Activation of Steroidogenesis, Anti-Apoptotic Activity, and Proliferation in Porcine Granulosa Cells by RUNX1 Is Negatively Regulated by H3K27me3 Transcriptional Repression
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Porcine Granulosa Cell Culture
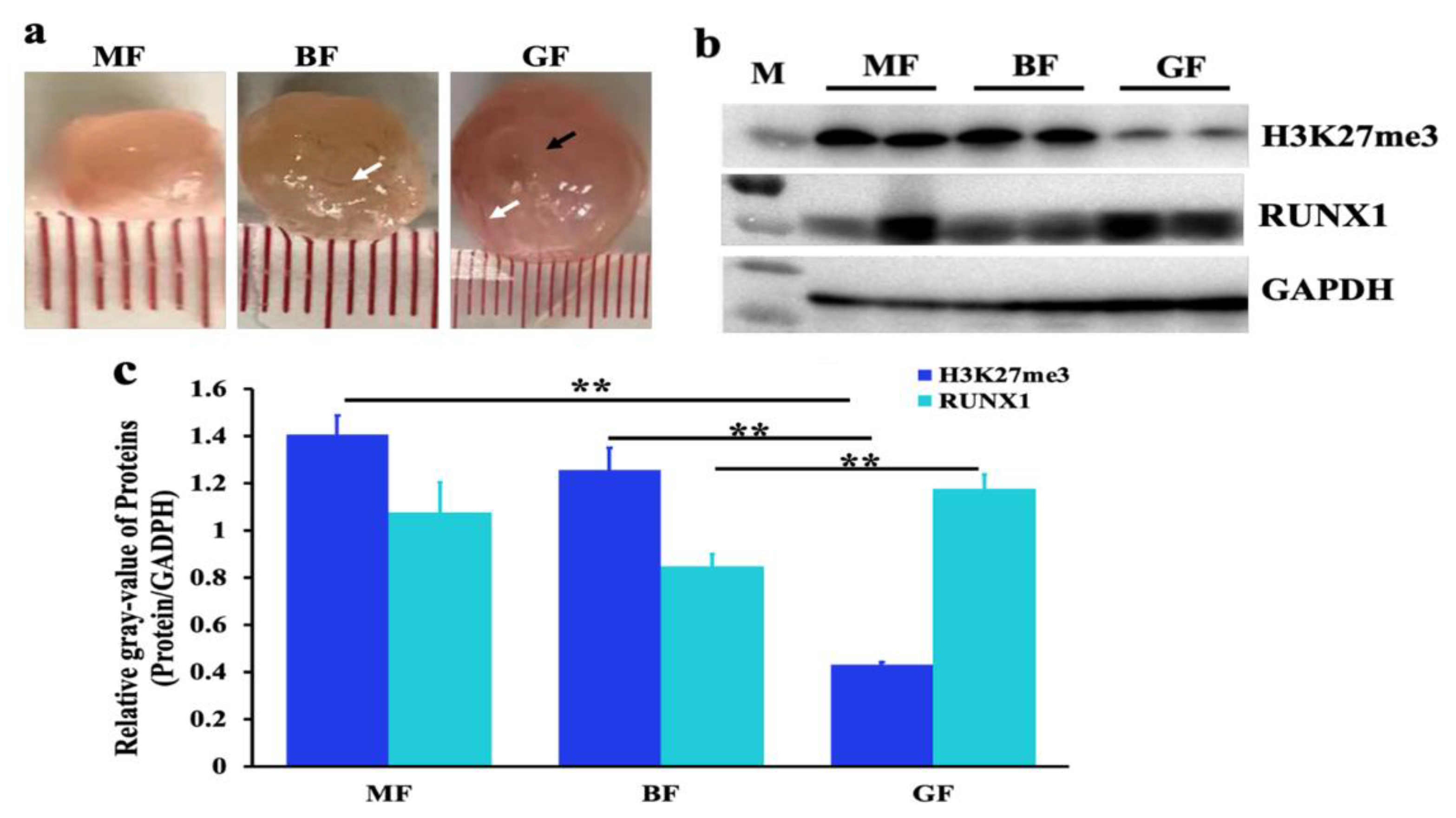
2.3. Expression Profiles of H3K27me3 in Different Stage Follicles
2.4. H3K27me3 Inhibitor and Activator
2.5. Reconstructed Vectors and RNAi Fragments of RUNX1
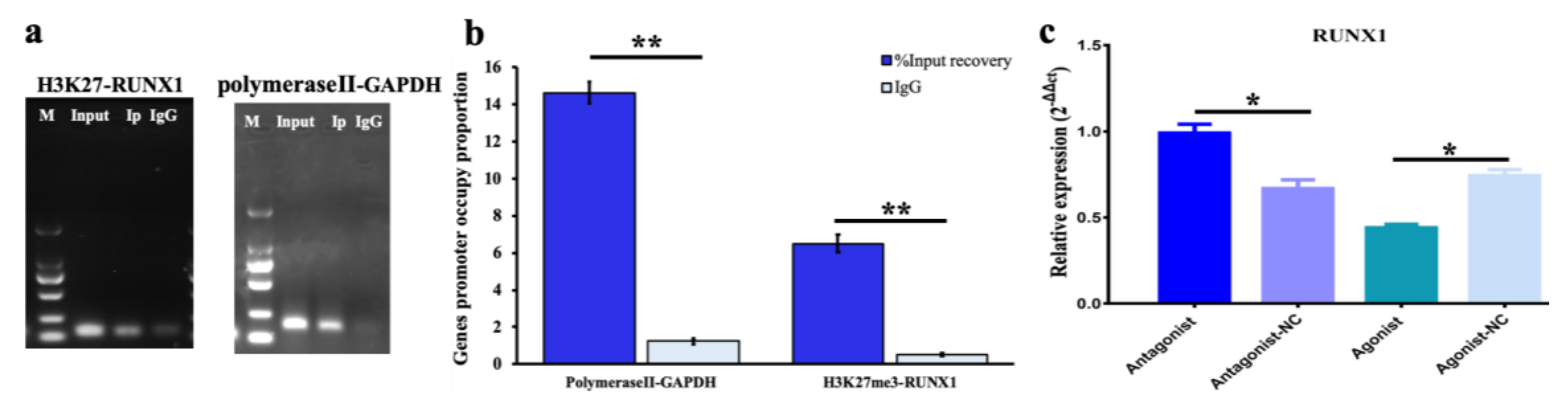
2.6. Bioinformation Prediction and Chromatin Immunoprecipitation (ChIP)
2.7. qPCR
2.8. Western Blotting
2.9. Hormone Detection
2.10. Cell Apoptosis and Proliferation
2.11. Data Analysis
3. Result
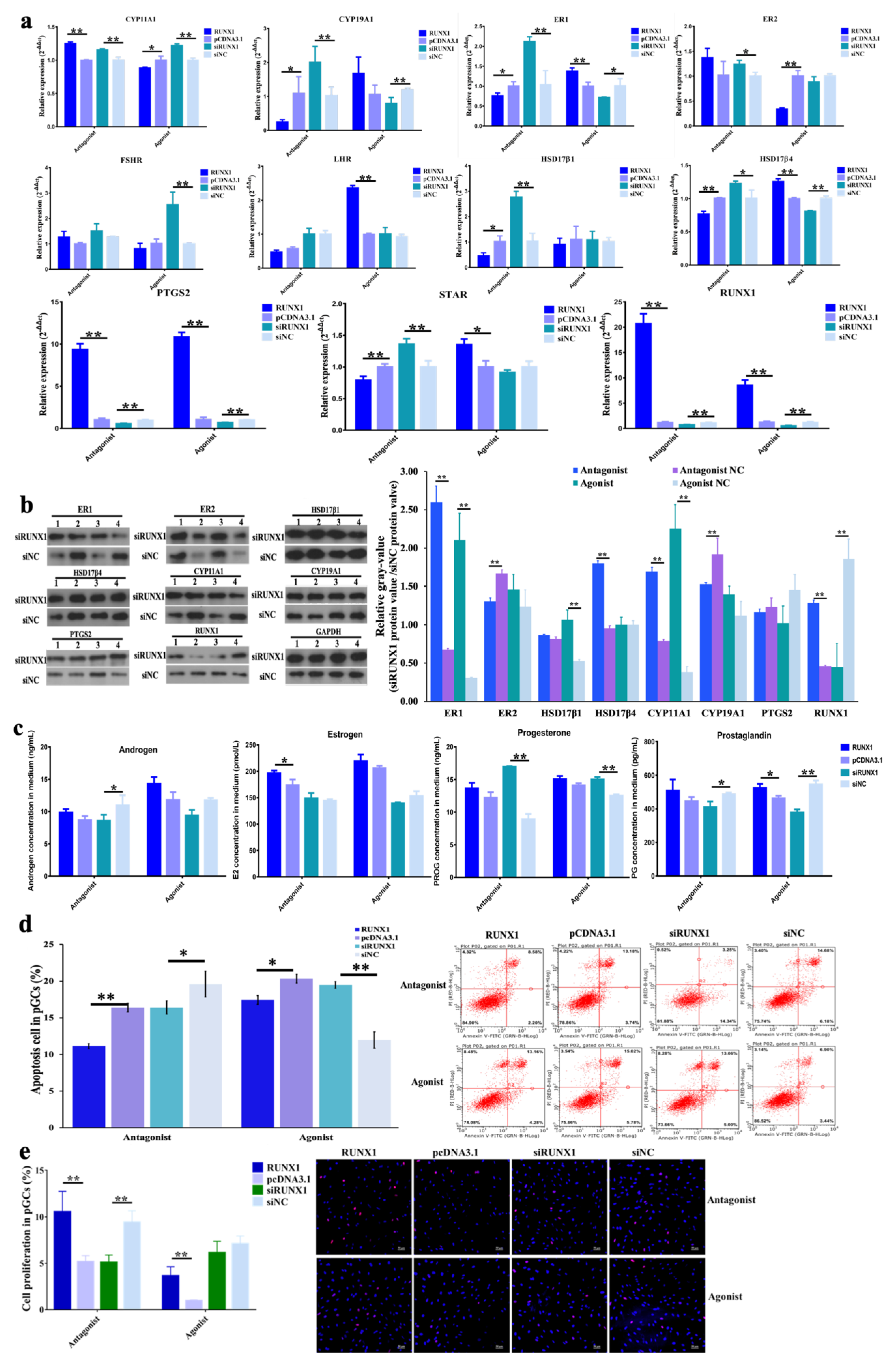
3.1. Function of H3K27me3 in Steroidogenesis of pGCs
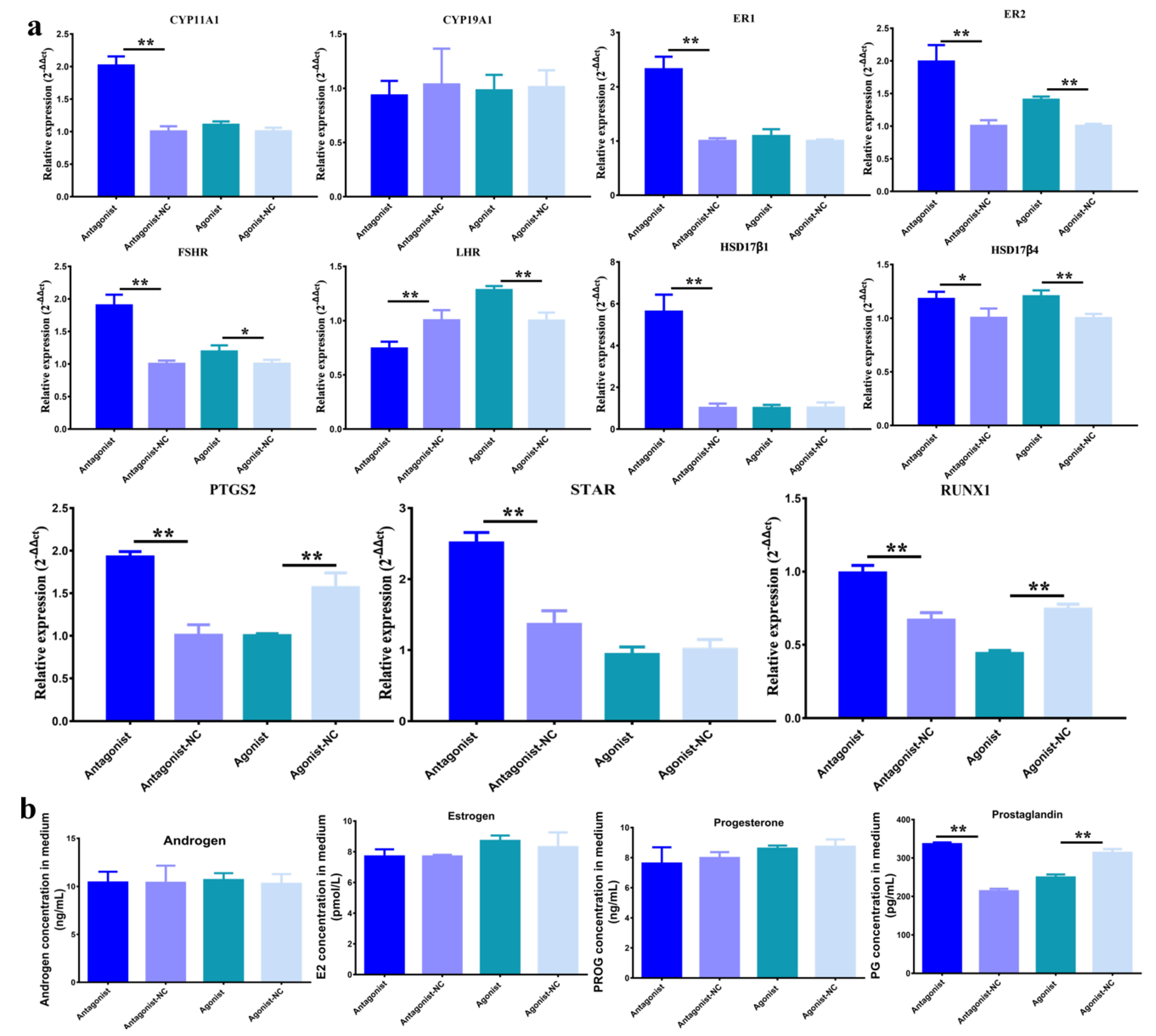
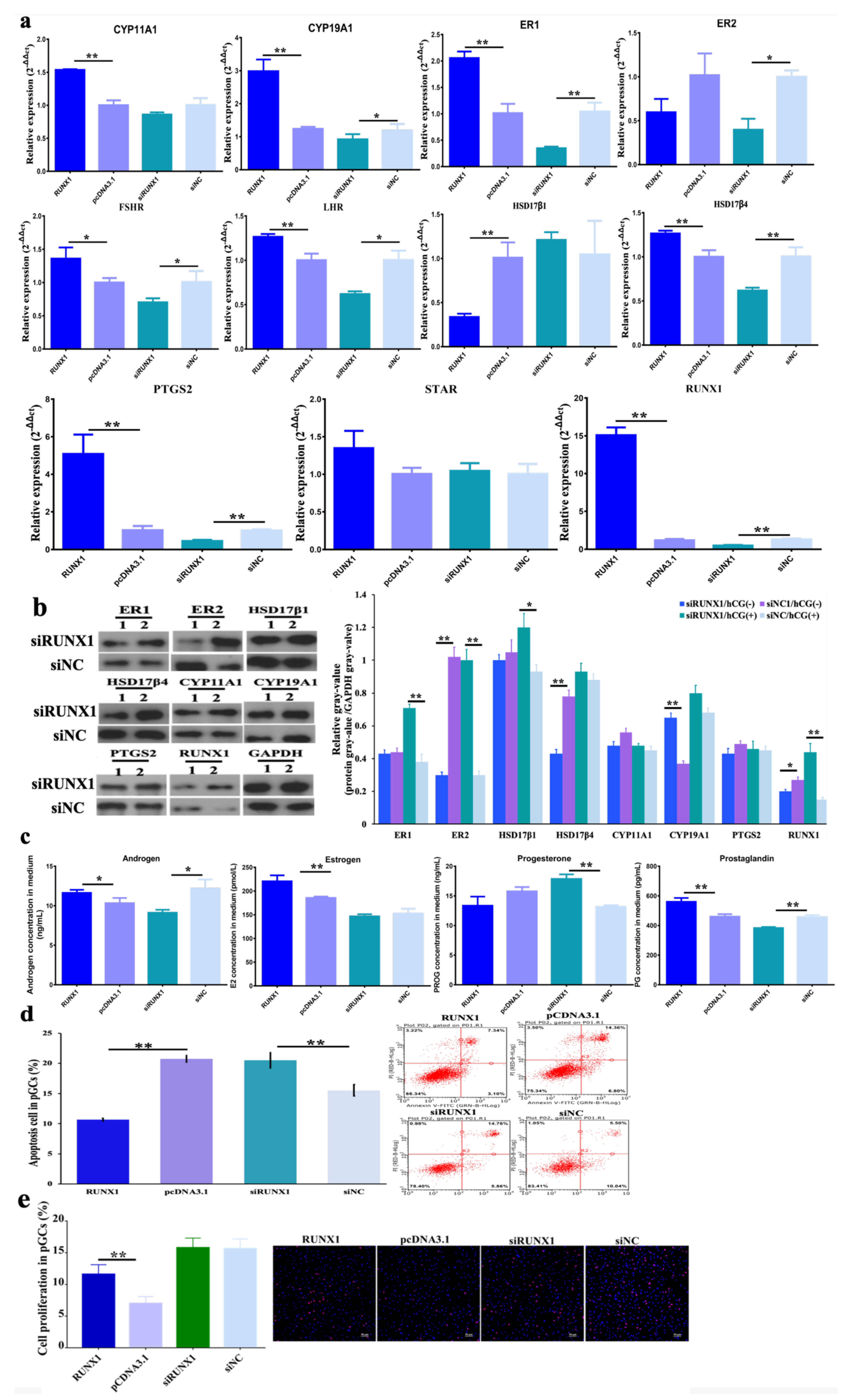
3.2. Function of H3K27me3 Target RUNX1for Steroidogenesis in pGCs
3.3. Effect of H3K27me3-RUNX1 Signals on Steroidogenesis in pGCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Matsuda, F.; Inoue, N.; Manabe, N.; Ohkura, S. Follicular growth and atresia in mammalian ovaries: Regulation by survival and death of granulosa cells. J. Reprod. Dev. 2012, 58, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Ciesiółka, S.; Budna, J.; Jopek, K.; Bryja, A.; Kranc, W.; Chachuła, A.; Borys, S.; Dyszkiewicz Konwińska, M.; Ziółkowska, A.; Antosik, P.; et al. Influence of Estradiol-17beta on Progesterone and Estrogen Receptor mRNA Expression in Porcine Follicular Granulosa Cells during Short-Term, in Vitro Real-Time Cell Proliferation. Biomed. Res. Int. 2016, 2016, 8431018. [Google Scholar] [CrossRef] [PubMed]
- Sekar, N.; Lavoie, H.A.; Veldhuis, J.D. Concerted regulation of steroidogenic acute regulatory gene expression by luteinizing hormone and insulin (or insulin-like growth factor I) in primary cultures of porcine granulosa-luteal cells. Endocrinology 2000, 141, 3983–3992. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alves, A.E.; Padilha-Nakaghi, L.C.; Pires-Butler, E.A.; Apparicio, M.; Silva, N.A.M.; Motheo, T.F.; Vicente, W.R.R.; Luvoni, G.C. Viability and growth of feline preantral follicles in vitro cultured with insulin growth factor and epidermal growth factor supplemented medium. Reprod. Domest. Anim. 2017, 52, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, F.; Inoue, N.; Maeda, A.; Cheng, Y.; Sai, T.; Gonda, H.; Goto, Y.; Sakamaki, K.; Manabe, N. Expression and function of apoptosis initiator FOXO3 in granulosa cells during follicular atresia in pig ovaries. J. Reprod. Dev. 2011, 57, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Fu, Y.H.; Han, J.; Shen, M.; Du, C.W.; Li, R.; Ma, X.S.; Liu, H.L. Changes in the expression of FoxO1 and death ligand genes during follicular atresia in porcine ovary. Genet. Mol. Res. 2014, 13, 6638–6645. [Google Scholar] [CrossRef]
- Cai, L.; Sun, A.; Li, H.; Tsinkgou, A.; Yu, J.; Ying, S.; Chen, Z.; Shi, Z. Molecular mechanisms of enhancing porcine granulosa cell proliferation and function by treatment in vitro with anti-inhibin alpha subunit antibody. Reprod. Biol. Endocrinol. 2015, 13, 1–10. [Google Scholar] [CrossRef]
- Terenina, E.; Fabre, S.; Bonnet, A.; Monniaux, D.; Robert-Granié, C.; Sancristobal, M.; Sarry, J.; Vignoles, F.; Gondret, F.; Monget, P.; et al. Differentially expressed genes and gene networks involved in pig ovarian follicular atresia. Physiol. Genom. 2017, 49, 67–80. [Google Scholar] [CrossRef]
- Hammond, J.M.; Baranao, J.L.S.; Skaleris, D.; Knight, A.B.; Romanus, J.A.; Rechler, M.M. Production of Insulin- Like Growth Factors by. Endocrinology 1985, 117, 2553–2555. [Google Scholar] [CrossRef]
- Medical, T.; Medical, T. Endothelin-1 is an autocrine/paracrine regulator of porcine granulosa cells Key-words. J. Endocrinol. Investig. 1993, 16, 425–431. [Google Scholar]
