Relevance of AKT and RAS Signaling Pathways for Antibody–Drug Conjugate Immunotherapies in Acute Lymphoblastic Leukemia
Abstract
1. Introduction
- Antibody-Based Immunotherapies and Their Great but Underutilized Potential in ALL
1.1. Current Challenges in ALL Treatment
1.2. Role of Antibody–Drug Conjugates
1.3. Key Signaling Pathways and Feedback Loops
2. The AKT and MAPK Pathway—Important Intracellular Regulatory Nodes
2.1. The AKT Pathway
2.2. The MAPK Pathway
3. Feedback Signaling of the PI3K/AKT and RAS/MAPK Pathways Influence the Regulation of Cell Surface Markers
4. Identification of ALL Subgroup-Specific Expression of Cell Surface Markers
| Subtype | Gene 1 | Protein ID | Normal Tissue Expression | Pathway | Gene 2 | Protein ID | Normal Tissue Expression | Pathway | Gene 3 | Protein ID | Normal Tissue Expression | Pathway |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KMT2A-r | CSPG4 | CSPG-4 | Intestine | MAPK; AKT | PSD2 | PSD-2 | Brain | n.a. | PCDHGC3 | PCDH-gamma-C3 | Brain | MAPK |
| ETV6-RUNX1 | DSC3 | DSC-3 | Esophagus, Skin | MAPK; AKT | LRRC4B | NGL-3 | Brain, Cervix, Pituitary gland | MAPK; AKT | PTPRK | R-PTP-kappa | Low tissue specificity | n.a. |
| TCF3-HLF | CLSTN2 | CLSTN2 | Brain, Ovary | n.a. | LPAR1 | LPA-1 | Brain | MAPK; AKT | F3 | TF/CD142 | Low tissue specificity | MAPK; AKT |
| Hyperdiploid (HeH) | EFNB1 | Ephrin-B1 | Low tissue specificity | MAPK | IL3RA | IL-3R-alpha/CD123 | Low tissue specificity | MAPK; AKT | IL13RA1 | IL-13R-alpha/CD213a1 | Low tissue specificity | MAPK; AKT |
| IKZF1-N159Y | TRPC4 | TrpC4 | Endometrium, Seminal vesicle, Smooth muscle | MAPK; AKT | ATP1A2 | ATP1A2 | Brain, Skeletal muscle, Tongue | n.a. | CACNG4 | TARP gamma-4 | Brain | n.a. |
| Low hyperdiploid (L-HD) | PDGFRB | PDGF-R-beta/CD140B | Low tissue specificity | MAPK; AKT | SLC6A9 | GlyT-1 | Brain, Skin | n.a. | DDR2 | CD167b | Low tissue specificity | MAPK; AKT |
| MEF2D | PTPRZ1 | R-PTP-zeta | Brain | AKT | TNFRSF13B | CD267 | Intestine, Lymphoid tissue, Skeletal muscle | n.a. | DCHS2 | Cadherin-27 | Brain, Intestine | n.a. |
| PAX5alt | ERBB2 | HER2/CD340 | Low tissue specificity | MAPK; AKT | VIPR2 | VIP-R-2 | Low tissue specificity | MAPK; AKT | MS4A1 | CD20 | Lymphoid tissue | MAPK; AKT |
| Ph-like | CRLF2 | CRLF-2 | Bone marrow, Gallbladder, Lymphoid tissue | AKT | TTYH2 | hTTY2 | Brain | n.a. | CHRNA1 | CHRNA-1 | Skeletal muscle | n.a. |
| PAX5-P80R | MEGF10 | MEGF-10 | Brain, Retina | n.a. | TNFRSF13B | CD267 | Intestine, Lymphoid tissue, Skeletal muscle | n.a. | PDGFRB | PDGF-R-beta/CD140B | Low tissue specificity | MAPK; AKT |
| TCF3-PBX1 | LRRC15 | LRRC-15 | Lymphoid tissue, Skin | n.a. | ITGA8 | ITG-alpha-8 | Prostate | AKT | SLAMF1 | IPO-3/CD150 | Lymphoid tissue | AKT |
| Ph-pos | IL2RA | CD25 | Adipose tissue, Lymphoid tissue, Urinary bladder | MAPK; AKT | TSPAN15 | Tspan-15 | Low tissue specificity | MAPK | OSMR | IL31RB | Low tissue specificity | MAPK; AKT |
| ZNF384 | XKR3 | XKR-3 | Testis | n.a. | SLC2A3 | GLUT-3 | Bone marrow | MAPK; AKT | CD226 | DNAM-1/CD226 | Lymphoid tissue, Parathyroid gland | MAPK; AKT |
