Single-Molecule Imaging and Super-Resolution Microscopy of Lipid Domains in Cell Membranes Using Lipid-Binding Proteins and Fluorophore-Conjugated Lipid Analogs
Abstract
1. Introduction
2. Observation of Lipid Probes Conjugated with Organic Dyes in the Outer Leaflet of the PM in Living Cells
2.1. Development of Lipid Probes Conjugated with Organic Dyes
2.2. Single-Molecule Tracking of Phospholipid Diffusion at High Temporal Resolution
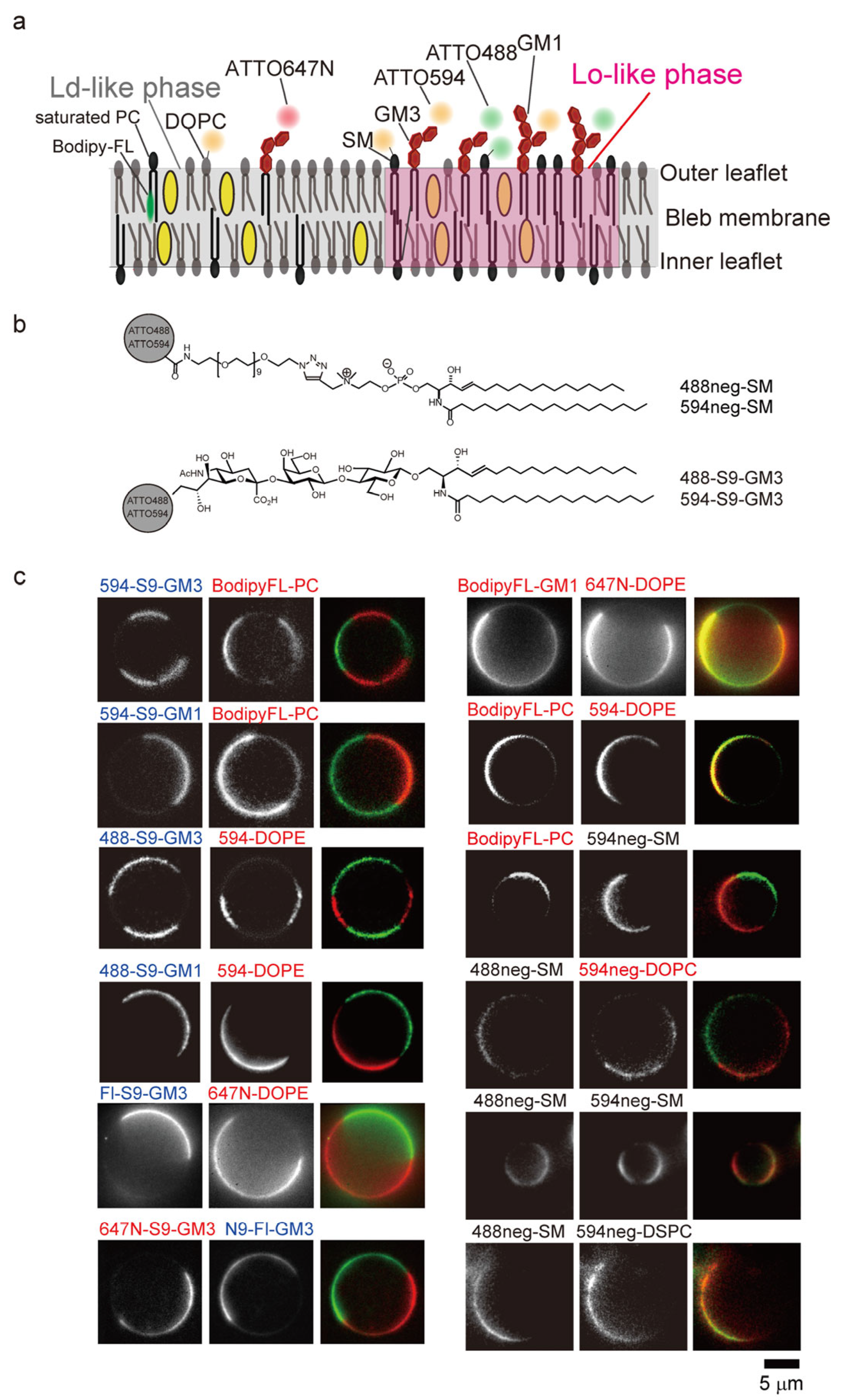
2.3. Development of Raftophilic Lipid Probes Tagged with Organic Fluorophores
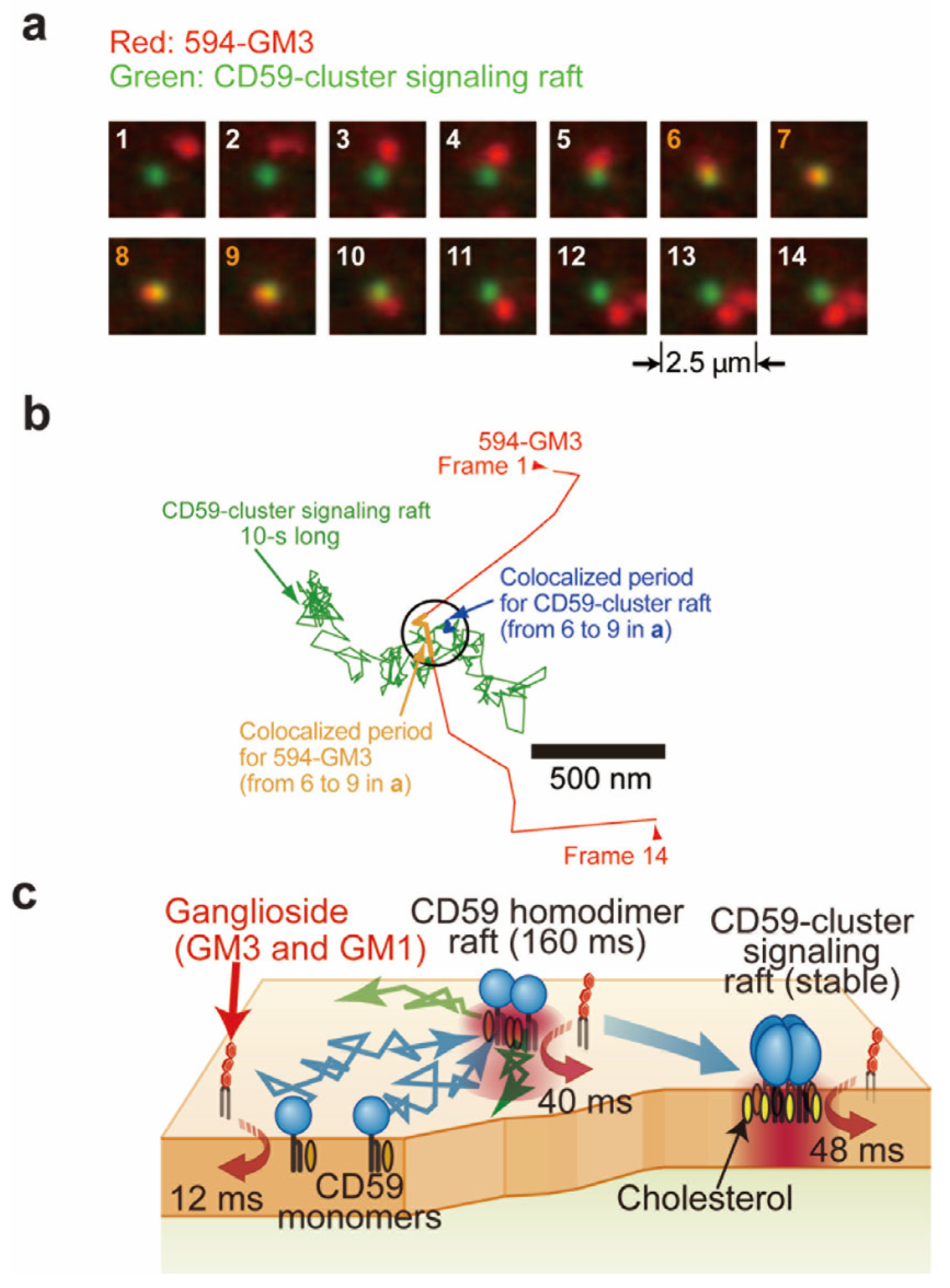
2.4. Microscopic Observation in the Outer Leaflet of the PM
3. Observation of Lipids in the Inner Leaflet of Cell PMs Using Lipid-Binding Proteins
3.1. Development of Phosphoinositide Probes Using Lipid-Binding Proteins
3.2. Development of Phosphatidylserine Probes in Cell Membranes
3.3. Development of SM-Binding Protein Probes in Cell Membranes
3.4. Development of Cholesterol-Binding Protein Probes in Cell Membranes
4. Advanced Microscopic Observation of Lipid-Binding Protein Probes in Cells
4.1. Observation in Fixed Cells
4.2. Observation in Living Cells
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lingwood, D.; Simons, K. Lipid rafts as a membrane-organizing principle. Science 2010, 327, 46–50. [Google Scholar] [CrossRef]
- Sezgin, E.; Levental, I.; Mayor, S.; Eggeling, C. The mystery of membrane organization: Composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Varshney, P.; Yadav, V.; Saini, N. Lipid rafts in immune signalling: Current progress and future perspective. Immunology 2016, 149, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Dinic, J.; Riehl, A.; Adler, J.; Parmryd, I. The T cell receptor resides in ordered plasma membrane nanodomains that aggregate upon patching of the receptor. Sci. Rep. 2015, 5, 10082. [Google Scholar] [CrossRef] [PubMed]
- Grassi, S.; Giussani, P.; Mauri, L.; Prioni, S.; Sonnino, S.; Prinetti, A. Lipid rafts and neurodegeneration: Structural and functional roles in physiologic aging and neurodegenerative diseases: Thematic Review Series: Biology of Lipid Rafts. J. Lipid Res. 2020, 61, 636–654. [Google Scholar] [CrossRef]
- Hulsmeier, A.J. Glycosphingolipids in neurodegeneration—Molecular mechanisms, cellular roles, and therapeutic perspectives. Neurobiol. Dis. 2025, 207, 106851. [Google Scholar] [CrossRef]
- Greenlee, J.D.; Subramanian, T.; Liu, K.; King, M.R. Rafting down the metastatic cascade: The role of lipid rafts in cancer metastasis, cell death, and clinical outcomes. Cancer Res. 2021, 81, 5–17. [Google Scholar] [CrossRef]
- Roy, A.; Patra, S.K. Lipid raft facilitated receptor organization and signaling: A functional rheostat in embryonic development, stem cell biology and cancer. Stem Cell Rev. Rep. 2023, 19, 2–25. [Google Scholar] [CrossRef]
- Kulkarni, R.; Wiemer, E.A.C.; Chang, W. Role of lipid rafts in pathogen-host interaction—A Mini Review. Front. Immunol. 2021, 12, 815020. [Google Scholar] [CrossRef]
- Fantini, J.; Yahi, N.; Azzaz, F.; Chahinian, H. Structural dynamics of SARS-CoV-2 variants: A health monitoring strategy for anticipating COVID-19 outbreaks. J. Infect. 2021, 83, 197–206. [Google Scholar] [CrossRef]
- Kusumi, A.; Suzuki, K. Toward understanding the dynamics of membrane-raft-based molecular interactions. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2005, 1746, 234–251. [Google Scholar] [CrossRef]
- Tanaka, K.A.; Suzuki, K.G.; Shirai, Y.M.; Shibutani, S.T.; Miyahara, M.S.; Tsuboi, H.; Yahara, M.; Yoshimura, A.; Mayor, S.; Fujiwara, T.K.; et al. Membrane molecules mobile even after chemical fixation. Nat. Methods 2010, 7, 865–866. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, T.; Wang, D.; Miyazawa, K.; Miyata, K.; Oshima, M.; Fukuma, T. Chemical fixation creates nanoscale clusters on the cell surface by aggregating membrane proteins. Commun. Biol. 2022, 5, 487. [Google Scholar] [CrossRef] [PubMed]
- Irgen-Gioro, S.; Yoshida, S.; Walling, V.; Chong, S. Fixation can change the appearance of phase separation in living cells. Elife 2022, 11, e79903. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, R.; Okada, T.; Ogura, T.; Ogura, T.; Baglioni, P. Direct observation of the effects of chemical fixation in MNT-1 cells: A SE-ADM and Raman study. Proc. Natl. Acad. Sci. USA 2023, 120, e2308088120. [Google Scholar] [CrossRef]
- Bhojoo, U.; Chen, M.; Zou, S. Temperature induced lipid membrane restructuring and changes in nanomechanics. Biochim. Biophys. Acta (BBA)—Biomembr. 2018, 1860, 700–709. [Google Scholar] [CrossRef]
- Ollila, O.H.S.; Risselada, H.J.; Louhivuori, M.; Lindahl, E.; Vattulainen, I.; Marrink, S.J. 3D Pressure Field in Lipid Membranes and Membrane-Protein Complexes. Phys. Rev. Lett. 2009, 102, 078101. [Google Scholar] [CrossRef]
