Transgenerational Inheritance of Environmentally Induced Epigenetic Alterations during Mammalian Development
Abstract
1. Introduction
2. The DNA Code-Based Genotype-Phenotype Relationship
3. Epigenetic Regulation via DNA Methylation
4. Epigenetic Regulation and Chromatin Proteins
5. Epigenetic Control Mediated by Short and Long Non-Coding RNAs
6. Germ Cell Specification and Reprograming
7. Retrotransposons Successfully Evade the Second Reprogramming Wave
8. Resetting of the Parental Epigenetic Makeup in Early Zygotes Reveals the Epigenome’s Plasticity
9. Epimutations Can Have Long-Term Consequences in Determining Phenotypes
10. Environmentally Induced Epigenetic Mutations
- Outbred models: Inbred strains are the best models to have an unbiased view of genetic and epigenetic variations following exposure to environmental factors. However, inbred strains are less susceptible to epigenetic transgenerational changes, and they do not reflect the complexity of the human genome. At the moment, most data are available on outbred strains.
- Biological replicates and statistical analyses: This is associated with the previous one. A large number of biological are required due to the high heterogeneity of outbred strains, and this is especially challenging when conducting studies up to three generations.
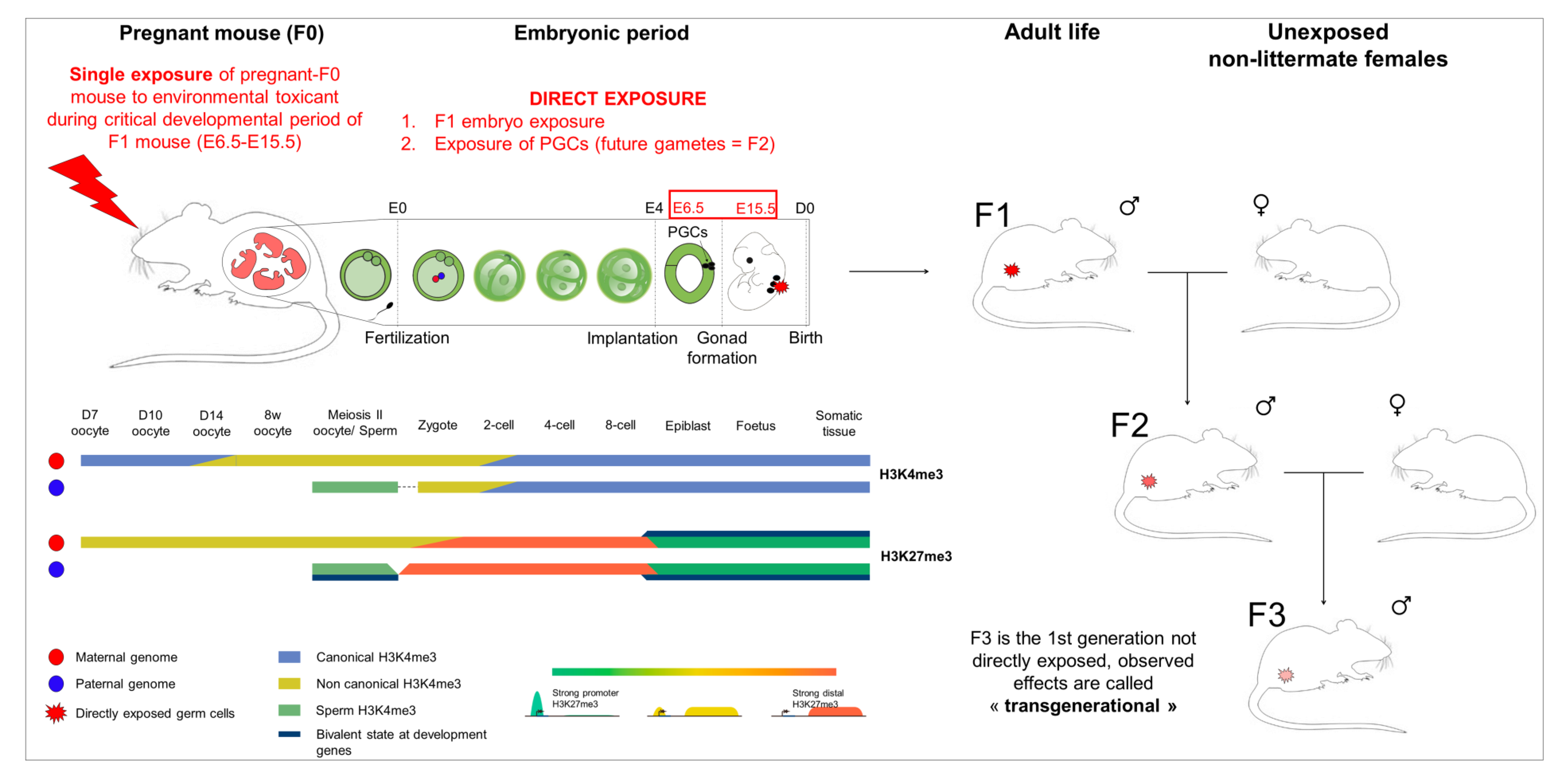
- Short exposure window: The developmental period is particularly susceptible to environmental factors due to epigenetic reprogramming, where the somatic-to-germline transition takes place. In mouse, this window is short, between E6–E15, and cannot be extended to avoid secondary effects associated with general toxicity.
- Choice of the molecule: For transgenerational studies, the environmental chemicals that are globally used or the ones that are of high-priority could be tested.
- Establishing dose-response: For studies involving environmental chemicals, the doses that induce transgenerational response without causing systemic toxicity needs to be experimentally established. Ideally, the minimal dose relevant to environmental doses where phenotypic effects are detected needs to be identified.
- Sex-specific transgenerational effects: Epidemiological studies have shown an association between various environmental factors and phenotypic changes in the offspring. However, it is difficult to study the exact mechanisms involved in maternal epigenetic inheritance due to the limited number of oocytes available in rodent models. Most studies that analyzed the epigenetic mechanisms in rodents to date focused on paternal inheritance.
