehner, J., McCloy, K.R., Strobl, J., Eds.; Goettinger Geographische Abhandlungen: Goettingen, Germany, 2006; pp. 13–28. [Google Scholar]
- Gitelson, A.A.; Zur, Y.; Chivkunova, O.B.; Merzlyak, M.N. Assessing Carotenoid Content in Plant Leaves with Reflectance Spectroscopy¶. Photochem. Photobiol. 2002, 75, 272. [Google Scholar] [CrossRef]
- Daughtry, C.; Hunt, E.; McMurtrey, J. Assessing crop residue cover using shortwave infrared reflectance. Remote Sens. Environ. 2004, 90, 126–134. [Google Scholar] [CrossRef]
- Mallick, D.I.J. A review of: “Image Interpretation in Geology ” by S. A. Drury. London: Allen & Unwin. Int. J. Remote Sens. 1987, 8, 1399–1400. [Google Scholar] [CrossRef]
- Pinty, B.; Verstraete, M. GEMI: A non-linear index to monitor global vegetation from satellites. Vegetatio 1992, 101, 15–20. [Google Scholar] [CrossRef]
- Segal, D. Theoretical Basis for Differentiation of Ferric-Iron Bearing Minerals, Using Landsat MSS Data. In Proceedings of the 2nd Thematic Conference on Remote Sensing for Exploratory Geology, Symposium for Remote Sensing of Environment, Fort Worth, TX, USA, 6–10 December 1982; pp. 949–951. [Google Scholar]
- Daughtry, C. Estimating Corn Leaf Chlorophyll Concentration from Leaf and Canopy Reflectance. Remote Sens. Environ. 2000, 74, 229–239. [Google Scholar] [CrossRef]
- Haboudane, D. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Serrano, L.; Penuelas, J.; Ustin, S.L. Remote sensing of nitrogen and lignin in Mediterranean vegetation from AVIRIS data. Remote Sens. Environ. 2002, 81, 355–364. [Google Scholar] [CrossRef]
- Fourty, T.; Baret, F.; Jacquemoud, S.; Schmuck, G.; Verdebout, J. Leaf optical properties with explicit description of its biochemical composition: Direct and inverse problems. Remote Sens. Environ. 1996, 56, 104–117. [Google Scholar] [CrossRef]
- Penuelas, J.; Filella, I.; Gamon, J.A. Assessment of photosynthetic radiation-use efficiency with spectral reflectance. New Phytol. 1995, 131, 291–296. [Google Scholar] [CrossRef]
- Gamon, J.A.; Serrano, L.; Surfus, J.S. The photochemical reflectance index: An optical indicator of photosynthetic radiation use efficiency across species, functional types, and nutrient levels. Oecologia 1997, 112, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Penuelas, J.; Baret, F.; Filella, I. Semi-Empirical Indices to Assess Carotenoids/Chlorophyll-a Ratio from Leaf Spectral Reflectance. Photosynthetica 1995, 31, 221–230. [Google Scholar]
- Broge, N.; Leblanc, E. Comparing prediction power and stability of broadband and hyperspectral vegetation indices for estimation of green leaf area index and canopy chlorophyll density. Remote Sens. Environ. 2001, 76, 156–172. [Google Scholar] [CrossRef]
- Wolf, A.F. Using WorldView-2 Vis-NIR multispectral imagery to support land mapping and feature extraction using normalized difference index ratios. Spie Def. Secur. Sens. 2012, 8390, 83900. [Google Scholar] [CrossRef]
- Mokarram, M.; Roshan, G.; Negahban, S. Landform classification using topography position index (case study: Salt dome of Korsia-Darab plain, Iran). Model. Earth Syst. Environ. 2015, 1, 40. [Google Scholar] [CrossRef]
- García-Rivero, A.E.; Olivera, J.; Salinas, E.; Yuli, R.A.; Bulege, W. Use of Hydrogeomorphic Indexes in SAGA-GIS for the Characterization of Flooded Areas in Madre de Dios, Peru. Int. J. Appl. Eng. Res. 2017, 12, 9078–9086. [Google Scholar]



| Habitat Class | ||||||
|---|---|---|---|---|---|---|
| Area | Acquisition | 6120 | 6440 | 6510 | Background (9999) | Total |
| BN | Spring | 144 | 492 | 105 | 722 | 1463 |
| Summer | 146 | 376 | 111 | 619 | 1252 | |
| Autumn | 141 | 315 | 74 | 550 | 1080 | |
| BG1 | Spring | 191 | - | 235 | 1036 | 1462 |
| Summer | 180 | - | 193 | 917 | 1290 | |
| Autumn | 192 | - | 249 | 996 | 1437 | |
| BG2 | Spring | 272 | 289 | - | 587 | 1148 |
| Summer | 268 | 224 | - | 586 | 1078 | |
| Objective | Objective | Input features | RFE | Feature nr. | Runs | Training/Validation Sampling |
|---|---|---|---|---|---|---|
| 1a | Spectral features of importance for selected habitats classification | SR | yes | 430 | 10 | 50/50, random manual selection |
| 1b | MNF | no | 30 | 10 | 50/50, random manual selection | |
| 2 | Best-to-use HS + ALS products and accuracy improvement | MNF + LiDAR + SI | yes | 188 | 10 | 50/50, random manual selection |
| 3a | Feature selection validation attempt | Area-dependent selection | no | 24-28 | 50 | 50/50, random automatic selection |