- Ghosh, S.; Wagenknecht-Wiesner, A.; Desai, S.; Vyphuis, J.; Ramos, M.S.; Grazul, J.L.; Baird, B.A. Synergy between membrane topography and domains to control signaling protein localization in mast cells facilitates their activation. Proc. Natl. Acad. Sci. USA 2025, 122, e2424427122. [Google Scholar] [CrossRef]
- Kot, E.F.; Goncharuk, S.A.; Franco, M.L.; McKenzie, D.M.; Arseniev, A.S.; Benito-Martínez, A.; Costa, M.; Cattaneo, A.; Hristova, K.; Vilar, M.; et al. Structural basis for the transmembrane signaling and antidepressant-induced activation of the receptor tyrosine kinase TrkB. Nat. Commun. 2024, 15, 9316. [Google Scholar] [CrossRef]
- Anishkin, A.; Loukin, S.H.; Teng, J.; Kung, C. Feeling the hidden mechanical forces in lipid bilayer is an original sense. Proc. Natl. Acad. Sci. USA 2014, 111, 7898–7905. [Google Scholar] [CrossRef]
- Suzuki, K.G.N.; Kusumi, A. Refinement of Singer-Nicolson fluid-mosaic model by microscopy imaging: Lipid rafts and actin-induced membrane compartmentalization. Biochim. Biophys. Acta (BBA)—Biomembr. 2023, 1865, 184093. [Google Scholar] [CrossRef] [PubMed]
- Aghaaminiha, M.; Farnoud, A.M.; Sharma, S. Quantitative relationship between cholesterol distribution and ordering of lipids in asymmetric lipid bilayers. Soft Matter 2021, 17, 2742–2752. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.K.; Takeuchi, S.; Kalay, Z.; Nagai, Y.; Tsunoyama, T.A.; Kalkbrenner, T.; Iwasawa, K.; Ritchie, K.P.; Suzuki, K.G.N.; Kusumi, A. Development of ultrafast camera-based single fluorescent-molecule imaging for cell biology. J. Cell Biol. 2023, 222, e202110160. [Google Scholar] [CrossRef] [PubMed]
- Hirosawa, K.M.; Sato, Y.; Kasai, R.S.; Yamaguchi, E.; Komura, N.; Ando, H.; Hoshino, A.; Yokota, Y.; Suzuki, K.G.N. Uptake of small extracellular vesicles by recipient cells is facilitated by paracrine adhesion signaling. Nat. Commun. 2025, 16, 2419. [Google Scholar] [CrossRef]
- Isogai, T.; Hirosawa, K.M.; Kanno, M.; Sho, A.; Kasai, R.S.; Komura, N.; Ando, H.; Furukawa, K.; Ohmi, Y.; Furukawa, K.; et al. Extracellular vesicles adhere to cells primarily by interactions of integrins and GM1 with laminin. J. Cell Biol. 2025, 224, e29249464. [Google Scholar] [CrossRef]
- Kinoshita, M.; Suzuki, K.G.; Matsumori, N.; Takada, M.; Ano, H.; Morigaki, K.; Abe, M.; Makino, A.; Kobayashi, T.; Hirosawa, K.M.; et al. Raft-based sphingomyelin interactions revealed by new fluorescent sphingomyelin analogs. J. Cell Biol. 2017, 216, 1183–1204. [Google Scholar] [CrossRef]
- Komura, N.; Suzuki, K.G.; Ando, H.; Konishi, M.; Koikeda, M.; Imamura, A.; Chadda, R.; Fujiwara, T.K.; Tsuboi, H.; Sheng, R.; et al. Raft-based interactions of gangliosides with a GPI-anchored receptor. Nat. Chem. Biol. 2016, 12, 402–410. [Google Scholar] [CrossRef]
- Asfia, S.; Seemann, R.; Fleury, J.-B. Phospholipids diffusion on the surface of model lipid droplets. Biochim. Biophys. Acta (BBA)—Biomembr. 2023, 1865, 184074. [Google Scholar] [CrossRef]
- Ruan, H.; Zou, C.; Xu, Y.; Fang, X.; Xia, T.; Shi, Y. N-(3-Oxododecanoyl) homoserine lactone is a generalizable plasma membrane lipid-ordered domain modifier. Front. Physiol. 2021, 12, 758458. [Google Scholar] [CrossRef]
- Eggeling, C.; Ringemann, C.; Medda, R.; Schwarzmann, G.; Sandhoff, K.; Polyakova, S.; Belov, V.N.; Hein, B.; von Middendorff, C.; Schonle, A.; et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 2009, 457, 1159–1162. [Google Scholar] [CrossRef]
- Honigmann, A.; Mueller, V.; Hell, S.W.; Eggeling, C. STED microscopy detects and quantifies liquid phase separation in lipid membranes using a new far-red emitting fluorescent phosphoglycerolipid analogue. Faraday Discuss. 2013, 161, 77–89; discussion 113–150. [Google Scholar] [CrossRef]
- Carten, J.D.; Bradford, M.K.; Farber, S.A. Visualizing digestive organ morphology and function using differential fatty acid metabolism in live zebrafish. Dev. Biol. 2011, 360, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Beuchat, M.H.; Chevallier, J.; Makino, A.; Mayran, N.; Escola, J.M.; Lebrand, C.; Cosson, P.; Kobayashi, T.; Gruenberg, J. Separation and characterization of late endosomal membrane domains. J. Biol. Chem. 2002, 277, 32157–32164. [Google Scholar] [CrossRef]
- Sezgin, E.; Levental, I.; Grzybek, M.; Schwarzmann, G.; Mueller, V.; Honigmann, A.; Belov, V.N.; Eggeling, C.; Coskun, U.; Simons, K.; et al. Partitioning, diffusion, and ligand binding of raft lipid analogs in model and cellular plasma membranes. Biochim. Biophys. Acta (BBA)—Biomembr. 2012, 1818, 1777–1784. [Google Scholar] [CrossRef]
- Suzuki, K.G.N.; Komura, N.; Ando, H. Recently developed glycosphingolipid probes and their dynamic behavior in cell plasma membranes as revealed by single-molecule imaging. Glycoconj. J. 2023, 40, 305–314. [Google Scholar] [CrossRef]