| DUX4 | PTPRM | R-PTP-mu | Low tissue specificity | MAPK; AKT | MCAM | MUC18/CD146 | Low tissue specificity | MAPK; AKT | CLEC12B | CLEC-12B | Bone marrow, Skin, Testis | MAPK; AKT |
| BCL2-MYC | CNR1 | CB-1 | Adipose tissue, Pituitary gland | MAPK; AKT | CD70 | CD70 | Lymphoid tissue | MAPK; AKT | BEST3 | Bestrophin-3 | Skeletal muscle, Tongue | MAPK |
| T-ALL | CD1B | CD1b | Lymphoid tissue | n.a. | NOTCH3 | Notch-3 | Low tissue specificity | MAPK; AKT | CCR9 | CCR-9/CD199 | Lymphoid tissue | MAPK; AKT |
| Subtype | Gene 1 | ADC | Reference | Gene 2 | ADC | Reference | Gene 3 | ADC | Reference |
|---|---|---|---|---|---|---|---|---|---|
| KMT2A-r | CSPG4 | anti-CSPG4-(PDD); anti-CSPG4(scFv)-SNAP-AURIF | [108,109], experimental | PSD2 | No | PCDHGC3 | No | ||
| ETV6-RUNX1 | DSC3 | No | LRRC4B | No | PTPRK | No | |||
| TCF3-HLF | CLSTN2 | No | LPAR1 | No | F3 | Tisotumab vedotin | [110], approved | ||
| Hyperdiploid (HeH) | EFNB1 | No | IL3RA | BAY-943 | [111], experimental | IL13RA1 | No | ||
| IKZF1-N159Y | TRPC4 | No | ATP1A2 | No | CACNG4 | No | |||
| Low hyperdiploid (L-HD) | PDGFRB | No | SLC6A9 | No | DDR2 | No | |||
| MEF2D | PTPRZ1 | No | TNFRSF13B | anti-TNFRSF13B (Tabalumab)-MC-Vc-PAB-MMAE | CreativeBiolabs, CAT#ADC-W-1907 experimental | DCHS2 | No | ||
| PAX5alt | ERBB2 | Trastuzumab deruxtecan; Trastuzumab emtansine | [112,113], approved | VIPR2 | No | MS4A1 | MRG001 | [114], NCT05155839 (Phase 1) | |
| Ph-like | CRLF2 | No | TTYH2 | No | CHRNA1 | No | |||
| PAX5-P80R | MEGF10 | No | TNFRSF13B | anti-TNFRSF13B (Tabalumab)-MC-Vc-PAB-MMAE | CreativeBiolabs, CAT#ADC-W-1907 experimental | PDGFRB | No | ||
| TCF3-PBX1 | LRRC15 | ABBV-085 | [115], NCT02565758 (Phase 1) | ITGA8 | No | SLAMF1 | No | ||
| Ph-pos | IL2RA | Camidanlumab tesirine | [116], NCT04052997 (Phase 2) | TSPAN15 | No | OSMR | No | ||
| ZNF384 | XKR3 | No | SLC2A3 | anti-SLC3A2 ADC (19G4-MMAE); | [117], experimental | CD226 | No | ||
| DUX4 | PTPRM | No | MCAM | AMT-253 | [118], NCT06209580 (Phase 1/2) | CLEC12B | No | ||
| BCL2-MYC | CNR1 | No | CD70 | PRO1160; Vorsetuzumab mafodotin (SGN-75) | [119], NCT05721222 (Phase 1/2), NCT01015911 (Phase 1) | BEST3 | No | ||
| T-ALL | CD1B | No | NOTCH3 | PF-06650808 | [120], NCT02129205 (Phase 1) | CCR9 | No |
5. Introduction of New ADC Target Genes
5.1. KMT2A-r (CSPG4, PSD2, PCDHGC3)
5.2. ETV6-RUNX1 (DSC3, LRRC4B, PTPRK)
5.3. TCF-HLF (CLSTN2, LPAR1, F3)
5.4. Hyperdiploid (EFNB1, IL3RA, IL13RA)
5.5. IKZF1-N159Y (TRPC4, ATP1A2, CACNG4)
5.6. Low Hyperdiploid (PDGFRB, SLC6A9, DDR2)
5.7. MEF2D (PTPRZ1, TNFRSF13B, DCHS2)
5.8. PAX5alt (ERBB2, VIPR2, MS4A1)
5.9. Ph-like (CRLF2, TTYH2, CHRNA1)
5.10. PAX5-P80R (MEGF10, PDGFRB, TNFRSF13B)
5.11. TCF-PBX1 (LRRC15, ITGA8, SLAMF1)
5.12. Ph-pos (IL2RA, TSPAN15, OSMR)
5.13. ZNF384 (XKR3, SLC2A3, CD226)
5.14. DUX4 (PTPRM, MCAM, CLEC12B)
5.15. BCL2-MYC (CNR1, CD70, BEST3)
5.16. T-ALL (CD1B, NOTCH3, CCR9)
6. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Pui, C.H.; Yang, J.J.; Bhakta, N.; Rodriguez-Galindo, C. Global efforts toward the cure of childhood acute lymphoblastic leukaemia. Lancet Child Adolesc. Health 2018, 2, 440–454, Erratum in Lancet Child Adolesc. Health 2018, 2, e25. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Berkman, A.M.; Andersen, C.R.; Cuglievan, B.; McCall, D.C.; Lupo, P.J.; Parsons, S.K.; DiNardo, C.D.; Short, N.J.; Jain, N.; Kadia, T.M.; et al. Long-Term Outcomes among Adolescent and Young Adult Survivors of Acute Leukemia: A Surveillance, Epidemiology, and End Results Analysis. Cancer Epidemiol. Biomark. Prev. 2022, 31, 1176–1184. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rheingold, S.R.; Bhojwani, D.; Ji, L.; Xu, X.; Devidas, M.; Kairalla, J.A.; Shago, M.; Heerema, N.A.; Carroll, A.J.; Breidenbach, H.; et al. Determinants of survival after first relapse of acute lymphoblastic leukemia: A Children’s Oncology Group study. Leukemia 2024, 38, 2382–2394. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Luskin, M.R. Acute lymphoblastic leukemia in older adults: Curtain call for conventional chemotherapy? Hematol. Am. Soc. Hematol. Educ. Program 2021, 2021, 7–14. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kantarjian, H.; Jabbour, E. Adult Acute Lymphoblastic Leukemia: 2025 Update on Diagnosis, Therapy, and Monitoring. Am. J. Hematol. 2025, 100, 1205–1231. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhu, H.H. Targeted treatment of T-cell acute lymphoblastic leukemia: Latest updates from the 2022 ASH Annual Meeting. Exp. Hematol. Oncol. 2023, 12, 30. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kuster, L.; Grausenburger, R.; Fuka, G.; Kaindl, U.; Krapf, G.; Inthal, A.; Mann, G.; Kauer, M.; Rainer, J.; Kofler, R.; et al. ETV6/RUNX1-positive relapses evolve from an ancestral clone and frequently acquire deletions of genes implicated in glucocorticoid signaling. Blood 2011, 117, 2658–2667. [Google Scholar] [CrossRef] [PubMed]
- Kantarjian, H.M.; DeAngelo, D.J.; Stelljes, M.; Martinelli, G.; Liedtke, M.; Stock, W.; Gökbuget, N.; O’Brien, S.; Wang, K.; Wang, T.; et al. Inotuzumab Ozogamicin versus Standard Therapy for Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2016, 375, 740–753. [Google Scholar] [CrossRef] [PubMed]
- Kantarjian, H.M.; Stein, A.; Gökbuget, N.; Fielding, A.K.; Schuh, A.C.; Ribera, J.-M.; Wei, A.; Dombret, H.; Foà, R.; Bassan, R.; et al. Blinatumomab versus Chemotherapy for Advanced Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2017, 376, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Tomuleasa, C.; Tigu, A.B.; Munteanu, R.; Moldovan, C.S.; Kegyes, D.; Onaciu, A.; Gulei, D.; Ghiaur, G.; Einsele, H.; Croce, C.M. Therapeutic advances of targeting receptor tyrosine kinases in cancer. Signal Transduct. Target. Ther. 2024, 9, 201. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gore, L.; Locatelli, F.; Zugmaier, G.; Handgretinger, R.; O’bRien, M.M.; Bader, P.; Bhojwani, D.; Schlegel, P.-G.; Tuglus, C.A.; von Stackelberg, A. Survival after blinatumomab treatment in pediatric patients with relapsed/refractory B-cell precursor acute lymphoblastic leukemia. Blood Cancer J. 2018, 8, 80. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- van der Sluis, I.M.; de Lorenzo, P.; Kotecha, R.S.; Attarbaschi, A.; Escherich, G.; Nysom, K.; Stary, J.; Ferster, A.; Brethon, B.; Locatelli, F.; et al. Blinatumomab Added to Chemotherapy in Infant Lymphoblastic Leukemia. N. Engl. J. Med. 2023, 388, 1572–1581. [Google Scholar] [CrossRef]
