Phenolic Compounds Inhibit 3T3-L1 Adipogenesis Depending on the Stage of Differentiation and Their Binding Affinity to PPARγ
Abstract
1. Introduction
2. Results and Discussion
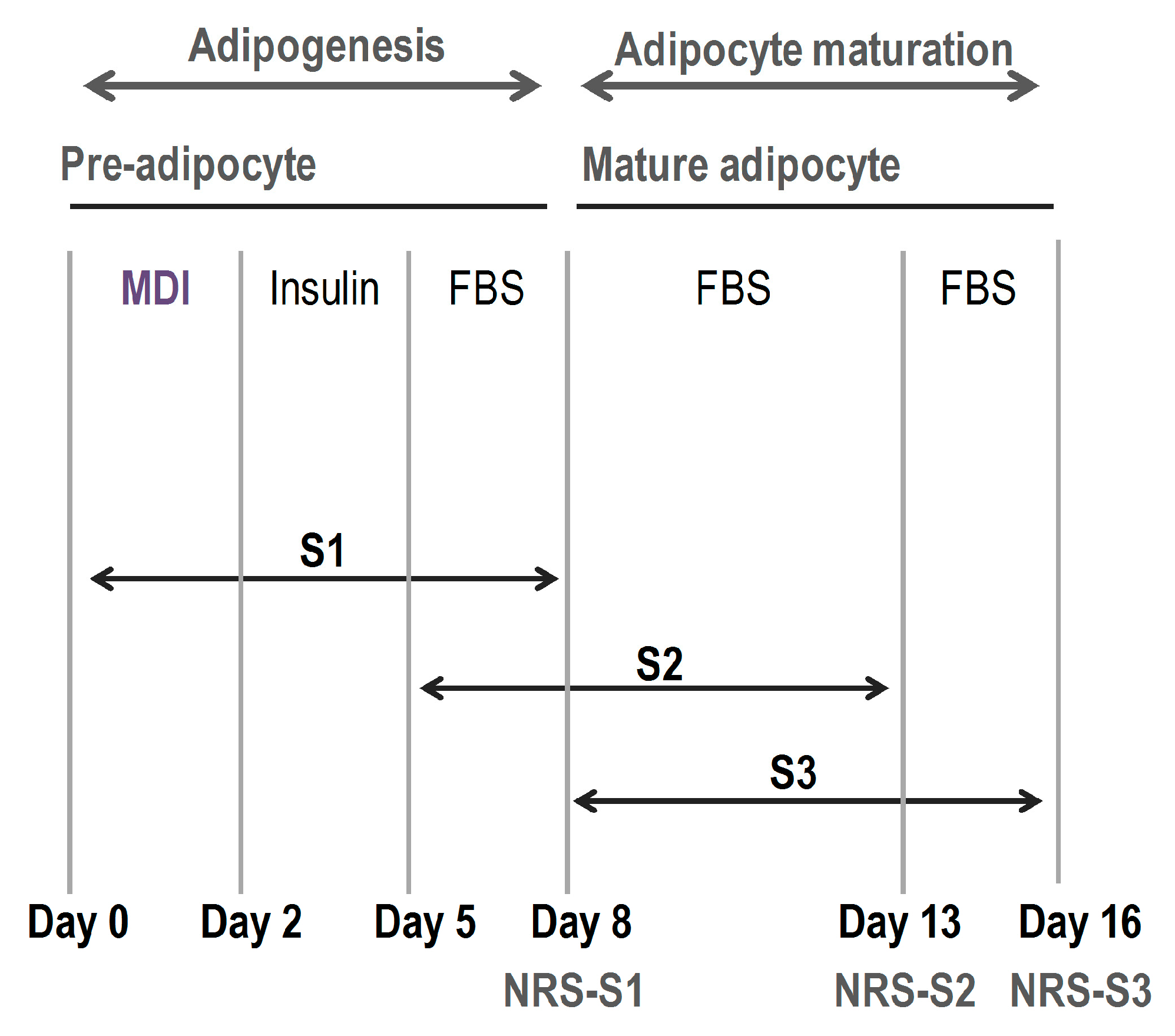
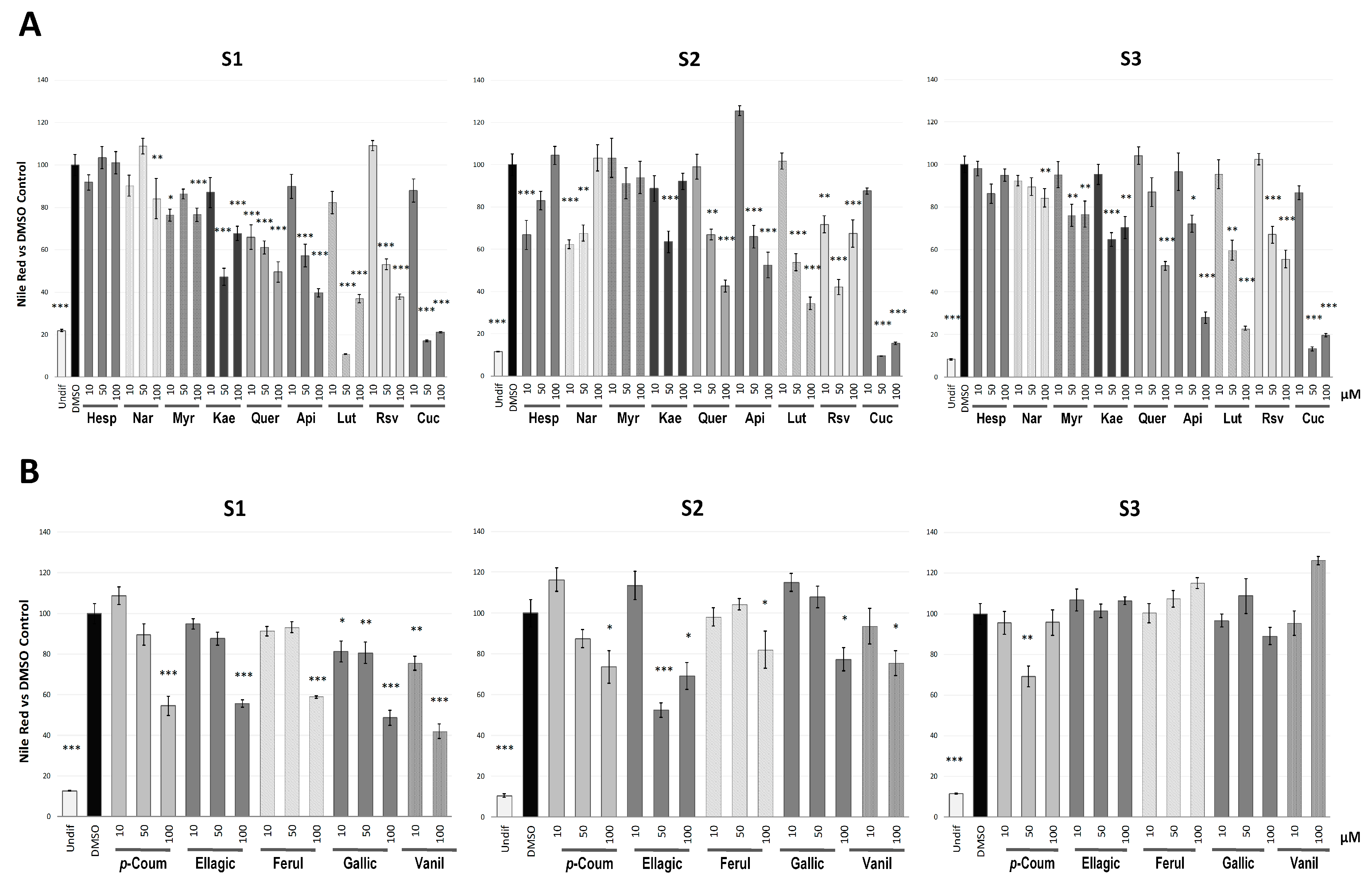
2.1. Phenolic Compounds Inhibited Adipogenesis with Different Intensity Depending on the Stage of the Differentiation Process
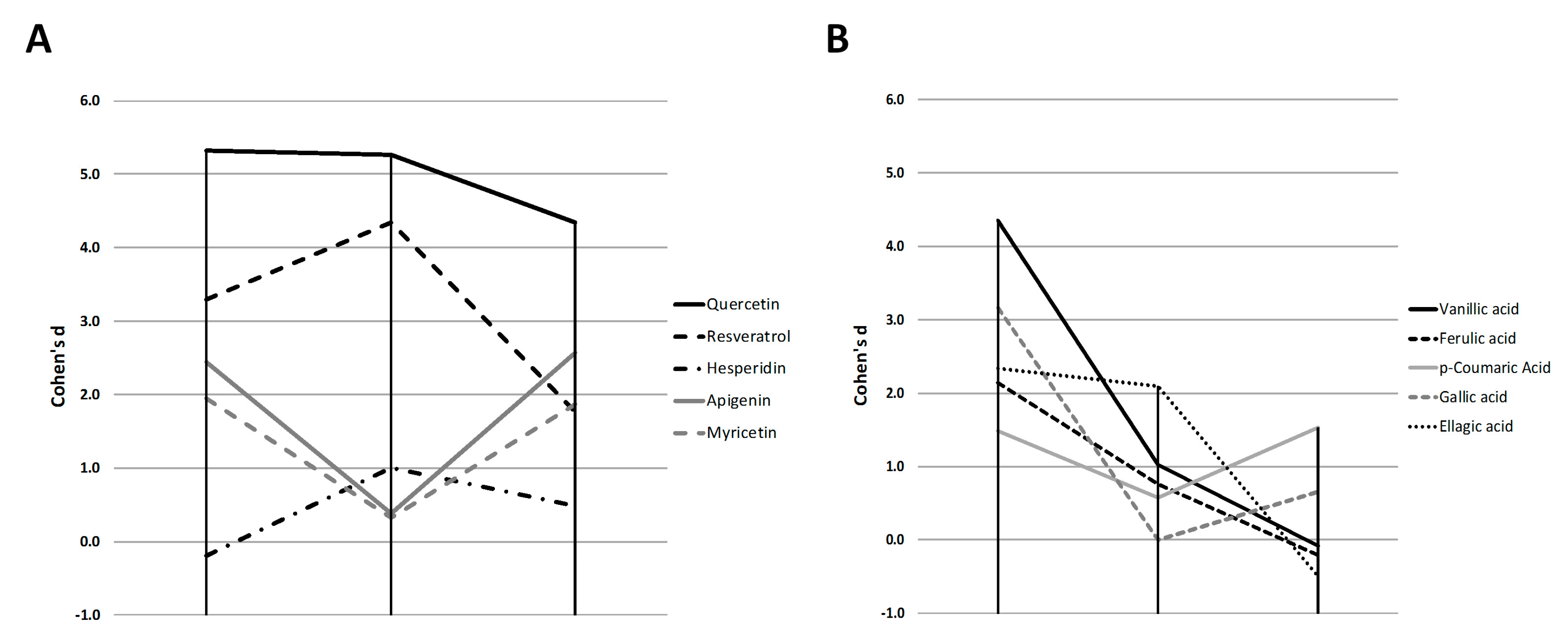
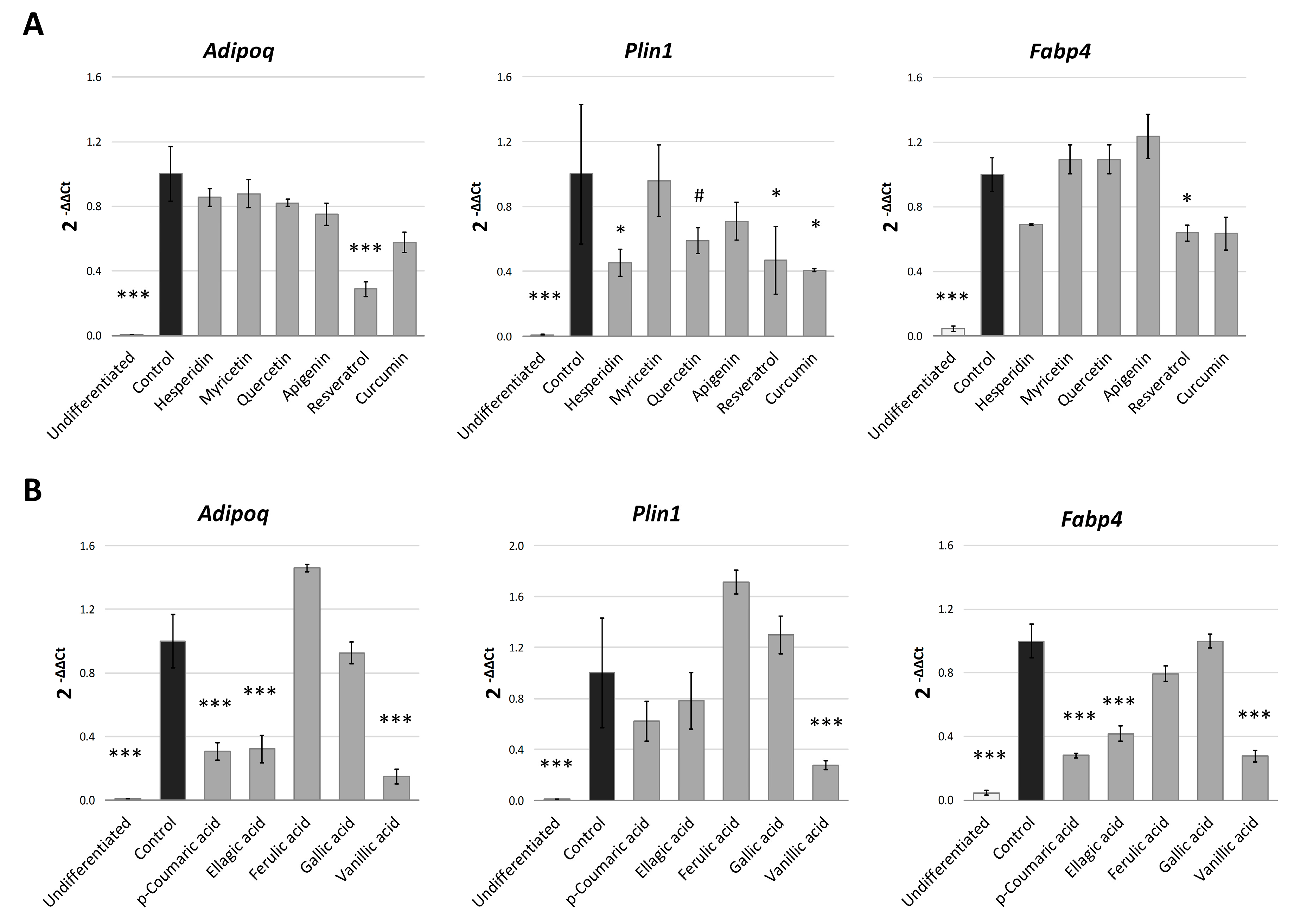
2.2. Quercetin, Resveratrol and Vanillic Acid were the Most Effective Phenolic Compounds Inhibiting Adipogenesis
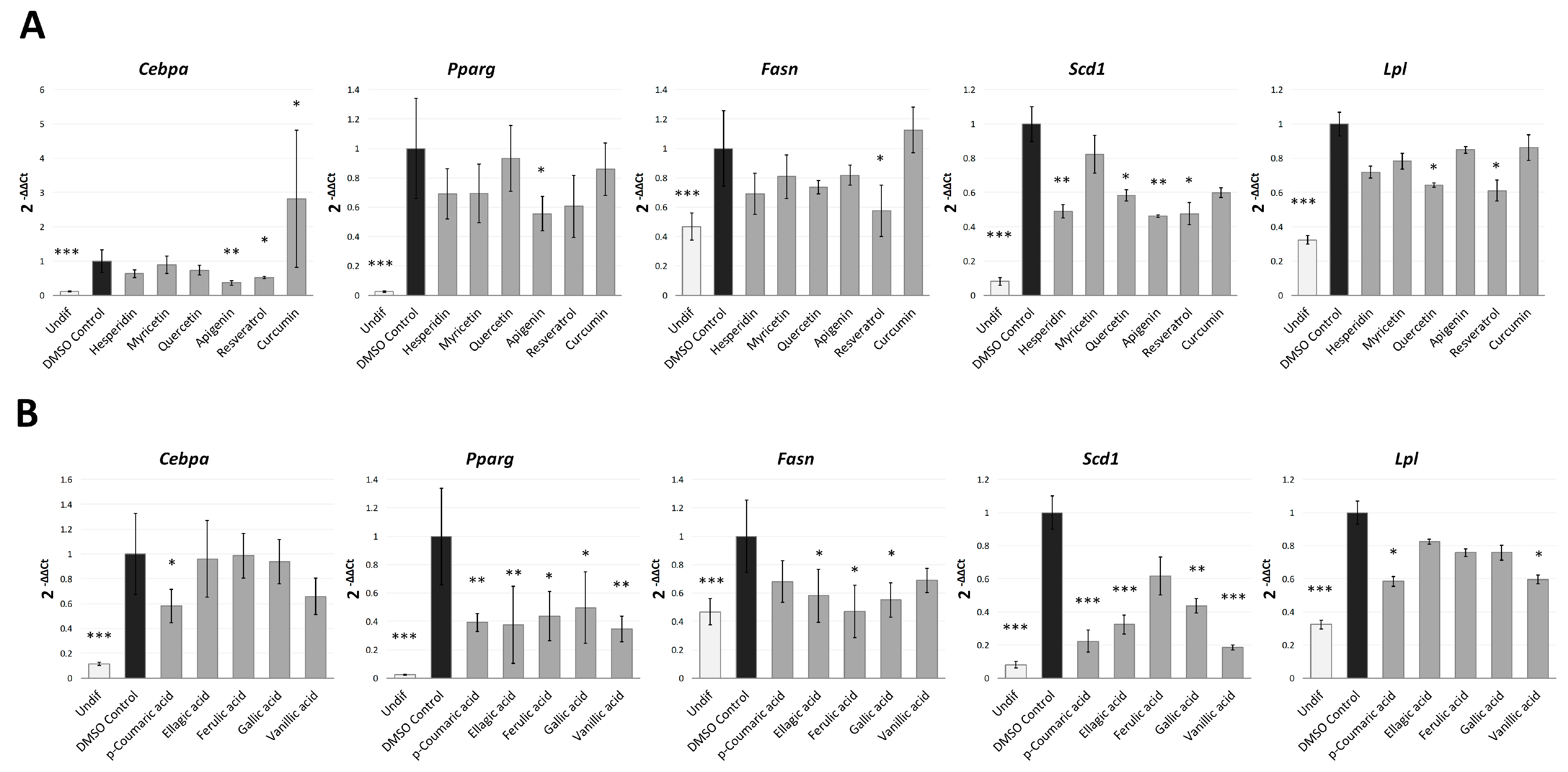
2.3. Phenolic Compounds Block the Early Stages of Adipogenesis by Regulating the Expression of Adipogenic Genes
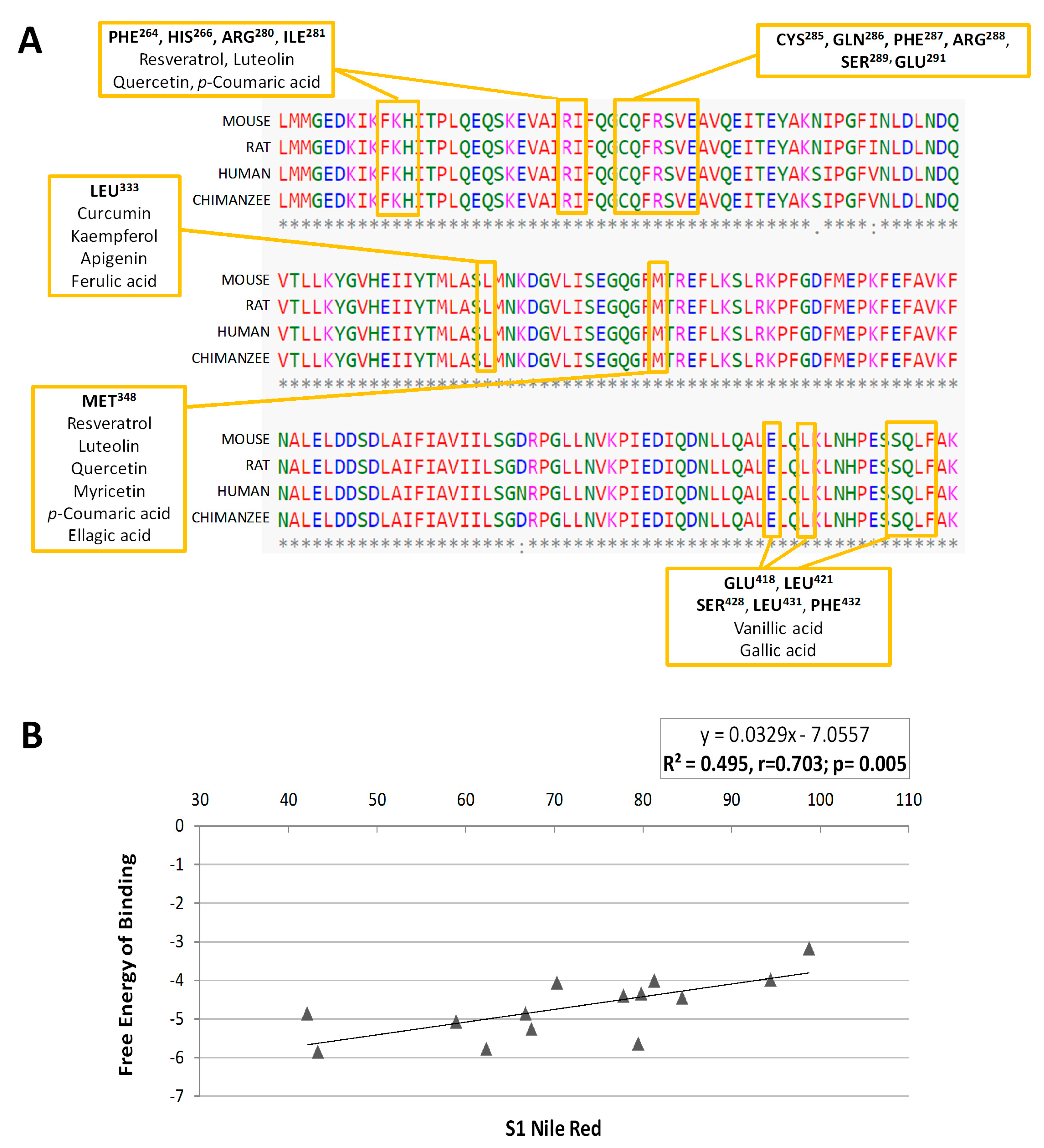
2.4. Molecular Docking Analysis Reveals that PPARγ Interaction is Crucial for the Inhibition of Adipogenesis
3. Materials and Methods
3.1. Reagents
3.2. Cell Culture
3.3. Cell Viability Assay
3.4. Effect of Phenolic Compounds on Adipogenesis
3.5. Intracellular Triglyceride Quantification
3.6. Cohen’s D Effect Size Calculation
3.7. Gene Expression Quantification (qPCR)
3.8. Molecular Docking Analysis: DockingServer
3.9. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Furuyashiki, T.; Nagayasu, H.; Aoki, Y.; Bessho, H.; Hashimoto, T.; Kanazawa, K.; Ashida, H. Tea catechin suppresses adipocyte differentiation accompanied by down-regulation of PPARgamma2 and C/EBPalpha in 3T3-L1 cells. Biosci. Biotechnol. Biochem. 2004, 68, 2353–2359. [Google Scholar] [CrossRef] [PubMed]
- Li, K.K.; Liu, C.L.; Shiu, H.T.; Wong, H.L.; Siu, W.S.; Zhang, C.; Han, X.Q.; Ye, C.X.; Leung, P.C.; Ko, C.H. Cocoa tea (Camellia ptilophylla) water extract inhibits adipocyte differentiation in mouse 3T3-L1 preadipocytes. Sci. Rep. 2016, 6, 20172. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Lee, M.; Lefevre, M.; Kim, H.R. Anthocyanins inhibit lipogenesis during adipocyte differentiation of 3T3-L1 preadipocytes. Plant Foods Hum. Nutr. 2014, 69, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Kersten, S. Mechanisms of nutritional and hormonal regulation of lipogenesis. EMBO Rep. 2001, 2, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Wafer, R.; Tandon, P.; Minchin, J.E.N. The role of peroxisome proliferator-activated receptor gamma (PPARG) in adipogenesis: Applying knowledge from the fish aquaculture industry to biomedical research. Front. Endocrinol. 2017, 8, 102. [Google Scholar] [CrossRef] [PubMed]