- Hull, K.L.; Harvey, S. Growth hormone: A reproductive endocrine-paracrine regulator? Rev. Reprod. 2000, 5, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Tsang, B.K.; Ainsworth, L.; Downey, B.R.; Marcus, G.J. Differential production of steroids by dispersed granulosa and theca interna cells from developing preovulatory follicles of pigs. J. Reprod. Fertil. 1985, 74, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Marcus, G.J.; Ainsworth, L.; Tsang, B.K. Prostaglandin Production Cells from by Dispersed Porcine Granulosa and Theca Interna Preovulatory. Biol. Reprod. 1984, 121, 115–121. [Google Scholar]
- Oxender, D.; Colenbrander, B. Ovarian Development in Fetal and Prepubertal Pigs retard. Biol. Reprod. 1979, 21, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Esbenshade, K.L.; Paterson, A.M.; Cantley, T.C.; Day, B.N. Changes in plasma hormone concentrations associated with the onset of puberty in the gilt. J. Anim. Sci. 1982, 54, 320–324. [Google Scholar] [CrossRef]
- Ainsworth, L.; Tsang, B.K.; Downey, B.R.; Marcus, G.J. The synthesis and actions of steroids and prostaglandins during follicular maturation in the pig. J. Reprod. Fertil. Suppl. 1990, 40, 137–150. [Google Scholar]
- Strott, C.A.; Yoshimi, T.; Ross, G.T.; Lipsett, M.B. Ovarian physiology: Relationship between plasma LH and steroidogenesis by the follicle and corpus luteum; effect of HCG. J. Clin. Endocrinol. Metab. 1969, 29, 1157–1167. [Google Scholar] [CrossRef]
- Winters, T.A.; Hanten, J.A.; Veldhuis, J.D. In situ amplification of the cytochrome P-450 cholesterol side-chain cleavage enzyme mRNA in single porcine granulosa cells by IGF-1 and FSH acting alone or in concert. Endocrine 1998, 9, 57–63. [Google Scholar] [CrossRef]
- Liu, Z.; Rudd, M.D.; Hernandez-Gonzalez, I.; Gonzalez-Robayna, I.; Fan, H.Y.; Zeleznik, A.J.; Richards, J.A.S. FSH and FOXO1 regulate genes in the sterol/steroid and lipid biosynthetic pathways in granulosa cells. Mol. Endocrinol. 2009, 23, 649–661. [Google Scholar] [CrossRef]
- LaVoie, H.A. Transcriptional control of genes mediating ovarian follicular growth, differentiation, and steroidogenesis in pigs. Mol. Reprod. Dev. 2017, 84, 788–801. