Abstract
Biofilms exert a profound impact on various facets of human life. Positive instances of biofilm usage involve their capacity to immobilize pollutants such as heavy metals, while adverse cases result in infections like urinary tract infections. Therefore, the study of biofilm engineering emerges as crucial. Employing a bibliographic research approach, this paper delves into biofilm engineering, identifying key species like Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus, among others. The investigation also unveils major research subjects and corresponding institutions dedicated to biofilm research. A comprehensive understanding of biofilm engineering holds profound implications for advancing knowledge in this domain.
1. Introduction
Biofilms, complex communities of microorganisms encapsulated in self-produced extracellular polymeric substances, pervade diverse realms of human existence [1]. Their influence spans from beneficial applications, such as the immobilization of environmental pollutants [2], to deleterious effects like the onset of infections [3]. This dual nature underscores the critical importance of delving into the intricacies of biofilm engineering [4].
On one hand, the positive aspects of biofilm engineering are exemplified by its role in environmental remediation. Notably, biofilms can serve as effective agents for immobilizing pollutants, including heavy metals, through intricate metabolic processes and structural adaptations [2]. This aspect not only aligns with environmental sustainability goals but also offers innovative solutions to mitigate the impact of industrial activities on ecosystems [5].
Conversely, the negative consequences of biofilm formation are starkly evident in the realm of human health [6]. Biofilms contribute significantly to the persistence and resistance of bacterial infections, exemplified by conditions such as urinary tract infections [7]. The resilient nature of biofilms shields bacterial communities from conventional antimicrobial treatments, posing a formidable challenge in healthcare settings [8]. Hence, an in-depth exploration of biofilm engineering becomes imperative to develop strategies that combat harmful biofilm formation while capitalizing on their constructive applications [9].
The methodological approach employed in this study hinges on bibliographic research, allowing for a comprehensive survey of existing literature on biofilm engineering [10]. Key microbial species, such as Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus, have been identified as pivotal players in biofilm engineering research [11,12,13,14]. Understanding their roles and behaviors in biofilm formation is paramount for tailoring engineering strategies that can either enhance beneficial biofilm functions [15] or curtail detrimental effects [9].
Furthermore, the investigation has unveiled the primary research subjects in the field of biofilm engineering and the corresponding research institutions. This mapping of research entities not only provides insight into the current landscape of biofilm research but also fosters potential collaborations and synergies among researchers and institutions working towards a common goal. The interconnectedness of biofilm engineering research, as highlighted by the diverse range of species studied and the multitude of research entities engaged, underscores the interdisciplinary nature of this field.
In conclusion, this paper serves as a stepping stone toward a comprehensive understanding of biofilm engineering. By identifying key microbial species, exploring their applications, and mapping the research landscape, the study contributes to the collective knowledge base. The implications extend beyond academic boundaries, offering valuable insights for environmental scientists, healthcare professionals, and researchers alike. As society grapples with the intricate dynamics of biofilms, unraveling their mysteries becomes not only a scientific endeavor but a pathway toward a sustainable and healthier coexistence with these omnipresent microbial communities.
2. Materials and Methods
In November 2023, an extensive data collection initiative was conducted, utilizing the reputable bibliographic database Web of Science, which comprises various subdatabases [16,17]. To enhance the interpretability of the bibliographic analysis, the study utilized the powerful data visualization tool VOSviewer (Version 1.6.19, Universiteit Leiden, Leiden, The Netherlands) [18]. Unless explicitly stated otherwise, the mapping process conducted with VOSviewer adhered to default settings, aligning with methodologies established in prior studies [19,20].
In the keyword analysis, a minimum keyword occurrence threshold of 25 was selected to focus on terms with significant relevance and prevalence. Similarly, in the country study, a minimum requirement of 10 documents from a particular country was imposed for inclusion, allowing for a concentrated examination of regions with substantial research contributions. Furthermore, in the organization study, a minimum of 5 documents from a specific organization was deemed adequate for analysis, providing insights into institutions actively contributing to the field of biofilm engineering. This meticulous approach ensured a comprehensive and nuanced exploration of the biofilm engineering landscape.
3. Results
Figure 1 provides a comprehensive overview of key themes and keywords within the research domain of biofilm engineering, shedding light on various aspects that define the breadth and depth of this biofilm engineering field. The depicted keywords encompass a spectrum of entities, including specific microbial species that play a pivotal role in biofilm formation and dynamics. Notable examples include “Escherichia coli”, “Pseudomonas aeruginosa”, and “Staphylococcus aureus”, reflecting the microorganisms under scrutiny within the context of biofilm engineering.
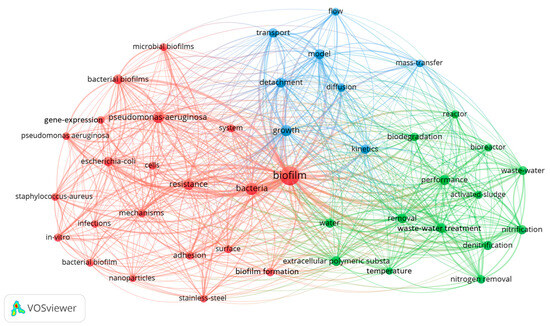
Figure 1.