- Encinar, J.A.; Fernández-Ballester, G.; Galiano-Ibarra, V.; Micol, V. In silico approach for the discovery of new PPARγ modulators among plant-derived polyphenols. Drug Des. Dev. Ther. 2015. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y.; Koh, M.; Bae, H.; An, D.R.; Im, H.N.; Kim, H.S.; Yoon, J.Y.; Yoon, H.J.; Han, B.W.; Park, S.B.; et al. Structural basis for differential activities of enantiomeric PPARγ agonists: Binding of S35 to the alternate site. Biochim. Biophys. Acta Proteins Proteom. 2017. [Google Scholar] [CrossRef]
- Wang, L.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Blunder, M.; Liu, X.; Malainer, C.; Blazevic, T.; Schwaiger, S.; Rollinger, J.M.; Heiss, E.H.; et al. Natural product agonists of peroxisome proliferator-activated receptor gamma (PPARgamma): A review. Biochem. Pharmacol. 2014, 92, 73–89. [Google Scholar] [CrossRef]
- Nolte, R.T.; Wisely, G.B.; Westin, S.; Cobb, J.E.; Lambert, M.H.; Kurokawa, R.; Rosenfeld, M.G.; Willson, T.M.; Glass, C.K.; Milburn, M. V Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 1998, 395, 137–143. [Google Scholar] [CrossRef]
- Puhl, A.C.; Bernardes, A.; Silveira, R.L.; Yuan, J.; Campos, J.L.; Saidemberg, D.M.; Palma, M.S.; Cvoro, A.; Ayers, S.D.; Webb, P.; et al. Mode of peroxisome proliferator-activated receptor gamma activation by luteolin. Mol. Pharmacol. 2012. [Google Scholar] [CrossRef]
- Puhl, A.C.; Milton, F.A.; Cvoro, A.; Sieglaff, D.H.; Filgueira, C.S.; Lindemann, J.L.; Deng, T.; Neves, F.A.R.; Polikarpov, I.; Webb, P. Mechanisms of peroxisome proliferator activated receptor γ regulation by non-steroidal anti-inflammatory drugs. Nucl. Recept. Signal. 2015. [Google Scholar] [CrossRef] [PubMed]
- Colitti, M.; Stefanon, B. Different anti-adipogenic effects of bio-compounds on primary visceral pre-adipocytes and adipocytes. EXCLI J. 2016, 15, 362–377. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Liu, X.; Pan, J.; Yang, H. Synergistic algicidal effect and mechanism of two diketopiperazines produced by Chryseobacterium sp. strain GLY-1106 on the harmful bloom-forming Microcystis aeruginosa. Sci. Rep. 2015. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.L.; Yen, G.C. Induction of cell apoptosis in 3T3-L1 pre-adipocytes by flavonoids is associated with their antioxidant activity. Mol. Nutr. Food Res. 2006. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.A.; Kang, K.; Lee, H.J.; Kim, M.; Kim, C.Y.; Nho, C.W. Apigenin isolated from Daphne genkwa Siebold et Zucc. inhibits 3T3-L1 preadipocyte differentiation through a modulation of mitotic clonal expansion. Life Sci. 2014. [Google Scholar] [CrossRef] [PubMed]