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin Modifications and Their Function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Reik, W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 2007, 447, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Lee, L.; Maekawa, R.; Sato, S.; Kajimura, T.; Shinagawa, M.; Tamura, I.; Taketani, T.; Asada, H.; Tamura, H.; et al. Epigenetic changes of the cyp11a1 promoter region in granulosa cells undergoing luteinization during ovulation in female rats. Endocrinology 2016, 157, 3344–3354. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Wang, P.; Peng, J.; Xue, J.; Chen, K.; Song, Y.; Wang, J.; Li, G.; An, X.; Cao, B. Regulation and function of runt-related transcription factors (RUNX1 and RUNX2) in goat granulosa cells. J. Steroid Biochem. Mol. Biol. 2018, 181, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Hayes, E.; Biswas, A.; Seger, C.; Prizant, H.; Hammes, S.R.; Sen, A. Androgens regulate ovarian gene expression through modulation of Ezh2 expression and activity. Endocrinology 2017, 158, 2944–2954. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.; Li, Z.; Zhong, Y.; Li, Q.; Wang, J.; Zhang, H.; Yuan, X.; Li, J.; Zhang, Z. KISS1 suppresses apoptosis and stimulates the synthesis of E2 in porcine ovarian granulosa cells. Animals 2019, 9, 54. [Google Scholar] [CrossRef]
- Marchal, R.; Vigneron, C.; Perreau, C.; Bali-Papp, A.; Mermillod, P. Effect of follicular size on meiotic and developmental competence of porcine oocytes. Theriogenology 2002, 57, 1523–1532. [Google Scholar] [CrossRef]
- Qi, W.; Zhao, K.; Gu, J.; Huang, Y.; Wang, Y.; Zhang, H.; Zhang, M.; Zhang, J.; Yu, Z.; Li, L.; et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat. Chem. Biol. 2017, 13, 381–388. [Google Scholar] [CrossRef]
- Hu, G.; Dong, B.; Zhang, J.; Zhai, W.; Xie, T.; Huang, B.; Huang, C.; Yao, X.; Zheng, J.; Che, J.; et al. The long noncoding RNA HOTAIR activates the Hippo pathway by directly binding to SAV1 in renal cell carcinoma. Oncotarget 2017, 8, 58654–58667. [Google Scholar] [CrossRef]
- Chinaranagari, S.; Sharma, P.; Chaudhary, J. EZH2 dependent H3K27me3 is involved in epigenetic silencing of ID4 in prostate cancer. Oncotarget 2014, 5, 7172–7182. [Google Scholar] [CrossRef]