11. Perspectives
Funding
Conflicts of Interest
References
- Wolffe, A.P.; Guschin, D. Review: Chromatin structural features and targets that regulate transcription. J. Struct. Biol. 2000, 129, 102–122. [Google Scholar] [CrossRef] [PubMed]
- Waddington, C.H. Towards a theoretical biology. Nature 1968, 218, 525. [Google Scholar] [CrossRef] [PubMed]
- Holliday, R. DNA methylation and epigenetic inheritance. Philosophical transactions of the Royal Society of London. Ser. B Biol. Sci. 1990, 326, 329–338. [Google Scholar] [CrossRef]
- Holliday, R. The inheritance of epigenetic defects. Science 1987, 238, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Jablonka, E.; Lamb, M.J. The inheritance of acquired epigenetic variations. J. Theor. Biol. 1989, 139, 69–83. [Google Scholar] [CrossRef]
- Jablonka, E.; Lamb, M.J. The changing concept of epigenetics. Ann. N. Y. Acad. Sci. 2002, 981, 82–96. [Google Scholar] [CrossRef]
- Heard, E.; Martienssen, R.A. Transgenerational epigenetic inheritance: Myths and mechanisms. Cell 2014, 157, 95–109. [Google Scholar] [CrossRef]
- Bollati, V.; Baccarelli, A. Environmental epigenetics. Heredity 2010, 105, 105–112. [Google Scholar] [CrossRef]
- Lowdon, R.F.; Zhang, B.; Bilenky, M.; Mauro, T.; Li, D.; Gascard, P.; Sigaroudinia, M.; Farnham, P.J.; Bastian, B.C.; Tlsty, T.D.; et al. Regulatory network decoded from epigenomes of surface ectoderm-derived cell types. Nat. Commun. 2014, 5, 5442. [Google Scholar] [CrossRef]
- Kim, K.; Doi, A.; Wen, B.; Ng, K.; Zhao, R.; Cahan, P.; Kim, J.; Aryee, M.J.; Ji, H.; Ehrlich, L.I.; et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010, 467, 285–290. [Google Scholar] [CrossRef]
- Whitelaw, N.C.; Whitelaw, E. Transgenerational epigenetic inheritance in health and disease. Curr. Opin. Genet. Dev. 2008, 18, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Iyama, T.; Wilson, D.M. DNA repair mechanisms in dividing and non-dividing cells. Dna Repair 2013, 12, 620–636. [Google Scholar] [CrossRef]
- Betti, M.; Casalone, E.; Ferrante, D.; Aspesi, A.; Morleo, G.; Biasi, A.; Sculco, M.; Mancuso, G.; Guarrera, S.; Righi, L.; et al. Germline mutations in DNA repair genes predispose asbestos-exposed patients to malignant pleural mesothelioma. Cancer Lett. 2017, 405, 38–45. [Google Scholar] [CrossRef]
- Costa, E.O.A.; Pinto, I.P.; Goncalves, M.W.; da Silva, J.F.; Oliveira, L.G.; da Cruz, A.S.; Silva, D.M.E.; da Silva, C.C.; Pereira, R.W.; da Cruz, A.D. Small de novo cnvs as biomarkers of parental exposure to low doses of ionizing radiation of caesium-137. Sci. Rep. 2018, 8, 5914. [Google Scholar] [CrossRef]
- McCarthy, M.I.; Abecasis, G.R.; Cardon, L.R.; Goldstein, D.B.; Little, J.; Ioannidis, J.P.; Hirschhorn, J.N. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat. Rev. Genet. 2008, 9, 356–369. [Google Scholar] [CrossRef]
- Hirschhorn, J.N.; Daly, M.J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005, 6, 95–108. [Google Scholar] [CrossRef]
- Mott, R.; Flint, J. Dissecting quantitative traits in mice. Annu. Rev. Genom. Hum. Genet. 2013, 14, 421–439. [Google Scholar] [CrossRef]
- Renaud, S.; Auffray, J.C.; de la Porte, S. Epigenetic effects on the mouse mandible: Common features and discrepancies in remodeling due to muscular dystrophy and response to food consistency. BMC Evol. Biol. 2010, 10, 28. [Google Scholar] [CrossRef]
- Chittka, A.; Chittka, L. Epigenetics of royalty. PLoS Biol. 2010, 8, e1000532. [Google Scholar] [CrossRef]
- Ralston, A.S. Environment controls gene expression: Sex determination and the onset of genetic disorders. Nat. Educ. 2008, 1, 203. [Google Scholar]
- Hunter, D.J. Gene-environment interactions in human diseases. Nat. Rev. Genet. 2005, 6, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Baarends, W.M.; van der Laan, R.; Grootegoed, J.A. DNA repair mechanisms and gametogenesis. Reproduction 2001, 121, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Zhang, Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a019133. [Google Scholar] [CrossRef] [PubMed]
- Holliday, R.; Pugh, J.E. DNA modification mechanisms and gene activity during development. Science 1975, 187, 226–232. [Google Scholar] [CrossRef]
- Riggs, A.D. X inactivation, differentiation, and DNA methylation. Cytogenet. Cell Genet. 1975, 14, 9–25. [Google Scholar] [CrossRef]
- Rakyan, V.K.; Preis, J.; Morgan, H.D.; Whitelaw, E. The marks, mechanisms and memory of epigenetic states in mammals. Biochem. J. 2001, 356, 1–10. [Google Scholar] [CrossRef]
- Duffie, R.; Ajjan, S.; Greenberg, M.V.; Zamudio, N.; del Arenal, M.E.; Iranzo, J.; Okamoto, I.; Barbaux, S.; Fauque, P.; Bourc’his, D. The gpr1/zdbf2 locus provides new paradigms for transient and dynamic genomic imprinting in mammals. Genes Dev. 2014, 28, 463–478. [Google Scholar] [CrossRef]
- Carninci, P.; Sandelin, A.; Lenhard, B.; Katayama, S.; Shimokawa, K.; Ponjavic, J.; Semple, C.A.; Taylor, M.S.; Engstrom, P.G.; Frith, M.C.; et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006, 38, 626–635. [Google Scholar] [CrossRef]
- Jang, H.S.; Shin, W.J.; Lee, J.E.; Do, J.T. Cpg and non-cpg methylation in epigenetic gene regulation and brain function. Genes 2017, 8. [Google Scholar]
- Laurent, L.; Wong, E.; Li, G.; Huynh, T.; Tsirigos, A.; Ong, C.T.; Low, H.M.; Kin Sung, K.W.; Rigoutsos, I.; Loring, J.; et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010, 20, 320–331. [Google Scholar] [CrossRef]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, A.; Ehrenhofer-Murray, A.; Jurkowski, T.P.; Lyko, F.; Reuter, G.; Ankri, S.; Nellen, W.; Schaefer, M.; Helm, M. Mechanism and biological role of dnmt2 in nucleic acid methylation. Rna Biol. 2017, 14, 1108–1123. [Google Scholar] [CrossRef] [PubMed]
- Easwaran, H.P.; Schermelleh, L.; Leonhardt, H.; Cardoso, M.C. Replication-independent chromatin loading of dnmt1 during g2 and m phases. Embo Rep. 2004, 5, 1181–1186. [Google Scholar] [CrossRef] [PubMed]
- Vandiver, A.R.; Idrizi, A.; Rizzardi, L.; Feinberg, A.P.; Hansen, K.D. DNA methylation is stable during replication and cell cycle arrest. Sci. Rep. 2015, 5, 17911. [Google Scholar] [CrossRef] [PubMed]
- Barau, J.; Teissandier, A.; Zamudio, N.; Roy, S.; Nalesso, V.; Herault, Y.; Guillou, F.; Bourc’his, D. The DNA methyltransferase dnmt3c protects male germ cells from transposon activity. Science 2016, 354, 909–912. [Google Scholar] [CrossRef]
- Elmhiri, G.; Gloaguen, C.; Grison, S.; Kereselidze, D.; Elie, C.; Tack, K.; Benderitter, M.; Lestaevel, P.; Legendre, A.; Souidi, M. DNA methylation and potential multigenerational epigenetic effects linked to uranium chronic low-dose exposure in gonads of males and females rats. Toxicol. Lett. 2018, 282, 64–70. [Google Scholar] [CrossRef]
- Lind, L.; Penell, J.; Luttropp, K.; Nordfors, L.; Syvanen, A.C.; Axelsson, T.; Salihovic, S.; van Bavel, B.; Fall, T.; Ingelsson, E.; et al. Global DNA hypermethylation is associated with high serum levels of persistent organic pollutants in an elderly population. Environ. Int. 2013, 59, 456–461. [Google Scholar] [CrossRef]
- Martin, E.M.; Fry, R.C. Environmental influences on the epigenome: Exposure-associated DNA methylation in human populations. Annu. Rev. Public Health 2018, 39, 309–333. [Google Scholar] [CrossRef]
- Hossain, K.; Suzuki, T.; Hasibuzzaman, M.M.; Islam, M.S.; Rahman, A.; Paul, S.K.; Tanu, T.; Hossain, S.; Saud, Z.A.; Rahman, M.; et al. Chronic exposure to arsenic, line-1 hypomethylation, and blood pressure: A cross-sectional study in bangladesh. Environ. Health A Glob. Access Sci. Source 2017, 16, 20. [Google Scholar] [CrossRef]
