Metabolic Reprogramming of Host Cells in Response to Enteroviral Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Cell Viability Determination
2.2. Virological Techniques
2.3. RNAi Knockdown
2.4. Plasmid DNA and DNA Transfection
2.5. Metabolomic Analysis
2.6. Processing of Metabolomics Data and Metabolite Validation
2.7. Immunoprecipitation and Proteomic Analysis
2.8. Western Blotting
2.9. Assessment of Cellular CAD Flux
2.10. Statistical Analyses
3. Results
3.1. Identification of Metabolic Changes in EV71-Infected Cells
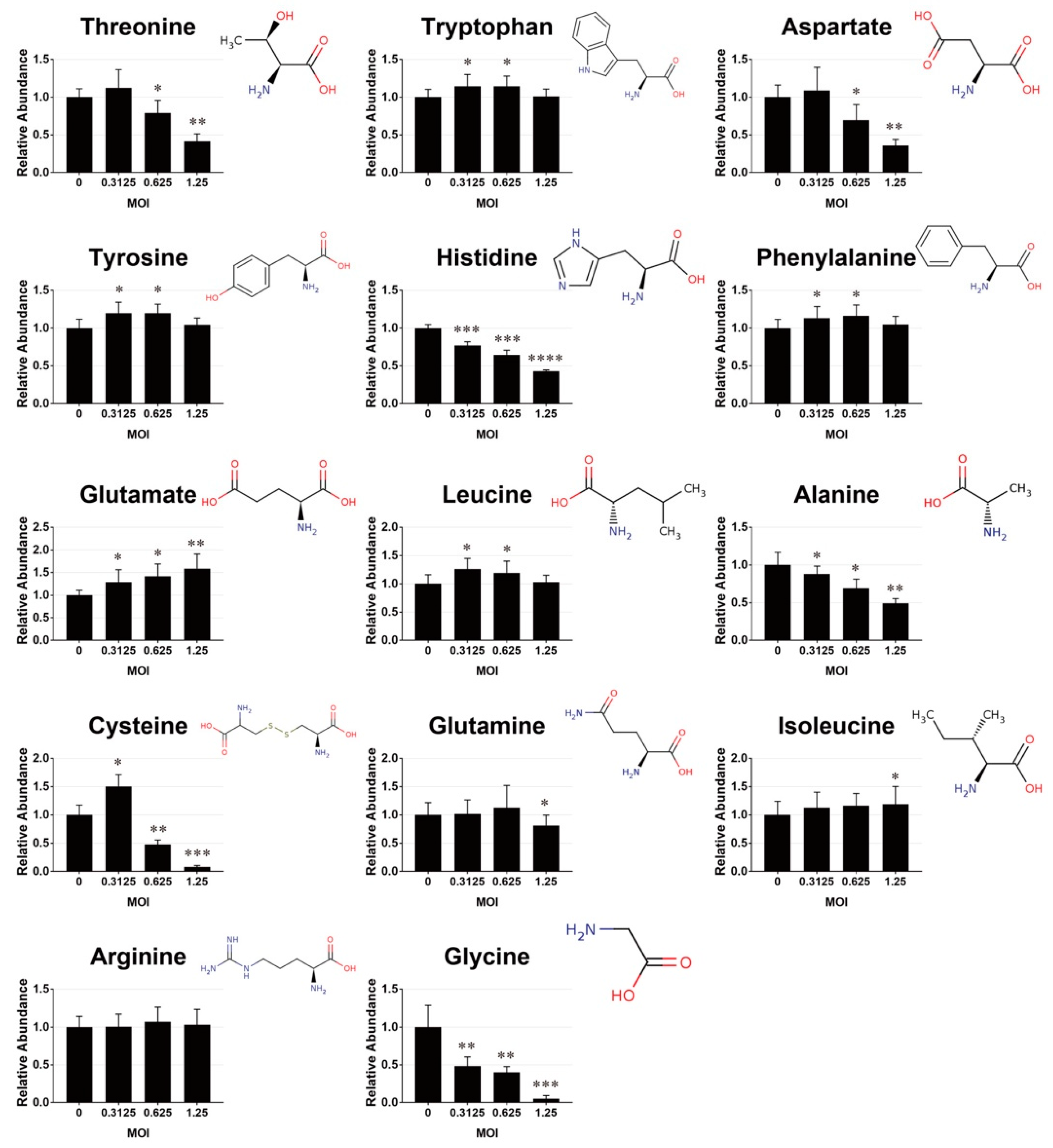
3.2. Changes in Amino Acid Pools in EV71-Infected Cells
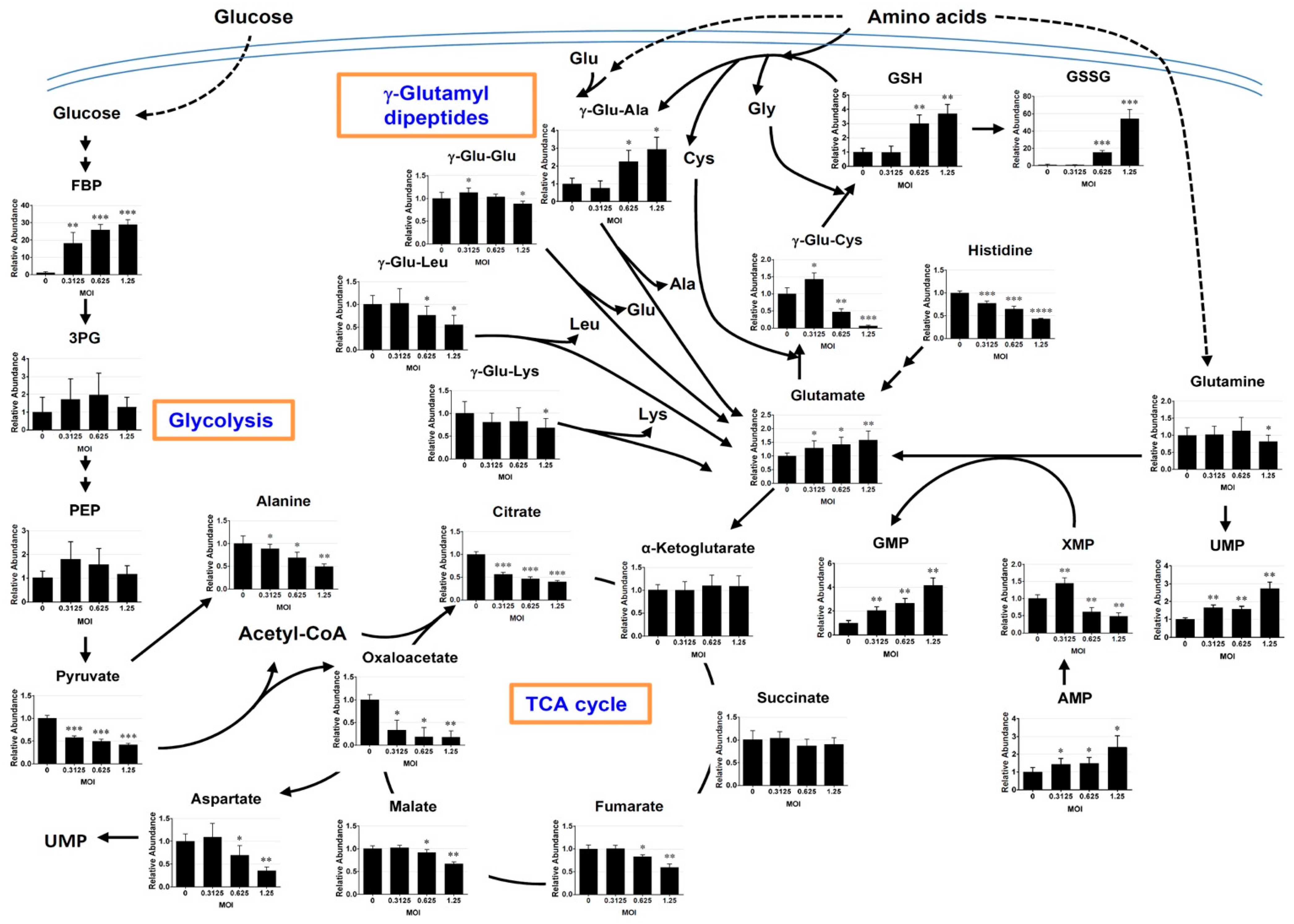
3.3. Mapping of Metabolites onto Metabolic Pathways
3.3.1. Changes in γ-Glutamyl Dipeptides
3.3.2. Changes in the Metabolites Related to Glutathione Metabolism
3.3.3. Changes in the Intermediates of Glycolysis and Citric Acid Cycle
3.3.4. Changes in the Purine and Pyrimidine Metabolism
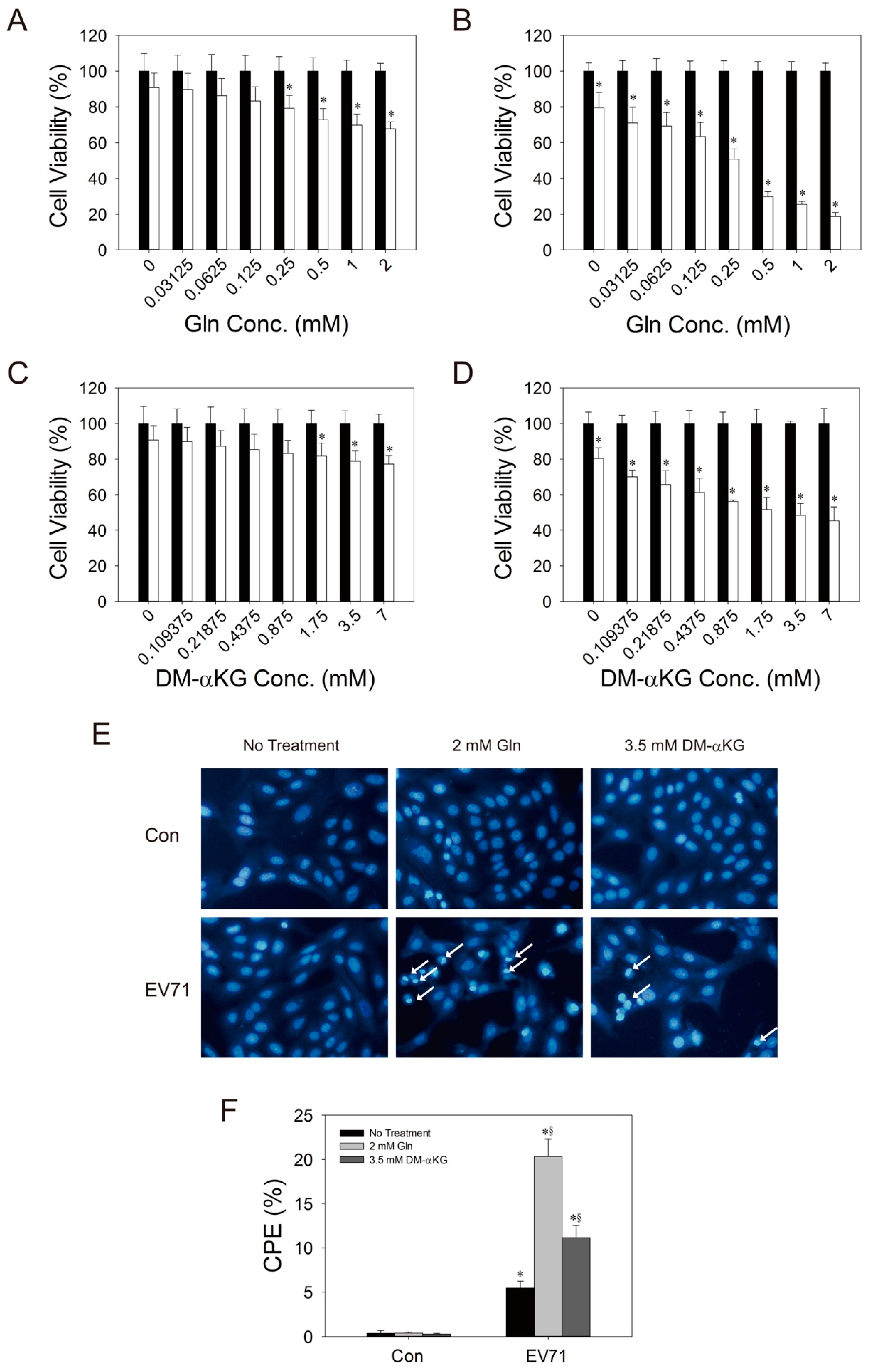
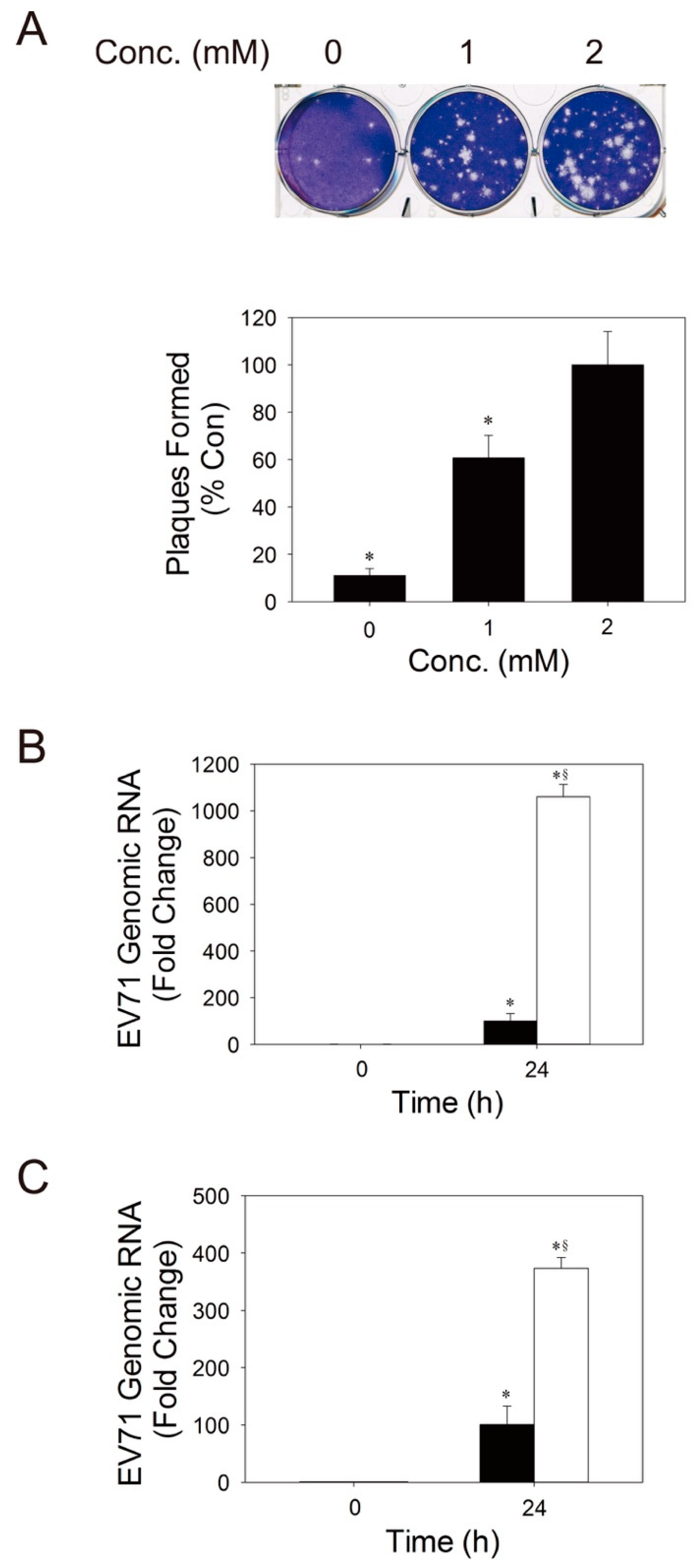
3.4. Glutamine is Essential to Viral Replication
3.5. Expression of GDH and CAD is Upregulated during EV71 Infection
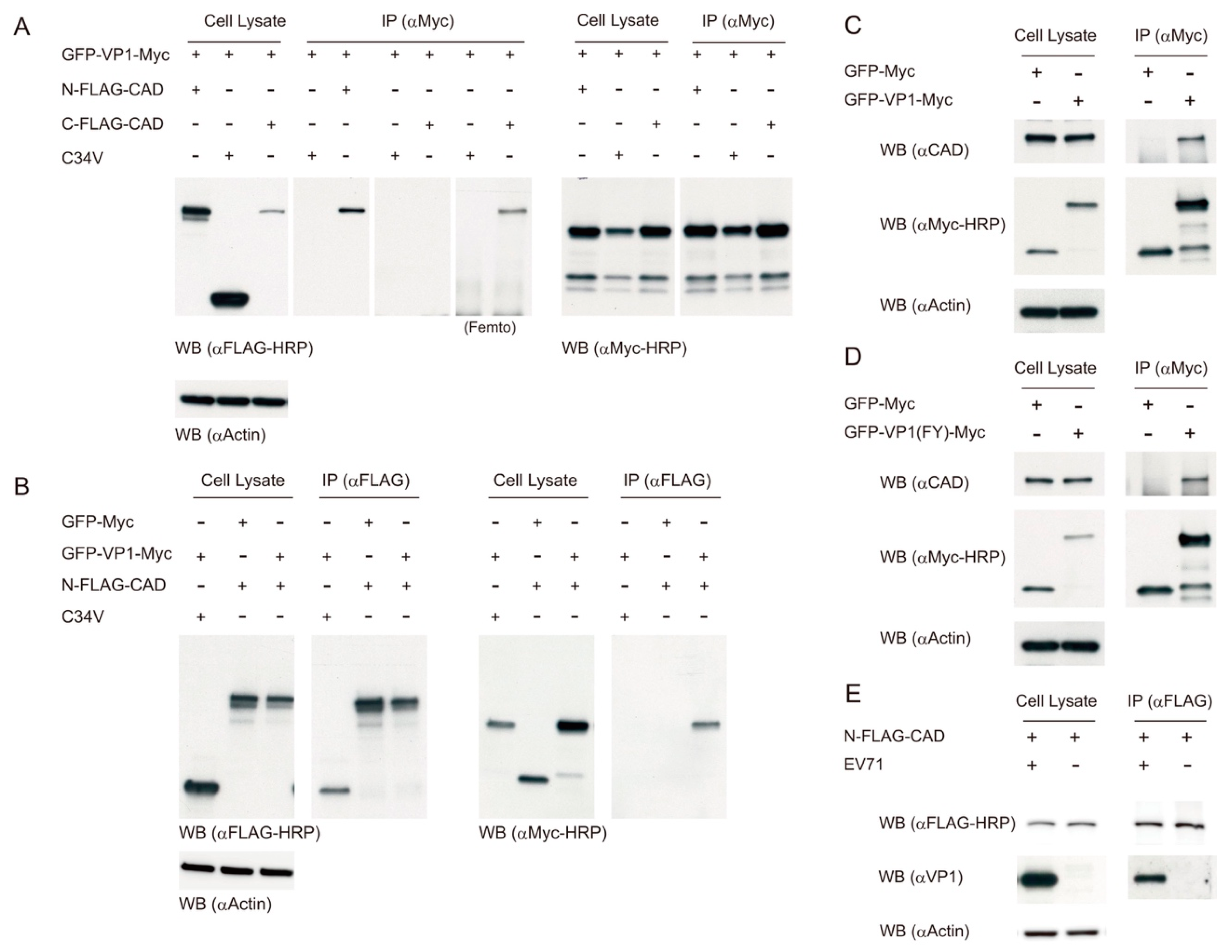
3.6. EV71 VP1 Protein Interacts with CAD
3.7. Exogenous VP1 Expression and Infection Increase the Flux of CAD-Catalyzed Reactions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Racaniello, V.R. Picornaviridae: the viruses and their replication. In Fields Virology; Fields, B., Knipe, D.M., Howley, P.M., Eds.; Lippincott-Williams & Wilkins: New York, NY, USA, 2006; pp. 795–838. [Google Scholar]
- Solomon, T.; Lewthwaite, P.; Perera, D.; Cardosa, M.J.; McMinn, P.; Ooi, M.H. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 2010, 10, 778–790. [Google Scholar] [CrossRef]
- Weng, K.F.; Chen, L.L.; Huang, P.N.; Shih, S.R. Neural pathogenesis of enterovirus 71 infection. Microbes Infect. 2010, 12, 505–510. [Google Scholar] [CrossRef]
- Yang, F.; Ren, L.; Xiong, Z.; Li, J.; Xiao, Y.; Zhao, R.; He, Y.; Bu, G.; Zhou, S.; Wang, J.; et al. Enterovirus 71 outbreak in the People’s Republic of China in 2008. J. Clin. Microbiol. 2009, 47, 2351–2352. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, T.; Hu, Y.; Wang, X.; Du, J.; Li, Y.; Sun, S.; Sun, X.; Li, Z.; Jin, Q. Survey of enterovirus infections from hand, foot and mouth disease outbreak in China, 2009. Virol J. 2011, 8, 508. