Abstract
Gout is a prevalent form of inflammatory arthritis caused by the crystallization of uric acid in the joints and soft tissues, leading to acute, painful attacks. Activation of the NLRP3 inflammasome in mononuclear cells, along with inflammasome-independent pathways, is responsible for the inflammatory phenotype in gout. Research into the different aspects of gout pathophysiology and potential treatment options is ongoing. This review highlights some of the basic research published in the 12 months following the 2022 Gout, Hyperuricemia, and Crystal-Associated Disease Network (G-CAN) conference and focuses on mechanisms of inflammation, encompassing pro- and anti-inflammatory pathways, as well as the exploration of various biological systems, such as single-cell transcriptomics, proteomics, metabolomics, and microbiome analyses.
1. Introduction
Gout is the most common inflammatory type of arthritis and is characterized by the rapid onset of painful flares. Uric acid, a metabolic byproduct resulting from the breakdown of purines, plays a central role in the pathogenesis of gout. Genetic and environmental factors can lead to hyperuricemia by overproduction, but most cases are due to inadequate excretion [1]. When serum urate concentration exceeds saturation point, monosodium urate (MSU) is able to crystallize and form deposits in joints and soft tissues. These crystal deposits trigger an innate immune response, causing inflammation and, thereafter, characteristic symptoms of gout [2].
A key element in gout-related inflammation is the activation of NLRP3 inflammasomes in mononuclear cells, which takes place in two steps: first, a “priming” signal promotes the expression of inflammasome components. These remain inactive precursors in the cytosol until the second “activating” signal prompts the formation of the inflammasome complex and proteolytic activation of caspase-1 [3], which, in turn, activates proIL-1β into bioactive IL-1β, one of the main proinflammatory cytokines in gout [4].
Concurrently, inflammasome-independent mechanisms also contribute to gouty inflammation. These involve direct cell stimulation, neutrophil recruitment, and the release of proinflammatory mediators, though neutrophils also have a role in the resolution of inflammation [5].
Though progress is being made each year, the characterization of the mechanisms of inflammation in gout remains an ongoing effort. This review summarizes research published in the 12 months after the 2022 Gout, Hyperuricemia, and Crystal-Associated Disease Network (G-CAN) Annual Meeting and follows the oral talk given at the G-CAN 2023 9th Annual Research Symposium in San Diego, California (7–8 November 2023).
2. Fundamental Studies on the Pathophysiology of Gouty Inflammation
Several studies have shed new light on various processes that either enhance or resolve inflammation in gout, and they identify targetable mechanisms in gouty inflammation. A schematic overview of the main insights developed in this section is represented in Figure 1 for proinflammatory mechanisms and in Figure 2 for anti-inflammatory and therapeutic strategies.
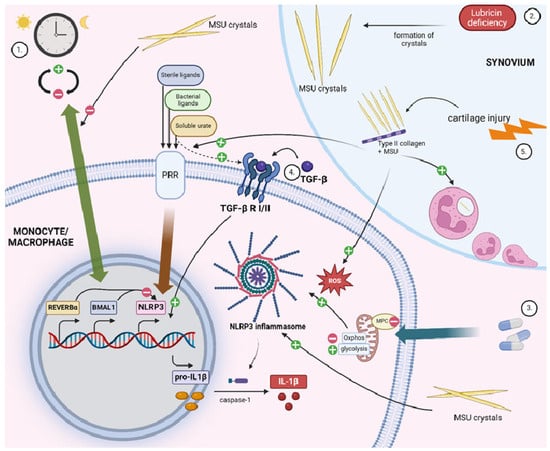
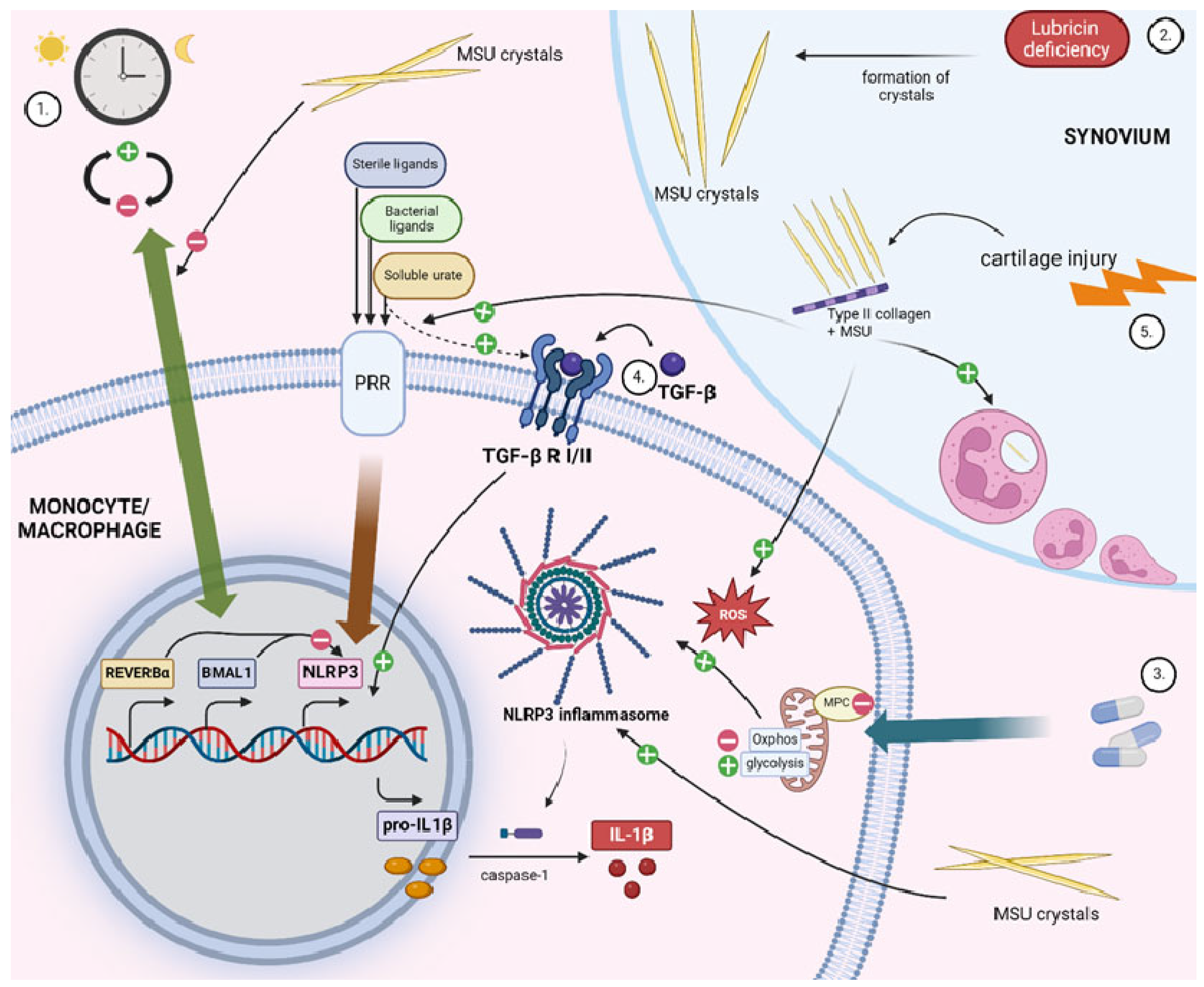
Figure 1.