- Hossain, M.B.; Vahter, M.; Concha, G.; Broberg, K. Low-level environmental cadmium exposure is associated with DNA hypomethylation in argentinean women. Environ. Health Perspect. 2012, 120, 879–884. [Google Scholar] [CrossRef]
- Breton, C.V.; Yao, J.; Millstein, J.; Gao, L.; Siegmund, K.D.; Mack, W.; Whitfield-Maxwell, L.; Lurmann, F.; Hodis, H.; Avol, E.; et al. Prenatal air pollution exposures, DNA methyl transferase genotypes, and associations with newborn line1 and alu methylation and childhood blood pressure and carotid intima-media thickness in the children’s health study. Environ. Health Perspect. 2016, 124, 1905–1912. [Google Scholar] [CrossRef] [PubMed]
- Tsai, P.C.; Glastonbury, C.A.; Eliot, M.N.; Bollepalli, S.; Yet, I.; Castillo-Fernandez, J.E.; Carnero-Montoro, E.; Hardiman, T.; Martin, T.C.; Vickers, A.; et al. Smoking induces coordinated DNA methylation and gene expression changes in adipose tissue with consequences for metabolic health. Clin. Epigenet. 2018, 10, 126. [Google Scholar] [CrossRef] [PubMed]
- Braithwaite, E.C.; Kundakovic, M.; Ramchandani, P.G.; Murphy, S.E.; Champagne, F.A. Maternal prenatal depressive symptoms predict infant nr3c1 1f and bdnf iv DNA methylation. Epigenetics 2015, 10, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Mansell, T.; Novakovic, B.; Meyer, B.; Rzehak, P.; Vuillermin, P.; Ponsonby, A.L.; Collier, F.; Burgner, D.; Saffery, R.; Ryan, J.; et al. The effects of maternal anxiety during pregnancy on igf2/h19 methylation in cord blood. Transl. Psychiatry 2016, 6, e765. [Google Scholar] [CrossRef]
- Voisin, S.; Eynon, N.; Yan, X.; Bishop, D.J. Exercise training and DNA methylation in humans. Acta Physiol. (Oxf.) 2015, 213, 39–59. [Google Scholar] [CrossRef] [PubMed]
- Nogues, P.; Dos Santos, E.; Jammes, H.; Berveiller, P.; Arnould, L.; Vialard, F.; Dieudonné, M.-N. Maternal obesity influences expression and DNA methylation of the adiponectin and leptin systems in human third-trimester placenta. Clin. Epigenet. 2019, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Weng, X.; Liu, F.; Zhang, H.; Kan, M.; Wang, T.; Dong, M.; Liu, Y. Genome-wide DNA methylation profiling in infants born to gestational diabetes mellitus. Diabetes Res. Clin. Pract. 2018, 142, 10–18. [Google Scholar] [CrossRef]
- Verdaasdonk, J.S.; Bloom, K. Centromeres: Unique chromatin structures that drive chromosome segregation. Nat. Rev. Mol. Cell Biol. 2011, 12, 320–332. [Google Scholar] [CrossRef]
- Limoli, C.L.; Giedzinski, E.; Bonner, W.M.; Cleaver, J.E. Uv-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, gamma -h2ax formation, and mre11 relocalization. Proc. Natl. Acad. Sci. USA 2002, 99, 233–238. [Google Scholar] [CrossRef]
- Fernandez-Capetillo, O.; Mahadevaiah, S.K.; Celeste, A.; Romanienko, P.J.; Camerini-Otero, R.D.; Bonner, W.M.; Manova, K.; Burgoyne, P.; Nussenzweig, A. H2ax is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev. Cell 2003, 4, 497–508. [Google Scholar] [CrossRef]
- Giaimo, B.D.; Ferrante, F.; Herchenrother, A.; Hake, S.B.; Borggrefe, T. The histone variant h2a.Z in gene regulation. Epigenet. Chromatin 2019, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Q.; McEachin, R.C.; Cavalcoli, J.D.; Yu, X. H2a.B facilitates transcription elongation at methylated cpg loci. Genome Res. 2014, 24, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, K.; Shindo, A.; Harada, A.; Kurumizaka, H.; Kimura, H.; Ohkawa, Y.; Matsuyama, H. Roles of histone h3.5 in human spermatogenesis and spermatogenic disorders. Andrology 2018, 6, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Fyodorov, D.V.; Zhou, B.R.; Skoultchi, A.I.; Bai, Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 2018, 19, 192–206. [Google Scholar] [CrossRef]
- Brockers, K.; Schneider, R. Histone h1, the forgotten histone. Epigenomics 2019, 11, 363–366. [Google Scholar] [CrossRef]
- Hayakawa, K.; Ohgane, J.; Tanaka, S.; Yagi, S.; Shiota, K. Oocyte-specific linker histone h1foo is an epigenomic modulator that decondenses chromatin and impairs pluripotency. Epigenetics 2012, 7, 1029–1036. [Google Scholar] [CrossRef]
- Cockerill, P.N. Structure and function of active chromatin and dnase i hypersensitive sites. Febs J. 2011, 278, 2182–2210. [Google Scholar] [CrossRef]
- Sapojnikova, N.; Thorne, A.; Myers, F.; Staynov, D.; Crane-Robinson, C. The chromatin of active genes is not in a permanently open conformation. J. Mol. Biol. 2009, 386, 290–299. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Suganuma, T.; Workman, J.L. Crosstalk among histone modifications. Cell 2008, 135, 604–607. [Google Scholar] [CrossRef]
- Ooi, L.; Wood, I.C. Chromatin crosstalk in development and disease: Lessons from rest. Nat. Rev. Genet. 2007, 8, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Hammond, C.M.; Stromme, C.B.; Huang, H.; Patel, D.J.; Groth, A. Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 2017, 18, 141–158. [Google Scholar] [CrossRef]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-resolution profiling of histone methylations in the human genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef]
- Liu, X.; Wang, C.; Liu, W.; Li, J.; Li, C.; Kou, X.; Chen, J.; Zhao, Y.; Gao, H.; Wang, H.; et al. Distinct features of h3k4me3 and h3k27me3 chromatin domains in pre-implantation embryos. Nature 2016, 537, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Huang, B.; Zhang, B.; Xiang, Y.; Du, Z.; Xu, Q.; Li, Y.; Wang, Q.; Ma, J.; Peng, X.; et al. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol. Cell 2016, 63, 1066–1079. [Google Scholar] [CrossRef] [PubMed]
- Lesch, B.J.; Dokshin, G.A.; Young, R.A.; McCarrey, J.R.; Page, D.C. A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. Proc. Natl. Acad. Sci. USA 2013, 110, 16061–16066. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K.; et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006, 125, 315–326. [Google Scholar] [CrossRef]
- Gaydos, L.J.; Wang, W.; Strome, S. Gene repression. H3k27me and prc2 transmit a memory of repression across generations and during development. Science 2014, 345, 1515–1518. [Google Scholar] [CrossRef]
- Schuettengruber, B.; Bourbon, H.M.; Di Croce, L.; Cavalli, G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef]
- Siklenka, K.; Erkek, S.; Godmann, M.; Lambrot, R.; McGraw, S.; Lafleur, C.; Cohen, T.; Xia, J.; Suderman, M.; Hallett, M.; et al. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science 2015, 350, aab2006. [Google Scholar] [CrossRef]
- Teperek, M.; Simeone, A.; Gaggioli, V.; Miyamoto, K.; Allen, G.E.; Erkek, S.; Kwon, T.; Marcotte, E.M.; Zegerman, P.; Bradshaw, C.R.; et al. Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Res. 2016, 26, 1034–1046. [Google Scholar] [CrossRef] [PubMed]
- Raposo, A.E.; Piller, S.C. Protein arginine methylation: An emerging regulator of the cell cycle. Cell Div. 2018, 13, 3. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, T.R.; Glenfield, K.; Jeyanthan, K.; Zhu, X.D. Arginine methylation regulates telomere length and stability. Mol. Cell. Biol. 2009, 29, 4918–4934. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, M.R.; Scherer, C.A.; Chen, J.; Roshon, M.J.; Ruley, H.E. Arginine n-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol. Cell. Biol. 2000, 20, 4859–4869. [Google Scholar] [CrossRef]