- Picton, H.M.; Campbell, B.K.; Hunter, M.G. Maintenance of oestradiol production and expression of cytochrome P450 aromatase enzyme mRNA in long-term serum-free cultures of pig granulosa cells. J. Reprod. Fertil. 1999, 115, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Piunti, A.; Shilatifard, A. Epigenetic balance of gene expression by polycomb and compass families. Science 2016, 352, aad9780. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Hyttel, P.; Hall, V.J. Regulation of H3K27me3 and H3K4me3 during early porcine embryonic development. Mol. Reprod. Dev. 2010, 77, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K.; et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell 2006, 125, 315–326. [Google Scholar] [CrossRef]
- Wei, G.; Wei, L.; Zhu, J.; Zang, C.; Hu-Li, J.; Yao, Z.; Cui, K.; Kanno, Y.; Roh, T.Y.; Watford, W.T.; et al. Global Mapping of H3K4me3 and H3K27me3 Reveals Specificity and Plasticity in Lineage Fate Determination of Differentiating CD4+ T Cells. Immunity 2009, 30, 155–167. [Google Scholar] [CrossRef]
- Lomniczi, A.; Loche, A.; Castellano, J.M.; Ronnekleiv, O.K.; Bosch, M.; Kaidar, G.; Knoll, J.G.; Wright, H.; Pfeifer, G.P.; Ojeda, S.R. Epigenetic control of female puberty. Nat. Neurosci. 2013, 16, 281–289. [Google Scholar] [CrossRef]
- Ross, P.J.; Ragina, N.P.; Rodriguez, R.M.; Iager, A.E.; Siripattarapravat, K.; Lopez-Corrales, N.; Cibelli, J.B. Polycomb gene expression and histone H3 lysine 27 trimethylation changes during bovine preimplantation development. Reproduction 2008, 136, 777–785. [Google Scholar] [CrossRef]
- Fang, L.; Chang, H.M.; Cheng, J.C.; Leung, P.C.K.; Sun, Y.P. TGF-β1 induces COX-2 expression and PGE2 production in human granulosa cells through Smad signaling pathways. J. Clin. Endocrinol. Metab. 2014, 99, 1217–1226. [Google Scholar] [CrossRef]
- Rekawiecki, R.; Nowik, M.; Kotwica, J. Stimulatory effect of LH, PGE2 and progesterone on StAR protein, cytochrome P450 cholesterol side chain cleavage and 3β hydroxysteroid dehydrogenase gene expression in bovine luteal cells. Prostaglandins Other Lipid Mediat. 2005, 78, 169–184. [Google Scholar] [CrossRef]