- Chinnapen, D.J.; Hsieh, W.T.; Te Welscher, Y.M.; Saslowsky, D.E.; Kaoutzani, L.; Brandsma, E.; D’Auria, L.; Park, H.; Wagner, J.S.; Drake, K.R.; et al. Lipid sorting by ceramide structure from plasma membrane to ER for the cholera toxin receptor ganglioside GM1. Dev. Cell 2012, 23, 573–586. [Google Scholar] [CrossRef]
- Arumugam, S.; Schmieder, S.; Pezeshkian, W.; Becken, U.; Wunder, C.; Chinnapen, D.; Ipsen, J.H.; Kenworthy, A.K.; Lencer, W.; Mayor, S.; et al. Ceramide structure dictates glycosphingolipid nanodomain assembly and function. Nat. Commun. 2021, 12, 3675. [Google Scholar] [CrossRef]
- Asano, S.; Pal, R.; Tanaka, H.; Imamura, A.; Ishida, H.; Suzuki, K.G.; Ando, H. Development of fluorescently labeled SSEA-3, SSEA-4, and Globo-H glycosphingolipids for elucidating molecular interactions in the cell membrane. Int. J. Mol. Sci. 2019, 20, 6187. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Komura, N.; Yoshida, Y.; Yamaguchi, E.; Hasegawa, A.; Tanaka, H.; Imamura, A.; Ishida, H.; Suzuki, K.G.; Ando, H. Development of lacto-series ganglioside fluorescent probe using late-stage sialylation and behavior analysis with single-molecule imaging. RSC Chem. Biol. 2022, 3, 868–885. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, E.; Komura, N.; Tanaka, H.; Imamura, A.; Ishida, H.; Groux-Degroote, S.; Muhlenhoff, M.; Suzuki, K.G.; Ando, H. Fluorescent GD2 analog for single-molecule imaging. Glycoconj. J. 2023, 40, 247–257. [Google Scholar] [CrossRef]
- Konishi, M.; Komura, N.; Hirose, Y.; Suganuma, Y.; Tanaka, H.; Imamura, A.; Ishida, H.; Suzuki, K.G.; Ando, H. Development of fluorescent ganglioside GD3 and GQ1b analogs for elucidation of raft-associated interactions. J. Org. Chem. 2020, 85, 15998–16013. [Google Scholar] [CrossRef]
- Shaw, J.E.; Epand, R.F.; Epand, R.M.; Li, Z.; Bittman, R.; Yip, C.M. Correlated fluorescence-atomic force microscopy of membrane domains: Structure of fluorescence probes determines lipid localization. Biophys. J. 2006, 90, 2170–2178. [Google Scholar] [CrossRef]
- Ariola, F.S.; Li, Z.; Cornejo, C.; Bittman, R.; Heikal, A.A. Membrane fluidity and lipid order in ternary giant unilamellar vesicles using a new bodipy-cholesterol derivative. Biophys. J. 2009, 96, 2696–2708. [Google Scholar] [CrossRef]
- Hiramoto-Yamaki, N.; Tanaka, K.A.; Suzuki, K.G.; Hirosawa, K.M.; Miyahara, M.S.; Kalay, Z.; Tanaka, K.; Kasai, R.S.; Kusumi, A.; Fujiwara, T.K. Ultrafast diffusion of a fluorescent cholesterol analog in compartmentalized plasma membranes. Traffic 2014, 15, 583–612. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Zha, X.; Tabas, I.; Maxfield, F.R. Cholesterol distribution in living cells: Fluorescence imaging using dehydroergosterol as a fluorescent cholesterol analog. Biophys. J. 1998, 75, 1915–1925. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, M.; Halder, S.; Reinholdt, P.; Bashawat, M.; Scheidt, H.A.; Leopold, J.; Schiller, J.; Prima, D.d.; Akkerman, V.; Szomek, M.; et al. Synthesis and Characterization of a novel intrinsically fluorescent analog of cholesterol with improved photophysical properties. Anal. Chem. 2024, 96, 18596–18604. [Google Scholar] [CrossRef]
- Ritchie, K.; Shan, X.Y.; Kondo, J.; Iwasawa, K.; Fujiwara, T.; Kusumi, A. Detection of non-Brownian diffusion in the cell membrane in single molecule tracking. Biophys. J. 2005, 88, 2266–2277. [Google Scholar] [CrossRef]
- Ploetz, E.; Ambrose, B.; Barth, A.; Börner, R.; Erichson, F.; Kapanidis, A.N.; Kim, H.D.; Levitus, M.; Lohman, T.M.; Mazumder, A.; et al. A new twist on PIFE: Photoisomerisation-related fluorescence enhancement. Methods Appl. Fluoresc. 2023, 12, 012001. [Google Scholar] [CrossRef]
- Pagano, R.E.; Martin, O.C.; Kang, H.C.; Haugland, R.P. A novel fluorescent ceramide analogue for studying membrane traffic in animal cells: Accumulation at the Golgi apparatus results in altered spectral properties of the sphingolipid precursor. J. Cell Biol. 1991, 113, 1267–1279. [Google Scholar] [CrossRef]
- Yoon, S.J.; Nakayama, K.; Hikita, T.; Handa, K.; Hakomori, S.I. Epidermal growth factor receptor tyrosine kinase is modulated by GM3 interaction with N-linked GlcNAc termini of the receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 18987–18991. [Google Scholar] [CrossRef]
- Patry, R.T.; Stahl, M.; Perez-Munoz, M.E.; Nothaft, H.; Wenzel, C.Q.; Sacher, J.C.; Coros, C.; Walter, J.; Vallance, B.A.; Szymanski, C.M. Bacterial AB(5) toxins inhibit the growth of gut bacteria by targeting ganglioside-like glycoconjugates. Nat. Commun. 2019, 10, 1390. [Google Scholar] [CrossRef]