Highlights of inflammatory mechanisms in gout. In monocytes/macrophages, inflammation in response to urate and monosodium urate crystals is driven by proinflammatory cytokines, mainly IL-1β. IL-1β activation is controlled by two main steps: (a) transcriptional regulation and production of proIL-1β and (b) post-translational processing by the inflammasome complex. In the joint, neutrophil influx dominates inflammatory phases initiated by IL-1β signaling. Several players have been identified to modulate these processes, as outlined in the manuscript (see text for details): (1) Expression of clock genes oscillate within the circadian rhythm. Clock components BMAL1 and REV-ERB-α, which are involved in NLRP3 repression, are shown to have decreased expression in MSU-exposed macrophages. (2) Lubricin deficiency promotes urate crystal precipitation. (3) Oral antidiabetic drug pioglitazone inhibits mitochondrial pyruvate carrier (MPC) and, as a result, promotes NLRP3 inflammasome hyperactivation. (4) TGF-β mediates a proinflammatory response in the process of soluble urate-induced reprogramming of myeloid cells. (5) Type 2 collagen (CII) is more abundant in synovial fluid from patients with cartilage injuries and forms complexes with MSU crystals. These CII-MSU complexes lead to higher activation of TLR2/4- NF-κB signal pathway in macrophages, higher ROS production, and higher neutrophil recruitment. Figure created with BioRender.com.
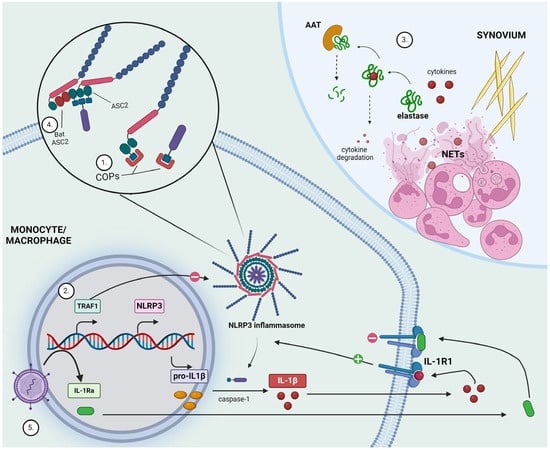
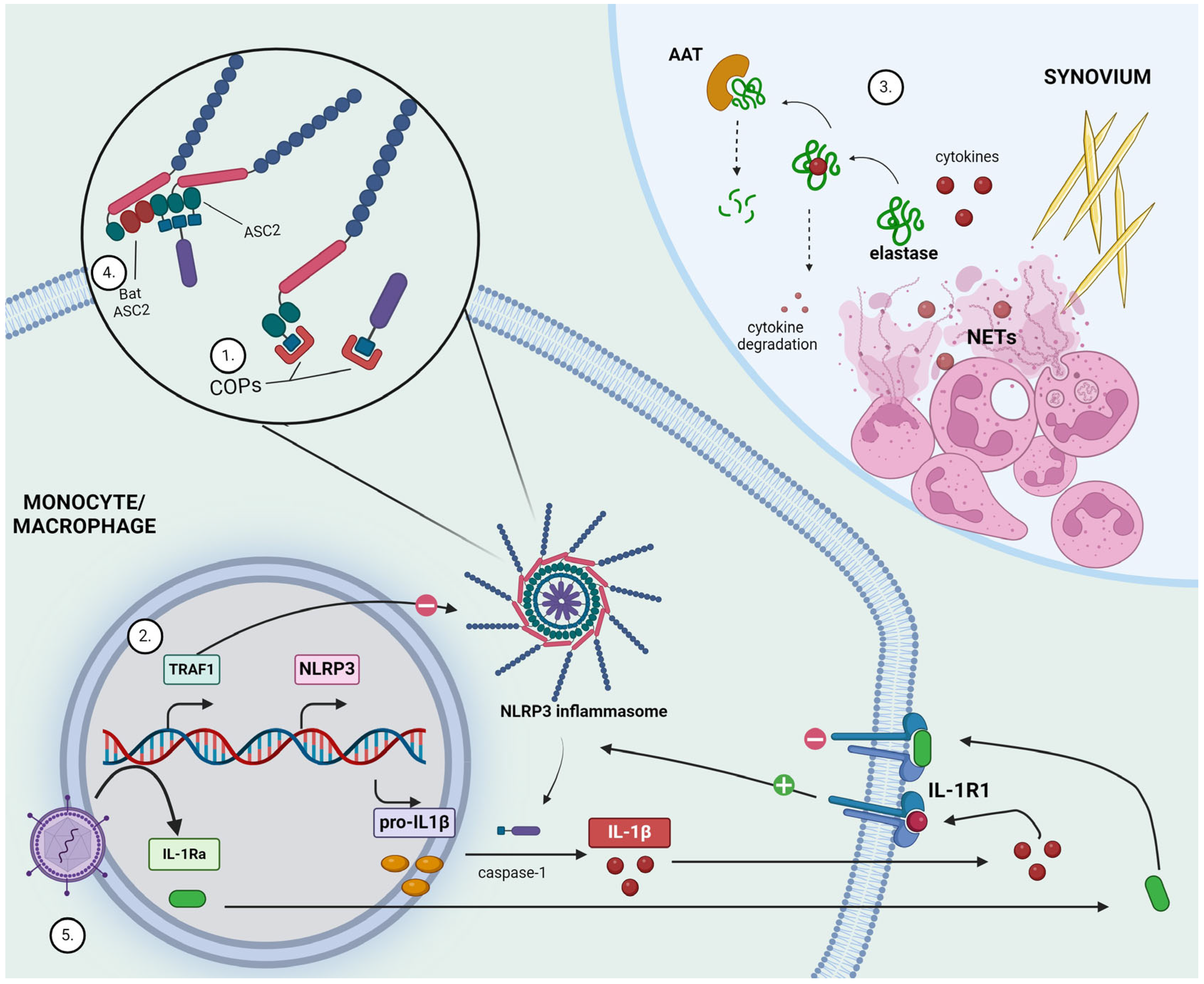
Figure 2.
Highlights of anti-inflammatory mechanisms and potential new therapeutic development in gout. (1) CARD-only proteins (COPs) competitively bind to caspase recruitment domains (CARD) and hinder interactions needed for inflammasome activation. (2) TNFR-associated factor 1 (TRAF1) acts as a negative regulator of inflammasome activation. (3) Monosodium urate (MSU) crystals formed within the synovial space trigger neutrophil recruitment and activation, eventually leading to the release of neutrophil extracellular traps (NETs). Neutrophil elastase is capable of degrading cytokines, thereby limiting inflammatory response. α1-antitrypsin (AAT) counteracts this by degrading elastase. (4) Bat ASC2, lacking the caspase-recruiting-domain, potently inhibits NLRP3 inflammasome activity. (5) Lentivirus-mediated gene transfer generates stable interleukin-1 receptor antagonist (IL-1Ra) production as a means to suppress IL-1R1 receptor activation. Figure created with BioRender.com.
2.1. New Insights into Proinflammatory Mechanisms in Gout
2.1.1. Circadian Clock
While NLRP3 inflammasome activation is recognized as a pivotal mechanism in gouty inflammation, the specific reason why flares occur disproportionately during night-time or early morning remains unknown. Popov et al. [6] sought to investigate whether MSU crystals induce changes in the circadian clock of THP1 macrophages in vitro. Clocks were synchronized within the cell population by serum starvation followed by re-feeding with serum-supplemented media (in the presence or absence of MSU crystals). MSU-exposed THP1 macrophages exhibited increased expression of the negative limb components PER2 and CRY1. The positive limb component BMAL1 and auxiliary component REV-ERBα, which have a role in NLRP3 repression, also showed an altered expression pattern, and although this was not as consistent throughout all time points, overall, BMAL1 and REV-ERBα protein levels were decreased across the 24 h period.
Next, the authors demonstrated a time-of-day-dependent effect of MSU on NLRP3 activation by pointing out that caspase-1 activity and IL-1β protein levels both showed a greater increase when MSU was introduced in the first 12 h following re-feeding, rather than in the 12–24 h period. The role of BMAL1 variation in these time-of-day differences was validated using adenoviral vectors to either over- or under-express BMAL1. The knockdown of BMAL1 led to an increase in caspase-1 activity and pro-IL-1β when MSU was introduced in the 12–24 h interval, but there was no effect in the 0–12 h interval. The overexpression of BMAL1 repressed caspase-1 and pro-IL-1β regardless of the time point.