The VOSviewer displays the main keywords. Colors in these networks indicate different clusters, while the lines indicate the interconnections between keywords. Larger circles indicate a higher frequency of mentioned keywords.
Moreover, the figure reveals a cluster of process-related keywords that elucidate critical biochemical and physicochemical mechanisms integral to biofilm engineering research. This encompasses terms such as “nitrification”, “denitrification”, “nitrogen removal”, “kinetics”, “diffusion”, “detachment”, and “mass transfer.” These terms collectively underscore the intricate processes involved in the development, maintenance, and functional attributes of biofilms, providing a nuanced understanding of the dynamics at play.
Further delving into the research targets within the biofilm engineering paradigm, the figure delineates keywords associated with specific areas of investigation. Noteworthy examples include “wastewater”, a crucial focus in the context of environmental and water treatment applications, and “activated sludge”, a key component in many wastewater treatment processes. Additionally, the figure encapsulates terms such as “extracellular polymeric substance” and “nanoparticles”, indicating the diverse materials and substances under exploration for their influence on biofilm properties and behavior.
In essence, Figure 1 serves as an intricate map, navigating through the multifaceted landscape of biofilm engineering research. It not only highlights specific microbial species, crucial processes, and research targets but also lays the groundwork for a more profound understanding of the interconnectedness of these elements. This comprehensive visualization serves as a valuable tool for scholars, researchers, and practitioners in the field of biofilm engineering. It offers a nuanced insight into the keywords that define and influence the progress of advancements within biofilm engineering. In this field, scientists focus on understanding the process where bacteria attach onto material surfaces and produce extracellular polymeric substances (including proteins, polysaccharides, extracellular DNA, etc.), similar to the construction of a house by bacteria [21].
Figure 2 delineates the primary countries and regions actively engaged in the expansive domain of biofilm engineering research, elucidating the global landscape of contributions to this dynamic field. At the forefront of this international endeavor are the United States and China, both assuming pivotal roles and making substantial strides in advancing biofilm engineering knowledge and applications. These two influential nations emerge as central pillars, driving innovation, and spearheading research initiatives within the biofilm engineering spectrum. Furthermore, the figure unveils a constellation of other significant contributors to biofilm engineering research, underscoring the truly global nature of collaboration in this scientific domain. The United Kingdom, Canada, Australia, Sweden, Denmark, Portugal, The Netherlands, and Singapore emerge as key players, each contributing unique perspectives, expertise, and research outcomes to the broader discourse on biofilm engineering. Noteworthy in this panorama is the emphasis on international collaboration as a driving force behind advancements in biofilm engineering research. Unlike endeavors that are solely reliant on the efforts of individual countries, the research depicted in Figure 2 highlights the interconnectedness and synergy achieved through collaborative initiatives spanning multiple nations and regions. This collaborative ethos enhances the diversity of insights, methodologies, and approaches brought to the table, fostering a richer and more comprehensive understanding of biofilm engineering dynamics. The collaborative efforts showcased in the figure underscore the importance of a collective, global approach to addressing the multifaceted challenges and opportunities within biofilm engineering. This interconnected network of nations not only shares knowledge and resources but also cultivates a truly international perspective, enriching the overall quality and impact of biofilm engineering research outcomes. In essence, Figure 2 serves as a visual testament to the collaborative spirit permeating the biofilm engineering research landscape. It not only identifies key players such as the USA and China but also emphasizes the collective contributions of a diverse array of countries and regions. In the realm of biofilm engineering, which significantly impacts human health and industry, international collaboration is particularly crucial. This collaborative effort mirrors a shared dedication to advancing the understanding of biofilm engineering on a global scale.
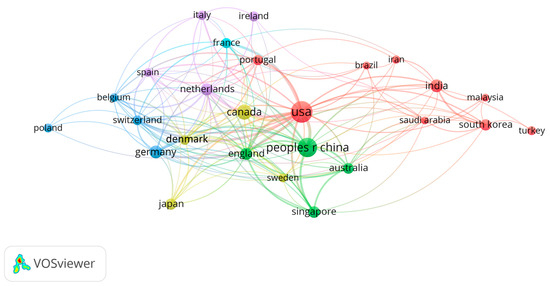
Figure 2.
The VOSviewer displays the main countries or regions working in the biofilm engineering field. Colors in these networks indicate different clusters, while the lines indicate international collaboration. Larger circles signify a greater number of publications.