- Mosqueda-Solís, A.; Lasa, A.; Gómez-Zorita, S.; Eseberri, I.; Picó, C.; Portillo, M.P. Screening of potential anti-adipogenic effects of phenolic compounds showing different chemical structure in 3T3-L1 preadipocytes. Food Funct. 2017. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.Y.; Le, T.T.; Chen, C.; Cheng, J.X.; Kim, K.H. Curcumin inhibits adipocyte differentiation through modulation of mitotic clonal expansion. J. Nutr. Biochem. 2011. [Google Scholar] [CrossRef]
- Hsu, C.L.; Yen, G.C. Effects of flavonoids and phenolic acids on the inhibition of adipogenesis in 3T3-L1 adipocytes. J. Agric. Food Chem. 2007. [Google Scholar] [CrossRef]
- Hsu, C.L.; Lo, W.H.; Yen, G.C. Gallic acid induces apoptosis in 3T3-L1 pre-adipocytes via a Fas- and mitochondrial-mediated pathway. J. Agric. Food Chem. 2007, 55, 7359–7365. [Google Scholar] [CrossRef]
- Oh, D.R.; Kim, Y.; Choi, E.J.; Lee, H.; Jung, M.A.; Bae, D.; Jo, A.; Kim, Y.R.; Kim, S. Antiobesity effects of unripe Rubus coreanus Miquel and its constituents: An in vitro and in vivo characterization of the underlying mechanism. Evid.-Based Complement. Altern. Med. 2016. [Google Scholar] [CrossRef]
- Prabhakar, P.K.; Doble, M. Effect of Natural Products on Commercial Oral Antidiabetic Drugs in Enhancing 2-Deoxyglucose Uptake by 3T3-L1 Adipocytes. Ther. Adv. Endocrinol. Metab. 2011, 2, 103–114. [Google Scholar] [CrossRef]
- Prabhakar, P.K.; Doble, M. Interaction of cinnamic acid derivatives with commercial hypoglycemic drugs on 2-deoxyglucose uptake in 3T3-L1 adipocytes. J. Agric. Food Chem. 2011. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.W.; Kang, S.I.; Shin, H.S.; Yoon, S.A.; Kim, J.H.; Ko, H.C.; Kim, S.J. Sasa quelpaertensis Nakai extract and its constituent p-coumaric acid inhibit adipogenesis in 3T3-L1 cells through activation of the AMPK pathway. Food Chem. Toxicol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Islam, M.; Shilpi, J.A.; Hasan, S. Inhibition of VEGF: A novel mechanism to control angiogenesis by Withania somnifera’s key metabolite Withaferin A. Silico Pharmacol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Lehrke, M.; Lazar, M.A. The many faces of PPARgamma. Cell 2005. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, S.; O’Driscoll, L. Metabolic syndrome: A closer look at the growing epidemic and its associated pathologies. Obes. Rev. 2015. [Google Scholar] [CrossRef] [PubMed]
- Rupasinghe, H.P.; Sekhon-Loodu, S.; Mantso, T.; Panayiotidis, M.I. Phytochemicals in regulating fatty acid beta-oxidation: Potential underlying mechanisms and their involvement in obesity and weight loss. Pharmacol. Ther. 2016, 165, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Trigueros, L.; Pena, S.; Ugidos, A.V.; Sayas-Barbera, E.; Pérez-Álvarez, J.A.; Sendra, E. Food ingredients as anti-obesity agents: A review. Crit. Rev. Food Sci. Nutr. 2013. [Google Scholar] [CrossRef]