- Dumontet, C.; Reichert, J.M.; Senter, P.D.; Lambert, J.M.; Beck, A. Antibody-drug conjugates come of age in oncology. Nat. Rev. Drug Discov. 2023, 22, 641–661. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Gillespie, E.; Naqvi, A.S.; Hayer, K.E.; Ang, Z.; Torres-Diz, M.; Quesnel-Vallières, M.; Hottman, D.A.; Bagashev, A.; Chukinas, J.; et al. Modulation of CD22 Protein Expression in Childhood Leukemia by Pervasive Splicing Aberrations: Implications for CD22-Directed Immunotherapies. Blood Cancer Discov. 2022, 3, 103–115. [Google Scholar] [CrossRef]
- Talleur, A.C.; Pui, C.H.; Karol, S.E. What is Next in Pediatric B-cell Precursor Acute Lymphoblastic Leukemia. Lymphatics 2023, 1, 34–44. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lu, Y.; Zhao, F. Strategies to overcome tumour relapse caused by antigen escape after CAR T therapy. Mol. Cancer 2025, 24, 126. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bosi, C.; Bartha, Á.; Galbardi, B.; Notini, G.; Naldini, M.M.; Licata, L.; Viale, G.; Mariani, M.; Pistilli, B.; Ali, H.R.; et al. Pan-cancer analysis of antibody-drug conjugate targets and putative predictors of treatment response. Eur. J. Cancer 2023, 195, 113379. [Google Scholar] [CrossRef] [PubMed]
- Razzaghdoust, A.; Rahmatizadeh, S.; Mofid, B.; Muhammadnejad, S.; Parvin, M.; Torbati, P.M.; Basiri, A. Data-Driven Discovery of Molecular Targets for Antibody-Drug Conjugates in Cancer Treatment. BioMed. Res. Int. 2021, 2021, 2670573. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kathad, U.; Biyani, N.; Peru YColón De Portugal, R.L.; Zhou, J.; Kochat, H.; Bhatia, K. Expanding the repertoire of Antibody Drug Conjugate (ADC) targets with improved tumor selectivity and range of potent payloads through in-silico analysis. PLoS ONE 2024, 19, e0308604. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xie, J.; Weiskirchen, R. What Does the “AKT” Stand for in the Name “AKT Kinase”? Some Historical Comments. Front. Oncol. 2020, 10, 1329. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hassan, D.; Menges, C.W.; Testa, J.R.; Bellacosa, A. AKT kinases as therapeutic targets. J. Exp. Clin. Cancer Res. 2024, 43, 313. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Corre, E.; Soum, C.; Pfeifer, R.; Bessière, C.; Dailhau, S.; Marbœuf, C.; Meggetto, F.; Touriol, C.; Récher, C.; Bousquet, M.; et al. Differential prognostic values of the three AKT isoforms in acute myeloid leukemia. Sci. Rep. 2024, 14, 7070. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Geißert, R.; Lammert, A.; Wirth, S.; Hönig, R.; Lohfink, D.; Unger, M.; Pek, D.; Schlüter, K.; Scheftschik, T.; Smit, D.J.; et al. K-Ras(V12) differentially affects the three Akt isoforms in lung and pancreatic carcinoma cells and upregulates E-cadherin and NCAM via Akt3. Cell Commun. Signal. 