| 3b | Common selection | no | 24 | 50 | 50/50, random automatic selection |
| Exp. 3a | Exp. 3b | ||||||
|---|---|---|---|---|---|---|---|
| BN | BG1 | BG2 | ALL | ||||
| LiDAR OPALS: | Category: | Input Layers: | Ref.: | ||||
| ALL_Amplitude_mean | Morphology | DTM | - | ||||
| DSM_sigma0 | Morphology | DSM | - | ||||
| DTM_sigma0 | Morphology | DTM | - | ||||
| LiDAR SAGA: | |||||||
| DiffI (Diffuse Insolation) | Light availability | DSM | [97,98] | ||||
| DurI (Duration of Insolation) | Light availability | DSM | [97,98] | ||||
| MRRTF (Multiresolution Index of the Ridge Top Flatness) | Morphology | DTM | [99] | ||||
| MRVBF (Multiresolution Index of Valley Bottom Flatness) | Morphology | DTM | [99] | ||||
| TPI (Topographic Position Index) | Morphology | DTM | [98,100] | ||||
| MCA (Modified Catchment Area) | Wetness | DTM | [101] | ||||
| TWI (Topographic Wetness Index) | Wetness | DTM | [101] | ||||
| Spectral Indices: | |||||||
| 5-CRI1: Carotenoid Reflectance Index 1 | Leaf Pigments | 510, 550 | [102] | ||||
| 7-CAI: Cellulose Absorption Index | Dry or Senescent Carbon | 2000, 2200, 2100 | [103] | ||||
| 8-CM: Clay Minerals Ratio | Geology Indices | 1550–1750, 2080–2350 | [104] | ||||
| 12-GEMI: Global Environmental Monitoring Index | Broadband Greenness | 650, 850 | [105] | ||||
| 19-IO: Iron Oxide Ratio | Geology Indices | 450–520, 630–690 | [104,106] | ||||
| 21-MCARI: Modified Chlorophyll Absorption Ratio Index | Narrowband Greenness | 550, 670, 700 | [107] | ||||
| 22-MCARI2: Modified Chlorophyll Absorption Ratio Index Improved | Narrowband Greenness | 550, 670, 800 | [108] | ||||
| 28-MTVI: Modified Triangular Vegetation Index | Narrowband Greenness | 550, 670, 800 | [108] | ||||
| 35-NDLI: Normalized Difference Lignin Index | Dry or Senescent Carbon | 1680, 1754 | [109,110] | ||||
| 37-NDNI: Normalized Difference Nitrogen Index | Canopy Nitrogen | 1510, 1680 | [109,110] | ||||
| 43-PRI: Photochemical Reflectance Index | Light Use Efficiency | 531, 570 | [111,112] | ||||
| 51-SIPI: Structure Insensitive Pigment Index | Light Use Efficiency | 445, 680, 800 | [113] | ||||
| 53-TCARI: Transformed Chlorophyll Absorption Reflectance Index | Narrowband Greenness | 550, 670, 700 | [108] | ||||
| 55-TVI: Triangular Vegetation Index | Narrowband Greenness | 550, 670, 750 | [114] | ||||
| 60-WV-BI: WorldView Built-Up Index | Other | 450, 730 | [115] | ||||
| 62-WV-II: WorldView New Iron Index | Geology Indices | 550, 600, 470 | [115] | ||||
| 63-WVNHFD: WorldView Non-Homogeneous Feature Difference | Other | 450, 730 | [115] | ||||
| 65-WV-WI: WorldView Water Index | Other | 450, 870–1040 | [115] | ||||
| MNF components: | |||||||
| MNF 1, 3, 5, 6, 7, 9 | - | - | - | ||||
| MNF 2, 4 | - | - | - | ||||
| MNF 8, 10 | - | - | - | ||||
| MNF 11 | - | - | - | ||||
| MNF 13, 15 | - | - | - | ||||
| MNF 16 | - | - | - | ||||
| Total number of used features | 28 | 26 | 24 | 24 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demarchi, L.; Kania, A.; Ciężkowski, W.; Piórkowski, H.; Oświecimska-Piasko, Z.; Chormański, J. Recursive Feature Elimination and Random Forest Classification of Natura 2000 Grasslands in Lowland River Valleys of Poland Based on Airborne Hyperspectral and LiDAR Data Fusion. Remote Sens. 2020, 12, 1842. https://doi.org/10.3390/rs12111842
Demarchi L, Kania A, Ciężkowski W, Piórkowski H, Oświecimska-Piasko Z, Chormański J. Recursive Feature Elimination and Random Forest Classification of Natura 2000 Grasslands in Lowland River Valleys of Poland Based on Airborne Hyperspectral and LiDAR Data Fusion. Remote Sensing. 2020; 12(11):1842. https://doi.org/10.3390/rs12111842
Chicago/Turabian StyleDemarchi, Luca, Adam Kania, Wojciech Ciężkowski, Hubert Piórkowski, Zuzanna Oświecimska-Piasko, and Jarosław Chormański. 2020. "Recursive Feature Elimination and Random Forest Classification of Natura 2000 Grasslands in Lowland River Valleys of Poland Based on Airborne Hyperspectral and LiDAR Data Fusion" Remote Sensing 12, no. 11: 1842. https://doi.org/10.3390/rs12111842
APA StyleDemarchi, L., Kania, A., Ciężkowski, W., Piórkowski, H., Oświecimska-Piasko, Z., & Chormański, J. (2020). Recursive Feature Elimination and Random Forest Classification of Natura 2000 Grasslands in Lowland River Valleys of Poland Based on Airborne Hyperspectral and LiDAR Data Fusion. Remote Sensing, 12(11), 1842. https://doi.org/10.3390/rs12111842