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.X.; Wu, B.; Bao, W.X.; Han, F.A.; Xu, L.; Ge, Q.J.; Yang, J.; Yuan, Z.H.; Miao, C.H.; Huang, X.X.; et al. Epidemiology of hand, foot, and mouth disease and genotype characterization of Enterovirus 71 in Jiangsu, China. J. Clin. Virol. 2010, 49, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Munger, J.; Bajad, S.U.; Coller, H.A.; Shenk, T.; Rabinowitz, J.D. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2006, 2, e132. [Google Scholar] [CrossRef] [PubMed]
- Ripoli, M.; D’Aprile, A.; Quarato, G.; Sarasin-Filipowicz, M.; Gouttenoire, J.; Scrima, R.; Cela, O.; Boffoli, D.; Heim, M.H.; Moradpour, D.; et al. Hepatitis C virus-linked mitochondrial dysfunction promotes hypoxia-inducible factor 1 alpha-mediated glycolytic adaptation. J. Virol. 2010, 84, 647–660. [Google Scholar] [CrossRef] [PubMed]
- Vysochan, A.; Sengupta, A.; Weljie, A.M.; Alwine, J.C.; Yu, Y. ACSS2-mediated acetyl-CoA synthesis from acetate is necessary for human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 2017, 114, E1528–E1535. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Liu, N.; Yang, Z.; Song, W.; Wang, P.; Chen, H.; Lucio, M.; Schmitt-Kopplin, P.; Chen, G.; Cai, Z. GC/MS-based metabolomics reveals fatty acid biosynthesis and cholesterol metabolism in cell lines infected with influenza A virus. Talanta 2010, 83, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Ritter, J.B.; Wahl, A.S.; Freund, S.; Genzel, Y.; Reichl, U. Metabolic effects of influenza virus infection in cultured animal cells: Intra- and extracellular metabolite profiling. BMC Syst. Biol. 2010, 4, 61. [Google Scholar] [CrossRef] [PubMed]
- Smallwood, H.S.; Duan, S.; Morfouace, M.; Rezinciuc, S.; Shulkin, B.L.; Shelat, A.; Zink, E.E.; Milasta, S.; Bajracharya, R.; Oluwaseum, A.J.; et al. Targeting Metabolic Reprogramming by Influenza Infection for Therapeutic Intervention. Cell reports 2017, 19, 1640–1653. [Google Scholar] [CrossRef] [PubMed]
- Thai, M.; Graham, N.A.; Braas, D.; Nehil, M.; Komisopoulou, E.; Kurdistani, S.K.; McCormick, F.; Graeber, T.G.; Christofk, H.R. Adenovirus E4ORF1-induced MYC activation promotes host cell anabolic glucose metabolism and virus replication. Cell Metab. 2014, 19, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Jordan, T.X.; Randall, G. Flavivirus modulation of cellular metabolism. Curr. Opin. Virol. 