Lastly, the influence of REV-ERBα on MSU crystal-induced inflammasome activation was explored using a pharmacological modulator. Heme, a REV-ERBα agonist, suppressed caspase-1 activity, IL-1β secretion, and NLRP3 expression, while co-treatment with an antagonist blocked these effects. Unlike BMAL1, however, the REV-ERBα effects were not dependent on time points.
Overall, these findings contribute to our understanding of the underlying mechanisms leading to nocturnal gout flares, implicating BMAL1’s low expression in aggravated NLRP3-mediated inflammation in response to MSU crystals. Some limitations need to be acknowledged; notably, these findings are restricted to THP1 cells, and more importantly, the circadian rhythm of cells grown in vitro cannot be considered interchangeable with the day-night cycle experienced in vivo.
2.1.2. Src Family Kinases
Futosi et al. [7] investigated the role of the myeloid Src family kinases Hck, Fgr, and Lyn in gouty inflammation as well as systemic inflammation in motheaten viable (Ptpn6me-v/me-v) mice. Isolated neutrophils from mice lacking Hck, Fgr, and Lyn failed to produce the respiratory burst and cytokine release responses showed by WT mouse neutrophils following MSU stimulation. Macrophages from mutant mice also showed reduced ROS production, though IL-1β and MIP-2 were not significantly affected. In vivo, triple knockout mice also showed reduced inflammation and pain, a lack of MPO and NADPH-oxidase activity, reduced neutrophil and monocyte/macrophage infiltration, and reduced proinflammatory mediators at the site of MSU crystal injection, including IL-1β, MIP-2, and LTB4. In response to MSU crystals, neutrophils lacking the Src family kinases also had a reduced capacity for phagocytosis and reduced activation of ERK and p38 MAPK pathways, but migration and NET formation were unaffected. Human neutrophils treated with dasatinib, an inhibitor of Src family kinases, performed in a similar manner, and further experiments using the MSU-induced gouty arthritis murine model revealed that dasatinib was effective both at preventing and treating crystal-induced inflammation. To summarize, this paper identifies a role for myeloid Src family kinases in autoinflammatory diseases, including gout, and suggests targeting tyrosine phosphorylation pathways as a potential therapeutic strategy for managing conditions characterized by heightened neutrophil activity.
2.1.3. Immunometabolic Reprogramming through Mitochondrial Pyruvate Carrier (MPC)
Immunometabolic reprogramming, and in particular, the shift in rates of glycolysis versus oxidative phosphorylation in response to stressors, is a recognized mechanism of inflammation through the transcriptional upregulation of inflammatory genes [8,9]. Recent studies have also shown that immunometabolic reprogramming takes place in gout [10]. The role of glycolytic reprogramming and its effects in NLRP3-mediated inflammation and the risk of gout has recently been described by Chen et al. [11]. This study investigated the role of the mitochondrial pyruvate carrier (MPC) in NLRP3 activation. MPC deficiency, achieved through genetic knockout in mice or pharmacological inhibition with antidiabetic drug pioglitazone in human THP1 cells, led to elevated IL-1β secretion in response to NLRP3 agonists in macrophages. Additionally, MPC inactivation increased caspase-1 activation, IL-1β maturation, and ASC oligomerization, indicating enhanced NLRP3 inflammasome activation. Combining insulin and MPC inactivation enhanced lactic acid fermentation and promoted NLRP3 inflammasome hyperactivation in response to MSU stimulation both in vitro and in diabetic mice, with the latter also showing increased neutrophil recruitment.
Finally, the authors performed a retrospective analysis and found that patients using both pioglitazone and insulin exhibited a significantly higher risk of developing gout compared to those using either medication alone or none. Multivariate analyses revealed increased risks for specific gout subtypes, and higher cumulative doses of pioglitazone and insulin were associated with elevated gout risk.
In conclusion, this report studies the relationship between the MPC and NLRP3 activation in the context of pioglitazone treatment in diabetes mellitus and highlights that glycolytic reprogramming upon MPC modulation recapitulates the mechanism of NLRP3 activation and increases gout risk. These results seem to be somewhat in contrast with other studies, which found that pioglitazone reduces NLRP3 activation in certain cell types, including renal cells [12,13] and cardiomyocytes [14]. The retrospective nature of this study also means that a cause-and-effect relationship between pioglitazone use and an increased risk of gout cannot be established.
2.1.4. Transforming Growth Factor β (TGF-β)
Another report published last year looked at the transforming growth factor β (TGF-β), a known pleiotropic cytokine typically associated with anti-inflammatory roles during the resolution phase of gout flares. Klück et al. [15] investigated the involvement of TGF-β in the context of gout and hyperuricemia and the reprogramming of myeloid cells induced by urate.
In monocytes obtained from individuals with gout, TGFB1 mRNA expression and its downstream target ITGAV were elevated compared to healthy controls and correlated positively with serum urate levels. In a larger cohort analysis, however, ITGAV expression showed no significant difference, but instead, an increased MMP9 expression was present in gout patients. Protein levels of TGF-β-LAP, LIF, and VEGFA were significantly higher in hyperuricemic individuals compared to controls, and moreover, LIF was significantly higher in gout flares than in intercritical gout. Serum TGF-β-LAP positively correlated with urate levels across all cohorts. Further in vitro studies using human primary monocytes isolated from healthy volunteers showed no elevation of TGFB1 mRNA or TGF-β1 protein production 24h after urate treatment; however, the upregulation of MMP9, known to activate latent TGF-β and the downregulation of SMAD7, a negative regulator of TGF-β signaling, were noted. Additionally, both urate and TGF-β induced phosphorylation of SMAD2, indicating increased intracellular TGF-β signaling. Pretreatment of cells with urate and TGF-β, followed by stimulation with LPS, led to increased IL-1β and IL-6 release. The addition of anti-TGF receptor II antibodies partially reversed this inflammatory phenotype, as did the inhibition of TGFβ I receptor. This pinpoints the fact that, in the context of gout, TGF-β could play dual roles: while anti-inflammatory in the resolution of flares driven by MSU crystals, it mediates a proinflammatory response in the process of the soluble urate-induced reprogramming of myeloid cells. The study’s limitations include unexplored intracellular signaling pathways and a lack of in vivo assessment of TGF-β involvement in inflammatory priming by urate.
2.1.5. Lubricin Deficiency
Another study showcased a rare hyperuricemia-independent inflammatory mechanism. Elsaid et al. [16] reported on a case of a young female normouricemic patient presenting with classic features of gout, including inflammatory arthritis flares, extensive crystal deposition, and erosive joint lesions, as well as increased levels of acute-phase reactants. Whole genome sequencing revealed that the proband was heterozygous for several NLRP3 variants, one of which is associated with significantly higher rates of stimulated monocyte IL-1β release. Additionally, the patient was heterozygous for an ITIH3 variant predicted to be deleterious and homozygous for a SERPINB3 variant associated with antiapoptotic and proinflammatory effects. Pathway analysis of upregulated genes in the proband’s whole-blood transcripts showed increased interferon and cytokine signaling, potential disruptions in anti-inflammatory pathways, and potentially impaired cell homeostasis, consistent with an inflammatory phenotype.
Most notably, the patient had decreased serum levels of lubricin or proteoglycan 4 (PRG4), an important cartilage lubricant [17], but no decreased levels of whole-blood corresponding PRG4 mRNA, suggesting that the cause for this alteration was a heightened capacity for lubricin degradation. This was compatible with increased activity of lubricin-degrading cathepsin G (CTSG), brought on by decreased levels of CTSG inhibitors, which were also found in proband serum. Furthermore, the authors showed that lubricin inhibited urate crystal precipitation in a time- and dose-dependent manner. In addition to this, higher xanthine oxidase positive and more M1 synovial resident macrophages were observed in the knee joint of mice of the PRG4 gene trap colony compared to PRG4-reconstituted mice, and this was observed both at baseline and after IL-1β injection.
