Effects of Ionic Liquid Alkyl Chain Length on Denaturation of Myoglobin by Anionic, Cationic, and Zwitterionic Detergents
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Sample Preparation
2.3. Absorbance Measurements
2.4. Fluorescence Spectroscopy
2.5. Circular Dichroism Spectroscopy
2.6. Calculation of Gibbs Free Energy of Dissociation
3. Results
3.1. Critical Micelle Concentration (CMC) Analysis
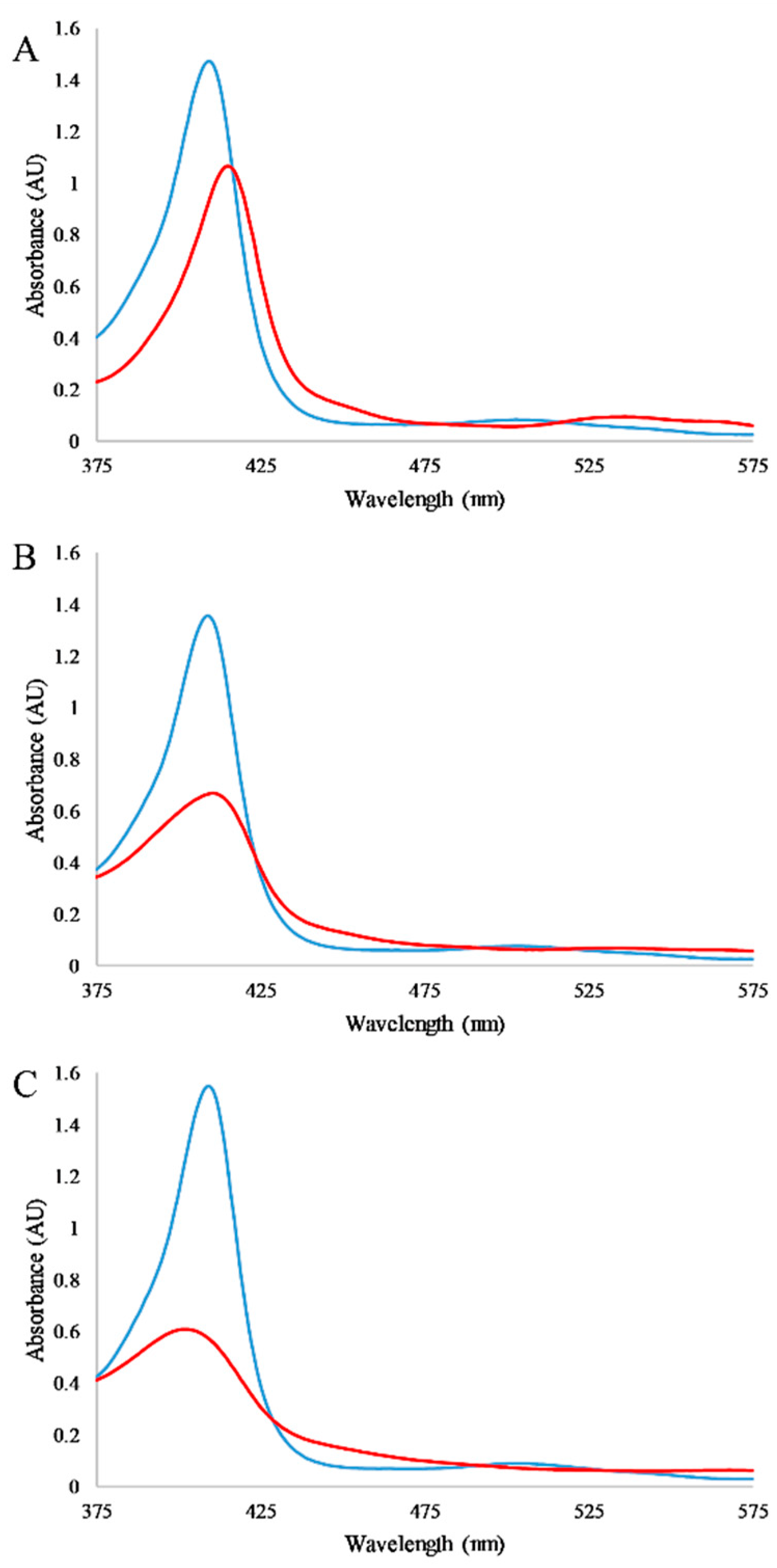
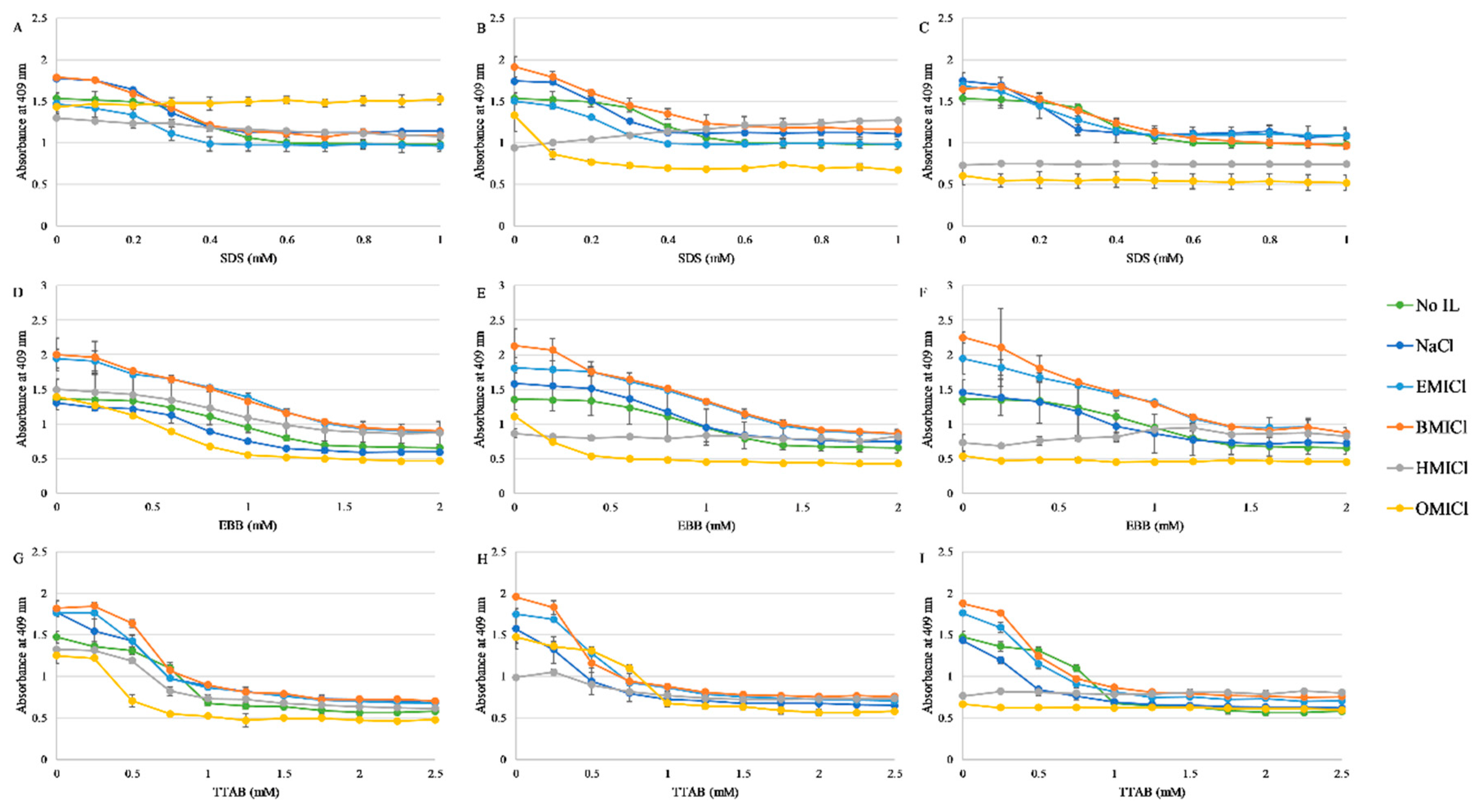
3.2. Absorbance Spectroscopy
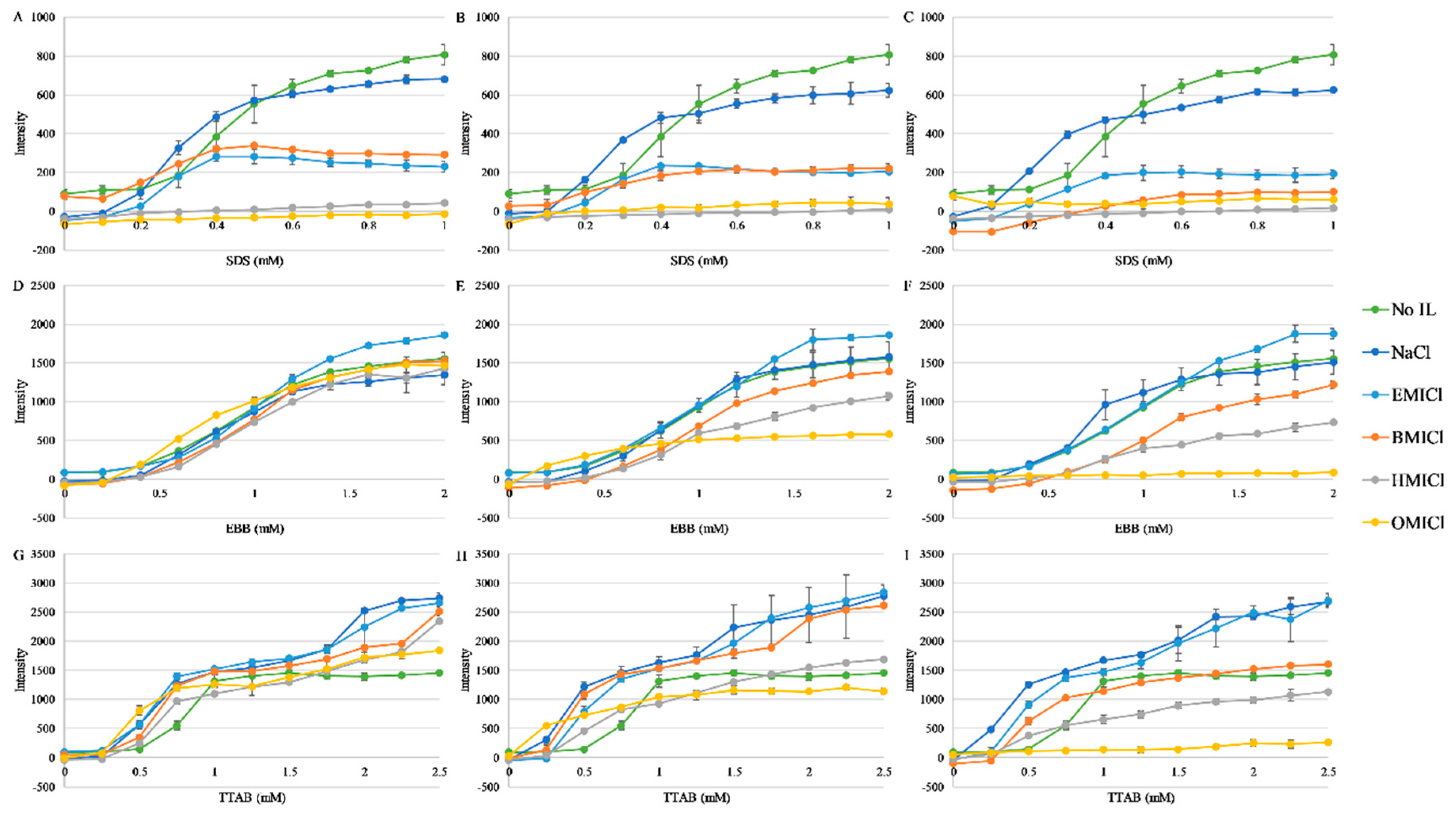
3.3. Fluorescence Spectroscopy
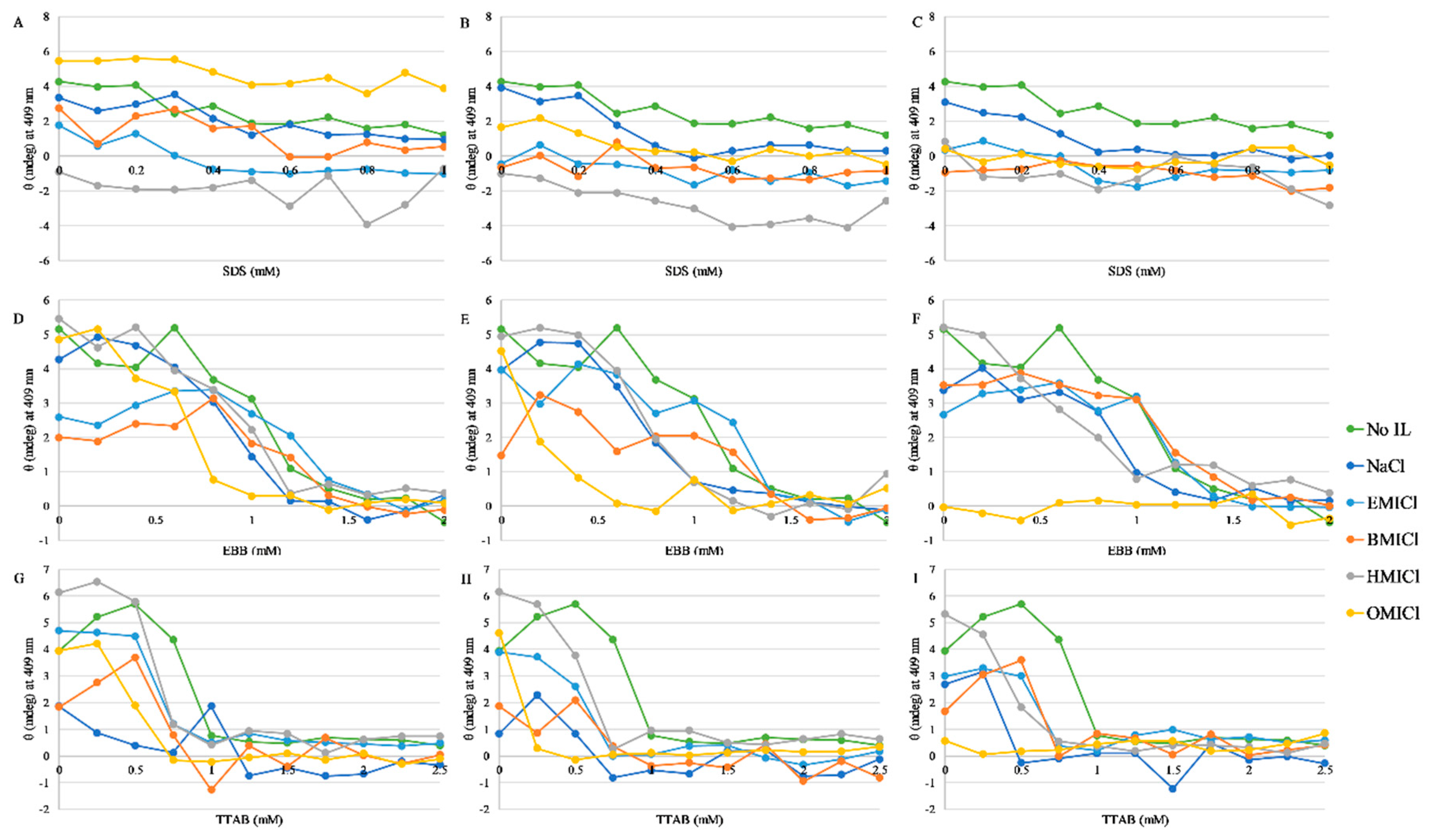
3.4. Circular Dichroism Spectroscopy
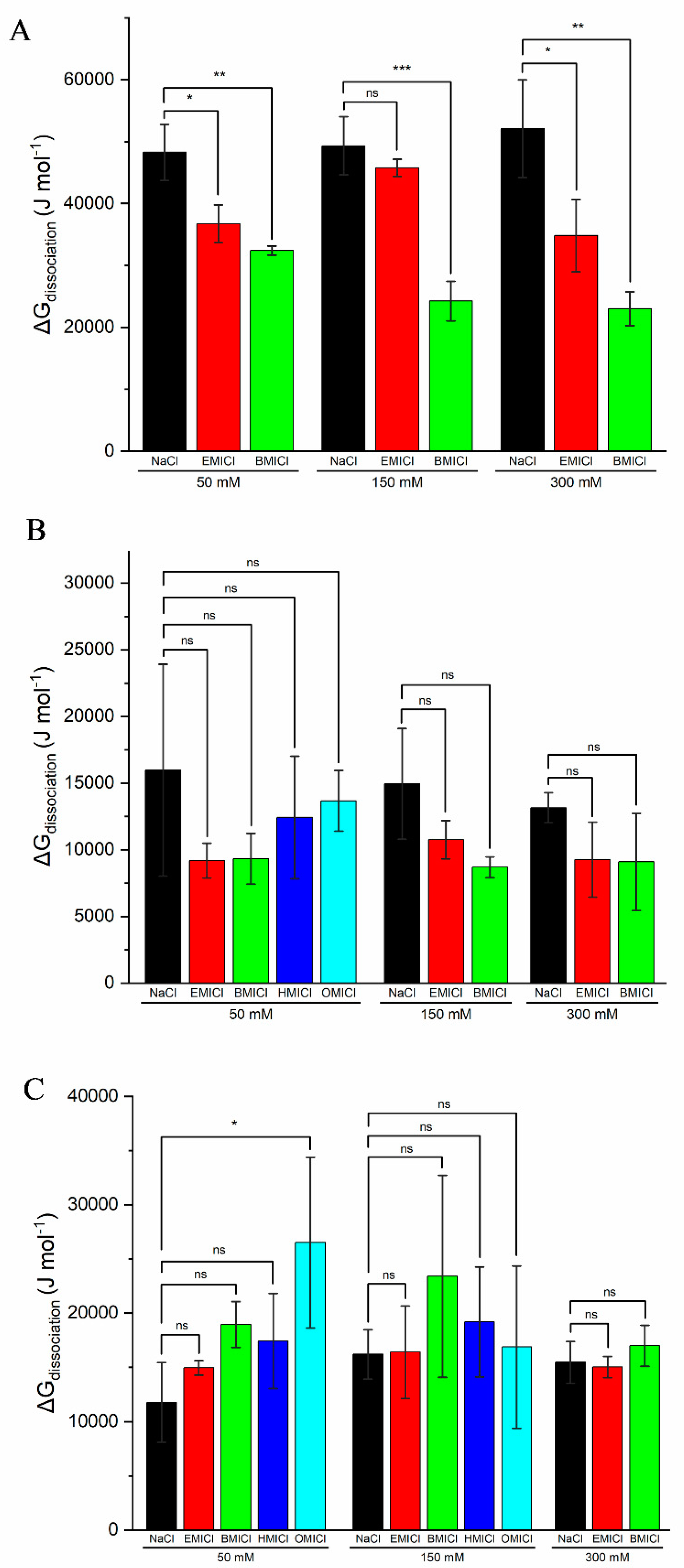
3.5. Thermodynamic Analysis of Heme Dissociation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marsh, K.N.; Boxall, J.A.; Lichtenthaler, R. Room temperature ionic liquids and their mixtures—A review. Fluid Phase Equilibr. 2004, 219, 93–98. [Google Scholar] [CrossRef]
- Salihu, A.; Alam, M.Z. Solvent tolerant lipases: A review. Process Biochem. 2015, 50, 86–96. [Google Scholar] [CrossRef]
- Troter, D.Z.; Todorović, Z.B.; Đokić-Stojanović, D.R.; Stamenković, O.S.; Veljković, V.B. Application of ionic liquids and deep eutectic solvents in biodiesel production: A review. Renew. Sustain. Energy Rev. 2016, 61, 473–500. [Google Scholar] [CrossRef]
- Vekariya, R.L. A review of ionic liquids: Applications towards catalytic organic transformations. J. Mol. Liquids 2017, 227, 44–60. [Google Scholar] [CrossRef]