2016, 19, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Shrinet, J.; Shastri, J.S.; Gaind, R.; Bhavesh, N.S.; Sunil, S. Serum metabolomics analysis of patients with chikungunya and dengue mono/co-infections reveals distinct metabolite signatures in the three disease conditions. Sci. Rep. 2016, 6, 36833. [Google Scholar] [CrossRef]
- Thaker, S.K.; Chapa, T.; Garcia, G., Jr.; Gong, D.; Schmid, E.W.; Arumugaswami, V.; Sun, R.; Christofk, H.R. Differential Metabolic Reprogramming by Zika Virus Promotes Cell Death in Human versus Mosquito Cells. Cell Metab. 2019, 29, 1206–1216 e1204. [Google Scholar] [CrossRef]
- Rodgers, M.A.; Saghatelian, A.; Yang, P.L. Identification of an overabundant cholesterol precursor in hepatitis B virus replicating cells by untargeted lipid metabolite profiling. J. Am. Chem. Soc. 2009, 131, 5030–5031. [Google Scholar] [CrossRef]
- Li, H.; Zhu, W.; Zhang, L.; Lei, H.; Wu, X.; Guo, L.; Chen, X.; Wang, Y.; Tang, H. The metabolic responses to hepatitis B virus infection shed new light on pathogenesis and targets for treatment. Sci. Rep. 2015, 5, 8421. [Google Scholar] [CrossRef]
- Shi, Y.X.; Huang, C.J.; Yang, Z.G. Impact of hepatitis B virus infection on hepatic metabolic signaling pathway. World J. Gastroenterol. 2016, 22, 8161–8167. [Google Scholar] [CrossRef]
- Roe, B.; Kensicki, E.; Mohney, R.; Hall, W.W. Metabolomic profile of hepatitis C virus-infected hepatocytes. PLoS ONE 2011, 6, e23641. [Google Scholar] [CrossRef]
- Sugiyama, K.; Ebinuma, H.; Nakamoto, N.; Sakasegawa, N.; Murakami, Y.; Chu, P.S.; Usui, S.; Ishibashi, Y.; Wakayama, Y.; Taniki, N.; et al. Prominent steatosis with hypermetabolism of the cell line permissive for years of infection with hepatitis C virus. PLoS ONE 2014, 9, e94460. [Google Scholar] [CrossRef]
- Tiwari, S.; Dhole, T.N. Metabolomics of Rhabdomyosarcoma Cell During Echovirus 30 Infection. Virol J. 2017, 14, 144. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.L.; Weng, S.F.; Kuo, C.H.; Ho, H.Y. Enterovirus 71 induces mitochondrial reactive oxygen species generation that is required for efficient replication. PLoS ONE 2014, 9, e113234. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Maguire, T.G.; Alwine, J.C. ChREBP, a glucose-responsive transcriptional factor, enhances glucose metabolism to support biosynthesis in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA 2014, 111, 1951–1956. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Maguire, T.G.; Alwine, J.C. Human cytomegalovirus activates glucose transporter 4 expression to increase glucose uptake during infection. J. Virol. 2011, 85, 1573–1580. [Google Scholar] [CrossRef]