- Ancelin, K.; Lange, U.C.; Hajkova, P.; Schneider, R.; Bannister, A.J.; Kouzarides, T.; Surani, M.A. Blimp1 associates with prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 2006, 8, 623–630. [Google Scholar] [CrossRef]
- Klosin, A.; Casas, E.; Hidalgo-Carcedo, C.; Vavouri, T.; Lehner, B. Transgenerational transmission of environmental information in c. Elegans. Science 2017, 356, 320–323. [Google Scholar] [CrossRef]
- Becker, J.S.; Nicetto, D.; Zaret, K.S. H3k9me3-dependent heterochromatin: Barrier to cell fate changes. Trends Genet. 2016, 32, 29–41. [Google Scholar] [CrossRef]
- Takada, Y.; Naruse, C.; Costa, Y.; Shirakawa, T.; Tachibana, M.; Sharif, J.; Kezuka-Shiotani, F.; Kakiuchi, D.; Masumoto, H.; Shinkai, Y.; et al. Hp1gamma links histone methylation marks to meiotic synapsis in mice. Development 2011, 138, 4207–4217. [Google Scholar] [CrossRef]
- Shi, M.; Whorton, A.E.; Sekulovski, N.; MacLean, J.A.; Hayashi, K. Prenatal exposure to bisphenol a, e and s induces transgenerational effects on male reproductive functions in mice. Toxicol. Sci. Off. J. Soc. Toxicol. 2019. [Google Scholar] [CrossRef]
- Milutinovic, S.; D’Alessio, A.C.; Detich, N.; Szyf, M. Valproate induces widespread epigenetic reprogramming which involves demethylation of specific genes. Carcinogenesis 2007, 28, 560–571. [Google Scholar] [CrossRef]
- Ooi, S.K.; Qiu, C.; Bernstein, E.; Li, K.; Jia, D.; Yang, Z.; Erdjument-Bromage, H.; Tempst, P.; Lin, S.P.; Allis, C.D.; et al. Dnmt3l connects unmethylated lysine 4 of histone h3 to de novo methylation of DNA. Nature 2007, 448, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Otani, J.; Nankumo, T.; Arita, K.; Inamoto, S.; Ariyoshi, M.; Shirakawa, M. Structural basis for recognition of h3k4 methylation status by the DNA methyltransferase 3a atrx-dnmt3-dnmt3l domain. Embo Rep. 2009, 10, 1235–1241. [Google Scholar] [CrossRef] [PubMed]
- Hill, P.W.S.; Leitch, H.G.; Requena, C.E.; Sun, Z.; Amouroux, R.; Roman-Trufero, M.; Borkowska, M.; Terragni, J.; Vaisvila, R.; Linnett, S.; et al. Epigenetic reprogramming enables the transition from primordial germ cell to gonocyte. Nature 2018, 555, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Cooper, S.; Brockdorff, N. The interplay of histone modifications—Writers that read. Embo Rep. 2015, 16, 1467–1481. [Google Scholar] [CrossRef]
- Beermann, J.; Piccoli, M.T.; Viereck, J.; Thum, T. Non-coding rnas in development and disease: Background, mechanisms, and therapeutic approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef]
- Diamantopoulos, M.A.; Tsiakanikas, P.; Scorilas, A. Non-coding rnas: The riddle of the transcriptome and their perspectives in cancer. Ann. Transl. Med. 2018, 6, 241. [Google Scholar] [CrossRef]
- Wei, J.W.; Huang, K.; Yang, C.; Kang, C.S. Non-coding rnas as regulators in epigenetics (review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef]
- Kanhere, A.; Viiri, K.; Araujo, C.C.; Rasaiyaah, J.; Bouwman, R.D.; Whyte, W.A.; Pereira, C.F.; Brookes, E.; Walker, K.; Bell, G.W.; et al. Short rnas are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol. Cell 2010, 38, 675–688. [Google Scholar] [CrossRef]
- Jinek, M.; Doudna, J.A. A three-dimensional view of the molecular machinery of rna interference. Nature 2009, 457, 405–412. [Google Scholar] [CrossRef]
- Morris, K.V.; Chan, S.W.; Jacobsen, S.E.; Looney, D.J. Small interfering rna-induced transcriptional gene silencing in human cells. Science 2004, 305, 1289–1292. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, J.; Man, W.Y.; Zhang, Q.W.; Xu, W.G. Sirna silencing ezh2 reverses cisplatin-resistance of human non-small cell lung and gastric cancer cells. Asian Pac. J. Cancer Prev. 2015, 16, 2425–2430. [Google Scholar] [CrossRef] [PubMed]
- Verdel, A.; Vavasseur, A.; Le Gorrec, M.; Touat-Todeschini, L. Common themes in sirna-mediated epigenetic silencing pathways. Int. J. Dev. Biol. 2009, 53, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Volpe, T.A.; Kidner, C.; Hall, I.M.; Teng, G.; Grewal, S.I.; Martienssen, R.A. Regulation of heterochromatic silencing and histone h3 lysine-9 methylation by rnai. Science 2002, 297, 1833–1837. [Google Scholar] [CrossRef] [PubMed]
- Luteijn, M.J.; Ketting, R.F. Piwi-interacting rnas: From generation to transgenerational epigenetics. Nat. Rev. Genet. 2013, 14, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Buckley, B.A.; Burkhart, K.B.; Gu, S.G.; Spracklin, G.; Kershner, A.; Fritz, H.; Kimble, J.; Fire, A.; Kennedy, S. A nuclear argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 2012, 489, 447–451. [Google Scholar] [CrossRef]
- Marczylo, E.L.; Amoako, A.A.; Konje, J.C.; Gant, T.W.; Marczylo, T.H. Smoking induces differential mirna expression in human spermatozoa: A potential transgenerational epigenetic concern? Epigenetics 2012, 7, 432–439. [Google Scholar] [CrossRef]
- Wagner, K.D.; Wagner, N.; Ghanbarian, H.; Grandjean, V.; Gounon, P.; Cuzin, F.; Rassoulzadegan, M. Rna induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev. Cell 2008, 14, 962–969. [Google Scholar] [CrossRef]
- Cuzin, F.; Rassoulzadegan, M. Non-mendelian epigenetic heredity: Gametic rnas as epigenetic regulators and transgenerational signals. Essays Biochem. 2010, 48, 101–106. [Google Scholar]
- Gapp, K.; Jawaid, A.; Sarkies, P.; Bohacek, J.; Pelczar, P.; Prados, J.; Farinelli, L.; Miska, E.; Mansuy, I.M. Implication of sperm rnas in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci. 2014, 17, 667–669. [Google Scholar] [CrossRef]
- Necsulea, A.; Soumillon, M.; Warnefors, M.; Liechti, A.; Daish, T.; Zeller, U.; Baker, J.C.; Grutzner, F.; Kaessmann, H. The evolution of lncrna repertoires and expression patterns in tetrapods. Nature 2014, 505, 635–640. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding rna biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Sarropoulos, I.; Marin, R.; Cardoso-Moreira, M.; Kaessmann, H. Developmental dynamics of lncrnas across mammalian organs and species. Nature 2019, 571, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Hollerer, I.; Higdon, A.; Brar, G.A. Strategies and challenges in identifying function for thousands of sorf-encoded peptides in meiosis. Proteomics 2018, 18, e1700274. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Zhang, J.; Lian, X.; Sun, L.; Meng, K.; Chen, Y.; Sun, Z.; Yin, X.; Li, Y.; Zhao, J.; et al. A hidden human proteome encoded by ‘non-coding’ genes. Nucleic Acids Res. 2019, 47, 8111–8125. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.R.; Makarewich, C.A.; Anderson, D.M.; Winders, B.R.; Troupes, C.D.; Wu, F.; Reese, A.L.; McAnally, J.R.; Chen, X.; Kavalali, E.T.; et al. A peptide encoded by a transcript annotated as long noncoding rna enhances serca activity in muscle. Science 2016, 351, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.W.; Wang, Y.; Chen, L.L. Cellular functions of long noncoding rnas. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef]
- Sahakyan, A.; Yang, Y.; Plath, K. The Role of Xist in X-Chromosome Dosage Compensation. Trends Cell Biol. 2018, 28, 999–1013. [Google Scholar] [CrossRef]
- Fang, H.; Disteche, C.M.; Berletch, J.B. X Inactivation and Escape: Epigenetic and Structural Features. Front. Cell Dev. Biol. 2019, 7. [Google Scholar] [CrossRef]