Figure 3 delineates the primary organizations at the forefront of biofilm engineering research, offering insights into the collaborative landscape and key contributors shaping advancements in this field. In contrast to the traditional biological powerhouses like Harvard, Stanford, and Cambridge, the central contributors to biofilm engineering are Montana State University in the USA and Nanyang Technological University in Singapore. There are two research centers, the Center for Biofilm Engineering at Montana State University and the Singapore Centre for Environmental Life Sciences Engineering primarily at Nanyang Technological University, both receiving specialized funding and focusing on research related to biofilm engineering. These institutions play pivotal roles in advancing biofilm engineering knowledge and applications, highlighting the diverse research avenues and expertise characterizing the global biofilm engineering landscape. Expanding the panorama, other prominent players in this field include Washington State University, Arizona State University, and Northwestern University in the United States, along with Tsinghua University, Tianjin University, Shenzhen University, and Tongji University in China. Additionally, esteemed institutions such as the Harbin Institute of Technology, University of Porto, Technical University of Denmark, Waseda University, University of Alberta, University of Ghent, Delft University of Technology, Korea University, and Seoul National University contribute significantly to the biofilm engineering research milieu. While Figure 3 underscores the pivotal role played by individual organizations, it also emphasizes the collaborative nature of biofilm engineering research. Biofilm engineering, as a complex and multidisciplinary field, benefits from the rich tapestry of perspectives and approaches that arise from the collaboration between different organizations. In essence, the figure serves as a visual representation of the intricate network of organizations propelling biofilm engineering research forward.
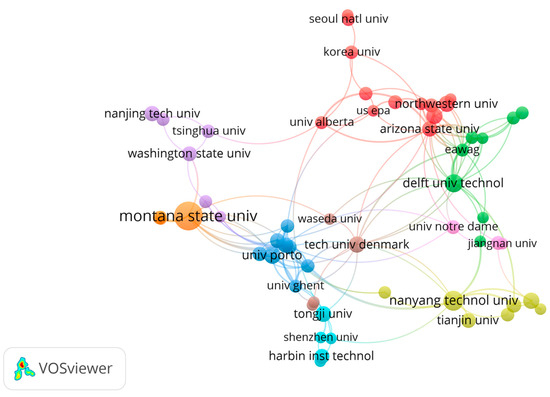
Figure 3.
The VOSviewer displays the main organizations working in the biofilm engineering field. Colors in these networks indicate different clusters, while the lines indicate the collaboration between different organizations. Larger circles signify a greater number of publications.
4. Discussion
4.1. Insights from Bibliographic Results: Recent Advances in Biofilm Engineering
After conducting bibliographic results, the authors identified the most representative recent papers in the field of biofilm engineering, which are listed in Table 1.
The investigation of biofilm engineering across different bacterial species has yielded substantial knowledge with wide-ranging implications for various applications. Particularly, in the field of water purification, Escherichia coli stands out by spearheading the development of a light-responsive, quorum-quenching biofilm engineered to mitigate biofouling on membranes [11]. This inventive strategy not only highlights the flexibility of biofilm engineering approaches but also offers solutions to practical challenges in water treatment technologies. The scientists also have the potential to modify Escherichia coli to achieve the light purpose [22] and microbial cell factories [23].
Shifting the focus to Pseudomonas aeruginosa, a multifaceted investigation delves into the manipulation of cyclic diguanylate (c-di-GMP) to generate and characterize biofilm-dispersed cells. This study employs both in vitro and in vivo methods, providing a comprehensive understanding of the dynamics of P. aeruginosa biofilm formation and dispersion [23]. Furthermore, the intriguing observation of functional amyloid formation within Pseudomonas aeruginosa biofilms adds a layer of complexity to our comprehension of biofilm architecture and functionality [24].
Expanding our understanding of Pseudomonas aeruginosa biofilms, evidence supporting the effects of iron override on quorum sensing and the regulation of biofilm-specific genes underscores the intricate regulatory mechanisms at play in biofilm development [25]. These findings contribute valuable insights into the environmental adaptability of Pseudomonas aeruginosa and its ability to modulate biofilm formation in response to external cues.
Turning our attention to Staphylococcus aureus, a critical role for the cidA murein hydrolase regulator is highlighted. This regulator contributes significantly to DNA release and biofilm development in Staphylococcus aureus, shedding light on the genetic and molecular factors influencing biofilm formation in this bacterial species [12].
In the domain of microbial fuel cell technology, Shewanella oneidensis emerges as a key player. The engineered Shewanella oneidensis-reduced graphene oxide biohybrid, with its enhanced biosynthesis and transport of flavins, stands out for its remarkable achievement in attaining the highest bioelectricity output in microbial fuel cells [26]. This not only showcases the potential of biofilm engineering in energy applications but also emphasizes the importance of understanding and manipulating electron transfer processes in microbial biofilms.
Expanding on the applications of Shewanella oneidensis, the promotion of bioelectricity generation is further observed with the enhanced biofilm of Shewanella oneidensis [27]. This finding reinforces the significance of biofilm architecture and composition in influencing microbial electrochemical activity [15]. Additionally, the efficient degradation of Bisphenol A achieved through the engineering of Shewanella oneidensis underscores the environmental remediation potential of biofilm engineering strategies [28].