In summary, this rare case of erosive gout in the absence of hyperuricemia led to the mechanistic implication of lubricin deficiency in gout pathogenesis via the diminished inhibition of urate crystal formation or diminished inhibition of M1 polarization and xanthine oxidase activity in synovial macrophages.
2.1.6. Type II Collagen (CII)
Xu et al. [18] examined synovial fluid from chronic gout patients with or without cartilage damage and found that type II collagen (CII) was higher in patients with cartilage destruction, and collagen in cartilage injuries was highly associated with MSU crystals. Next, they investigated the influence of CII on the crystallization process and pointed out that while pure synthetized crystals and crystals obtained from the synovial fluid of gout patients varied in length and straightness, macrophage-phagocyted crystals were homogenous, and crystals synthetized with CII were more similar to them. Using the label-free imaging of RAW264.7 macrophages, the study found that CII-MSU complexes increased lipid droplet accumulation, cell elongation, and phagocytosis rate compared to MSU crystals alone. CII-MSU also led to higher reactive oxygen species (ROS) production and increased levels of the proinflammatory cytokines IL-1β and TNF-α. RNA-Seq analysis showed that while MSU induced changes in over 8000 genes, CII-MSU primarily affected genes related to intercellular interactions and anabolism, suggesting that CII-MSU complexes significantly enhance MSU-induced inflammation. Gene enrichment analysis showed that CII-MSU complexes significantly upregulated genes related to leukocyte migration. In vitro, CII-MSU increased the secretion of CCL2 and CXCL2, which are key chemokines for recruiting macrophages and neutrophils, and migration assays confirmed that CII-MSU supernatants promoted greater macrophage and neutrophil migration compared to MSU alone. In vivo, the air-pouch model revealed that CII-MSU induced a higher percentage of CCL2+ and CXCL2+ cells and increased the recruitment of F4/80+ macrophages and MPO+ neutrophils to the inflammation site. Additionally, they showed that CII-MSU upregulated pattern recognition receptors, especially TLR2 and TLR4, leading to increased inflammatory responses and knocking down ITGB1 in macrophages, decreased TLR2/4 expression and the associated inflammatory responses. Using the air-pouch model, they showed that blocking TLR2/4 decreased the migration of neutrophils and macrophages, as well as the recruitment of CCR2+ and CXCR2+ cells in response to CII-MSU. This study had several limitations. The patient cohort was small, and the nature of their cartilage injuries was not specified, limiting the ability to generalize the findings to other gout patients with different types of cartilage damage. Additionally, the cohort comprised relatively young individuals (ages 20–33), which is not typical for gout and raises questions about underlying factors contributing to the early onset of the disease in these patients.
Finally, this report showcased how cartilage fragments, in particular, type II collagen, can enhance the proinflammatory effects of MSU crystals, which may exacerbate gout flares by increasing macrophage activation, inflammatory responses, and immune cell recruitment.
2.2. New Insights into Anti-Inflammatory Mechanisms in Gout
2.2.1. CARD-Only Proteins (COPs)
As mentioned above, in gout, a crucial inflammatory mechanism involves the NLRP3 inflammasome. The release of caspase-1 is initiated by pattern recognition receptors (PRRs) and the subsequent polymerization of proteins containing pyrin domains (PYDs) and caspase recruitment domains (CARDs). Small proteins known as PYD-only proteins (POPs) and CARD-only proteins (COPs) modulate inflammasome activation by competing with crucial CARD interactions, thereby limiting inflammation.
COPs are the main focus of a paper published last year by Devi et al. [19], in which they investigate their salutary effects on gout inflammation. In THP-1 macrophages treated with MSU crystals, CARD16 and CARD17 were significantly decreased, and a slight increase was observed in CARD18. Transgenic mice expressing human CARD16, 17, and 18 were further injected with MSU crystals using the subcutaneous air pouch model. In wild-type mice, this resulted in inflammation, as shown by the in vivo imaging of neutrophils and quantified by myeloperoxidase activity. However, the transgenic mice had an ameliorated response with reduced cellular infiltrates, similar to caspase-1-KO control mice. Lavage fluid of the pouch of transgenic mice also contained less IL-1β, caspase-1, and NLRP3 but not reduced levels of CXCL1 and 2, which are inflammasome-independent.
In bone-marrow-derived macrophages (BMDMs) isolated from COP-expressing transgenic mice, ELISA, following stimulation with crystalline and soluble NLRP3 activators, detected a significantly reduced release of IL-1β and IL-18 but not TNF, which is independent of NLRP3. The same results were obtained for human THP-1 cells. Moreover, the inhibitory effect of COPs was shown to not be exclusive to the NLRP3 inflammasome but also present for AIM2, NLRP1b, NLRC4, and NLRP1 inflammasomes. In immunoblot assays, BMDMs expressing CARD16, CARD17, and CARD18 and primed with MSU were shown to release less caspase-1. In line with this, gasdermin D, which is activated by caspase-1, was also reduced. Finally, employing coimmunoprecipitation, the authors sought to reveal how COPs inhibit inflammasome activation. Each COP displayed unique binding characteristics during macrophage activation, with CARD16 and CARD17 interacting after NLRP3 inflammasome activation, while CARD18 is constitutively bound to caspase-1. Despite these differences, what all COPs had in common was that they inhibited caspase-1 activation by interfering with its dimerization and polymerization, essential for inflammasome responses, therefore demonstrating their protective effect in gout inflammation. A major limitation is the use of human CARD16, CARD17, and CARD18 expression in mice, which may not perfectly mimic human processes. While the results indicate that COP expression alleviates gout severity in mice, more studies are required to validate the findings in human models. The study observed decreased CARD16 and increased CARD18 expression in gout patients, but the role of these expression changes in gout pathology remains unclear.
2.2.2. TNFR-Associated Factor 1 (TRAF1)
Another anti-inflammatory mechanism in gout was provided by Mirzaesmaeili et al. [20] in a paper focusing on TNFR-associated factor 1 (TRAF1). Using human THP1 cells transduced with short hairpin RNA targeting TRAF1, they showed that reduced expression of TRAF1 led to elevated IL-1β production and increased activation of multiple inflammasomes, including NLRP3, following stimulation. The transduced cells showed higher ASC oligomerization and displayed enhanced linear ubiquitination of ASC compared with control cells. In vivo, MSU-induced peritonitis was exacerbated in TRAF1-deficient mice, which also demonstrated higher IL-1β release and inflammatory cell recruitment. TRAF1-KO mice also displayed more pronounced joint inflammation and higher serum IL-1β in a model of MSU crystal-induced arthritis when compared to wild-type mice. However, one unaddressed limitation is how the use of short hairpin RNA might cause off-target effects or unintended alterations in gene expression, which could influence the observed results.
In summary, this paper described the role of TRAF1 in restricting inflammasome activation and IL-1β production by negatively regulating ASC ubiquitination and oligomer formation.
2.2.3. Neutrophil Extracellular Trap (NET)-Derived Elastase
During the initiation of urate-lowering therapy (ULT), patients often experience acute flares and consequently show low compliance to treatment. Guidelines for the treatment of gout recommend that prophylaxis against flares is necessary during the first several months of ULT [21,22]. Liu et al. [23] assessed the mechanism behind increased rates of gout flares during the initiation of ULT, specifically the role of neutrophil extracellular trap (NET)-derived elastase and α1-antitrypsin (AAT) in the inflammatory response to unstable tophi. Increased concentrations of cell-free DNA (cfDNA), a component of NETs, were found in serum samples of patients with tophaceous gout experiencing frequent flares, both in the acute phase and the intermittent phase, and these levels rose further after initiation of ULT. Patients with tophi also had significantly higher serum AAT levels; however, this was only observed in the acute phase, affirming its role in acute flares of tophaceous gout triggered by ULT.