- Hong, S.H.; Kwon, J.T.; Kim, J.; Jeong, J.; Kim, J.; Lee, S.; Cho, C. Profiling of testis-specific long noncoding rnas in mice. BMC Genom. 2018, 19, 539. [Google Scholar] [CrossRef]
- Rolland, A.D.; Evrard, B.; Darde, T.A.; Le Beguec, C.; Le Bras, Y.; Bensalah, K.; Lavoue, S.; Jost, B.; Primig, M.; Dejucq-Rainsford, N.; et al. Rna profiling of human testicular cells identifies syntenic lncrnas associated with spermatogenesis. Hum. Reprod. 2019, 34, 1278–1290. [Google Scholar] [CrossRef]
- Svoboda, P. Long and small noncoding rnas during oocyte-to-embryo transition in mammals. Biochem. Soc. Trans. 2017, 45, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- Sendler, E.; Johnson, G.D.; Mao, S.; Goodrich, R.J.; Diamond, M.P.; Hauser, R.; Krawetz, S.A. Stability, delivery and functions of human sperm rnas at fertilization. Nucleic Acids Res. 2013, 41, 4104–4117. [Google Scholar] [CrossRef] [PubMed]
- Ostermeier, G.C.; Miller, D.; Huntriss, J.D.; Diamond, M.P.; Krawetz, S.A. Reproductive biology: Delivering spermatozoan rna to the oocyte. Nature 2004, 429, 154. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding rnas in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mrna methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]
- Jacob, R.; Zander, S.; Gutschner, T. The dark side of the epitranscriptome: Chemical modifications in long non-coding rnas. Int. J. Mol. Sci. 2017, 18, 2387. [Google Scholar] [CrossRef]
- Hochedlinger, K. Balancing work and life: A conversation with konrad hochedlinger. Interview by majlinda lako and susan daher. Stem Cells 2009, 27, 991–992. [Google Scholar]
- Rhind, S.M.; Rae, M.T.; Brooks, A.N. Environmental influences on the fetus and neonate—Timing, mechanisms of action and effects on subsequent adult function. Domest. Anim. Endocrinol. 2003, 25, 3–11. [Google Scholar] [CrossRef]
- Hayashi, K.; de Sousa Lopes, S.M.; Surani, M.A. Germ cell specification in mice. Science 2007, 316, 394–396. [Google Scholar] [CrossRef]
- Eguizabal, C.; Herrera, L.; De Onate, L.; Montserrat, N.; Hajkova, P.; Izpisua Belmonte, J.C. Characterization of the epigenetic changes during human gonadal primordial germ cells reprogramming. Stem Cells 2016, 34, 2418–2428. [Google Scholar] [CrossRef]
- Magnusdottir, E.; Dietmann, S.; Murakami, K.; Gunesdogan, U.; Tang, F.; Bao, S.; Diamanti, E.; Lao, K.; Gottgens, B.; Azim Surani, M. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat. Cell Biol. 2013, 15, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Gkountela, S.; Li, Z.; Vincent, J.J.; Zhang, K.X.; Chen, A.; Pellegrini, M.; Clark, A.T. The ontogeny of ckit+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat. Cell Biol. 2013, 15, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Hackett, J.A.; Surani, M.A. Regulatory principles of pluripotency: From the ground state up. Cell Stem Cell 2014, 15, 416–430. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.; Smith, A. Naive and primed pluripotent states. Cell Stem Cell 2009, 4, 487–492. [Google Scholar] [CrossRef]
- Irie, N.; Weinberger, L.; Tang, W.W.; Kobayashi, T.; Viukov, S.; Manor, Y.S.; Dietmann, S.; Hanna, J.H.; Surani, M.A. Sox17 is a critical specifier of human primordial germ cell fate. Cell 2015, 160, 253–268. [Google Scholar] [CrossRef]
- Greber, B.; Wu, G.; Bernemann, C.; Joo, J.Y.; Han, D.W.; Ko, K.; Tapia, N.; Sabour, D.; Sterneckert, J.; Tesar, P.; et al. Conserved and divergent roles of fgf signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell 2010, 6, 215–226. [Google Scholar] [CrossRef]
- de Koning, A.P.; Gu, W.; Castoe, T.A.; Batzer, M.A.; Pollock, D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011, 7, e1002384. [Google Scholar] [CrossRef]
- Jern, P.; Coffin, J.M. Effects of retroviruses on host genome function. Annu. Rev. Genet. 2008, 42, 709–732. [Google Scholar] [CrossRef]
- Qin, C.; Wang, Z.; Shang, J.; Bekkari, K.; Liu, R.; Pacchione, S.; McNulty, K.A.; Ng, A.; Barnum, J.E.; Storer, R.D. Intracisternal a particle genes: Distribution in the mouse genome, active subtypes, and potential roles as species-specific mediators of susceptibility to cancer. Mol. Carcinog. 2010, 49, 54–67. [Google Scholar] [CrossRef]
- Maksakova, I.A.; Romanish, M.T.; Gagnier, L.; Dunn, C.A.; van de Lagemaat, L.N.; Mager, D.L. Retroviral elements and their hosts: Insertional mutagenesis in the mouse germ line. PLoS Genet. 2006, 2, e2. [Google Scholar] [CrossRef]
- Bourc’his, D.; Bestor, T.H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking dnmt3l. Nature 2004, 431, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Hajkova, P.; Ancelin, K.; Waldmann, T.; Lacoste, N.; Lange, U.C.; Cesari, F.; Lee, C.; Almouzni, G.; Schneider, R.; Surani, M.A. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 2008, 452, 877–881. [Google Scholar] [CrossRef] [PubMed]
- Lane, N.; Dean, W.; Erhardt, S.; Hajkova, P.; Surani, A.; Walter, J.; Reik, W. Resistance of iaps to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 2003, 35, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.M.; Goyal, P.; Maksakova, I.A.; Bilenky, M.; Leung, D.; Tang, J.X.; Shinkai, Y.; Mager, D.L.; Jones, S.; Hirst, M.; et al. DNA methylation and setdb1/h3k9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mescs. Cell Stem Cell 2011, 8, 676–687. [Google Scholar] [CrossRef] [PubMed]
- Elsasser, S.J.; Noh, K.M.; Diaz, N.; Allis, C.D.; Banaszynski, L.A. Histone h3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 2015, 522, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Rowe, H.M.; Jakobsson, J.; Mesnard, D.; Rougemont, J.; Reynard, S.; Aktas, T.; Maillard, P.V.; Layard-Liesching, H.; Verp, S.; Marquis, J.; et al. Kap1 controls endogenous retroviruses in embryonic stem cells. Nature 2010, 463, 237–240. [Google Scholar] [CrossRef]
- Maksakova, I.A.; Thompson, P.J.; Goyal, P.; Jones, S.J.; Singh, P.B.; Karimi, M.M.; Lorincz, M.C. Distinct roles of kap1, hp1 and g9a/glp in silencing of the two-cell-specific retrotransposon mervl in mouse es cells. Epigenet. Chromatin 2013, 6, 15. [Google Scholar] [CrossRef]
- Leung, D.; Du, T.; Wagner, U.; Xie, W.; Lee, A.Y.; Goyal, P.; Li, Y.; Szulwach, K.E.; Jin, P.; Lorincz, M.C.; et al. Regulation of DNA methylation turnover at ltr retrotransposons and imprinted loci by the histone methyltransferase setdb1. Proc. Natl. Acad. Sci. USA 2014, 111, 6690–6695. [Google Scholar] [CrossRef]
- Liu, X.; Gao, Q.; Li, P.; Zhao, Q.; Zhang, J.; Li, J.; Koseki, H.; Wong, J. Uhrf1 targets dnmt1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated h3k9. Nat. Commun. 2013, 4, 1563. [Google Scholar] [CrossRef]
- Hackett, J.A.; Sengupta, R.; Zylicz, J.J.; Murakami, K.; Lee, C.; Down, T.A.; Surani, M.A. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 2013, 339, 448–452. [Google Scholar] [CrossRef]
- Seisenberger, S.; Andrews, S.; Krueger, F.; Arand, J.; Walter, J.; Santos, F.; Popp, C.; Thienpont, B.; Dean, W.; Reik, W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 2012, 48, 849–862. [Google Scholar] [CrossRef] [PubMed]
- Gatewood, J.M.; Cook, G.R.; Balhorn, R.; Schmid, C.W.; Bradbury, E.M. Isolation of four core histones from human sperm chromatin representing a minor subset of somatic histones. J. Biol. Chem. 1990, 265, 20662–20666. [Google Scholar] [PubMed]