Bringing these insights together, a comprehensive review highlights the challenges and opportunities associated with electrochemically active biofilms (EABs) in environmental bioelectrochemical sensors. The limitations in sensitivity, specificity, and stability of EABs necessitate thoughtful engineering strategies for improved biosensor performance [13,14]. This review, focusing on extracellular electron transfer, development, matrix, and applications of EAB-enabled biosensors, provides a roadmap for addressing current challenges and advancing the field toward more robust and efficient sensor technologies. In conclusion, the collective findings across Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Shewanella oneidensis contribute to a nuanced understanding of biofilm engineering, offering valuable insights for applications ranging from water purification to energy production and environmental remediation.

Table 1.
Main species working in the biofilm engineering field.
Table 1.
Main species working in the biofilm engineering field.
| Species | Main Contribution | References |
|---|---|---|
| Escherichia coli | The engineering of a light-responsive, quorum-quenching biofilm is undertaken to mitigate biofouling on water purification membranes. | [11] |
| Escherichia coli | The study modified Escherichia coli to see the light. | [22] |
| Escherichia coli | The paper engineered Escherichia coli to achieve microbial cell factories. | |
| Pseudomonas aeruginosa | Manipulating c-di-GMP, this study focuses on generating and characterizing Pseudomonas aeruginosa biofilm-dispersed cells through in vitro and in vivo methods. | [23] |
| Pseudomonas aeruginosa | Formation of functional amyloid in Pseudomonas aeruginosa biofilms is observed. | [24] |
| Pseudomonas aeruginosa | Iron override effects on quorum sensing and the regulation of biofilm-specific genes are supported by evidence. | [25] |
| Staphylococcus aureus | The cidA murein hydrolase regulator plays a role in contributing to DNA release and biofilm development in Staphylococcus aureus. | [12] |
| Shewanella oneidensis | The engineered Shewanella oneidensis-reduced graphene oxide biohybrid, with enhanced biosynthesis and transport of flavins, enables the highest bioelectricity output in microbial fuel cells. | [26] |
| Shewanella oneidensis | Promotion of bioelectricity generation is observed with the enhanced biofilm of Shewanella. | [15] |
| Shewanella oneidensis | Efficient degradation of Bisphenol A is established through the engineering of Shewanella oneidensis. | [28] |
| Shewanella oneidensis | Silver nanoparticles enhance charge-extraction efficiency in Shewanella oneidensis microbial fuel cells. | [27] |
| Pseudomonas aeruginosa, Shewanella oneidensis, Geobacter metallireducens | Limitations in sensitivity, specificity, and stability hinder the potential of electrochemically active biofilms (EABs) in environmental bioelectrochemical sensors, necessitating engineering strategies for improved biosensor performance, as discussed in this review focusing on extracellular electron transfer, development, matrix, and applications. | [13,14] |
4.2. Microbial Marvels: Biofilm Reactors in Environmental Engineering
Biofilm reactors, situated at the fascinating intersection of environmental engineering and microbiology, stand as dynamic and adaptive systems, representing a technological paradigm with unparalleled versatility and sustainability. Their multifaceted applications span a spectrum of critical environmental domains, showcasing their efficacy in addressing pressing challenges, from heavy metal contamination to the generation of electricity. These reactors operate on the principle of harnessing the collective power of microbial communities, intricately embedded in a matrix of extracellular polymeric substances, fostering the development of resilient biofilms that adhere tenaciously to diverse surfaces [29].
In the realm of heavy metal removal, biofilm reactors emerge as formidable allies, demonstrating extraordinary efficiency through intricate mechanisms such as biosorption, precipitation, and complexation [2]. These mechanisms underscore their potential as potent tools for mitigating metal contamination across a myriad of environmental matrices, including the complex effluents of industrial processes and municipal wastewater. The adaptability and robustness of biofilm reactors become particularly evident in their ability to operate efficiently in diverse environmental conditions, making them versatile and scalable solutions for addressing metal pollution challenges on a broad scale.
Expanding their reach, biofilm reactors transition seamlessly into the sphere of water cleaning, where their dynamic microbial consortia play a pivotal role in catalyzing the breakdown of organic pollutants and reducing nutrient loads [30]. This transformative capability positions biofilm reactors as sustainable alternatives for wastewater treatment, aligning with the global quest for eco-friendly and resource-efficient solutions. The intricate interplay of microorganisms within biofilms contributes to the purification of water resources, promoting ecological balance and mitigating the environmental impact of anthropogenic activities.
Furthermore, the innovative prowess of biofilm reactors extends to the realm of electricity generation through the fascinating realm of microbial fuel cells [15,26]. By capitalizing on the metabolic activities of microorganisms, these reactors convert organic matter into electrical energy, offering a unique and sustainable approach to power generation. This dual functionality underscores the versatility of biofilm reactors, positioning them as integral components in the pursuit of environmentally conscious and resource-efficient technologies.