2024, 22, 85. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cho, H.; Thorvaldsen, J.L.; Chu, Q.; Feng, F.; Birnbaum, M.J. Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J. Biol. Chem. 2001, 276, 38349–38352. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Mu, J.; Kim, J.K.; Thorvaldsen, J.L.; Chu, Q.; Crenshaw EB3rd Kaestner, K.H.; Bartolomei, M.S.; Shulman, G.I.; Birnbaum, M.J. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta). Science 2001, 292, 1728–1731. [Google Scholar] [CrossRef]
- Tschopp, O.; Yang, Z.Z.; Brodbeck, D.; Dummler, B.A.; Hemmings-Mieszczak, M.; Watanabe, T.; Michaelis, T.; Frahm, J.; Hemmings, B.A. Essential role of protein kinase B gamma (PKB gamma/Akt3) in postnatal brain development but not in glucose homeostasis. Development 2005, 132, 2943–2954. [Google Scholar] [CrossRef]
- Ehm, P.; Grottke, A.; Bettin, B.; Jücker, M. Investigation of the function of the PI3-Kinase / AKT signaling pathway for leukemogenesis and therapy of acute childhood lymphoblastic leukemia (ALL). Cell Signal. 2022, 93, 110301. [Google Scholar] [CrossRef] [PubMed]
- Szymonowicz, K.; Oeck, S.; Malewicz, N.M.; Jendrossek, V. New Insights into Protein Kinase B/Akt Signaling: Role of Localized Akt Activation and Compartment-Specific Target Proteins for the Cellular Radiation Response. Cancers 2018, 10, 78. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Carpten, J.D.; Faber, A.L.; Horn, C.; Donoho, G.P.; Briggs, S.L.; Robbins, C.M.; Hostetter, G.; Boguslawski, S.; Moses, T.Y.; Savage, S.; et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 2007, 448, 439–444. [Google Scholar] [CrossRef]
- Silva, A.; Yunes, J.A.; Cardoso, B.A.; Martins, L.R.; Jotta, P.Y.; Abecasis, M.; Nowill, A.E.; Leslie, N.R.; Cardoso, A.A.; Barata, J.T. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J. Clin. Investig. 2008, 118, 3762–3774. [Google Scholar] [CrossRef]
- Gomes, A.M.; Soares, M.V.; Ribeiro, P.; Caldas, J.; Póvoa, V.; Martins, L.R.; Melão, A.; Serra-Caetano, A.; de Sousa, A.B.; Lacerda, J.F.; et al. Adult B-cell acute lymphoblastic leukemia cells display decreased PTEN activity and constitutive hyperactivation of PI3K/Akt pathway despite high PTEN protein levels. Haematologica 2014, 99, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Ehm, P.A.H.; Horn, S.; Hoffer, K.; Kriegs, M.; Horn, M.; Giehler, S.; Nalaskowski, M.; Rehbach, C.; Horstmann, M.A.; Jücker, M. Ikaros sets the threshold for negative B-cell selection by regulation of the signaling strength of the AKT pathway. Cell Commun. Signal. 2024, 22, 360. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Riggio, M.; Perrone, M.C.; Polo, M.L.; Rodriguez, M.J.; May, M.; Abba, M.; Lanari, C.; Novaro, V. AKT1 and AKT2 isoforms play distinct roles during breast cancer progression through the regulation of specific downstream proteins. Sci. Rep. 2017, 7, 44244. [Google Scholar] [CrossRef]