- Hammoud, S.S.; Nix, D.A.; Zhang, H.; Purwar, J.; Carrell, D.T.; Cairns, B.R. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009, 460, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Brykczynska, U.; Hisano, M.; Erkek, S.; Ramos, L.; Oakeley, E.J.; Roloff, T.C.; Beisel, C.; Schubeler, D.; Stadler, M.B.; Peters, A.H. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 2010, 17, 679–687. [Google Scholar] [CrossRef]
- Zhang, B.; Zheng, H.; Huang, B.; Li, W.; Xiang, Y.; Peng, X.; Ming, J.; Wu, X.; Zhang, Y.; Xu, Q.; et al. Allelic reprogramming of the histone modification h3k4me3 in early mammalian development. Nature 2016, 537, 553–557. [Google Scholar] [CrossRef]
- Katz, D.J.; Edwards, T.M.; Reinke, V.; Kelly, W.G. A c. Elegans lsd1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 2009, 137, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Nottke, A.C.; Beese-Sims, S.E.; Pantalena, L.F.; Reinke, V.; Shi, Y.; Colaiacovo, M.P. Spr-5 is a histone h3k4 demethylase with a role in meiotic double-strand break repair. Proc. Natl. Acad. Sci. USA 2011, 108, 12805–12810. [Google Scholar] [CrossRef]
- Stern, S.; Fridmann-Sirkis, Y.; Braun, E.; Soen, Y. Epigenetically heritable alteration of fly development in response to toxic challenge. Cell Rep. 2012, 1, 528–542. [Google Scholar] [CrossRef]
- Fullston, T.; Ohlsson Teague, E.M.; Palmer, N.O.; DeBlasio, M.J.; Mitchell, M.; Corbett, M.; Print, C.G.; Owens, J.A.; Lane, M. Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the f2 generation and alters the transcriptional profile of testis and sperm microrna content. Faseb J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2013, 27, 4226–4243. [Google Scholar] [CrossRef]
- Ren, J.; Cheng, Y.; Ming, Z.H.; Dong, X.Y.; Zhou, Y.Z.; Ding, G.L.; Pang, H.Y.; Rahman, T.U.; Akbar, R.; Huang, H.F.; et al. Intrauterine hyperglycemia exposure results in intergenerational inheritance via DNA methylation reprogramming on f1 pgcs. Epigenet. Chromatin 2018, 11, 20. [Google Scholar] [CrossRef]
- World Health Organization. Don’t Pollute My Future! The Impact of the Environment on the Children’s Health; WHO Press: Geneva, Switzerland, 2017. [Google Scholar]
- Anway, M.D.; Cupp, A.S.; Uzumcu, M.; Skinner, M.K. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 2005, 308, 1466–1469. [Google Scholar] [CrossRef] [PubMed]
- Manikkam, M.; Haque, M.M.; Guerrero-Bosagna, C.; Nilsson, E.E.; Skinner, M.K. Pesticide methoxychlor promotes the epigenetic transgenerational inheritance of adult-onset disease through the female germline. PLoS ONE 2014, 9, e102091. [Google Scholar] [CrossRef] [PubMed]
- Manikkam, M.; Tracey, R.; Guerrero-Bosagna, C.; Skinner, M.K. Pesticide and insect repellent mixture (permethrin and deet) induces epigenetic transgenerational inheritance of disease and sperm epimutations. Reprod. Toxicol. 2012, 34, 708–719. [Google Scholar] [CrossRef] [PubMed]
- Manikkam, M.; Tracey, R.; Guerrero-Bosagna, C.; Skinner, M.K. Plastics derived endocrine disruptors (bpa, dehp and dbp) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS ONE 2013, 8, e55387. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K.; Manikkam, M.; Tracey, R.; Guerrero-Bosagna, C.; Haque, M.; Nilsson, E.E. Ancestral dichlorodiphenyltrichloroethane (ddt) exposure promotes epigenetic transgenerational inheritance of obesity. BMC Med. 2013, 11, 228. [Google Scholar] [CrossRef]
- Tracey, R.; Manikkam, M.; Guerrero-Bosagna, C.; Skinner, M.K. Hydrocarbons (jet fuel jp-8) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. Reprod. Toxicol. 2013, 36, 104–116. [Google Scholar] [CrossRef]
- Diaz-Castillo, C.; Chamorro-Garcia, R.; Shioda, T.; Blumberg, B. Transgenerational self-reconstruction of disrupted chromatin organization after exposure to an environmental stressor in mice. Sci. Rep. 2019, 9, 13057. [Google Scholar] [CrossRef]
- Gely-Pernot, A.; Hao, C.; Legoff, L.; Multigner, L.; D’Cruz, S.C.; Kervarrec, C.; Jegou, B.; Tevosian, S.; Smagulova, F. Gestational exposure to chlordecone promotes transgenerational changes in the murine reproductive system of males. Sci. Rep. 2018, 8, 10274. [Google Scholar] [CrossRef]
- Hao, C.; Gely-Pernot, A.; Kervarrec, C.; Boudjema, M.; Becker, E.; Khil, P.; Tevosian, S.; Jegou, B.; Smagulova, F. Exposure to the widely used herbicide atrazine results in deregulation of global tissue-specific rna transcription in the third generation and is associated with a global decrease of histone trimethylation in mice. Nucleic Acids Res. 2016, 44, 9784–9802. [Google Scholar] [CrossRef]
- WHO/UNEP. State of the Science of Endocrine Disrupting Chemicals—2012; WHO Press: Geneva, Switzerland, 2013; pp. 1–296. [Google Scholar]
- Nieuwenhuijsen, M.J.; Dadvand, P.; Grellier, J.; Martinez, D.; Vrijheid, M. Environmental risk factors of pregnancy outcomes: A summary of recent meta-analyses of epidemiological studies. Environ. Health A Glob. Access Sci. Source 2013, 12, 6. [Google Scholar] [CrossRef]
- DiVall, S.A. The influence of endocrine disruptors on growth and development of children. Curr. Opin. Endocrinol. Diabetes Obes. 2013, 20, 50–55. [Google Scholar] [CrossRef] [PubMed]
- de Cock, M.; Maas, Y.G.; van de Bor, M. Does perinatal exposure to endocrine disruptors induce autism spectrum and attention deficit hyperactivity disorders? Review. Acta Paediatr. 2012, 101, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, H.E.; Koskenniemi, J.J.; Sundqvist, E.; Main, K.M.; Kiviranta, H.; Tuomisto, J.T.; Tuomisto, J.; Viluksela, M.; Vartiainen, T.; Skakkebaek, N.E.; et al. Associations between congenital cryptorchidism in newborn boys and levels of dioxins and pcbs in placenta. Int. J. Androl. 2012, 35, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, H.E.; Adamsson, A. Cryptorchidism and endocrine disrupting chemicals. Mol. Cell. Endocrinol. 2012, 355, 208–220. [Google Scholar] [CrossRef]
- Bromer, J.G.; Wu, J.; Zhou, Y.; Taylor, H.S. Hypermethylation of homeobox a10 by in utero diethylstilbestrol exposure: An epigenetic mechanism for altered developmental programming. Endocrinology 2009, 150, 3376–3382. [Google Scholar] [CrossRef][Green Version]
- Bibbo, M.; Al-Naqeeb, M.; Baccarini, I.; Gill, W.; Newton, M.; Sleeper, K.M.; Sonek, R.N.; Wied, G.L. Follow-up study of male and female offspring of des-treated mothers a preliminary report. J. Reprod. Med. 1975, 15, 29–32. [Google Scholar]
- Herbst, A.L. Summary of the changes in the human female genital tract as a consequence of maternal diethylstilbestrol therapy. J. Toxicol. Environ. Health Suppl. 1976, 1, 13–20. [Google Scholar]
- Ozgyin, L.; Erdos, E.; Bojcsuk, D.; Balint, B.L. Nuclear receptors in transgenerational epigenetic inheritance. Prog. Biophys. Mol. Biol. 2015, 118, 34–43. [Google Scholar] [CrossRef]
- Drobna, Z.; Henriksen, A.D.; Wolstenholme, J.T.; Montiel, C.; Lambeth, P.S.; Shang, S.; Harris, E.P.; Zhou, C.; Flaws, J.A.; Adli, M.; et al. Transgenerational effects of bisphenol a on gene expression and DNA methylation of imprinted genes in brain. Endocrinology 2018, 159, 132–144. [Google Scholar] [CrossRef]
- Lombo, M.; Fernandez-Diez, C.; Gonzalez-Rojo, S.; Navarro, C.; Robles, V.; Herraez, M.P. Transgenerational inheritance of heart disorders caused by paternal bisphenol a exposure. Environ. Pollut. 2015, 206, 667–678. [Google Scholar] [CrossRef]
- Wolstenholme, J.T.; Goldsby, J.A.; Rissman, E.F. Transgenerational effects of prenatal bisphenol a on social recognition. Horm. Behav. 2013, 64, 833–839. [Google Scholar] [CrossRef]