In a murine model of MSU-induced peritonitis, IL-1β and IL-6 concentrations rose locally and spontaneously resolved after 8h. The application of AAT during the resolution phase led to a relapse of inflammation and an increase in IL-1β and IL-6 levels, demonstrating the serin-protease-dependent resolution of inflammation. This role of elastase and its counteraction by AAT is not new in the context of gout; however, this report shows their involvement in the initial phases of ULT.
2.2.4. ABCG2
ABCG2 is a known urate transporter involved in the control of serum urate and associated with the transition from hyperuricemia to gout [24,25].
Recently, Zhao et al. [26] pointed out the role of ABCG2 present in intestinal epithelial cells in decreasing serum uric acid following a flare. After inducing gout in a mouse model and stimulating Caco2 cells with IL-1β, they noted that ABCG2 expression and UA transport were increased, and the pharmacological inhibition and genetic knockout of ABCG2 resulted in decreased UA transport and excretion, respectively. The upregulation of ABCG2 and uric acid transport was inhibited in vitro by blocking the phosphoinositide 3-kinase (PI3K)/Akt pathway. The ABCG2 transporter is also present in the kidney, and another recent study [27] revealed that during gout flares, urinary excretion of UA also increases. A recent study by Notsu et al. found that both the genetic and pharmacological inhibition of ABCG2 enhanced IL-1β production in macrophages in response to MSU crystals, suggesting that ABCG2 may limit inflammation in response to MSU crystals beyond its effect on urate levels [28].
2.3. Potential New Therapeutic Development in Gout Inflammation
2.3.1. Bat ASC2
Continuing the theme of the inhibitory effects on the inflammasome with potential therapeutic implications, Ahn et al. [29] assessed the ASC2 protein originating in bats and described its anti-inflammatory properties. Bats have increased resistance to highly pathogenic viruses and increased lifespan through unclear mechanisms. Previous studies show altered inflammasomes in bats, including dampened NLRP3 and lower IL-1β. ASC2 is a pyrin domain-only containing protein (POP), shown to inhibit ASC polymerization and, therefore, inhibit inflammasome activation in a competitive manner. The overexpression of bat ASC2 in HEK293T cells had an inhibitory effect on human NLRP3, bat NLRP3, and AIM2 inflammasome activation. Moreover, bat ASC2 was found to be significantly more potent than human ASC2.
The authors generated the first transgenic mice expressing a bat gene, and these mice (expressing human ASC2) showed less IL-1β and less cell death measured by LDH release in response to activators of several inflammasomes, such as NLRP3, AIM2, or NLRC4. With relevance to gout, bat ASC2 transgenic mice were injected with MSU intraperitoneally. Increased inflammatory monocytes and increased neutrophil counts were observed in the peritoneal lavage at 4 h post-MSU injection, and this was dramatically reduced in ASC2 transgenic mice. Moreover, IL-1β was diminished to control levels by bat ASC2, and ASC speck formation was significantly reduced in both monocyte and neutrophil infiltrating cells. This paper indicates that bat ASC2 is a very potent inflammasome inhibitor, which interferes with ASC speck formation and inhibits cytokines and pyroptosis. This report has implications for a wide range of stimuli and several inflammasomes, including MSU-induced inflammation, which was dramatically blocked by bat ASC2. Insights from bat ASC2 could help develop new treatments for autoimmune diseases and chronic inflammation and extend our understanding of longevity and resistance to viruses, though this study focused on acute inflammation using transgenic mice, not addressing chronic conditions or aging, and the applicability of these findings in humans remains to be explored.
2.3.2. Constitutive Interleukin-1 Receptor Antagonist (IL-1Ra)
Another therapeutic approach is described by Colantuoni et al. [30], who utilized lentivirus-mediated gene transfer into transplanted autologous hematopoietic stem/progenitor cells (HSPCs) to produce increased constitutive interleukin-1 receptor antagonist (IL-1Ra), a strategy which ensured stable and systemic IL-1 inhibition while also preserving the functional capacities of HSPCs. The efficacy of this method was evaluated in three distinct models of IL-1-dependent inflammation: in gout, where it hindered neutrophil recruitment; in cryopyrin-associated periodic syndromes, where it prevented systemic inflammation; and in experimental autoimmune encephalomyelitis associated with multiple sclerosis, where it reduced disease severity. Importantly, no signs of toxicity were observed, and IL-1RA expression did not adversely affect the homing, engraftment, or differentiation of HSPCs into immune cell subsets. However, potential limitations include the need for myeloablative conditioning for optimal engraftment and the need to confirm the absence of anti-IL-1RA autoantibodies in a clinical setting. Additionally, while the strategy was tested in mice before disease development, its effectiveness in human patients with pre-existing conditions remains to be determined. Nevertheless, this innovative approach holds promise as a therapeutic measure for suppressing IL-1-driven inflammation.
3. Omics Studies in Gout
Recent years have seen a surge in omics studies in the field of gout and hyperuricemia. In 2023, several biological data systems were assessed individually, as described below (and summarized in Table 1), and it is an exciting prospect that multi-omics integrative approaches will become possible with respect to inflammation in gout.

Table 1.
Significant targets identified in omics studies in human samples.
3.1. Single-Cell Transcriptomics and Immune Cell Phenotyping
3.1.1. Single-Cell Transcriptomics in Peripheral Blood and Synovial Fluid in Acute Gout
The first study to use single-cell RNA sequencing in order to characterize immune cell populations in gout was performed by Chang et al. [31]. They describe increased levels of naïve CD4 T cells as well as classical monocytes in the peripheral blood of patients with acute gout compared to healthy controls. They also found that plasmacytoid dendritic cells and intermediate monocytes were more abundant in healthy controls, but no significant differences in the presence of B cells, megakaryocytes, progenitor cells, low-density neutrophils, and low-density basophils were found between groups. A major advantage of this study was that in addition to peripheral mononuclear cells, it also characterized cell populations present in the synovial fluid of three patients during a gout flare. Synovial fluid was enriched in macrophages and myeloid dendritic cells, Th1/Th17 cells, effector memory CD8 T cells, and mucosal-associated invariant T cells and was depleted in classical monocytes, central memory CD8 T cells, and NK cells.
3.1.2. Single-Cell Transcriptomics in Intercritical and Advanced Gout
While the previously mentioned study focused on acute gout, Gu et al. [32] employed single-cell RNA sequencing to compare patients with intercritical gout, patients with advanced disease, and healthy controls (three donors per group). Patients included in this study are referred to as “in remission” because they do not display clinical inflammation, their serum urate is below 360 mM, and their C-reactive protein levels are normal. Proportions of monocytes, dendritic cells, T- and NK cells, and B cells were not significantly different between the three groups. Among monocytes, however, one subcluster, HLA-DQA1high, was shown to be more prevalent in advanced gout patients, and this was validated through flow cytometry in a separate cohort. Differentially expressed gene (DEG) analysis in this cell subtype found upregulated genes related to antigen processing and presentation, osteoclast differentiation, and parathyroid hormone synthesis. Through Single-cell Regulatory Network Inference and Clustering (SCENIC) analysis, transcription factors (TF) highly expressed in HLA-DQA1high were found to be enhanced in advanced gout compared to intercritical gout and normal controls. Several TFs implicated in bone metabolism (with many promoting osteoclast differentiation) and others known to be key regulators of the type I interferon (IFN) signaling pathway showed higher expression in intercritical gout than in advanced gout. Furthermore, the study identified 23 interferon-stimulated genes (ISGs) differentially expressed between intercritical and advanced gout, some of which displayed higher expression in intercritical gout, suggesting a contribution to systemic inflammatory responses during this phase.