- McArdle, J.; Moorman, N.J.; Munger, J. HCMV targets the metabolic stress response through activation of AMPK whose activity is important for viral replication. PLoS Pathog. 2012, 8, e1002502. [Google Scholar] [CrossRef]
- Yu, Y.; Maguire, T.G.; Alwine, J.C. Human cytomegalovirus infection induces adipocyte-like lipogenesis through activation of sterol regulatory element binding protein 1. J. Virol. 2012, 86, 2942–2949. [Google Scholar] [CrossRef]
- Yu, Y.; Pierciey, F.J., Jr.; Maguire, T.G.; Alwine, J.C. PKR-like endoplasmic reticulum kinase is necessary for lipogenic activation during HCMV infection. PLoS Pathog. 2013, 9, e1003266. [Google Scholar] [CrossRef]
- Heaton, N.S.; Perera, R.; Berger, K.L.; Khadka, S.; Lacount, D.J.; Kuhn, R.J.; Randall, G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 17345–17350. [Google Scholar] [CrossRef]
- Ho, H.Y.; Cheng, M.L.; Weng, S.F.; Chang, L.; Yeh, T.T.; Shih, S.R.; Chiu, D.T. Glucose-6-phosphate dehydrogenase deficiency enhances enterovirus 71 infection. J. Gen. Virol. 2008, 89, 2080–2089. [Google Scholar] [CrossRef]
- Ho, H.Y.; Cheng, M.L.; Chiu, H.Y.; Weng, S.F.; Chiu, D.T. Dehydroepiandrosterone induces growth arrest of hepatoma cells via alteration of mitochondrial gene expression and function. Int. J. Oncol. 2008, 33, 969–977. [Google Scholar]
- Chen, S.G.; Cheng, M.L.; Chen, K.H.; Horng, J.T.; Liu, C.C.; Wang, S.M.; Sakurai, H.; Leu, Y.L.; Wang, S.D.; Ho, H.Y. Antiviral activities of Schizonepeta tenuifolia Briq. against enterovirus 71 in vitro and in vivo. Sci. Rep. 2017, 7, 935. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.G.; Leu, Y.L.; Cheng, M.L.; Ting, S.C.; Liu, C.C.; Wang, S.D.; Yang, C.H.; Hung, C.Y.; Sakurai, H.; Chen, K.H.; et al. Anti-enterovirus 71 activities of Melissa officinalis extract and its biologically active constituent rosmarinic acid. Sci. Rep. 2017, 7, 12264. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.H.; Liang, C.T.; Jiang, S.T.; Chen, K.H.; Yang, C.C.; Cheng, M.L.; Ho, H.Y. A Novel Murine Model Expressing a Chimeric mSCARB2/hSCARB2 Receptor Is Highly Susceptible to Oral Infection with Clinical Isolates of Enterovirus 71. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.J.; Puhl, H.L., 3rd; Ikeda, S.R. Rapid modification of proteins using a rapamycin-inducible tobacco etch virus protease system. PLoS ONE 2009, 4, e7474. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.L.; Lin, J.F.; Huang, C.Y.; Li, G.J.; Shih, L.M.; Chiu, D.T.; Ho, H.Y. Sedoheptulose-1,7-bisphospate Accumulation and Metabolic Anomalies in Hepatoma Cells Exposed to Oxidative Stress. Oxid. Med. Cell. Longev. 2019, 2019, 5913635. [Google Scholar] [CrossRef] [PubMed]
- Tautenhahn, R.; Cho, K.; Uritboonthai, W.; Zhu, Z.; Patti, G.J.; Siuzdak, G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat. Biotechnol. 2012, 30, 826–828. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–807. [Google Scholar] [CrossRef]
- Franco, R.; Schoneveld, O.J.; Pappa, A.; Panayiotidis, M.I. The central role of glutathione in the pathophysiology of human diseases. Arch. Physiol. Biochem. 2007, 113, 234–258. [Google Scholar] [CrossRef]
- Guo, D.; Zhu, Q.; Zhang, H.; Sun, D. Proteomic analysis of membrane proteins of vero cells: exploration of potential proteins responsible for virus entry. DNA Cell Biol. 2014, 33, 20–28. [Google Scholar] [CrossRef]
- Chambers, J.W.; Maguire, T.G.; Alwine, J.C. Glutamine metabolism is essential for human cytomegalovirus infection. J. Virol. 2010, 84, 1867–1873. [Google Scholar] [CrossRef]
- Levy, P.L.; Duponchel, S.; Eischeid, H.; Molle, J.; Michelet, M.; Diserens, G.; Vermathen, M.; Vermathen, P.; Dufour, J.F.; Dienes, H.P.; et al. Hepatitis C virus infection triggers a tumor-like glutamine metabolism. Hepatology (Baltimore, Md.) 2017, 65, 789–803. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, E.L.; Carroll, P.A.; Thalhofer, A.B.; Lagunoff, M. Latent KSHV Infected Endothelial Cells Are Glutamine Addicted and Require Glutaminolysis for Survival. PLoS Pathog. 2015, 11, e1005052. [Google Scholar] [CrossRef] [PubMed]
- Owen, O.E.; Kalhan, S.C.; Hanson, R.W. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002, 277, 30409–30412. [Google Scholar] [CrossRef]
- Frenkel, R. Regulation and physiological functions of malic enzymes. Curr. Top. Cell. Regul. 1975, 9, 157–181. [Google Scholar] [PubMed]