- Wahlström, R.M.; Suurnäkki, A. Enzymatic hydrolysis of lignocellulosic polysaccharides in the presence of ionic liquids. Green Chem. 2015, 17, 694–714. [Google Scholar] [CrossRef]
- Adawiyah, N.; Moniruzzaman, M.; Hawatulaila, S.; Goto, M. Ionic liquids as a potential tool for drug delivery systems. Med. Chem. Comm. 2016, 7, 1881–1897. [Google Scholar] [CrossRef]
- Egorova, K.S.; Gordeev, E.G.; Ananikov, V.P. Biological activity of ionic liquids and their application in pharmaceutics and medicine. Chem. Rev. 2017, 117, 7132–7189. [Google Scholar] [CrossRef] [PubMed]
- Sahbaz, Y.; Williams, H.D.; Nguyen, T.-H.; Saunders, J.; Ford, L.; Charman, S.A.; Scammells, P.J.; Porter, C.J.H. Transformation of poorly water-soluble drugs into lipophilic ionic liquids enhances oral drug exposure from lipid based formulations. Mol. Pharm. 2015, 12, 1980–1991. [Google Scholar] [CrossRef] [PubMed]
- Yabre, M.; Ferey, L.; Some, I.T.; Gaudin, K. Greening reversed-phase liquid chromatography methods using alternative solvents for pharmaceutical analysis. Molecules 2018, 23, 1065. [Google Scholar] [CrossRef]
- Fiebig, O.C.; Mancini, E.; Caputo, G.; Vaden, T.D. Quantitative evaluation of myoglobin unfolding in the presence of guanidinium hydrochloride and ionic liquids in solution. J. Phys. Chem. B 2014, 118, 406–412. [Google Scholar] [CrossRef]