As we contemplate the far-reaching implications of biofilm reactors, it becomes evident that their promise extends beyond singular applications. These reactors hold the potential to revolutionize diverse fields, from environmental remediation and water management to renewable energy. Their adaptability, efficiency, and sustainability mark them as transformative agents in the ongoing quest for a harmonious coexistence between technological progress and environmental stewardship. As research endeavors continue to unravel the intricacies of biofilm reactor dynamics, the horizon of possibilities for their applications in creating a greener and more sustainable future appears boundless.
4.3. Revolutionizing Biofilm Engineering: The Impact of Big Data and Machine Learning
Recent developments in big data and machine learning have opened up new and promising avenues for advancements in biofilm engineering. The widespread application of big data and machine learning across various domains, such as the development of autonomous vehicles [31], facial recognition technology [32], prediction of biological species distribution [33], and forecasting student performance in education [34], is widely acknowledged as a rapid and effective approach.
In the context of biofilm engineering, the increasing integration of big data and machine learning presents exciting opportunities [35]. Both big data and machine learning have proven to be valuable tools in processing and analyzing large datasets, providing insights and predictions that were previously challenging to attain [36]. This discussion explores the potential impact of big data and machine learning on biofilm engineering, focusing on the generation of experimental data and the subsequent enhancement of biofilm-related processes [37].
One significant aspect is the generation of substantial experimental data in biofilm engineering [38]. Experiments conducted in bioreactors, for instance, generate a wealth of data related to pollutant removal rates [39]. Similarly, microbial fuel cells produce data on electricity generation [40]. These datasets, when properly leveraged, can serve as a foundation for the application of machine learning algorithms [41]. By utilizing these datasets, we can develop machine learning models to gain a deeper understanding of biofilm behavior and performance in various environments [42].
The integration of machine learning into biofilm engineering holds great potential for advancing our understanding and optimizing processes [43]. One key application lies in predicting biofilm behavior and performance based on experimental data [44]. Machine learning models can analyze the relationships between different variables, such as environmental conditions, microbial composition, and biofilm characteristics, to predict outcomes [45]. For example, a machine learning model could predict the pollutant removal efficiency of a bioreactor under specific conditions or forecast the electricity generation of a microbial fuel cell based on various parameters [46].
Moreover, machine learning can facilitate the identification of patterns and trends within biofilm-related datasets that may not be immediately apparent through traditional analytical methods [47]. This ability to uncover hidden patterns can contribute to a more comprehensive understanding of biofilm dynamics [48]. For instance, machine learning algorithms could reveal correlations between specific microbial species and enhanced biofilm performance, leading to targeted strategies for biofilm improvement [49].
The application of big data and machine learning in biofilm engineering can also streamline the optimization of bioprocesses [50]. Traditional trial-and-error approaches to improve biofilm-related technologies can be time-consuming and resource-intensive [51]. Machine learning models, on the other hand, can efficiently analyze vast datasets and identify optimal conditions for enhanced biofilm performance [52]. This approach enables a more systematic and targeted approach to biofilm engineering, potentially accelerating the development of more efficient and sustainable biofilm-based technologies [53].
Furthermore, the integration of big data and machine learning in biofilm engineering can contribute to the design of adaptive and responsive systems [54]. As biofilm-related processes often operate in dynamic environments, the ability to predict and adapt to changing conditions is crucial [55]. Machine learning models can continuously learn and adjust based on real-time data, allowing for the development of biofilm systems that can respond to environmental changes and fluctuations in performance [56].
5. Conclusions
In conclusion, the impact of biofilms on human life is vast, ranging from the positive contributions of biofilm reactors in removing heavy metal pollutants from water to the negative consequences of biofilm-induced infections. The significance of biofilm engineering in navigating these dual effects cannot be overstated. Utilizing a bibliographic research approach, this study delved into the pivotal aspects of biofilm engineering, with a specific emphasis on bacteria species that are well-explored within this domain.
Moreover, in the era of big data and machine learning, this study underscored how these technological advancements can propel the field of biofilm engineering forward. By harnessing the power of big data, researchers can efficiently analyze large datasets generated from biofilm experiments, leading to deeper insights and more informed decision making. Machine learning, on the other hand, offers the potential to predict biofilm behavior, optimize processes, and uncover hidden patterns within complex datasets.
In essence, this exploration into biofilm engineering not only revealed the current state of research but also highlighted the immense potential for growth and innovation. The interdisciplinary nature of this field, coupled with global collaborations and advancements in data science, positions biofilm engineering at the forefront of scientific progress. As we continue to unlock the mysteries of biofilms and leverage cutting-edge technologies, the future of biofilm engineering holds great promise for addressing environmental challenges, improving human health, and advancing sustainable technologies.
Author Contributions
Conceptualization, Y.D.; methodology, S.C. and Y.D.; software, S.C. and Y.D.; validation, Y.D.; formal analysis, Y.D.; investigation, Y.D.; resources, Y.D.; data curation, Y.D.; writing—original draft preparation, Y.D.; writing—review and editing, Y.D.; visualization, S.C.; supervision, Y.D.; project administration, Y.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Dataset available on request from the authors.