- Du, J.; Yanagida, A.; Knight, K.; Engel, A.L.; Vo, A.H.; Jankowski, C.; Sadilek, M.; Tran, V.T.; Manson, M.A.; Ramakrishnan, A.; et al. Reductive carboxylation is a major metabolic pathway in the retinal pigment epithelium. Proc. Natl. Acad. Sci. USA 2016, 113, 14710–14715. [Google Scholar] [CrossRef]
- Thai, M.; Thaker, S.K.; Feng, J.; Du, Y.; Hu, H.; Ting Wu, T.; Graeber, T.G.; Braas, D.; Christofk, H.R. MYC-induced reprogramming of glutamine catabolism supports optimal virus replication. Nat. Commun. 2015, 6, 8873. [Google Scholar] [CrossRef]
- Ratuszny, D.; Suhs, K.W.; Novoselova, N.; Kuhn, M.; Kaever, V.; Skripuletz, T.; Pessler, F.; Stangel, M. Identification of Cerebrospinal Fluid Metabolites as Biomarkers for Enterovirus Meningitis. Int. J. Mol. Sci. 2019, 20, 337. [Google Scholar] [CrossRef]
- Orlowski, M.; Wilk, S. Intermediates of the gamma-glutamyl cycle in mouse tissues. Influence of administration of amino acids on pyrrolidone carboxylate and gamma-glutamyl amino acids. Eur. J. Biochem. 1975, 53, 581–590. [Google Scholar] [CrossRef]
- Moore, W.R.; Meister, A. Enzymatic synthesis of novel glutathione analogs. Anal. Biochem. 1987, 161, 487–493. [Google Scholar] [CrossRef]
- Dunkel, A.; Koster, J.; Hofmann, T. Molecular and sensory characterization of gamma-glutamyl peptides as key contributors to the kokumi taste of edible beans (Phaseolus vulgaris L.). J. Agric. Food Chem. 2007, 55, 6712–6719. [Google Scholar] [CrossRef]
- Roudot-Algaron, F.; Kerhoas, L.; Le Bars, D.; Einhorn, J.; Gripon, J.C. Isolation of gamma-Glutamyl Peptides from Comte Cheese. J. Dairy Sci. 1994, 77, 1161–1166. [Google Scholar] [CrossRef]
- Toelstede, S.; Dunkel, A.; Hofmann, T. A series of kokumi peptides impart the long-lasting mouthfulness of matured Gouda cheese. J. Agric. Food Chem. 2009, 57, 1440–1448. [Google Scholar] [CrossRef] [PubMed]
- Priolo, C.; Khabibullin, D.; Reznik, E.; Filippakis, H.; Ogorek, B.; Kavanagh, T.R.; Nijmeh, J.; Herbert, Z.T.; Asara, J.M.; Kwiatkowski, D.J.; et al. Impairment of gamma-glutamyl transferase 1 activity in the metabolic pathogenesis of chromophobe renal cell carcinoma. Proc. Natl. Acad. Sci. USA 2018, 115, E6274–E6282. [Google Scholar] [CrossRef] [PubMed]
- Soga, T.; Sugimoto, M.; Honma, M.; Mori, M.; Igarashi, K.; Kashikura, K.; Ikeda, S.; Hirayama, A.; Yamamoto, T.; Yoshida, H.; et al. Serum metabolomics reveals gamma-glutamyl dipeptides as biomarkers for discrimination among different forms of liver disease. J. Hepatol. 2011, 55, 896–905. [Google Scholar] [CrossRef]