- Yu, R.K.; Usuki, S.; Ariga, T. Ganglioside molecular mimicry and its pathological roles in Guillain-Barré syndrome and related diseases. Infect. Immun. 2006, 74, 6517–6527. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, K.; Odaka, A.; Suzuki, N.; Ihara, Y. GM1 ganglioside–bound amyloid β–protein (Aβ): A possible form of preamyloid in Alzheimer’s disease. Nat. Med. 1995, 1, 1062–1066. [Google Scholar] [CrossRef] [PubMed]
- Kabbani, A.M.; Raghunathan, K.; Lencer, W.I.; Kenworthy, A.K.; Kelly, C.V. Structured clustering of the glycosphingolipid GM1 is required for membrane curvature induced by cholera toxin. Proc. Natl. Acad. Sci. USA 2020, 117, 14978–14986. [Google Scholar] [CrossRef] [PubMed]
- Sahl, S.J.; Leutenegger, M.; Hilbert, M.; Hell, S.W.; Eggeling, C. Fast molecular tracking maps nanoscale dynamics of plasma membrane lipids. Proc. Natl. Acad. Sci. USA 2010, 107, 6829–6834. [Google Scholar] [CrossRef]
- Honigmann, A.; Mueller, V.; Ta, H.; Schoenle, A.; Sezgin, E.; Hell, S.W.; Eggeling, C. Scanning STED-FCS reveals spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells. Nat. Commun. 2014, 5, 5412. [Google Scholar] [CrossRef]
- Suzuki, K.G.; Kasai, R.S.; Hirosawa, K.M.; Nemoto, Y.L.; Ishibashi, M.; Miwa, Y.; Fujiwara, T.K.; Kusumi, A. Transient GPI-anchored protein homodimers are units for raft organization and function. Nat. Chem. Biol. 2012, 8, 774–783. [Google Scholar] [CrossRef]
- Suzuki, K.G.; Fujiwara, T.K.; Sanematsu, F.; Iino, R.; Edidin, M.; Kusumi, A. GPI-anchored receptor clusters transiently recruit Lyn and G alpha for temporary cluster immobilization and Lyn activation: Single-molecule tracking study 1. J. Cell Biol. 2007, 177, 717–730. [Google Scholar] [CrossRef]
- Suzuki, K.G.; Fujiwara, T.K.; Edidin, M.; Kusumi, A. Dynamic recruitment of phospholipase C gamma at transiently immobilized GPI-anchored receptor clusters induces IP3-Ca2+ signaling: Single-molecule tracking study 2. J. Cell Biol. 2007, 177, 731–742. [Google Scholar] [CrossRef]
- Falck, E.; Patra, M.; Karttunen, M.; Hyvonen, M.T.; Vattulainen, I. Lessons of slicing membranes: Interplay of packing, free area, and lateral diffusion in phospholipid/cholesterol bilayers. Biophys. J. 2004, 87, 1076–1091. [Google Scholar] [CrossRef]
- Stauffer, T.P.; Ahn, S.; Meyer, T. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 1998, 8, 343–346. [Google Scholar] [CrossRef]
- Várnai, P.; Balla, T. Visualization of phosphoinositides that bind pleckstrin homology domains: Calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 1998, 143, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Knight, J.D.; Falke, J.J. Single-molecule fluorescence studies of a PH domain: New insights into the membrane docking reaction. Biophys. J. 2009, 96, 566–582. [Google Scholar] [CrossRef]
- Mashanov, G.I.; Tacon, D.; Peckham, M.; Molloy, J.E. The spatial and temporal dynamics of pleckstrin homology domain binding at the plasma membrane measured by imaging single molecules in live mouse myoblasts. J. Biol. Chem. 2004, 279, 15274–15280. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Niki, T.; Uchida, Y.; Mukai, K.; Kuchitsu, Y.; Kishimoto, T.; Sakai, S.; Makino, A.; Kobayashi, T.; Arai, H.; et al. A non-toxic equinatoxin-II reveals the dynamics and distribution of sphingomyelin in the cytosolic leaflet of the plasma membrane. Sci. Rep. 2024, 14, 16872. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Makino, A.; Murate, M.; Hullin-Matsuda, F.; Yanagawa, M.; Sako, Y.; Kobayashi, T. PMP2/FABP8 induces PI(4,5)P(2)-dependent transbilayer reorganization of sphingomyelin in the plasma membrane. Cell Rep. 2021, 37, 109935. [Google Scholar] [CrossRef]
- Abe, M.; Yanagawa, M.; Hiroshima, M.; Kobayashi, T.; Sako, Y. Bilateral regulation of EGFR activity and local PI(4,5)P(2) dynamics in mammalian cells observed with superresolution microscopy. Elife 2024, 13, e101652. [Google Scholar] [CrossRef]
- Lemmon, M.A.; Ferguson, K.M.; O’Brien, R.; Sigler, P.B.; Schlessinger, J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. USA 1995, 92, 10472–10476. [Google Scholar] [CrossRef]
- Garcia, P.; Gupta, R.; Shah, S.; Morris, A.J.; Rudge, S.A.; Scarlata, S.; Petrova, V.; McLaughlin, S.; Rebecchi, M.J. The pleckstrin homology domain of phospholipase C-delta 1 binds with high affinity to phosphatidylinositol 4,5-bisphosphate in bilayer membranes. Biochemistry 1995, 34, 16228–16234. [Google Scholar] [CrossRef]
- Thallmair, V.; Schultz, L.; Evers, S.; Jolie, T.; Goecke, C.; Leitner, M.G.; Thallmair, S.; Oliver, D. Localization of the tubby domain, a PI(4,5)P2 biosensor, to E-Syt3-rich endoplasmic reticulum-plasma membrane junctions. J. Cell Sci. 2023, 136, jcs260848. [Google Scholar] [CrossRef]
- Goulden, B.D.; Pacheco, J.; Dull, A.; Zewe, J.P.; Deiters, A.; Hammond, G.R.V. A high-avidity biosensor reveals plasma membrane PI(3,4)P(2) is predominantly a class I PI3K signaling product. J. Cell Biol. 2019, 218, 1066–1079. [Google Scholar] [CrossRef]