Acknowledgments
The authors acknowledge the computation support from the School of Geography and the Environment, University of Oxford.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Ding, Y.; Peng, N.; Du, Y.; Ji, L.; Cao, B. Disruption of putrescine biosynthesis in Shewanella oneidensis enhances biofilm cohesiveness and performance in Cr (VI) immobilization. Appl. Environ. Microbiol. 2014, 80, 1498–1506. [Google Scholar] [CrossRef]
- Yang, L.; Liu, Y.; Wu, H.; Song, Z.; Høiby, N.; Molin, S.; Givskov, M. Combating biofilms. FEMS Immunol. Med. Microbiol. 2012, 65, 146–157. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Mishra, S.; Huang, Y.; Li, J.; Wu, X.; Zhou, Z.; Lei, Q.; Bhatt, P.; Chen, S. Biofilm-mediated bioremediation is a powerful tool for the removal of environmental pollutants. Chemosphere 2022, 294, 133609. [Google Scholar] [CrossRef] [PubMed]
- Schulze, A.; Mitterer, F.; Pombo, J.P.; Schild, S. Biofilms by bacterial human pathogens: Clinical relevance-development, composition and regulation-therapeutical strategies. Microb. Cell 2021, 8, 28. [Google Scholar] [CrossRef]
- Tenke, P.; Köves, B.; Nagy, K.; Hultgren, S.J.; Mendling, W.; Wullt, B.; Grabe, M.; Wagenlehner, F.M.E.; Cek, M.; Pickard, R. Update on biofilm infections in the urinary tract. World J. Urol. 2012, 30, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, D.; Von Holy, A. Bacterial biofilms within the clinical setting: What healthcare professionals should know. J. Hosp. Infect. 2006, 64, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Xu, S.; Wang, Y.; Zhang, Y.; Chou, S.-H.; Galperin, M.Y.; He, J. Ways to control harmful biofilms: Prevention, inhibition, and eradication. Crit. Rev. Microbiol. 2021, 47, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, J.J.; Reng, J.; Wang, S.; Zhang, R.; Wang, B. Global trends of Pseudomonas aeruginosa biofilm research in the past two decades: A bibliometric study. MicrobiologyOpen 2020, 9, 1102–1112. [Google Scholar] [CrossRef]
- Mukherjee, M.; Hu, Y.; Tan, C.H.; Rice, S.A.; Cao, B. Engineering a light-responsive, quorum quenching biofilm to mitigate biofouling on water purification membranes. Sci. Adv. 2018, 4, eaau1459. [Google Scholar] [CrossRef]
- Rice, K.C.; Mann, E.E.; Endres, J.L.; Weiss, E.C.; Cassat, J.E.; Smeltzer, M.S.; Bayles, K.W. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2007, 104, 8113–8118. [Google Scholar] [CrossRef]
- Shi, L.; Dong, H.; Reguera, G.; Beyenal, H.; Lu, A.; Liu, J.; Yu, H.-Q.; Fredrickson, J.K. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Microbiol. 2016, 14, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Han, X.; Shi, L.; Cao, B. Electrochemically active biofilm-enabled biosensors: Current status and opportunities for biofilm engineering. Electrochim. Acta 2022, 428, 140917. [Google Scholar] [CrossRef]
- Liu, T.; Yu, Y.Y.; Deng, X.P.; Ng, C.K.; Cao, B.; Wang, J.Y.; Rice, S.A.; Kjelleberg, S.; Song, H. Enhanced Shewanella biofilm promotes bioelectricity generation. Biotechnol. Bioeng. 2015, 112, 2051–2059. [Google Scholar] [CrossRef] [PubMed]
- Pranckutė, R. Web of Science (WoS) and Scopus: The titans of bibliographic information in today’s academic world. Publications 2021, 9, 12. [Google Scholar] [CrossRef]
- Archambault, É.; Campbell, D.; Gingras, Y.; Larivière, V. Comparing bibliometric statistics obtained from the Web of Science and Scopus. J. Am. Soc. Inf. Sci. Technol. 2009, 60, 1320–1326. [Google Scholar] [CrossRef]
- Xie, L.; Chen, Z.; Wang, H.; Zheng, C.; Jiang, J. Bibliometric and visualized analysis of scientific publications on atlantoaxial spine surgery based on Web of Science and VOSviewer. World Neurosurg. 2020, 137, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ding, Y. Tackling Heavy Metal Pollution: Evaluating Governance Models and Frameworks. Sustainability 2023, 15, 15863. [Google Scholar] [CrossRef]
- Chen, S.; Ding, Y. A bibliography study of Shewanella oneidensis biofilm. FEMS Microbiol. Ecol. 2023, 99, fiad124. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Neu, T.R.; Wozniak, D.J. The EPS matrix: The “house of biofilm cells”. J. Bacteriol. 2007, 189, 7945–7947. [Google Scholar] [CrossRef] [PubMed]