- Hanna, S.L.; Huang, J.L.; Swinton, A.J.; Caputo, G.A.; Vaden, T.D. Synergistic effects of polymyxin and ionic liquids on lipid vesicle membrane stability and aggregation. Biophys. Chem. 2017, 227, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kohn, E.M.; Lee, J.Y.; Calabro, A.; Vaden, T.D.; Caputo, G.A. Heme dissociation from myoglobin in the presence of the zwitterionic detergent N,N-Dimethyl-N-Dodecylglycine betaine: Effects of ionic liquids. Biomolecules 2018, 8, 126. [Google Scholar] [CrossRef] [PubMed]
- Takada, A.; Kadokawa, J. Fabrication and characterization of polysaccharide ion gels with ionic liquids and their further conversion into value-added sustainable materials. Biomolecules 2015, 5, 244–262. [Google Scholar] [CrossRef] [PubMed]
- Borrell, K.L.; Cancglin, C.; Stinger, B.L.; DeFrates, K.G.; Caputo, G.A.; Wu, C.; Vaden, T.D. An experimental and molecular dynamics study of red fluorescent protein mcherry in novel aqueous amino acid ionic liquids. J. Phys. Chem. B 2017, 121, 4823–4832. [Google Scholar] [CrossRef] [PubMed]
- Irimescu, R.; Kato, K. Lipase-catalyzed enantioselective reaction of amines with carboxylic acids under reduced pressure in non-solvent system and in ionic liquids. Tetrah. Lett. 2004, 45, 523–525. [Google Scholar] [CrossRef]
- Gromiha, M.M. Protein stability. In Protein Bioinformatics; Gromiha, M.M., Ed.; Academic Press: Singapore, 2010; Chapter 6; pp. 209–245. [Google Scholar]
- Pace, C.N. Conformational stability of globular proteins. Trends Biochem. Sci. 1990, 15, 14–17. [Google Scholar] [CrossRef]
- Elgharbawy, A.A.; Alam, M.Z.; Moniruzzaman, M.; Goto, M. Ionic liquid pretreatment as emerging approaches for enhanced enzymatic hydrolysis of lignocellulosic biomass. Biochem. Eng. J. 2016, 109, 252–267. [Google Scholar] [CrossRef]