- Liu, P.P.; Chang, H.M.; Cheng, J.C.; Leung, P.C.K. Activin a upregulates PTGS2 expression and increases PGE2 production in human granulosa-lutein cells. Reproduction 2016, 152, 655–664. [Google Scholar] [CrossRef]
- Jo, M.; Curry, T.E. Luteinizing hormone-induced RUNX1 regulates the expression of genes in granulosa cells of rat periovulatory follicles. Mol. Endocrinol. 2006, 20, 2156–2172. [Google Scholar] [CrossRef] [PubMed]
- Huyen, L.V.N. Effects of varying doses of HCG on the evolution of preovulatory rabbit follicles and oocytes Sign in Oxford Academic account Sign in via your Institution. Hum. Reprod. 1989, 4, 636–642. [Google Scholar]
- Li, Y.; Jin, C.; Bai, H.; Gao, Y.; Sun, S.; Chen, L.; Qin, L.; Liu, P.P.; Cheng, L.; Wang, Q.F. Human NOTCH4 is a key target of RUNX1 in megakaryocytic differentiation. Blood 2018, 131, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Luo, M.; Cai, W.; Zhou, S.; Feng, D.; Xu, C.; Wang, H. Runt-Related Transcription Factor 1 (RUNX1) Promotes TGF-β-Induced Renal Tubular Epithelial-to-Mesenchymal Transition (EMT) and Renal Fibrosis through the PI3K Subunit p110δ. EBioMedicine 2018, 31, 217–225. [Google Scholar] [CrossRef]
- Słomczyñska, M.; Tabarowski, Z. Localization of androgen receptor and cytochrome P450 aromatase in the follicle and corpus luteum of the porcine ovary. Anim. Reprod. Sci. 2001, 65, 127–134. [Google Scholar] [CrossRef]
- Schams, D.; Berisha, B. Steroids as local regulators of ovarian activity in domestic animals. Domest. Anim. Endocrinol. 2002, 23, 53–65. [Google Scholar] [CrossRef]
- Armstrong, D.T. Prostaglandins and follicular functions. J. Reprod. Fertil. 1981, 62, 283–291. [Google Scholar] [CrossRef]
- McNatty, K.P.; Henderson, K.M.; Sawers, R.S. Effects of prostaglandin F(2α) and E2 on the production of progesterone by human granulosa cells in tissue culture. J. Endocrinol. 1975, 67, 231–240. [Google Scholar] [CrossRef]