- Wolstenholme, J.T.; Edwards, M.; Shetty, S.R.; Gatewood, J.D.; Taylor, J.A.; Rissman, E.F.; Connelly, J.J. Gestational exposure to bisphenol a produces transgenerational changes in behaviors and gene expression. Endocrinology 2012, 153, 3828–3838. [Google Scholar] [CrossRef]
- Doherty, L.F.; Bromer, J.G.; Zhou, Y.; Aldad, T.S.; Taylor, H.S. In utero exposure to diethylstilbestrol (des) or bisphenol-a (bpa) increases ezh2 expression in the mammary gland: An epigenetic mechanism linking endocrine disruptors to breast cancer. Horm. Cancer 2010, 1, 146–155. [Google Scholar] [CrossRef]
- Krebs, M.; Sakurai, R.; Torday, J.S.; Rehan, V.K. Evidence for in vivo nicotine-induced alveolar interstitial fibroblast-to-myofibroblast transdifferentiation. Exp. Lung Res. 2010, 36, 390–398. [Google Scholar] [CrossRef]
- Rehan, V.K.; Liu, J.; Sakurai, R.; Torday, J.S. Perinatal nicotine-induced transgenerational asthma. Am. J. Physiol. Lung Cell. Mol. Physiol. 2013, 305, L501–L507. [Google Scholar] [CrossRef]
- Greer, E.L.; Maures, T.J.; Ucar, D.; Hauswirth, A.G.; Mancini, E.; Lim, J.P.; Benayoun, B.A.; Shi, Y.; Brunet, A. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 2011, 479, 365–371. [Google Scholar] [CrossRef]
- Dumasia, K.; Kumar, A.; Deshpande, S.; Balasinor, N.H. Estrogen, through estrogen receptor 1, regulates histone modifications and chromatin remodeling during spermatogenesis in adult rats. Epigenetics 2017, 12, 953–963. [Google Scholar] [CrossRef]
- Andersen, H.R.; Andersson, A.M.; Arnold, S.F.; Autrup, H.; Barfoed, M.; Beresford, N.A.; Bjerregaard, P.; Christiansen, L.B.; Gissel, B.; Hummel, R.; et al. Comparison of short-term estrogenicity tests for identification of hormone-disrupting chemicals. Environ. Health Perspect. 1999, 107, 89–108. [Google Scholar] [CrossRef]
- Okada, H.; Tokunaga, T.; Liu, X.; Takayanagi, S.; Matsushima, A.; Shimohigashi, Y. Direct evidence revealing structural elements essential for the high binding ability of bisphenol a to human estrogen-related receptor-gamma. Environ. Health Perspect. 2008, 116, 32–38. [Google Scholar] [CrossRef]
- Lemaire, G.; Mnif, W.; Mauvais, P.; Balaguer, P.; Rahmani, R. Activation of alpha- and beta-estrogen receptors by persistent pesticides in reporter cell lines. Life Sci. 2006, 79, 1160–1169. [Google Scholar] [CrossRef]
- Sado, T.; Sakaguchi, T. Species-specific differences in x chromosome inactivation in mammals. Reproduction 2013, 146, R131–R139. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.F.; Lin, R.C.; Laybutt, D.R.; Barres, R.; Owens, J.A.; Morris, M.J. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature 2010, 467, 963–966. [Google Scholar] [CrossRef]
- Carone, B.R.; Fauquier, L.; Habib, N.; Shea, J.M.; Hart, C.E.; Li, R.; Bock, C.; Li, C.; Gu, H.; Zamore, P.D.; et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 2010, 143, 1084–1096. [Google Scholar] [CrossRef]
- Hardikar, A.A.; Satoor, S.N.; Karandikar, M.S.; Joglekar, M.V.; Puranik, A.S.; Wong, W.; Kumar, S.; Limaye, A.; Bhat, D.S.; Januszewski, A.S.; et al. Multigenerational undernutrition increases susceptibility to obesity and diabetes that is not reversed after dietary recuperation. Cell Metab. 2015, 22, 312–319. [Google Scholar] [CrossRef]
- Masuyama, H.; Mitsui, T.; Eguchi, T.; Tamada, S.; Hiramatsu, Y. The effects of paternal high-fat diet exposure on offspring metabolism with epigenetic changes in the mouse adiponectin and leptin gene promoters. Am. J. Physiol. Endocrinol. Metab. 2016, 311, E236–E245. [Google Scholar] [CrossRef]
- Nowacka-Woszuk, J.; Szczerbal, I.; Malinowska, A.M.; Chmurzynska, A. Transgenerational effects of prenatal restricted diet on gene expression and histone modifications in the rat. PLoS ONE 2018, 13, e0193464. [Google Scholar] [CrossRef]
- Franklin, T.B.; Russig, H.; Weiss, I.C.; Graff, J.; Linder, N.; Michalon, A.; Vizi, S.; Mansuy, I.M. Epigenetic transmission of the impact of early stress across generations. Biol. Psychiatry 2010, 68, 408–415. [Google Scholar] [CrossRef]
- Dong, E.; Dzitoyeva, S.G.; Matrisciano, F.; Tueting, P.; Grayson, D.R.; Guidotti, A. Brain-derived neurotrophic factor epigenetic modifications associated with schizophrenia-like phenotype induced by prenatal stress in mice. Biol. Psychiatry 2015, 77, 589–596. [Google Scholar] [CrossRef]
- Zucchi, F.C.; Yao, Y.; Ward, I.D.; Ilnytskyy, Y.; Olson, D.M.; Benzies, K.; Kovalchuk, I.; Kovalchuk, O.; Metz, G.A. Maternal stress induces epigenetic signatures of psychiatric and neurological diseases in the offspring. PLoS ONE 2013, 8, e56967. [Google Scholar] [CrossRef]
- Dias, B.G.; Ressler, K.J. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat. Neurosci. 2014, 17, 89–96. [Google Scholar] [CrossRef]
- Gapp, K.; Soldado-Magraner, S.; Alvarez-Sanchez, M.; Bohacek, J.; Vernaz, G.; Shu, H.; Franklin, T.B.; Wolfer, D.; Mansuy, I.M. Early life stress in fathers improves behavioural flexibility in their offspring. Nat. Commun. 2014, 5, 5466. [Google Scholar] [CrossRef]
- Bogdarina, I.; Welham, S.; King, P.J.; Burns, S.P.; Clark, A.J. Epigenetic modification of the renin-angiotensin system in the fetal programming of hypertension. Circ. Res. 2007, 100, 520–526. [Google Scholar] [CrossRef]
- Gregory, D.J.; Kobzik, L.; Yang, Z.; McGuire, C.C.; Fedulov, A.V. Transgenerational transmission of asthma risk after exposure to environmental particles during pregnancy. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017, 313, L395–L405. [Google Scholar] [CrossRef]
- Skinner, M.K.; Anway, M.D.; Savenkova, M.I.; Gore, A.C.; Crews, D. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoS ONE 2008, 3, e3745. [Google Scholar] [CrossRef]
- Stouder, C.; Paoloni-Giacobino, A. Transgenerational effects of the endocrine disruptor vinclozolin on the methylation pattern of imprinted genes in the mouse sperm. Reproduction 2010, 139, 373–379. [Google Scholar] [CrossRef]
- Singh, S.P.; Chand, H.S.; Langley, R.J.; Mishra, N.; Barrett, T.; Rudolph, K.; Tellez, C.; Filipczak, P.T.; Belinsky, S.; Saeed, A.I.; et al. Gestational exposure to sidestream (secondhand) cigarette smoke promotes transgenerational epigenetic transmission of exacerbated allergic asthma and bronchopulmonary dysplasia. J. Immunol. 2017, 198, 3815–3822. [Google Scholar] [CrossRef]
- McBirney, M.; King, S.E.; Pappalardo, M.; Houser, E.; Unkefer, M.; Nilsson, E.; Sadler-Riggleman, I.; Beck, D.; Winchester, P.; Skinner, M.K. Atrazine induced epigenetic transgenerational inheritance of disease, lean phenotype and sperm epimutation pathology biomarkers. PLoS ONE 2017, 12, e0184306. [Google Scholar] [CrossRef]
| Epigenetic Mark that Was Affected | Environmental Factor Involved | Organ/Matrices Studied | Animal Model | Associated Health Issue in Offspring | Reference | |
|---|---|---|---|---|---|---|
| Metabolic effects | Hypomethylation of Il13ra2 (interleukin 13 receptor subunit alpha 2), altered expression of 642 pancreatic islet genes in female F1 offspring following paternal high-fat diet. | Paternal high-fat diet | Pancreatic islets | Sprague–Dawley rats | Impaired glucose-insulin homeostatsis (Type 2 diabetes) | [184] |
| 20% change in cytosine methylation along with methylation at the enhancer of a key lipid regulator, PPARα, in the liver of F1 offspring following paternal low-protein diet. | Paternal low-protein diet | Liver | C57/Bl6 mice | Impaired cholesterol and lipid metabolism | [185] | |