- Bisht, M.; Venkatesu, P. Influence of cholinium-based ionic liquids on the structural stability and activity of α-chymotrypsin. New J. Chem. 2017, 41, 13902–13911. [Google Scholar] [CrossRef]
- Vicente, F.A.; Lario, L.D.; Pessoa, A.; Ventura, S.P.M. Recovery of bromelain from pineapple stem residues using aqueous micellar two-phase systems with ionic liquids as co-surfactants. Process Biochem. 2016, 51, 528–534. [Google Scholar] [CrossRef]
- Constantinescu, D.; Weingartner, H.; Herrmann, C. Protein denaturation by ionic liquids and the Hofmeister series: A case study of aqueous solutions of ribonuclease A. Angew. Chem. Int. Ed. Engl. 2007, 46, 8887–8889. [Google Scholar] [CrossRef]
- Zhao, H. Protein stabilization and enzyme activation in ionic liquids: Specific ion effects. J. Chem. Technol. Biotechnol. 2016, 91, 25–50. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.C.; Hanna, S.L.; DeFrates, K.G.; Fiebig, O.C.; Vaden, T.D. Kinetics and mass spectrometric measurements of myoglobin unfolding in aqueous ionic liquid solutions. Int. J. Biol. Macromol. 2016, 85, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Jha, I.; Kumar, A.; Venkatesu, P. The overriding roles of concentration and hydrophobic effect on structure and stability of heme protein induced by imidazolium-based ionic liquids. J. Phys. Chem. B 2015, 119, 8357–8368. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Zhang, L.; Xie, J.; Guo, R. Binding characteristics and molecular mechanism of interaction between ionic liquid and DNA. J. Phys. Chem. B 2010, 114, 2033–2043. [Google Scholar] [CrossRef]