- Fowkes, R.C.; Chandras, C.; Chin, E.C.; Okolo, S.; Abayasekara, D.R.E.; Michael, A.E. Relationship between the production of prostaglandins and progesterone by luteinizing human granulosa cells. J. Endocrinol. 2001, 171, 455–462. [Google Scholar] [CrossRef]
- Haney, A.F.; Schomberg, D.W. Steroidal Modulation of Progesterone Secretion by Granulosa Cells from Large Porcine Follicles: A Role for Androgens and Estrogens in Controlling Steroidogenesis. Biol. Reprod. 1978, 19, 242–248. [Google Scholar] [CrossRef]
- Fritsche-Guenther, R.; Witzel, F.; Sieber, A.; Herr, R.; Schmidt, N.; Braun, S.; Brummer, T.; Sers, C.; Blüthgen, N. Strong negative feedback from Erk to Raf confers robustness to MAPK signalling. Mol. Syst. Biol. 2011, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, A.; Leigh-Brown, S.; Thybert, D.; Stefflova, K.; Turro, E.; Flicek, P.; Brazma, A.; Odom, D.T.; Marioni, J.C. Extensive compensatory cis-trans regulation in the evolution of mouse gene expression. Genome Res. 2012, 22, 2376–2384. [Google Scholar] [CrossRef] [PubMed]
- Barron, N.; Kumar, N.; Sanchez, N.; Doolan, P.; Clarke, C.; Meleady, P.; O’Sullivan, F.; Clynes, M. Engineering CHO cell growth and recombinant protein productivity by overexpression of miR-7. J. Biotechnol. 2011, 151, 204–211. [Google Scholar] [CrossRef]
- De Amicis, F.; Thirugnansampanthan, J.; Cui, Y.; Selever, J.; Beyer, A.; Parra, I.; Weigel, N.L.; Herynk, M.H.; Tsimelzon, A.; Lewis, M.T.; et al. Androgen receptor overexpression induces tamoxifen resistance in human breast cancer cells. Breast Cancer Res. Treat. 2010, 121, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Becker, A.; Zimmer, A.; Lu, J.; Buettner, R.; Kirfel, J. SNAI1-Mediated Epithelial-Mesenchymal Transition Confers Chemoresistance and Cellular Plasticity by Regulating Genes Involved in Cell Death and Stem Cell Maintenance. PLoS ONE 2013, 8, e66558. [Google Scholar] [CrossRef] [PubMed]