- Levskaya, A.; Chevalier, A.A.; Tabor, J.J.; Simpson, Z.B.; Lavery, L.A.; Levy, M.; Davidson, E.A.; Scouras, A.; Ellington, A.D.; Marcotte, E.M. Engineering Escherichia coli to see light. Nature 2005, 438, 441–442. [Google Scholar] [CrossRef] [PubMed]
- Chua, S.L.; Hultqvist, L.D.; Yuan, M.; Rybtke, M.; Nielsen, T.E.; Givskov, M.; Tolker-Nielsen, T.; Yang, L. In vitro and in vivo generation and characterization of Pseudomonas aeruginosa biofilm–dispersed cells via c-di-GMP manipulation. Nat. Protoc. 2015, 10, 1165–1180. [Google Scholar] [CrossRef] [PubMed]
- Bleem, A.; Christiansen, G.; Madsen, D.J.; Maric, H.; Strømgaard, K.; Bryers, J.D.; Daggett, V.; Meyer, R.L.; Otzen, D.E. Protein engineering reveals mechanisms of functional amyloid formation in Pseudomonas aeruginosa biofilms. J. Mol. Biol. 2018, 430, 3751–3763. [Google Scholar] [CrossRef] [PubMed]
- Bollinger, N.; Hassett, D.J.; Iglewski, B.H.; Costerton, J.W.; McDermott, T.R. Gene expression in Pseudomonas aeruginosa: Evidence of iron override effects on quorum sensing and biofilm-specific gene regulation. J. Bacteriol. 2001, 183, 1990–1996. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Ding, W.; Sun, L.; Wang, L.; Liu, C.-G.; Song, H. Engineered Shewanella oneidensis-reduced graphene oxide biohybrid with enhanced biosynthesis and transport of flavins enabled a highest bioelectricity output in microbial fuel cells. Nano Energy 2018, 50, 639–648. [Google Scholar] [CrossRef]
- Cao, B.; Zhao, Z.; Peng, L.; Shiu, H.-Y.; Ding, M.; Song, F.; Guan, X.; Lee, C.K.; Huang, J.; Zhu, D. Silver nanoparticles boost charge-extraction efficiency in Shewanella microbial fuel cells. Science 2021, 373, 1336–1340. [Google Scholar] [CrossRef]
- Zhou, J.; Hong, S.H. Establishing Efficient Bisphenol A Degradation by Engineering Shewanella oneidensis. Ind. Eng. Chem. Res. 2021, 60, 16864–16873. [Google Scholar] [CrossRef]
- Nguyen, P.Q.; Courchesne, N.M.D.; Duraj-Thatte, A.; Praveschotinunt, P.; Joshi, N.S. Engineered living materials: Prospects and challenges for using biological systems to direct the assembly of smart materials. Adv. Mater. 2018, 30, 1704847. [Google Scholar] [CrossRef]
- Edwards, S.J.; Kjellerup, B.V. Applications of biofilms in bioremediation and biotransformation of persistent organic pollutants, pharmaceuticals/personal care products, and heavy metals. Appl. Microbiol. Biotechnol. 2013, 97, 9909–9921. [Google Scholar] [CrossRef] [PubMed]
- Navarro, P.J.; Fernandez, C.; Borraz, R.; Alonso, D. A machine learning approach to pedestrian detection for autonomous vehicles using high-definition 3D range data. Sensors 2016, 17, 18. [Google Scholar] [CrossRef] [PubMed]
- Andrejevic, M.; Selwyn, N. Facial recognition technology in schools: Critical questions and concerns. Learn. Media Technol. 2020, 45, 115–128. [Google Scholar] [CrossRef]
- Chen, S.; Ding, Y. Machine Learning and Its Applications in Studying the Geographical Distribution of Ants. Diversity 2022, 14, 706. [Google Scholar] [CrossRef]
- Chen, S.; Ding, Y. A Machine Learning Approach to Predicting Academic Performance in Pennsylvania’s Schools. Soc. Sci. 2023, 12, 118. [Google Scholar] [CrossRef]
- Zhou, L.; Pan, S.; Wang, J.; Vasilakos, A.V. Machine learning on big data: Opportunities and challenges. Neurocomputing 2017, 237, 350–361. [Google Scholar] [CrossRef]
- Elshawi, R.; Sakr, S.; Talia, D.; Trunfio, P. Big data systems meet machine learning challenges: Towards big data science as a service. Big Data Res. 2018, 14, 1–11. [Google Scholar] [CrossRef]
- Highmore, C.J.; Melaugh, G.; Morris, R.J.; Parker, J.; Direito, S.O.L.; Romero, M.; Soukarieh, F.; Robertson, S.N.; Bamford, N.C. Translational challenges and opportunities in biofilm science: A BRIEF for the future. Npj Biofilms Microbiomes 2022, 8, 68. [Google Scholar] [CrossRef] [PubMed]
- Volk, E.; Iden, S.C.; Furman, A.; Durner, W.; Rosenzweig, R. Biofilm effect on soil hydraulic properties: Experimental investigation using soil-grown real biofilm. Water Resour. Res. 2016, 52, 5813–5828. [Google Scholar] [CrossRef]