- Jungnickel, C.; Łuczak, J.; Ranke, J.; Fernández, J.F.; Müller, A.; Thöming, J. Micelle formation of imidazolium ionic liquids in aqueous solution. Colloid. Surf. A Physicochem. Eng. Asp. 2008, 316, 278–284. [Google Scholar] [CrossRef]
- Chattopadhyay, A.; London, E. Fluorimetric determination of critical micelle concentration avoiding interference from detergent charge. Anal. Biochem. 1984, 139, 408–412. [Google Scholar] [CrossRef]
- Dutkiewicz, E.; Jakubowska, A. Effect of electrolytes on the physicochemical behaviour of sodium dodecyl sulphate micelles. Colloid Polym. Sci. 2002, 280, 1009–1014. [Google Scholar]
- Andersen, K.K.; Westh, P.; Otzen, D.E. Global study of myoglobin−surfactant interactions. Langmuir 2008, 24, 399–407. [Google Scholar] [CrossRef]
- Hansen, J.H.; Petersen, S.V.; Andersen, K.K.; Enghild, J.J.; Damhus, T.; Otzen, D. Stable intermediates determine proteins’ primary unfolding sites in the presence of surfactants. Biopolymers 2009, 91, 221–231. [Google Scholar] [CrossRef]
- Gudiksen, K.L.; Gitlin, I.; Whitesides, G.M. Differentiation of proteins based on characteristic patterns of association and denaturation in solutions of SDS. Proc. Nat. Acad. Sci. USA 2006, 103, 7968. [Google Scholar] [CrossRef]