- Ciesiółka, S.; Budna, J.; Jopek, K.; Bryja, A.; Kranc, W.; Borys, S.; Jeseta, M.; Chachuła, A.; Ziółkowska, A.; Antosik, P.; et al. Time- and Dose-Dependent Effects of 17 Beta-Estradiol on Short-Term, Real-Time Proliferation and Gene Expression in Porcine Granulosa Cells. BioMed Res. Int. 2017, 2017, 9738640. [Google Scholar] [CrossRef] [PubMed]
- Duda, M.; Durlej, M.; Knet, M.; Knapczyk-Stwora, K.; Tabarowski, Z.; Slomczynska, M. Does 2-hydroxyflutamide inhibit apoptosis in porcine granulosa cells? An in vitro study. J. Reprod. Dev. 2012, 58, 438–444. [Google Scholar] [CrossRef]
- Blaha, M.; Prochazka, R.; Adamkova, K.; Nevoral, J.; Nemcova, L. Prostaglandin E2 stimulates the expression of cumulus expansion-related genes in pigs: The role of protein kinase B. Prostaglandins Other Lipid Mediat. 2017, 130, 38–46. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, Y.; Li, L.; He, Y.; He, B.; Li, Z.; Zhang, Z.; Zhang, H.; Yuan, X.; Li, J. Activation of Steroidogenesis, Anti-Apoptotic Activity, and Proliferation in Porcine Granulosa Cells by RUNX1 Is Negatively Regulated by H3K27me3 Transcriptional Repression. Genes 2020, 11, 495. https://doi.org/10.3390/genes11050495
Zhong Y, Li L, He Y, He B, Li Z, Zhang Z, Zhang H, Yuan X, Li J. Activation of Steroidogenesis, Anti-Apoptotic Activity, and Proliferation in Porcine Granulosa Cells by RUNX1 Is Negatively Regulated by H3K27me3 Transcriptional Repression. Genes. 2020; 11(5):495. https://doi.org/10.3390/genes11050495
Chicago/Turabian StyleZhong, Yuyi, Liying Li, Yingting He, Bo He, Zhonghui Li, Zhe Zhang, Hao Zhang, Xiaolong Yuan, and Jiaqi Li. 2020. "Activation of Steroidogenesis, Anti-Apoptotic Activity, and Proliferation in Porcine Granulosa Cells by RUNX1 Is Negatively Regulated by H3K27me3 Transcriptional Repression" Genes 11, no. 5: 495. https://doi.org/10.3390/genes11050495
APA StyleZhong, Y., Li, L., He, Y., He, B., Li, Z., Zhang, Z., Zhang, H., Yuan, X., & Li, J. (2020). Activation of Steroidogenesis, Anti-Apoptotic Activity, and Proliferation in Porcine Granulosa Cells by RUNX1 Is Negatively Regulated by H3K27me3 Transcriptional Repression. Genes, 11(5), 495. https://doi.org/10.3390/genes11050495