- Healy, M.G.; Ibrahim, T.G.; Lanigan, G.J.; Serrenho, A.J.; Fenton, O. Nitrate removal rate, efficiency and pollution swapping potential of different organic carbon media in laboratory denitrification bioreactors. Ecol. Eng. 2012, 40, 198–209. [Google Scholar] [CrossRef]
- Mohan, Y.; Kumar, S.M.M.; Das, D. Electricity generation using microbial fuel cells. Int. J. Hydrogen Energy 2008, 33, 423–426. [Google Scholar] [CrossRef]
- Paullada, A.; Raji, I.D.; Bender, E.M.; Denton, E.; Hanna, A. Data and its (dis)contents: A survey of dataset development and use in machine learning research. Patterns 2021, 2, 100336. [Google Scholar] [CrossRef] [PubMed]
- Long, F.; Wang, L.; Cai, W.; Lesnik, K.; Liu, H. Predicting the performance of anaerobic digestion using machine learning algorithms and genomic data. Water Res. 2021, 199, 117182. [Google Scholar] [CrossRef] [PubMed]
- Helleckes, L.M.; Hemmerich, J.; Wiechert, W.; von Lieres, E.; Grünberger, A. Machine learning in bioprocess development: From promise to practice. Trends Biotechnol. 2022, 41, 817–835. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.A.; Harnisch, F.; Kapadnis, B.; Schröder, U. Electroactive mixed culture biofilms in microbial bioelectrochemical systems: The role of temperature for biofilm formation and performance. Biosens. Bioelectron. 2010, 26, 803–808. [Google Scholar] [CrossRef]
- Lesnik, K.L.; Liu, H. Predicting microbial fuel cell biofilm communities and bioreactor performance using artificial neural networks. Environ. Sci. Technol. 2017, 51, 10881–10892. [Google Scholar] [CrossRef]
- Tsipa, A.; Varnava, C.K.; Grenni, P.; Ferrara, V.; Pietrelli, A. Bio-electrochemical system depollution capabilities and monitoring applications: Models, applicability, advanced bio-based concept for predicting pollutant degradation and microbial growth kinetics via gene regulation modelling. Processes 2021, 9, 1038. [Google Scholar] [CrossRef]
- Capatina, D.; Feier, B.; Hosu, O.; Tertis, M.; Cristea, C. Analytical methods for the characterization and diagnosis of infection with Pseudomonas aeruginosa: A critical review. Anal. Chim. Acta 2022, 1204, 339696. [Google Scholar] [CrossRef]
- Shade, A.; Gilbert, J.A. Temporal patterns of rarity provide a more complete view of microbial diversity. Trends Microbiol. 2015, 23, 335–340. [Google Scholar] [CrossRef]
- Artini, M.; Patsilinakos, A.; Papa, R.; Božović, M.; Sabatino, M.; Garzoli, S.; Vrenna, G.; Tilotta, M.; Pepi, F.; Ragno, R. Antimicrobial and antibiofilm activity and machine learning classification analysis of essential oils from different mediterranean plants against Pseudomonas aeruginosa. Molecules 2018, 23, 482. [Google Scholar] [CrossRef]
- Wainaina, S.; Taherzadeh, M.J. Automation and artificial intelligence in filamentous fungi-based bioprocesses: A review. Bioresour. Technol. 2022, 369, 128421. [Google Scholar] [CrossRef]
- Mountcastle, S.E.; Vyas, N.; Villapun, V.M.; Cox, S.C.; Jabbari, S.; Sammons, R.L.; Shelton, R.M.; Walmsley, A.D.; Kuehne, S.A. Biofilm viability checker: An open-source tool for automated biofilm viability analysis from confocal microscopy images. Npj Biofilms Microbiomes 2021, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Shaban, T.F.; Alkawareek, M.Y. Prediction of qualitative antibiofilm activity of antibiotics using supervised machine learning techniques. Comput. Biol. Med. 2022, 140, 105065. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-W.; Chen, S. Potential of biofilm-based biofuel production. Appl. Microbiol. Biotechnol. 2009, 83, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-T.; Kristiani, E.; Leong, Y.K.; Chang, J.-S. Big Data and Machine Learning Driven Bioprocessing-Recent trends and critical analysis. Bioresour. Technol. 2023, 372, 128625. [Google Scholar] [CrossRef]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The biofilm life cycle: Expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef]
- Yu, J.; Tang, S.N.; Lee, P.K.H. Universal Dynamics of Microbial Communities in Full-Scale Textile Wastewater Treatment Plants and System Prediction by Machine Learning. Environ. Sci. Technol. 2023, 57, 3345–3356. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).