- Andersen, K.K.; Otzen, D.E. Denaturation of α-lactalbumin and myoglobin by the anionic biosurfactant rhamnolipid. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2014, 1844, 2338–2345. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.K.; Chen, L.C.; Yu, Z.; Nonami, H.; Erra-Balsells, R.; Hiraoka, K. Detection of protein from detergent solutions by probe electrospray ionization mass spectrometry (PESI-MS). J. Mass Spectrom. 2011, 46, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Urner, L.H.; Maier, Y.B.; Haag, R.; Pagel, K. Exploring the potential of dendritic oligoglycerol detergents for protein mass spectrometry. J. Am. Soc. Mass Spectrom. 2019, 30, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Tofani, L.; Feis, A.; Snoke, R.E.; Berti, D.; Baglioni, P.; Smulevich, G. Spectroscopic and interfacial properties of myoglobin/surfactant complexes. Biophys. J. 2004, 87, 1186–1195. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Ghosh, S. Effect of curcumin on the binding of cationic, anionic and nonionic surfactants with myoglobin. J. Mol. Struct. 2017, 1134, 292–297. [Google Scholar] [CrossRef]
- Moulik, S.P.; Haque, M.E.; Jana, P.K.; Das, A.R. Micellar properties of cationic surfactants in pure and mixed states. J. Phys. Chem. 1996, 100, 701–708. [Google Scholar] [CrossRef]
- Fujita, K.; MacFarlane, D.R.; Forsyth, M. Protein solubilising and stabilising ionic liquids. Chem. Commun. 2005, 38, 4804–4806. [Google Scholar] [CrossRef]
- Weingartner, H.; Cabrele, C.; Herrmann, C. How ionic liquids can help to stabilize native proteins. Phys. Chem. Chem. Phys. 2012, 14, 415–426. [Google Scholar] [CrossRef]