Next, the authors investigated T- and NK cell subsets and found that there was a gradual decrease in naïve CD8+ T cells from advanced gout to intercritical to healthy controls, while cytotoxic CD8+ T cells followed the opposite trend. They validated the lower proportion of naïve CD8+ T cells in advanced gout patients using flow cytometry on another cohort. Additionally, they observed higher proportions of effective memory and senescent CD8+ T cells and lower proportions of functional CD8+ T cells in intercritical and advanced gout patients compared to healthy volunteers, with no significant differences between intercritical and advanced gout patients. When comparing the DEGs per various cell types, upregulated genes in advanced gout patients were enriched in pathways related to osteoclast differentiation, PTH synthesis, antigen processing, NF-kB signaling, leukocyte migration, cytokine interaction, and chemokine signaling. Cell–cell communication analysis revealed more numerous and stronger interactions among immune cells in gout patients compared to healthy controls, and advanced gout patients exhibited the highest interactions.
Lastly, the researchers noted that gene expression analysis revealed increased COX-2 in monocytes of advanced gout patients, while plasma analysis on a different cohort confirmed elevated arachidonic acid levels in intercritical and advanced gout, with specific metabolites showing higher levels in advanced gout and lower levels in intercritical gout compared to healthy controls.
3.1.3. Single-Cell Mass Cytometry in Acute Gout
Wang et al. [33] used single-cell mass cytometry and compared PBMCs from five people without gout to seven people with gout during a flare and the same seven people after the flare. A significant decrease in the percentage of T cells was observed in gout (acute and intercritical). Additionally, multiple immune cell subsets expressing CCR4 and OX40 were increased in gout. In line with this, expression levels of CCL17 and CCL22, which are known to be specific chemokines that can recruit cells expressing CCR4, were increased in the plasma of flaring patients compared to controls and intercritical patients.
Altogether, the importance of these studies consists in characterizing immune cell populations, revealing differences in immune cell abundances in gout-related phenotypes. The observed transcriptional signatures identified by these studies give links to new players that could impact inflammatory signaling in gout. These studies also indicate that other immune cell populations, in addition to monocyte/macrophages, warrant further investigation in the context of gout as lymphoid cell subsets are differentially abundant in gout and show expression profiles that could pathogenetically contribute to inflammatory mechanisms. However, a common limitation of these reports is the small sample size; therefore, replicative studies and further validation of identified hits are important.
3.2. Proteome
Using a targeted proteomic assay through the Olink Target 96 Inflammation Panel™, Cabău et al. [34] characterized inflammatory proteomic signatures in gout and asymptomatic hyperuricemia. While no differentially expressed proteins were found in gout patients when compared to non-gout controls (both hyperuricemic and normouricemic), when they compared individuals with asymptomatic hyperuricemia to normouricemic individuals, a distinct inflammatory signature emerged, featuring 58 significantly differentially expressed proteins. Among these were cytokines, chemokines, growth factors, and immunoregulatory proteins, with the most upregulated proteins being the mTOR effector 4E-BP1 and IL-18R1. When stratifying patients with gout based on serum urate levels, specific proteins, such as FGF-21 and CCL25, were significantly overexpressed in hyperuricemic gout. Further experiments using PBMCs demonstrated that rhFGF-21 treatment reduced the production of IL-1β and IL-1Ra in cells stimulated with palmitate and MSU, pointing toward a role in the modulation of gout-related inflammation. Serum proteomes of flaring and non-flaring gout patients also revealed several differences, and certain proteins correlated to the extent of joint involvement.
Lastly, in patients treated with urate-lowering therapy and who reached the target serum urate after three months, eight proteins associated with hyperuricemia were significantly downregulated, indicating a reversal of inflammatory effects.
This report shows that the hyperuricemia status is associated with a broad inflammatory signature in individuals with gout and non-gout controls. It identifies markers associated with gout sub-phenotypes and prompts future studies into the causal roles of asymptomatic hyperuricemia for inflammatory complications.
3.3. Metabolome
A noteworthy paper published by Wang et al. [35] assessed differentially abundant metabolites and pathways to discover differences between patients with frequent gout flares versus infrequent flaring patients. The authors performed untargeted metabolomics in the sera of patients in the discovery cohort, followed by targeted metabolomics in a validation cohort. In total, 116 metabolites were upregulated, and 323 were downregulated. A quantitative enrichment analysis highlighted 57 pathways with significant alterations, primarily in carbohydrate, amino acid, and nucleotide metabolism. The network propagation-based algorithm, FELLA, identified the crosstalk between purine and caffeine metabolism as the most altered sub-network, emphasizing the role of xanthine dehydrogenase (XDH), which was upregulated in the frequent gout flare group. Additionally, connections between a number of amino acid sub-networks and bile acid biosynthesis suggested interactions involving gut bacteria in inflammation. Drug analysis showed limited effects on metabolomic outcomes, with key metabolic alterations remaining significant after adjusting for clinical parameters. Finally, the authors used machine learning algorithms to construct and validate a prediction model centered on a selection of six metabolites, which performed well in discriminating between frequent versus infrequent gout flare groups based on a prediction score (with an AUROC of 0.88 in the discovery cohort and 0.67 in the validation cohort). This report has implications for patient stratification and risk management based on metabolite features.
3.4. Microbiome
Two comprehensive studies have investigated the role of gut microbiota in urate homeostasis.
3.4.1. Gut Anaerobic Bacteria Are Implicated in Urate Metabolism and Risk of Gout
Liu et al. [36] reported identifying numerous uric-acid-consuming gut bacteria across four phyla and pointed out two possible routes for urate metabolism: conversion to xanthine or short-chain fatty acids. Subsequently, the authors used RNA-sequencing analysis to identify a set of uric-acid-inducible genes shared among three phylogenetically distinct bacteria and found that mutations in these genes block urate metabolism. Furthermore, the authors inferred that the genes identified in their study were likely to be specific to anaerobic metabolism. In mice, cecal contents were shown to consume urate in vitro, and the ablation of gut microflora in urate oxidase-deficient mice led to severe hyperuricemia and eventually acute kidney injury, leading to the supposition that gut bacteria might compensate for the loss of uricase in mice.
Seeking to explore the relationship between gut microflora and urate metabolism in humans, they further decided to investigate whether antibiotics that target anaerobic bacteria increase the risk of gout. This was examined via a retrospective study comparing gout incidence over a period of four years in two groups of patients with similar characteristics treated with either an antibiotic that targets aerobic bacteria (trimethoprim/sulfamethoxazole) or one that targets both aerobic and anaerobic microbes (clindamycin). Their findings suggest that the disruption of anaerobic flora might indeed increase the risk of gout.
3.4.2. Atherosclerosis Burden Depends on Gut Microbiome and Is Associated with Urate Levels
Another significant step toward defining the importance of gut bacteria purine metabolism in host health was made by Kasahara et al. [37], who investigated the role of purine-degrading bacteria in the development of atherosclerosis. They hypothesized that the variation in disease burden observed among the strains of the Hybrid Mouse Diversity Panel (HMDP) could be, in part, due to gut microbiome differences. To test this, they colonized germ-free ApoE knockout mice with microbial samples obtained from mouse strains with differing atherosclerosis phenotypes: two of the strains had large lesions, while another two showed few signs of disease. Their findings support their hypothesis as, after 8 weeks, disease progression in the two groups of recipient mice was different, with those receiving bacteria from the more atherosclerosis-prone donors having more advanced lesions. Furthermore, after performing metabolome analysis of plasma samples, the authors found purine metabolites that positively correlated with atherosclerotic lesion size. These metabolites included xanthine, xanthosine, inosine, and uric acid.
In a study examining the relationships between urate, atherosclerosis, and gut bacteria in a human cohort, their analysis revealed higher urate levels in individuals with coronary artery calcification. Using XGBoost regression, the study identified Clostridia-class gut bacteria negatively associated with urate, suggesting an influence of the gut microbiome on urate levels.