- Rayalam, S.; Yang, J.Y.; Ambati, S.; Della-Fera, M.A.; Baile, C.A. Resveratrol induces apoptosis and inhibits adipogenesis in 3T3-L1 adipocytes. Phytother. Res. 2008, 22, 1367–1371. [Google Scholar] [CrossRef]
- Wang, P.; Mariman, E.; Keijer, J.; Bouwman, F.; Noben, J.P.; Robben, J.; Renes, J. Profiling of the secreted proteins during 3T3-L1 adipocyte differentiation leads to the identification of novel adipokines. Cell. Mol. Life Sci. 2004. [Google Scholar] [CrossRef]
- Ilavenil, S.; Arasu, M.V.; Lee, J.C.; Kim, D.H.; Roh, S.G.; Park, H.S.; Choi, G.J.; Mayakrishnan, V.; Choi, K.C. Trigonelline attenuates the adipocyte differentiation and lipid accumulation in 3T3-L1 cells. Phytomedicine 2014. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Castejon, M.; Garcia-Carrasco, B.; Fernandez-Dacosta, R.; Davalos, A.; Rodriguez-Casado, A. Reduction of adipogenesis and lipid accumulation by Taraxacum officinale (Dandelion) extracts in 3T3L1 adipocytes: An in vitro study. Phytother. Res. 2014, 28, 745–752. [Google Scholar] [CrossRef]
- Ono, M.; Fujimori, K. Antiadipogenic effect of dietary apigenin through activation of AMPK in 3T3-L1 cells. J. Agric. Food Chem. 2011, 59, 13346–13352. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.L.; Huang, S.L.; Yen, G.C. Inhibitory effect of phenolic acids on the proliferation of 3T3-L1 preadipocytes in relation to their antioxidant activity. J. Agric. Food Chem. 2006. [Google Scholar] [CrossRef] [PubMed]
- Woo, M.S.; Choi, H.S.; Seo, M.J.; Jeon, H.J.; Lee, B.Y. Ellagic acid suppresses lipid accumulation by suppressing early adipogenic events and cell cycle arrest. Phyther. Res. 2015. [Google Scholar] [CrossRef]
- Rosen, E.D. The transcriptional basis of adipocyte development. Prostaglandins. Leukot. Essent. Fatty Acids 2005, 73, 31–34. [Google Scholar] [CrossRef]
- Bouvy-Liivrand, M.; Heinäniemi, M.; John, E.; Schneider, J.G.; Sauter, T.; Sinkkonen, L. Combinatorial regulation of lipoprotein lipase by microRNAs during mouse adipogenesis. RNA Biol. 2014. [Google Scholar] [CrossRef]
- Zhang, X.H.; Huang, B.; Choi, S.K.; Seo, J.S. Anti-obesity effect of resveratrol-amplified grape skin extracts on 3T3-L1 adipocytes differentiation. Nutr. Res. Pract. 2012, 6, 286–293. [Google Scholar] [CrossRef]
- Giles, E.D.; Steig, A.J.; Jackman, M.R.; Higgins, J.A.; Johnson, G.C.; Lindstrom, R.C.; MacLean, P.S. Exercise decreases lipogenic gene expression in adipose tissue and alters adipocyte cellularity during weight regain after weight loss. Front. Physiol. 2016. [Google Scholar] [CrossRef]