- Attri, P.; Venkatesu, P.; Kumar, A. Activity and stability of α-chymotrypsin in biocompatible ionic liquids: Enzyme refolding by triethyl ammonium acetate. Phys. Chem. Chem. Phys. 2011, 13, 2788–2796. [Google Scholar] [CrossRef]
- Greaves, T.L.; Drummond, C.J. Ionic liquids as amphiphile self-assembly media. Chem. Soc. Rev. 2008, 37, 1709–1726. [Google Scholar] [CrossRef]
- Tariq, M.; Freire, M.G.; Saramago, B.; Coutinho, J.A.P.; Lopes, J.N.C.; Rebelo, L.P.N. Surface tension of ionic liquids and ionic liquid solutions. Chem. Soc. Rev. 2012, 41, 829–868. [Google Scholar] [CrossRef] [PubMed]
- Santos, D. Micelle formation of protic ionic liquids in aqueous solution. J. Chem. Eng. Data 2018, 63, 1480–1487. [Google Scholar] [CrossRef]
- Jaeger, V.; Burney, P.; Pfaendtner, J. Comparison of three ionic liquid-tolerant cellulases by molecular dynamics. Biophys. J. 2015, 108, 880–892. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, A.M.; Sardinha, J.; Moore, G.R.; Cabrita, E.J. Protein destabilisation in ionic liquids: The role of preferential interactions in denaturation. Phys. Chem. Chem. Phys. 2013, 15, 19632–19643. [Google Scholar] [CrossRef] [PubMed]
- Micaelo, N.M.; Soares, C.M. Protein structure and dynamics in ionic liquids. Insights from molecular dynamics simulation studies. J. Phys. Chem. B 2008, 112, 2566–2572. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q. On the influence of hydrated imidazolium-based ionic liquid on protein structure stability: A molecular dynamics simulation study. J. Chem. Phys. 2013, 139, 115102. [Google Scholar] [CrossRef] [PubMed]