| Altered epigenetic signatures in the insulin-2 gene promoter region and inefficient binding of transcription factor PDX1 at the insulin-2 promotor region following undernutrition for 50 generations. | Undernutrition (protein and caloric restriction) | Pancreas | Wistar rats | Adiposity/Type 2 diabetes | [186] | |
| Decrease in acetyl H3K9 and increase in dimethyl H3K9 levels in adiponectin and leptin gene promotor region in the offspring when mothers were fed high-fat diet for multiple generations. | Maternal high-fat diet | Adipose tissue | ICR outbred mice | Impaired glucose homeostasis and obesity | [187] | |
| Differentially methylated genes enriched for obesity/diabetic and metabolic changes in F2, not F3 generation males on intrauterine exposure to hyperglycemia. | Intrauterine hyperglycemia | Primordial germ cells | ICR mice | Obesity and insulin resistance | [150] | |
| Paternal diet restriction significantly changed the DNA methyltransferase, Dnmt1 and the transcript of methyl CpG binding protein 2 (Mecp2) in F1, not in F2 and F3 generations. An increase in the expression of histone modification gene, histone deacetylase 1 (Hdac1) in fetus liver was found in F1 and F2. An increased H3 acetylation in fetuses was also detected in F2 generation. | Diet restriction | Liver, adipose and muscle | Wistar rats | Metabolic changes | [188] | |
| Neurological effects | Hypo- and hyper-methylation of several candidate genes including MeCP2, cannabinoid receptor 1 (CB1), corticotropin-releasing hormone 2 (CRFR2) genes in the sperm of males and the brain of F2 females. | Stress (chronic maternal separation) | Brain, Sperm | C57Bl6/J mice | Depressive-like behavior | [189] |
| An increase in DNA methyltransferase 1 (DNMT1) and ten-eleven translocation hydroxylases (TET1) in the frontal cortex and hippocampus of offspring along with a decrease in 5-methylcytosine and 5-hydroxylmethylcytosine levels at Bdnf gene regulatory regions following prenatal stress. | Stress (restraining movement in pregnant dams for 45 min from gestation day 7 until delivery) | Brain | Swiss albino mice | Schizophrenia-like phenotype | [190] | |
| Upregulation of miR-103 and downregulation of its target gene Ptplb, downregulation of mIR-145 (a marker of multiple sclerosis), upregulation of miR-323, miR-98 (involved in inflammatory responses in brain), and miR-219 that targets the gene Dazap1 (marker of schizophrenia and bipolar disorders) was observed in the offspring following induction of stress to pregnant mothers. | Stress (Pregnant dams were forced to swim for 5 min and restrained body movement for 20 min from gestational day 12 to 18) | Brain | Long-Evans rats | Brain diseases (genes involved in multiple sclerosis, schizophrenia and bipolar disorder) | [99,191] | |
| CpG hypomethylation in the olfactory Olfr151 gene in the sperm of F1 generation whose parents underwent olfactory fear conditioning with acetophenone. | Olfactory fear conditioning with acetophenone or propanol | Sperm | C57BL/6J and M71-LacZ transgenic mice | Fear/behavioral sensitivity | [192] | |
| Acetylation of H4K5 and H3K14, dimethylation of H3K4, and trimethylation of H3K36 (H3K36me3) were significantly decreased in mineralocorticoid receptor (MR) gene in the F2 hippocampus following maternal separation in F0 generation offspring. Sperm DNA methylation was significantly increased at several CpGs across the MR promoter. In a follow-up study, miR-375 was found to be upregulated in the hippocampus of F1 and F2 offspring. | Maternal separation and maternal stress for 2 weeks (F0) | Hippocampus Sperm | C57BL/6 mice | Traumatic stress/depressive anxiety-like behavior | [193] | |
| Cardio-vascular disorders | Decreased methylation of the AT1b angiotensin receptor gene in the offspring following maternal low protein diet. | Maternal low protein diet | Adrenal gland | Wistar rats | Hypertension (renin-angiotensin system) | [194] |
| Respiratory diseases | Altered methylation of 14480 individual CpG loci in F1, 9413 loci in F2 and 6239 in F3 generations in dendritic cell methylome following maternal exposure to intranasal instillation of environmental particles. | Maternal intranasal instillation of environmental particles | Dendrite cells | BALB/C mice | Asthma | [195] |
| Environmental toxicants/factors | Transgenerational differential expression of 92 genes in the hippocampus and 276 genes in amygdala in males, and 1301 genes in hippocampus and 172 in the amygdala in females following exposure to vinclozolin, an endocrine-disrupting chemical. | Maternal exposure to vinclozolin (100 mg/kg/day from gestational day 8–14) | Brain | Sprague–Dawley rats | Anxiety-like behavior | [196] |
| The number of methylated CpG in H19 and Gtl2 genes (paternally methylated) decreased while Peg1, Snrpn, and Peg3 (maternally methylated) increased in F1 male offspring. These effects were not significant in F2 and F3 generations. | Maternal exposure to vinclozolin (intraperitoneal injection at a dose of 50 mg/kg/day) from gestation day 10–18 | Sperm | FVB/N mice | Decrease in sperm concentration | [197] | |
| Lower levels of microRNA, miR-130a, and increased levels of miR-16 and miR-221 along with a decreased expression of HIF-1α and other biochemical and histological changes in the lungs (F1 and F2 generations) where mothers were exposed to second-hand cigarette smoke (mice). | Maternal exposure to second-hand cigarette smoke | Lungs | BALB/C mice | Asthma and Bronchopulmonary dysplasia | [198] | |
| A significant increase in DNA methylation regions in sperm (also called epimutations) was observed in F1, F2, and F3 generations when pregnant rats were administered atrazine, a herbicide. | Maternal exposure to atrazine (intraperitoneal injection at a dose of 25 mg/kg body weight/day) from gestational day 8 to 14. | Sperm | Sprague–Dawley rats | Lean phenotype and hyperactivity | [199] | |
| Differentially methylated DNA methylation regions in sperm (epimutations) in 197 different promoters in the F3 generation were observed following the administration of a plastic compound mixture to pregnant rats. | Maternal exposure to plastic mixture (intraperitoneal injection of a mixture of bisphenol A 50 mg/kg BW/day, DEHP 750 mg/kg BW/day and DBP 66 mg/kg/BW/day) from gestational day 8 to 14. | Sperm | Sprague–Dawley rats | Obesity and sperm abnormalities | [155] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Legoff, L.; D’Cruz, S.C.; Tevosian, S.; Primig, M.; Smagulova, F. Transgenerational Inheritance of Environmentally Induced Epigenetic Alterations during Mammalian Development. Cells 2019, 8, 1559. https://doi.org/10.3390/cells8121559
Legoff L, D’Cruz SC, Tevosian S, Primig M, Smagulova F. Transgenerational Inheritance of Environmentally Induced Epigenetic Alterations during Mammalian Development. Cells. 2019; 8(12):1559. https://doi.org/10.3390/cells8121559
Chicago/Turabian StyleLegoff, Louis, Shereen Cynthia D’Cruz, Sergei Tevosian, Michael Primig, and Fatima Smagulova. 2019. "Transgenerational Inheritance of Environmentally Induced Epigenetic Alterations during Mammalian Development" Cells 8, no. 12: 1559. https://doi.org/10.3390/cells8121559
APA StyleLegoff, L., D’Cruz, S. C., Tevosian, S., Primig, M., & Smagulova, F. (2019). Transgenerational Inheritance of Environmentally Induced Epigenetic Alterations during Mammalian Development. Cells, 8(12), 1559. https://doi.org/10.3390/cells8121559