- Cohen, P.; Friedman, J.M. Leptin and the control of metabolism: Role for stearoyl-CoA desaturase-1 (SCD-1). J. Nutr. 2004. [Google Scholar] [CrossRef]
- Xia, L.; Fan, C.; Dong, H.; Wang, C.; Lu, Y.; Deckelbaum, R.J.; Qi, K. Particle size determines effects of lipoprotein lipase on the catabolism of n-3 triglyceride-rich particles. Clin. Nutr. 2015. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-C.; Lin, K.-Y.; Peng, K.-Y.; Day, Y.-J.; Hung, L.-M. Resveratrol exerts anti-obesity effects in high-fat diet obese mice and displays differential dosage effects on cytotoxicity, differentiation, and lipolysis in 3T3-L1 cells. Endocr. J. 2016. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Della-Fera, M.A.; Rayalam, S.; Ambati, S.; Hartzell, D.L.; Park, H.J.; Baile, C.A. Enhanced inhibition of adipogenesis and induction of apoptosis in 3T3-L1 adipocytes with combinations of resveratrol and quercetin. Life Sci. 2008, 82, 1032–1039. [Google Scholar] [CrossRef] [PubMed]
- Beales, P.E.; Liddi, R.; Giorgini, A.E.; Signore, A.; Procaccini, E.; Batchelor, K.; Pozzilli, P. Troglitazone prevents insulin dependent diabetes in the non-obese diabetic mouse. Eur. J. Pharmacol. 1998, 357, 221–225. [Google Scholar] [CrossRef]
- Hazai, E.; Kovács, S.; Demkó, L.; Bikádi, Z. DockingServer: Molecular docking calculations online. Acta Pharm. Hung. 2009, 79, 17–21. [Google Scholar]
- Bikadi, Z.; Hazai, E. Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J. Cheminform. 2009. [Google Scholar] [CrossRef] [PubMed]
- Djanic, M.; Pavlovic, N.; Stanimirov, B.; Stojancevic, T.; Golocorbin-Kon, S.; Bojic, G.; Mikov, M. Docking-based preliminary study on the interactions of bile acids with drugs at the transporter level in intestinal bacteria. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 553–560. [Google Scholar] [PubMed]
- Jacob, R.B.; Andersen, T.; McDougal, O.M. Accessible high-throughput virtual screening molecular docking software for students and educators. PLoS Comput. Biol. 2012. [Google Scholar] [CrossRef]
- Park, H.S.; Kim, S.H.; Kim, Y.S.; Ryu, S.Y.; Hwang, J.T.; Yang, H.J.; Kim, G.H.; Kwon, D.Y.; Kim, M.S. Luteolin inhibits adipogenic differentiation by regulating PPARc activation. BioFactors 2009. [Google Scholar] [CrossRef]
- Calleri, E.; Pochetti, G.; Dossou, K.S.S.; Laghezza, A.; Montanari, R.; Capelli, D.; Prada, E.; Loiodice, F.; Massolini, G.; Bernier, M.; et al. Resveratrol and its metabolites bind to PPARs. ChemBioChem 2014. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.W.; Ntambi, J.M. Peroxisome proliferators induce mouse liver stearoyl-CoA desaturase 1 gene expression. Proc. Natl. Acad. Sci. USA 2002. [Google Scholar] [CrossRef]