- Hammond, G.R.; Machner, M.P.; Balla, T. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol. 2014, 205, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, M.; Fairn, G.D. Complementary probes reveal that phosphatidylserine is required for the proper transbilayer distribution of cholesterol. J. Cell Sci. 2015, 128, 1422–1433. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, J.; Guo, X.; Tong, T.; Shi, X.; Li, L.; Qi, M.; Wang, Y.; Cai, M.; Jiang, J.; et al. Regulation of EGFR nanocluster formation by ionic protein-lipid interaction. Cell Res. 2014, 24, 959–976. [Google Scholar] [CrossRef] [PubMed]
- Raut, P.; Obeng, B.; Waters, H.; Zimmerberg, J.; Gosse, J.A.; Hess, S.T. Phosphatidylinositol 4,5-bisphosphate mediates the co-distribution of influenza A hemagglutinin and matrix protein M1 at the plasma membrane. Viruses 2022, 14, 2509. [Google Scholar] [CrossRef]
- Sztacho, M.; Červenka, J.; Šalovská, B.; Antiga, L.; Hoboth, P.; Hozák, P. The RNA-dependent association of phosphatidylinositol 4,5-bisphosphate with intrinsically disordered proteins contribute to nuclear compartmentalization. PLoS Genet. 2024, 20, e1011462. [Google Scholar] [CrossRef]
- Ji, C.; Lou, X. Single-molecule Super-resolution imaging of phosphatidylinositol 4,5-bisphosphate in the plasma membrane with novel fluorescent probes. J. Vis. Exp. 2016, 116, e54466. [Google Scholar] [CrossRef]
- Kuramoto, R.; Ikuta, T.; Carino, C.M.C.; Kawakami, K.; Kushiro, M.; Watanabe, C.; Uchida, Y.; Abe, M.; Sako, Y.; Taguchi, T.; et al. Membrane-domain compartmentalization of active GPCRs by β-arrestins through PtdIns(4,5)P2 binding. Nat. Chem. Biol. 2025. [Google Scholar] [CrossRef]
- Posor, Y.; Eichhorn-Gruenig, M.; Puchkov, D.; Schöneberg, J.; Ullrich, A.; Lampe, A.; Müller, R.; Zarbakhsh, S.; Gulluni, F.; Hirsch, E.; et al. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature 2013, 499, 233–237. [Google Scholar] [CrossRef]
- He, K.; Marsland, R., III; Upadhyayula, S.; Song, E.; Dang, S.; Capraro, B.R.; Wang, W.; Skillern, W.; Gaudin, R.; Ma, M.; et al. Dynamics of phosphoinositide conversion in clathrin-mediated endocytic traffic. Nature 2017, 552, 410–414. [Google Scholar] [CrossRef]
- Zhou, Y.; Prakash, P.; Liang, H.; Cho, K.J.; Gorfe, A.A.; Hancock, J.F. Lipid-sorting specificity encoded in K-Ras membrane anchor regulates signal output. Cell 2017, 168, 239–251.e16. [Google Scholar] [CrossRef] [PubMed]
- Thallmair, V.; Schultz, L.; Zhao, W.; Marrink, S.J.; Oliver, D.; Thallmair, S. Two cooperative binding sites sensitize PI(4,5)P(2) recognition by the tubby domain. Sci. Adv. 2022, 8, eabp9471. [Google Scholar] [CrossRef] [PubMed]
- Kimber, W.A.; Trinkle-Mulcahy, L.; Cheung, P.C.; Deak, M.; Marsden, L.J.; Kieloch, A.; Watt, S.; Javier, R.T.; Gray, A.; Downes, C.P.; et al. Evidence that the tandem-pleckstrin-homology-domain-containing protein TAPP1 interacts with Ptd(3,4)P2 and the multi-PDZ-domain-containing protein MUPP1 in vivo. Biochem. J. 2002, 361, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Vines, J.H.; Maib, H.; Buckley, C.M.; Gueho, A.; Zhu, Z.; Soldati, T.; Murray, D.H.; King, J.S. A PI(3,5)P2 reporter reveals PIKfyve activity and dynamics on macropinosomes and phagosomes. J. Cell Biol. 2023, 222, e202209077. [Google Scholar] [CrossRef]
- Rizalar, F.S.; Lucht, M.T.; Petzoldt, A.; Kong, S.; Sun, J.; Vines, J.H.; Telugu, N.S.; Diecke, S.; Kaas, T.; Bullmann, T.; et al. Phosphatidylinositol 3,5-bisphosphate facilitates axonal vesicle transport and presynapse assembly. Science 2023, 382, 223–230. [Google Scholar] [CrossRef]
- James, S.R.; Downes, C.P.; Gigg, R.; Grove, S.J.; Holmes, A.B.; Alessi, D.R. Specific binding of the Akt-1 protein kinase to phosphatidylinositol 3,4,5-trisphosphate without subsequent activation. Biochem. J. 1996, 315, 709–713. [Google Scholar] [CrossRef]
- Park, W.S.; Heo, W.D.; Whalen, J.H.; O’Rourke, N.A.; Bryan, H.M.; Meyer, T.; Teruel, M.N. Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. Sapiens by model prediction and live imaging. Mol. Cell 2008, 30, 381–392. [Google Scholar] [CrossRef]
- Singh, N.; Reyes-Ordoñez, A.; Compagnone, M.A.; Moreno, J.F.; Leslie, B.J.; Ha, T.; Chen, J. Redefining the specificity of phosphoinositide-binding by human PH domain-containing proteins. Nat. Commun. 2021, 12, 4339. [Google Scholar] [CrossRef]
- Lietzke, S.E.; Bose, S.; Cronin, T.; Klarlund, J.; Chawla, A.; Czech, M.P.; Lambright, D.G. Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol. Cell 2000, 6, 385–394. [Google Scholar] [CrossRef]
- Salim, K.; Bottomley, M.J.; Querfurth, E.; Zvelebil, M.J.; Gout, I.; Scaife, R.; Margolis, R.L.; Gigg, R.; Smith, C.I.; Driscoll, P.C.; et al. Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton’s tyrosine kinase. EMBO J. 1996, 15, 6241–6250. [Google Scholar] [CrossRef]