- Chee, M.E.; Majumder, K.; Mine, Y. Intervention of Dietary Dipeptide Gamma-l-Glutamyl-l-Valine (gamma-EV) Ameliorates Inflammatory Response in a Mouse Model of LPS-Induced Sepsis. J. Agric. Food Chem. 2017, 65, 5953–5960. [Google Scholar] [CrossRef]
- Zhang, H.; Kovacs-Nolan, J.; Kodera, T.; Eto, Y.; Mine, Y. gamma-Glutamyl cysteine and gamma-glutamyl valine inhibit TNF-alpha signaling in intestinal epithelial cells and reduce inflammation in a mouse model of colitis via allosteric activation of the calcium-sensing receptor. Biochim. Biophys. Acta 2015, 1852, 792–804. [Google Scholar] [CrossRef]
- Wu, Y.H.; Chiu, D.T.; Lin, H.R.; Tang, H.Y.; Cheng, M.L.; Ho, H.Y. Glucose-6-Phosphate Dehydrogenase Enhances Antiviral Response through Downregulation of NADPH Sensor HSCARG and Upregulation of NF-kappaB Signaling. Viruses 2015, 7, 6689–6706. [Google Scholar] [CrossRef]
- Evans, D.R.; Guy, H.I. Mammalian pyrimidine biosynthesis: fresh insights into an ancient pathway. J. Biol. Chem. 2004, 279, 33035–33038. [Google Scholar] [CrossRef]
- Deans, R.M.; Morgens, D.W.; Okesli, A.; Pillay, S.; Horlbeck, M.A.; Kampmann, M.; Gilbert, L.A.; Li, A.; Mateo, R.; Smith, M.; et al. Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat. Chem. Biol. 2016, 12, 361–366. [Google Scholar] [CrossRef]
- Huang, M.; Graves, L.M. De novo synthesis of pyrimidine nucleotides; emerging interfaces with signal transduction pathways. Cell. Mol. Life Sci. 2003, 60, 321–336. [Google Scholar] [CrossRef]
- Boyd, K.E.; Farnham, P.J. Myc versus USF: discrimination at the cad gene is determined by core promoter elements. Mol. Cell. Biol. 1997, 17, 2529–2537. [Google Scholar] [CrossRef]
- Graves, L.M.; Guy, H.I.; Kozlowski, P.; Huang, M.; Lazarowski, E.; Pope, R.M.; Collins, M.A.; Dahlstrand, E.N.; Earp, H.S., 3rd; Evans, D.R. Regulation of carbamoyl phosphate synthetase by MAP kinase. Nature 2000, 403, 328–332. [Google Scholar] [CrossRef]
- Hann, S.R. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin. Cancer Biol. 2006, 16, 288–302. [Google Scholar] [CrossRef]
- Wong, W.R.; Chen, Y.Y.; Yang, S.M.; Chen, Y.L.; Horng, J.T. Phosphorylation of PI3K/Akt and MAPK/ERK in an early entry step of enterovirus 71. Life Sci. 2005, 78, 82–90. [Google Scholar] [CrossRef]