- Bogacka, I.; Xie, H.; Bray, G.A.; Smith, S.R. The effect of pioglitazone on peroxisome proliferator-activated receptor-γ target genes related to lipid storage in vivo. Diabetes Care 2004. [Google Scholar] [CrossRef]
- Jeong, Y.S.; Hong, J.H.; Cho, K.H.; Jung, H.K. Grape skin extract reduces adipogenesis- and lipogenesis-related gene expression in 3T3-L1 adipocytes through the peroxisome proliferator-activated receptor-γ signaling pathway. Nutr. Res. 2012, 32, 514–521. [Google Scholar] [CrossRef]
- Gregoire, F.M.; Smas, C.M.; Sul, H.S. Understanding Adipocyte Differentiation. Physiol. Rev. 2017. [Google Scholar] [CrossRef]
- Arimura, N.; Horiba, T.; Imagawa, M.; Shimizu, M.; Sato, R. The Peroxisome Proliferator-activated Receptor γ Regulates Expression of the Perilipin Gene in Adipocytes. J. Biol. Chem. 2004. [Google Scholar] [CrossRef]
- Fu, Y.; Luo, N.; Klein, R.L.; Garvey, W.T. Adiponectin promotes adipocyte differentiation, insulin sensitivity, and lipid accumulation. J. Lipid Res. 2005. [Google Scholar] [CrossRef]
- Park, E.; Kim, J.; Yeo, S.; Kim, G.; Ko, E.H.; Lee, S.W.; Li, W.Y.; Choi, C.W.; Jeong, S.Y. Antiadipogenic effects of loganic acid in 3T3-L1 preadipocytes and ovariectomized mice. Molecules 2018. [Google Scholar] [CrossRef]
- Amin, R.H.; Mathews, S.T.; Camp, H.S.; Ding, L.; Leff, T. Selective activation of PPARgamma in skeletal muscle induces endogenous production of adiponectin and protects mice from diet-induced insulin resistance. Am. J. Physiol. Endocrinol. Metab. 2010. [Google Scholar] [CrossRef]
- Pelton, P.D.; Zhou, L.; Demarest, K.T.; Burris, T.P. PPARγ activation induces the expression of the adipocyte fatty acid binding protein gene in human monocytes. Biochem. Biophys. Res. Commun. 1999. [Google Scholar] [CrossRef]
- Boß, M.; Kemmerer, M.; Brüne, B.; Namgaladze, D. FABP4 inhibition suppresses PPARγ activity and VLDL-induced foam cell formation in IL-4-polarized human macrophages. Atherosclerosis 2015. [Google Scholar] [CrossRef]
- Aranaz, P.; Romo-Hualde, A.; Zabala, M.; Navarro-Herrera, D.; Ruiz De Galarreta, M.; Gil, A.G.; Martinez, J.A.; Milagro, F.I.; González-Navarro, C.J. Freeze-dried strawberry and blueberry attenuates diet-induced obesity and insulin resistance in rats by inhibiting adipogenesis and lipogenesis. Food Funct. 2017. [Google Scholar] [CrossRef]
- Pereira-Fernandes, A.; Demaegdt, H.; Vandermeiren, K.; Hectors, T.L.; Jorens, P.G.; Blust, R.; Vanparys, C. Evaluation of a screening system for obesogenic compounds: Screening of endocrine disrupting compounds and evaluation of the PPAR dependency of the effect. PLoS ONE 2013, 8, e77481. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Della-Fera, M.A.; Hartzell, D.L.; Nelson-Dooley, C.; Hausman, D.B.; Baile, C.A. Esculetin induces apoptosis and inhibits adipogenesis in 3T3-L1 cells. Obesity (Silver Spring) 2006, 14, 1691–1699. [Google Scholar] [CrossRef]