- Javadian, S.; Nasiri, F.; Heydari, A.; Yousefi, A.; Shahir, A.A. Modifying effect of imidazolium-based ionic liquids on surface activity and self-assembled nanostructures of sodium dodecyl sulfate. J. Phys. Chem. B 2014, 118, 4140–4150. [Google Scholar] [CrossRef]
- Yuan, J.; Bai, X.; Zhao, M.; Zheng, L. C12mimBr ionic liquid/SDS vesicle formation and use as template for the synthesis of hollow silica spheres. Langmuir 2010, 26, 11726–11731. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.Y.; Selfridge, K.M.; Kohn, E.M.; Vaden, T.D.; Caputo, G.A. Effects of Ionic Liquid Alkyl Chain Length on Denaturation of Myoglobin by Anionic, Cationic, and Zwitterionic Detergents. Biomolecules 2019, 9, 264. https://doi.org/10.3390/biom9070264
Lee JY, Selfridge KM, Kohn EM, Vaden TD, Caputo GA. Effects of Ionic Liquid Alkyl Chain Length on Denaturation of Myoglobin by Anionic, Cationic, and Zwitterionic Detergents. Biomolecules. 2019; 9(7):264. https://doi.org/10.3390/biom9070264
Chicago/Turabian StyleLee, Joshua Y., Katherine M. Selfridge, Eric M. Kohn, Timothy D. Vaden, and Gregory A. Caputo. 2019. "Effects of Ionic Liquid Alkyl Chain Length on Denaturation of Myoglobin by Anionic, Cationic, and Zwitterionic Detergents" Biomolecules 9, no. 7: 264. https://doi.org/10.3390/biom9070264
APA StyleLee, J. Y., Selfridge, K. M., Kohn, E. M., Vaden, T. D., & Caputo, G. A. (2019). Effects of Ionic Liquid Alkyl Chain Length on Denaturation of Myoglobin by Anionic, Cationic, and Zwitterionic Detergents. Biomolecules, 9(7), 264. https://doi.org/10.3390/biom9070264