Next, the researchers aimed to identify bacteria capable of anaerobic purine degradation, a process that had been discussed but not thoroughly identified in human or mouse gut isolates. Through anaerobic enrichments and screenings, they isolated strains from the Bacillota (Firmicutes) and Pseudomonadota (Proteobacteria) phyla, including Enterocloster bolteae and Escherichia coli. The experiment confirmed the anaerobic growth and uric acid utilization of these strains, highlighting differences in purine utilization among strains, and explored factors influencing purine utilization. They also found that the ability to use urate was diminished or eliminated in media containing glucose or fructose, indicating regulation by catabolite repression.
In order to assess the effect of purine-degrading bacteria (PDB) in vivo, Kasahara et al. [37] used two groups of germ-free mice, one of which was colonized solely with species that did not degrade purines, while the other received the same bacteria but with the addition of PDB. Analysis of cecal contents revealed that mice with the “core plus PDB” community had higher levels of urate but lower levels of other purines, while plasma analysis showed consistently lower levels of several purine metabolites, including UA, suggesting that PDB influenced purine levels locally in the gut and systemically in circulation.
The in vitro growth of PDB on a medium supplemented with urate promoted a higher expression of 51 genes (compared to the same species grown on xylose), including some involved in micronutrient transport, glycine cleavage, and Se-dependent hydrolytic reactions. A gene cluster with conserved elements in purine-fermenting organisms was identified. In vivo experiments with germ-free mice showed that colonization with a wild-type strain of E. coli MS 200-1 led to lower plasma urate levels compared to colonization with a deletion variant, suggesting the role of these genes in urate homeostasis in vivo.
Performing BLASTP of the NCBI RefSeq Genome Database, they identified 230 bacterial taxa, with five genes detected among all confirmed PDB. The abundance of these genes in the cecum of gnotobiotic mice correlated negatively with cecal levels of purine-related metabolites. The findings suggest the potential of these genes as biomarkers for purine breakdown in the gut, offering insights for potential interventions in conditions related to hyperuricemia.
Overall, these recent studies have given insights into the mechanisms of bacterial urate degradation in the gut, offering evidence that this activity plays a crucial role in reducing circulating urate levels. While further research is needed, these findings pave the way for future microbiome-focused studies aimed at preventing or treating gout, atherosclerosis, or other diseases associated with urate.
4. Closing Remarks
Recent work in the field of gout basic science has been marked by an array of original research assessing mechanisms of inflammation (both pro- and anti-inflammatory) or exploring biological systems (single-cell transcriptomics, proteomics, metabolomics, and microbiomes). The papers highlighted in this review contribute to our understanding of the pathophysiology of gout and reveal emerging treatment options. Omics approaches are increasingly being employed to comprehensively study molecular signatures associated with gout. As more datasets are available, multi-omics integrative approaches are expected, with exciting prospects for assessing the interplay of different molecular layers and interindividual differences in gout basic and translational research.
Author Contributions
Conceptualization, M.M. and T.O.C.; writing—original draft preparation, M.M.; writing—review and editing, L.A.B.J. and T.O.C.; supervision, L.A.B.J. and T.O.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Romania’s National Recovery and Resilience Plan grant from the Romanian Ministry of Investments and European Projects (PNRR-III-C9-2022-I8, CF 85/15.11.2022).
Data Availability Statement
Data sharing is not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Dalbeth, N.; Choi, H.K.; Joosten, L.A.; Khanna, P.P.; Matsuo, H.; Perez-Ruiz, F.; Stamp, L.K. Gout (primer). Nat. Rev. Dis. Primers 2019, 5, 69. [Google Scholar] [CrossRef]
- Choi, H.K.; Mount, D.B.; Reginato, A.M. Pathogenesis of Gout. 2005. Available online: www.annals.org (accessed on 20 March 2024).
- Szekanecz, Z.; Szamosi, S.; Kovács, G.E.; Kocsis, E.; Benkő, S. The NLRP3 inflammasome—Interleukin 1 pathway as a therapeutic target in gout. Arch. Biochem. Biophys. 2019, 670, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Klück, V.; Liu, R.; Joosten, L.A.B. The role of interleukin-1 family members in hyperuricemia and gout. Jt. Bone Spine 2020, 88, 105092. [Google Scholar] [CrossRef]
- Joosten, L.A.B.; Crişan, T.O.; Bjornstad, P.; Johnson, R.J. Asymptomatic hyperuricaemia: A silent activator of the innate immune system. Nat. Rev. Rheumatol. 2020, 16, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Popov, D.; Jain, L.; Alhilali, M.; Dalbeth, N.; Poulsen, R.C. Monosodium urate crystals alter the circadian clock in macrophages leading to loss of NLRP3 inflammasome repression: Implications for timing of the gout flare. FASEB J. 2023, 37, e22940. [Google Scholar] [CrossRef]
- Futosi, K.; Németh, T.; Horváth, Á.I.; Abram, C.L.; Tusnády, S.; Lowell, C.A.; Helyes, Z.; Mócsai, A. Myeloid Src-family kinases are critical for neutrophil-mediated autoinflammation in gout and motheaten models. J. Exp. Med. 2023, 220, e20221010. [Google Scholar] [CrossRef]
- Corcoran, S.E.; O’Neill, L.A.J. HIF1α and metabolic reprogramming in inflammation. J. Clin. Investig. 2016, 126, 3699–3707. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.J.; Kishton, R.J.; Rathmell, J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016, 16, 553–565. [Google Scholar] [CrossRef] [PubMed]
- Cobo, I.; Murillo-Saich, J.; Alishala, M.; Guma, M. Epigenetic and Metabolic Regulation of Macrophages during Gout. Gout Urate Cryst. Depos. Dis. 2023, 1, 137–151. [Google Scholar] [CrossRef]
- Chen, L.; Chen, Y.; Chien, W.; Tsai, K.; Chung, C.; Wang, J.; Chen, C.; Liao, N.; Shih, C.; Lin, Y.; et al. Inactivation of mitochondrial pyruvate carrier promotes NLRP3 inflammasome activation and gout development via metabolic reprogramming. Immunology 2023, 169, 271–291. [Google Scholar] [CrossRef]