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences; Lawrence Erlbaum Associates: Mahwah, NJ, USA, 1988. [Google Scholar]
- Nakagawa, S.; Cuthill, I.C. Effect size, confidence interval and statistical significance: A practical guide for biologists. Biol. Rev. 2007, 82, 591–605. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, J.; Saller, H. Calculation of the Charge Distribution in Conjugated Systems by a Quantification of the Resonance Concept. Angew. Chem. Int. Ed. Engl. 1985. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, J.; Suzek, T.O.; Zhang, J.; Wang, J.; Bryant, S.H. PubChem: A public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009, 37 (Suppl. 2), W623–W633. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Goodsell, D.S.; Morris, G.M.; Olson, A.J. Automated docking of flexible ligands: Applications of AutoDock. J. Mol. Recognit. 1996, 9, 1–5. [Google Scholar] [CrossRef]
Sample Availability: Not available. |
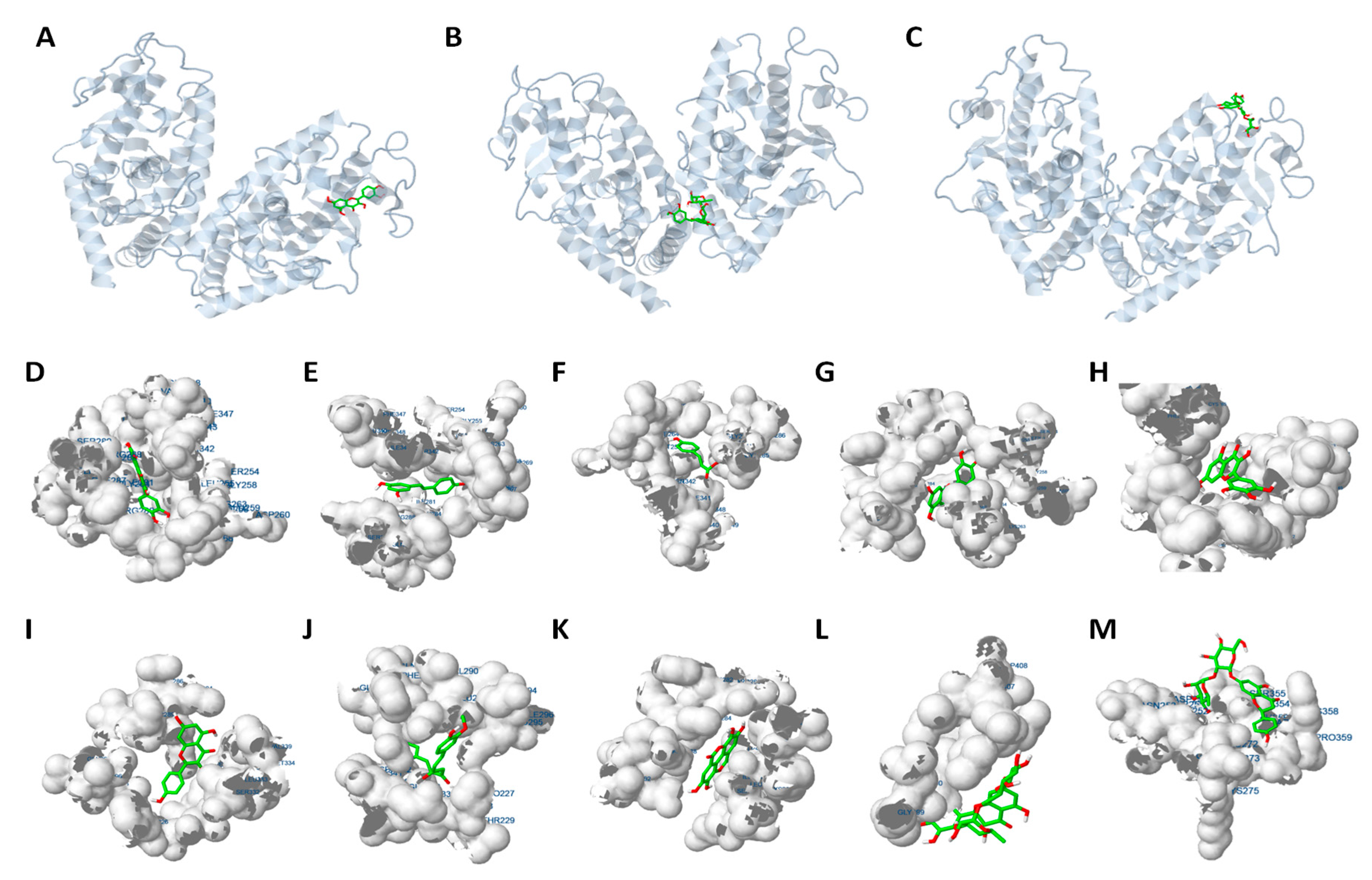
| Estimated Free Energy of Binding (kcal/mol) | Estimated Inhibition Constant, Ki (uM) | vdW + Hbond + desolv Energy (kcal/mol) | Total Intermolec. Energy (kcal/mol) | Interaction Surface | |
|---|---|---|---|---|---|
| Troglitazone | −7.23 | 5.00 | −8.83 | −8.93 | 1021.969 |
| Hesperetin | −5.95 | 43.34 | −6.38 | −6.42 | 942.649 |
| Luteolin | −5.84 | 52.07 | −6.07 | −6.32 | 667.019 |
| Apigenin | −5.78 | 57.62 | −6.30 | −6.32 | 774.163 |
| Naringenin | −5.77 | 58.80 | −6.27 | −6.30 | 897.003 |
| Ellagic acid | −5.64 | 73.75 | −5.59 | −5.70 | 871.940 |
| Kaempferol | −5.27 | 137.56 | −5.67 | −5.89 | 902.514 |
| Quercetin | −5.07 | 193.00 | −5.64 | −5.85 | 672.588 |
| Resveratrol | −4.86 | 273.59 | −6.04 | −6.36 | 614.078 |
| Curcumin | −4.86 | 272.69 | −6.70 | −6.95 | 1052.665 |
| p-Coumaric acid | −4.44 | 558.07 | −5.39 | −5.64 | 451.466 |
| Vanillic acid | −4.40 | 591.18 | −4.99 | −5.24 | 427.083 |
| Myricetin | −4.34 | 654.56 | −4.84 | −5.01 | 724.325 |
| Gallic acid | −4.05 | 1080 | −4.48 | −4.76 | 407.548 |
| Ferulic acid | −4.00 | 1180 | −5.04 | −5.11 | 567.571 |
| Naringin | −3.98 | 1200 | −5.84 | −5.85 | 657.646 |
| Hesperidin | −3.16 | 4810 | −4.94 | −5.07 | 561.726 |
| Troglitazone | Quercetin | Resveratrol | p-coumaric acid | Luteolin | Myricetin | Kaempferol | Curcumin | Ellagic acid | Apigenin | Ferulic acid | Gallic acid | Vanillic acid | Hesperetin * | Naringenin * | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leu228 | ● | ● | |||||||||||||