- Rameh, L.E.; Arvidsson, A.; Carraway, K.L., III; Couvillon, A.D.; Rathbun, G.; Crompton, A.; VanRenterghem, B.; Czech, M.P.; Ravichandran, K.S.; Burakoff, S.J.; et al. A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J. Biol. Chem. 1997, 272, 22059–22066. [Google Scholar] [CrossRef]
- Shin, J.J.H.; Liu, P.; Chan, L.J.; Ullah, A.; Pan, J.; Borchers, C.H.; Burke, J.E.; Stefan, C.; Smits, G.J.; Loewen, C.J.R. pH biosensing by PI4P regulates cargo sorting at the TGN. Dev. Cell 2020, 52, 461–476.e4. [Google Scholar] [CrossRef]
- Gillooly, D.J.; Morrow, I.C.; Lindsay, M.; Gould, R.; Bryant, N.J.; Gaullier, J.M.; Parton, R.G.; Stenmark, H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000, 19, 4577–4588. [Google Scholar] [CrossRef] [PubMed]
- Lawe, D.C.; Patki, V.; Heller-Harrison, R.; Lambright, D.; Corvera, S. The FYVE domain of early endosome antigen 1 is required for both phosphatidylinositol 3-phosphate and Rab5 binding. Critical role of this dual interaction for endosomal localization. J. Biol. Chem. 2000, 275, 3699–3705. [Google Scholar] [CrossRef] [PubMed]
- Burd, C.G.; Emr, S.D. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell 1998, 2, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Kutateladze, T.G.; Overduin, M. Structural mechanism of endosome docking by the FYVE domain. Science 2001, 291, 1793–1796. [Google Scholar] [CrossRef]
- Ketel, K.; Krauss, M.; Nicot, A.S.; Puchkov, D.; Wieffer, M.; Müller, R.; Subramanian, D.; Schultz, C.; Laporte, J.; Haucke, V. A phosphoinositide conversion mechanism for exit from endosomes. Nature 2016, 529, 408–412. [Google Scholar] [CrossRef]
- Yeung, T.; Gilbert, G.E.; Shi, J.; Silvius, J.; Kapus, A.; Grinstein, S. Membrane phosphatidylserine regulates surface charge and protein localization. Science 2008, 319, 210–213. [Google Scholar] [CrossRef]
- Kay, J.G.; Grinstein, S. Sensing phosphatidylserine in cellular membranes. Sensors 2011, 11, 1744–1755. [Google Scholar] [CrossRef]
- Koltsova, E.; Avilova, A.; Nikolaeva, E.; Kolchin, N.; Butov, K. Engineered Fluorescent Variants of Lactadherin C2 Domain for Phosphatidylserine Detection in Flow Cytometry. Biomolecules 2025, 15, 673. [Google Scholar] [CrossRef]
- Kay, J.G.; Koivusalo, M.; Ma, X.; Wohland, T.; Grinstein, S. Phosphatidylserine dynamics in cellular membranes. Mol. Biol. Cell 2012, 23, 2198–2212. [Google Scholar] [CrossRef]
- Uchida, Y.; Hasegawa, J.; Chinnapen, D.; Inoue, T.; Okazaki, S.; Kato, R.; Wakatsuki, S.; Misaki, R.; Koike, M.; Uchiyama, Y.; et al. Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes. Proc. Natl. Acad. Sci. USA 2011, 108, 15846–15851. [Google Scholar] [CrossRef]
- Koester, A.M.; Tao, K.; Szczepaniak, M.; Rames, M.J.; Nan, X. Nanoscopic spatial association between Ras and phosphatidylserine on the cell membrane studied with multicolor super resolution microscopy. Biomolecules 2022, 12, 1033. [Google Scholar] [CrossRef]
- Zhou, Y.; Wong, C.O.; Cho, K.J.; van der Hoeven, D.; Liang, H.; Thakur, D.P.; Luo, J.; Babic, M.; Zinsmaier, K.E.; Zhu, M.X.; et al. SIGNAL TRANSDUCTION. Membrane potential modulates plasma membrane phospholipid dynamics and K-Ras signaling. Science 2015, 349, 873–876. [Google Scholar] [CrossRef] [PubMed]
- Hirama, T.; Lu, S.M.; Kay, J.G.; Maekawa, M.; Kozlov, M.M.; Grinstein, S.; Fairn, G.D. Membrane curvature induced by proximity of anionic phospholipids can initiate endocytosis. Nat. Commun. 2017, 8, 1393. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, R.; Mori, T.; Hirosawa, K.M.; Kondo, R.; Taguchi, T.; Matsuhashi, N.; Suzuki, K.G.N. Single-molecule imaging quantifies oncogenic KRAS dynamics for enhanced accuracy of therapeutic efficacy assessment. iScience 2025, 28, 113374. [Google Scholar] [CrossRef] [PubMed]
- Kemmoku, H.; Takahashi, K.; Mukai, K.; Mori, T.; Hirosawa, K.M.; Kiku, F.; Uchida, Y.; Kuchitsu, Y.; Nishioka, Y.; Sawa, M.; et al. Single-molecule localization microscopy reveals STING clustering at the trans-Golgi network through palmitoylation-dependent accumulation of cholesterol. Nat. Commun. 2024, 15, 220. [Google Scholar] [CrossRef]
- Fernández-Golbano, I.M.; García, P.; Rebollo, E.; Geli, M.I.; Encinar Del Dedo, J. Use of the D4H Probe to Track Sterols in Yeast. Methods Mol. Biol. 2025, 2888, 35–52. [Google Scholar] [CrossRef]
- de Leeuw, S.M.; Nuriel, T. Intracellular cholesterol visualization in brain tissue using D4H*. STAR Protoc. 2024, 5, 102779. [Google Scholar] [CrossRef]
- Tomishige, N.; Murate, M.; Didier, P.; Richert, L.; Mély, Y.; Kobayashi, T. The use of pore-forming toxins to image lipids and lipid domains. Methods Enzym. 2021, 649, 503–542. [Google Scholar] [CrossRef]