- Ye, Z.; Zhang, J.; Xu, Z.; Li, Z.; Huang, G.; Tong, B.; Xia, P.; Shen, Y.; Hu, H.; Yu, P.; et al. Pioglitazone ameliorates ischemia/reperfusion-induced acute kidney injury via oxidative stress attenuation and NLRP3 inflammasome. Hum. Cell 2024, 37, 959–971. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, B.; Wang, L.; Yang, M.; Xia, Z.; Wei, W.; Zhang, F.; Yuan, X. Pioglitazone ameliorates glomerular NLRP3 inflammasome activation in apolipoprotein E knockout mice with diabetes mellitus. PLoS ONE 2017, 12, e0181248. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, C.; Zhang, D.; Zhu, Y. Pioglitazone Protects Against Hypoxia-Induced Cardiomyocyte Apoptosis Through Inhibiting NLRP3/Caspase-1 Pathway in vivo and in vitro. Int. Heart J. 2022, 63, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Klück, V.; Cabău, G.; Mies, L.; Bukkems, F.; Van Emst, L.; Bakker, R.; Van Caam, A.; Pop, I.V.; Popp, R.A.; Rednic, S.; et al. TGF-β is elevated in hyperuricemic individuals and mediates urate-induced hyperinflammatory phenotype in human mononuclear cells. Arthritis Res. Ther. 2023, 25, 30. [Google Scholar] [CrossRef] [PubMed]
- Elsaid, K.; Merriman, T.R.; Rossitto, L.; Liu-Bryan, R.; Karsh, J.; Phipps-Green, A.; Jay, G.D.; Elsayed, S.; Qadri, M.; Miner, M.; et al. Amplification of Inflammation by Lubricin Deficiency Implicated in Incident, Erosive Gout Independent of Hyperuricemia. Arthritis Rheumatol. 2023, 75, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Waller, K.A.; Zhang, L.X.; Elsaid, K.A.; Fleming, B.C.; Warman, M.L.; Jay, G.D. Role of lubricin and boundary lubrication in the prevention of chondrocyte apoptosis. Proc. Natl. Acad. Sci. USA 2013, 110, 5852–5857. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhang, B.; Chen, Y.; Zeng, F.; Wang, W.; Chen, Z.; Cao, L.; Shi, J.; Chen, J.; Zhu, X.; et al. Type II collagen facilitates gouty arthritis by regulating MSU crystallisation and inflammatory cell recruitment. Ann. Rheum. Dis. 2023, 82, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Devi, S.; Indramohan, M.; Jäger, E.; Carriere, J.; Chu, L.H.; De Almeida, L.; Greaves, D.R.; Stehlik, C.; Dorfleutner, A. CARD-only proteins regulate in vivo inflammasome responses and ameliorate gout. Cell Rep. 2023, 42, 112265. [Google Scholar] [CrossRef] [PubMed]
- Mirzaesmaeili, A.; Zangiabadi, S.; Raspanti, J.; Akram, A.; Inman, R.D.; Abdul-Sater, A.A. Cutting Edge: Negative Regulation of Inflammasome Activation by TRAF1 Can Limit Gout. J. Immunol. 2023, 210, 531–535. [Google Scholar] [CrossRef]
- Richette, P.; Doherty, M.; Pascual, E.; Barskova, V.; Becce, F.; Castañeda-Sanabria, J.; Coyfish, M.; Guillo, S.; Jansen, T.L.; Janssens, H.; et al. 2016 updated EULAR evidence-based recommendations for the management of gout. Ann. Rheum. Dis. 2017, 76, 29–42. [Google Scholar] [CrossRef]
- FitzGerald, J.D.; Dalbeth, N.; Mikuls, T.; Brignardello-Petersen, R.; Guyatt, G.; Abeles, A.M.; Gelber, A.C.; Harrold, L.R.; Khanna, D.; King, C.; et al. 2020 American College of Rheumatology Guideline for the Management of Gout. Arthritis Care Res. 2020, 72, 744–760. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Shan, L.; Wang, H.; Schauer, C.; Schoen, J.; Zhu, L.; Lu, C.; Wang, Z.; Xue, Y.; Wu, H.; et al. Neutrophil Extracellular Trap–Borne Elastase Prevents Inflammatory Relapse in Intercritical Gout. Arthritis Rheumatol. 2023, 75, 1039–1047. [Google Scholar] [CrossRef] [PubMed]
- Leask, M.P.; Merriman, T.R. The genetic basis of urate control and gout: Insights into molecular pathogenesis from follow-up study of genome-wide association study loci. Best Pract. Res. Clin. Rheumatol. 2021, 35, 101721. [Google Scholar] [CrossRef]
- Wrigley, R.; Phipps-Green, A.J.; Topless, R.K.; Major, T.J.; Cadzow, M.; Riches, P.; Tausche, A.K.; Janssen, M.; Joosten LA, B.; Jansen, T.L.; et al. Pleiotropic effect of the ABCG2 gene in gout: Involvement in serum urate levels and progression from hyperuricemia to gout. Arthritis Res. Ther. 2020, 22, 45. [Google Scholar] [CrossRef] [PubMed]
- Wrigley, R.; Phipps-Green, A.J.; Topless, R.K.; Major, T.J.; Cadzow, M.; Riches, P.; Tausche, A.-K.; Janssen, M.; Joosten, L.A.B.; Jansen, T.L.; et al. Intestinal uric acid excretion contributes to serum uric acid decrease during acute gout attack. Rheumatology 2023, 62, 3984–3992. [Google Scholar]
- Zhang, J.; Sun, W.; Gao, F.; Lu, J.; Li, K.; Xu, Y.; Li, Y.; Li, C.; Chen, Y. Changes of serum uric acid level during acute gout flare and related factors. Front. Endocrinol. 2023, 14, 1077059. [Google Scholar] [CrossRef] [PubMed]
- Notsu, T.; Kurata, Y.; Ninomiya, H.; Taufiq, F.; Komatsu, K.; Miake, J.; Sawano, T.; Tsuneto, M.; Shirayoshi, Y.; Hisatome, I. Inhibition of the uric acid efflux transporter ABCG2 enhances stimulating effect of soluble uric acid on IL-1β production in murine macrophage-like J774.1 cells. Hypertens. Res. 2023, 46, 2368–2377. [Google Scholar] [CrossRef] [PubMed]
- Ahn, M.; Chen, V.C.-W.; Rozario, P.; Ng, W.L.; Kong, P.S.; Sia, W.R.; Kang, A.E.Z.; Su, Q.; Nguyen, L.H.; Zhu, F.; et al. Bat ASC2 suppresses inflammasomes and ameliorates inflammatory diseases. Cell 2023, 186, 2144–2159.e22. [Google Scholar] [CrossRef]
- Colantuoni, M.; Jofra Hernandez, R.; Pettinato, E.; Basso-Ricci, L.; Magnani, L.; Andolfi, G.; Rigamonti, C.; Finardi, A.; Romeo, V.; Soldi, M.; et al. Constitutive IL-1RA Production by Modified Immune Cells Protects against IL-1-Mediated Inflammatory Disorders. 2023. Available online: https://www.science.org (accessed on 4 April 2024).
- Chang, J.-G.; Tu, S.-J.; Huang, C.-M.; Chen, Y.-C.; Chiang, H.-S.; Lee, Y.-T.; Yen, J.-C.; Lin, C.-L.; Chung, C.-C.; Liu, T.-C.; et al. Single-cell RNA sequencing of immune cells in patients with acute gout. Sci. Rep. 2022, 12, 22130. [Google Scholar] [CrossRef]
- Gu, H.; Yu, H.; Qin, L.; Yu, H.; Song, Y.; Chen, G.; Zhao, D.; Wang, S.; Xue, W.; Wang, L.; et al. MSU crystal deposition contributes to inflammation and immune responses in gout remission. Cell Rep. 2023, 42, 113139. [Google Scholar] [CrossRef]
- Wang, M.; Chen, W.; Zhang, X.; Mei, L.; Wu, X.; Chen, X.; Yang, Z.; Gao, K.; Huang, H.; Huang, R. Single-Cell Analysis in Blood Reveals Distinct Immune Cell Profiles in Gouty Arthritis. J. Immunol. 2023, 210, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Cabău, G.; Gaal, O.; Badii, M.; Nica, V.; Mirea, A.-M.; Hotea, I.; Pamfil, C.; Popp, R.A.; Netea, M.G.; Rednic, S.; et al. Hyperuricemia remodels the serum proteome toward a higher inflammatory state. iScience 2023, 26, 107909. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, R.; Qi, H.; Pang, L.; Cui, L.; Liu, Z.; Lu, J.; Wang, R.; Hu, S.; Liang, N.; et al. Metabolomics and Machine Learning Identify Metabolic Differences and Potential Biomarkers for Frequent Versus Infrequent Gout Flares. Arthritis Rheumatol. 2023, 75, 2252–2264. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jarman, J.B.; Low, Y.S.; Augustijn, H.E.; Huang, S.; Chen, H.; DeFeo, M.E.; Sekiba, K.; Hou, B.-H.; Meng, X.; et al. A widely distributed gene cluster compensates for uricase loss in hominids. Cell 2023, 186, 3400–3413.e20. [Google Scholar] [CrossRef]
- Kasahara, K.; Kerby, R.L.; Zhang, Q.; Pradhan, M.; Mehrabian, M.; Lusis, A.J.; Bergström, G.; Bäckhed, F.; Rey, F.E. Gut bacterial metabolism contributes to host global purine homeostasis. Cell Host Microbe 2023, 31, 1038–1053.e10. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).