- Zhu, M.; Duan, H.; Gao, M.; Zhang, H.; Peng, Y. Both ERK1 and ERK2 are required for enterovirus 71 (EV71) efficient replication. Viruses 2015, 7, 1344–1356. [Google Scholar] [CrossRef]
- Angeletti, P.C.; Engler, J.A. Adenovirus preterminal protein binds to the CAD enzyme at active sites of viral DNA replication on the nuclear matrix. J. Virol. 1998, 72, 2896–2904. [Google Scholar] [CrossRef]
- Garcia-Dorival, I.; Wu, W.; Dowall, S.; Armstrong, S.; Touzelet, O.; Wastling, J.; Barr, J.N.; Matthews, D.; Carroll, M.; Hewson, R.; et al. Elucidation of the Ebola virus VP24 cellular interactome and disruption of virus biology through targeted inhibition of host-cell protein function. J. Proteome Res. 2014, 13, 5120–5135. [Google Scholar] [CrossRef]
- Kang, S.M.; Choi, J.K.; Kim, S.J.; Kim, J.H.; Ahn, D.G.; Oh, J.W. Regulation of hepatitis C virus replication by the core protein through its interaction with viral RNA polymerase. Biochem. Biophys. Res. Commun. 2009, 386, 55–59. [Google Scholar] [CrossRef]
- Yi, G.; Letteney, E.; Kim, C.H.; Kao, C.C. Brome mosaic virus capsid protein regulates accumulation of viral replication proteins by binding to the replicase assembly RNA element. RNA (New York, N.Y.) 2009, 15, 615–626. [Google Scholar] [CrossRef]
- Nasheri, N.; Joyce, M.; Rouleau, Y.; Yang, P.; Yao, S.; Tyrrell, D.L.; Pezacki, J.P. Modulation of fatty acid synthase enzyme activity and expression during hepatitis C virus replication. Chem. Biol. 2013, 20, 570–582. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, M.-L.; Chien, K.-Y.; Lai, C.-H.; Li, G.-J.; Lin, J.-F.; Ho, H.-Y. Metabolic Reprogramming of Host Cells in Response to Enteroviral Infection. Cells 2020, 9, 473. https://doi.org/10.3390/cells9020473
Cheng M-L, Chien K-Y, Lai C-H, Li G-J, Lin J-F, Ho H-Y. Metabolic Reprogramming of Host Cells in Response to Enteroviral Infection. Cells. 2020; 9(2):473. https://doi.org/10.3390/cells9020473
Chicago/Turabian StyleCheng, Mei-Ling, Kun-Yi Chien, Chien-Hsueh Lai, Guan-Jie Li, Jui-Fen Lin, and Hung-Yao Ho. 2020. "Metabolic Reprogramming of Host Cells in Response to Enteroviral Infection" Cells 9, no. 2: 473. https://doi.org/10.3390/cells9020473
APA StyleCheng, M.-L., Chien, K.-Y., Lai, C.-H., Li, G.-J., Lin, J.-F., & Ho, H.-Y. (2020). Metabolic Reprogramming of Host Cells in Response to Enteroviral Infection. Cells, 9(2), 473. https://doi.org/10.3390/cells9020473