- Kiyokawa, E.; Baba, T.; Otsuka, N.; Makino, A.; Ohno, S.; Kobayashi, T. Spatial and functional heterogeneity of sphingolipid-rich membrane domains. J. Biol. Chem. 2005, 280, 24072–24084. [Google Scholar] [CrossRef]
- Mizuno, H.; Abe, M.; Dedecker, P.; Makino, A.; Rocha, S.; Ohno-Iwashita, Y.; Hofkens, J.; Kobayashi, T.; Miyawaki, A. Fluorescent probes for superresolution imaging of lipid domains on the plasma membrane. Chem. Sci. 2011, 2, 1548–1553. [Google Scholar] [CrossRef]
- Kwiatkowska, K.; Hordejuk, R.; Szymczyk, P.; Kulma, M.; Abdel-Shakor, A.B.; Płucienniczak, A.; Dołowy, K.; Szewczyk, A.; Sobota, A. Lysenin-His, a sphingomyelin-recognizing toxin, requires tryptophan 20 for cation-selective channel assembly but not for membrane binding. Mol. Membr. Biol. 2007, 24, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Kundu, R.; Mondal, S.; Kapadia, A.; Banerjee, A.A.; Kucherak, O.A.; Klymchenko, A.S.; Koushika, S.P.; Venkatramani, R.; Vaidya, V.A.; Datta, A. A cell-permeable fluorescent probe reveals temporally diverse PI(4,5)P2 dynamics evoked by distinct GPCR agonists in neurons. Chem. Sci. 2025, 16, 10970–10982. [Google Scholar] [CrossRef] [PubMed]
- Dowler, S.; Currie, R.A.; Campbell, D.G.; Deak, M.; Kular, G.; Downes, C.P.; Alessi, D.R. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 2000, 351, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, A.; Sekizawa, Y.; Emoto, K.; Sakuraba, H.; Inoue, K.; Kobayashi, H.; Umeda, M. Lysenin, a novel sphingomyelin-specific binding protein. J. Biol. Chem. 1998, 273, 5300–5306. [Google Scholar] [CrossRef]
- Anderluh, G.; Pungercar, J.; Strukelj, B.; Macek, P.; Gubensek, F. Cloning, sequencing, and expression of equinatoxin II. Biochem. Biophys. Res. Commun. 1996, 220, 437–442. [Google Scholar] [CrossRef]
- Makino, A.; Abe, M.; Ishitsuka, R.; Murate, M.; Kishimoto, T.; Sakai, S.; Hullin-Matsuda, F.; Shimada, Y.; Inaba, T.; Miyatake, H.; et al. A novel sphingomyelin/cholesterol domain-specific probe reveals the dynamics of the membrane domains during virus release and in Niemann-Pick type C. Faseb J. 2017, 31, 1301–1322. [Google Scholar] [CrossRef]
- Ohno-Iwashita, Y.; Iwamoto, M.; Ando, S.; Mitsui, K.; Iwashita, S. A modified theta-toxin produced by limited proteolysis and methylation: A probe for the functional study of membrane cholesterol. Biochim. Biophys. Acta (BBA)—Biomembr. 1990, 1023, 441–448. [Google Scholar] [CrossRef]
- Ohno-Iwashita, Y.; Iwamoto, M.; Mitsui, K.; Ando, S.; Iwashita, S. A cytolysin, theta-toxin, preferentially binds to membrane cholesterol surrounded by phospholipids with 18-carbon hydrocarbon chains in cholesterol-rich region. J. Biochem. 1991, 110, 369–375. [Google Scholar] [CrossRef]
- Nakamura, M.; Sekino, N.; Iwamoto, M.; Ohno-Iwashita, Y. Interaction of theta-toxin (perfringolysin O), a cholesterol-binding cytolysin, with liposomal membranes: Change in the aromatic side chains upon binding and insertion. Biochemistry 1995, 34, 6513–6520. [Google Scholar] [CrossRef] [PubMed]
- Carquin, M.; Conrard, L.; Pollet, H.; Van Der Smissen, P.; Cominelli, A.; Veiga-da-Cunha, M.; Courtoy, P.J.; Tyteca, D. Cholesterol segregates into submicrometric domains at the living erythrocyte membrane: Evidence and regulation. Cell. Mol. Life Sci. 2015, 72, 4633–4651. [Google Scholar] [CrossRef]
- Johnson, B.B.; Moe, P.C.; Wang, D.; Rossi, K.; Trigatti, B.L.; Heuck, A.P. Modifications in perfringolysin O domain 4 alter the cholesterol concentration threshold required for binding. Biochemistry 2012, 51, 3373–3382. [Google Scholar] [CrossRef]
- Venugopal, S.; Martinez-Arguelles, D.B.; Chebbi, S.; Hullin-Matsuda, F.; Kobayashi, T.; Papadopoulos, V. Plasma Membrane Origin of the Steroidogenic Pool of Cholesterol Used in Hormone-induced Acute Steroid Formation in Leydig Cells. J. Biol. Chem. 2016, 291, 26109–26125. [Google Scholar] [CrossRef]
- Andronov, L.; Orlov, I.; Lutz, Y.; Vonesch, J.L.; Klaholz, B.P. ClusterViSu, a method for clustering of protein complexes by Voronoi tessellation in super-resolution microscopy. Sci. Rep. 2016, 6, 24084. [Google Scholar] [CrossRef]
- Hirano, M.; Ando, R.; Shimozono, S.; Sugiyama, M.; Takeda, N.; Kurokawa, H.; Deguchi, R.; Endo, K.; Haga, K.; Takai-Todaka, R.; et al. A highly photostable and bright green fluorescent protein. Nat. Biotechnol. 2022, 40, 1132–1142. [Google Scholar] [CrossRef]
- Isogai, T.; Hirosawa, K.M.; Suzuki, K.G.N. Recent Advancements in Imaging Techniques for Individual Extracellular Vesicles. Molecules 2024, 29, 5828. [Google Scholar] [CrossRef]

| Lipid | Probe | Fluorophore | Partitioning | References |
|---|---|---|---|---|
| Phosphoethanolamine (PE) | Cy3-DOPE | Cy3 | - | [23] |
| Phosphoethanolamine (PE) | 594-DOPE | ATTO594 | Ld | [26] |