| Leu255 | ● | ● | ● | ● | |||||||||||
| Glu259 | ● | ● | ● | ● | |||||||||||
| Lys263 | ● | ||||||||||||||
| Phe264 | ● | ● | ● | ● | ● | ● | ● | ||||||||
| His266 | ● | ● | ● | ● | ● | ● | |||||||||
| Arg280 | ● | ● | ● | ● | ● | ||||||||||
| Ile281 | ● | ● | ● | ● | ● | ● | |||||||||
| Cys285 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ||||||
| Arg288 | ● | ● | ● | ● | ● | ● | |||||||||
| Ser289 | ● | ● | |||||||||||||
| Glu291 | ● | ● | ● | ● | ● | ● | |||||||||
| Glu295 | ● | ● | |||||||||||||
| Ile326 | ● | ● | |||||||||||||
| Tyr327 | ● | ● | |||||||||||||
| Leu330 | ● | ● | ● | ||||||||||||
| Leu333 | ● | ● | ● | ● | ● | ● | ● | ||||||||
| Val339 | ● | ● | |||||||||||||
| Ile341 | ● | ● | ● | ● | ● | ● | |||||||||
| Met348 | ● | ● | ● | ● | ● | ● | |||||||||
| Phe363 | ● | ● | |||||||||||||
| Glu418 | ● | ● | |||||||||||||
| Leu421 | ● | ● | |||||||||||||
| Ser428 | ● | ● | |||||||||||||
| Leu431 | ● | ● | |||||||||||||
| Phe432 | ● | ● | |||||||||||||
| His449 | ● | ● |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aranaz, P.; Navarro-Herrera, D.; Zabala, M.; Miguéliz, I.; Romo-Hualde, A.; López-Yoldi, M.; Martínez, J.A.; Vizmanos, J.L.; Milagro, F.I.; González-Navarro, C.J. Phenolic Compounds Inhibit 3T3-L1 Adipogenesis Depending on the Stage of Differentiation and Their Binding Affinity to PPARγ. Molecules 2019, 24, 1045. https://doi.org/10.3390/molecules24061045
Aranaz P, Navarro-Herrera D, Zabala M, Miguéliz I, Romo-Hualde A, López-Yoldi M, Martínez JA, Vizmanos JL, Milagro FI, González-Navarro CJ. Phenolic Compounds Inhibit 3T3-L1 Adipogenesis Depending on the Stage of Differentiation and Their Binding Affinity to PPARγ. Molecules. 2019; 24(6):1045. https://doi.org/10.3390/molecules24061045
Chicago/Turabian StyleAranaz, Paula, David Navarro-Herrera, María Zabala, Itziar Miguéliz, Ana Romo-Hualde, Miguel López-Yoldi, J. Alfredo Martínez, José Luis Vizmanos, Fermín I. Milagro, and Carlos Javier González-Navarro. 2019. "Phenolic Compounds Inhibit 3T3-L1 Adipogenesis Depending on the Stage of Differentiation and Their Binding Affinity to PPARγ" Molecules 24, no. 6: 1045. https://doi.org/10.3390/molecules24061045
APA StyleAranaz, P., Navarro-Herrera, D., Zabala, M., Miguéliz, I., Romo-Hualde, A., López-Yoldi, M., Martínez, J. A., Vizmanos, J. L., Milagro, F. I., & González-Navarro, C. J. (2019). Phenolic Compounds Inhibit 3T3-L1 Adipogenesis Depending on the Stage of Differentiation and Their Binding Affinity to PPARγ. Molecules, 24(6), 1045. https://doi.org/10.3390/molecules24061045