| Phosphoethanolamine (PE) | FITC-DOPE | Fluorescein (FITC) | - | [28] |
| Phosphoethanolamine (PE) | TMR-PE | TMR | - | [29] |
| Phosphoethanolamine (PE) | Atto647N-DPPE | ATTO647N | Ld | [30] |
| Phosphoethanolamine (PE) | DSPE-PEG-KK114 | KK114 | Lo | [31] |
| Phosphatidylcholine (PC) | 594neg-DOPC | ATTO594 | Ld | [26] |
| Phosphatidylcholine (PC) | 594neg-DSPC | ATTO594 | Lo | [26] |
| Phosphatidylcholine (PC) | BODIPY-FL-C5- or -C12-PC | BODIPY-FL | Ld | [32] |
| Phosphatidylcholine (PC) | β-BODIPY FLC12-HPC | BODIPY-FL | Ld | [33] |
| Sphingomylein (SM) | 488neg-SM | ATTO488 | Lo | [26] |
| Sphingomylein (SM) | 594neg-SM | ATTO594 | Lo | [26] |
| Sphingomylein (SM) | Atto647N SM | ATTO647N | Ld | [34] |
| GM1 | 488-S9-GM1 | ATTO488 | Lo | [27,35] |
| GM1 | 594-S9-GM1 | ATTO594 | Lo | [27,35] |
| GM1 | BODIPY-FL-GM1 | BODIPY-FL | Ld | [27] |
| GM1 | Atto647N GM1 | ATTO647N | Ld | [34] |
| GM1 | Alexa-GM1 | Alexa488, 568, 647 | - | [36] |
| GM1 | Alexa-peptide-GM1 | Alexa-488 | Lo | [37] |
| GM3 | SF650B-GM3 | SF650B | - | [24] |
| GM3 | 488-S9-GM3 | ATTO488 | Lo | [27,35] |
| GM3 | 594-S9-GM3 | ATTO594 | Lo | [27,35] |
| GM3 | 647N-S9-GM3 | ATTO647N | Ld | [27] |
| GM3 | FI-S9-GM3 | Fluorescein (FI) | Lo | [27] |
| GM3 | N9-FI-GM3 | Fluorescein (FI) | Lo | [27] |
| Stage-specific embryonic antigen-3 (SSEA-3) | 594-SSEA-3 | ATTO594 | Lo | [35,38] |
| Stage-specific embryonic antigen-4 (SSEA-4) | 594-SSEA-4 | ATTO594 | Lo | [35,38] |
| Globohexaosylceramide (Globo-H) | 594-Globo-H | ATTO594 | Lo | [35,38] |
| Sialyl-lactotetraosylceramide (NeuAcLc4Cer) | ATTO594-NeuAcLc4-Cer | ATTO594 | Lo | [35,39] |
| Lactotetraosylceramide (Lc4Cer) | ATTO594-Lc4Cer | ATTO594 | Lo | [35,39] |
| GD2 | ATTO594-GD2 | ATTO594 | Lo | [35,40] |
| GD3 | ATTO594-GD3 | ATTO594 | Lo | [35,41] |
| GQ1b | ATTO594-GQ1b | ATTO594 | Lo | [35,41] |
| Cholesterol | BODIPY-Chl 2 | BODIPY | Lo | [42] |
| Cholesterol | Bdp-Chol | BODIPY | Lo | [43,44] |
| Cholesterol | Dehydroergosterol (DHE) | - | - | [45] |
| Cholesterol | Cholesterol analog 5 | - | Lo/Ld | [46] |
| Lipid | Probe | Fluorophore | Subcellular Localization | References |
|---|---|---|---|---|
| PI(4,5)P2 | PH domain of PLCδ1 | FP */Halo/SNAP | PM inner leaflet | [61,62,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81] |
| PI(4,5)P2 | Tubby C-terminal domain | FP | PM inner leaflet | [70,82] |
| PI(3,4)P2 | C-terminal PH domain of TAPP1 | FP | PM inner leaflet | [71,79,80,83] |
| PI(3,5)P2 | Dictyostelium SnxA | FP | PM inner leaflet | [84] |
| PI(3,5)P2 | KIF1A PH domain | FP | Synaptic vesicle | [85] |
| PI(3,4,5)P3 | PKB/Akt PH domain | FP/Halo | PM inner leaflet | [62,71,81,86,87,88] |
| PI(3,4,5)P3 | GRP1 PH domain | FP | PM inner leaflet | [63,67,89] |
| PI(3,4,5)P3 | Btk1 PH | FP | PM inner leaflet | [62,79,80,90,91] |
| PI(4)P | FAPP1 | FP | Golgi, PM | [72,92] |
| PI(4)P | P4M domain (SidM/SidC) | FP | Golgi, PM | [71,72,80] |
| PI(3)P | EEA1 FYVE domain | FP | Endocytic compartment | [71,72,93,94,95,96] |
| PI(3)P | Hrs 2xFYVE domain | FP | Early endosome | [79,80,84,93,97] |
| Phosphatidylserine (PS) | Lactadherin C2 (Lact-C2) | FP/Halo | PM inner leaflet | [66,73,81,98,99,100,101,102,103,104,105] |
| Phosphatidylserine (PS) | Evectin-2-PH domain | FP | Endosomes | [65,67,102,106] |
| Cholesterol | D4H (domain 4 of perfringolysin O) | FP | PM inner leaflet | [65,73,81,107,108,109] |
| Sphingomylein (SM) | NT-lysenin (C-terminal residues 161–297) | FP | PM outer leaflet | [66,110,111,112] |
| Sphingomylein (SM) | W20A mutant | FP | PM outer leaflet | [66,113] |
| Sphingomylein (SM) | NT-EqtII (EqtII mutant) | FP/Halo | PM outer and inner leaflet | [65] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mori, T.; Suzuki, K.G.N. Single-Molecule Imaging and Super-Resolution Microscopy of Lipid Domains in Cell Membranes Using Lipid-Binding Proteins and Fluorophore-Conjugated Lipid Analogs. Membranes 2025, 15, 317. https://doi.org/10.3390/membranes15100317
Mori T, Suzuki KGN. Single-Molecule Imaging and Super-Resolution Microscopy of Lipid Domains in Cell Membranes Using Lipid-Binding Proteins and Fluorophore-Conjugated Lipid Analogs. Membranes. 2025; 15(10):317. https://doi.org/10.3390/membranes15100317
Chicago/Turabian StyleMori, Toshiki, and Kenichi G. N. Suzuki. 2025. "Single-Molecule Imaging and Super-Resolution Microscopy of Lipid Domains in Cell Membranes Using Lipid-Binding Proteins and Fluorophore-Conjugated Lipid Analogs" Membranes 15, no. 10: 317. https://doi.org/10.3390/membranes15100317
APA StyleMori, T., & Suzuki, K. G. N. (2025). Single-Molecule Imaging and Super-Resolution Microscopy of Lipid Domains in Cell Membranes Using Lipid-Binding Proteins and Fluorophore-Conjugated Lipid Analogs. Membranes, 15(10), 317. https://doi.org/10.3390/membranes15100317