- Wang, L.; Ji, X.B.; Wang, L.H.; Qiu, J.G.; Zhou, F.M.; Liu, W.J.; Wan, D.D.; Lin, M.C.; Liu, L.Z.; Zhang, J.Y.; et al. Regulation of MicroRNA-497-Targeting AKT2 Influences Tumor Growth and Chemoresistance to Cisplatin in Lung Cancer. Front. Cell Dev. Biol. 2020, 8, 840, Erratum in Front. Cell Dev. Biol. 2022, 10, 1008088. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dilawari, A.; Buturla, J.; Osgood, C.; Gao, X.; Chen, W.; Ricks, T.K.; Schaefer, T.; Avasarala, S.; Reyes Turcu, F.; Pathak, A.; et al. US Food and Drug Administration Approval Summary: Capivasertib With Fulvestrant for Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Locally Advanced or Metastatic Breast Cancer With PIK3CA/AKT1/PTEN Alterations. J. Clin. Oncol. 2024, 42, 4103–4113. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Whitley, M.J.; Tran, T.H.; Rigby, M.; Yi, M.; Dharmaiah, S.; Waybright, T.J.; Ramakrishnan, N.; Perkins, S.; Taylor, T.; Messing, S.; et al. Comparative analysis of KRAS4a and KRAS4b splice variants reveals distinctive structural and functional properties. Sci. Adv. 2024, 10, eadj4137. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nuevo-Tapioles, C.; Philips, M.R. The role of KRAS splice variants in cancer biology. Front. Cell Dev. Biol. 2022, 10, 1033348. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Holderfield, M.; Lee, B.J.; Jiang, J.; Tomlinson, A.; Seamon, K.J.; Mira, A.; Patrucco, E.; Goodhart, G.; Dilly, J.; Gindin, Y.; et al. Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy. Nature 2024, 629, 919–926. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Irving, J.; Matheson, E.; Minto, L.; Blair, H.; Case, M.; Halsey, C.; Swidenbank, I.; Ponthan, F.; Kirschner-Schwabe, R.; Groeneveld-Krentz, S.; et al. Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood 2014, 124, 3420–3430. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Qian, J.; Li, Z.; Pei, K.; Li, Z.; Li, C.; Yan, M.; Qian, M.; Song, Y.; Zhang, H.; He, Y. Effects of NRAS Mutations on Leukemogenesis and Targeting of Children With Acute Lymphoblastic Leukemia. Front. Cell Dev. Biol. 2022, 10, 712484. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Janes, M.R.; Zhang, J.; Li, L.S.; Hansen, R.; Peters, U.; Guo, X.; Chen, Y.; Babbar, A.; Firdaus, S.J.; Darjania, L.; et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018, 172, 578–589.e17. [Google Scholar] [CrossRef] [PubMed]
- Canon, J.; Rex, K.; Saiki, A.Y.; Mohr, C.; Cooke, K.; Bagal, D.; Gaida, K.; Holt, T.; Knutson, C.G.; Koppada, N.; et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019, 575, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Hallin, J.; Engstrom, L.D.; Hargis, L.; Calinisan, A.; Aranda, R.; Briere, D.M.; Sudhakar, N.; Bowcut, V.; Baer, B.R.; Ballard, J.A.; et al. The KRASG12C Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov. 2020, 10, 54–71. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jain, N.; Curran, E.; Iyengar, N.M.; Diaz-Flores, E.; Kunnavakkam, R.; Popplewell, L.; Kirschbaum, M.H.; Karrison, T.; Erba, H.P.; Green, M.; et al. Phase II study of the oral MEK inhibitor selumetinib in advanced acute myelogenous leukemia: A University of Chicago phase II consortium trial. Clin. Cancer Res. 2014, 20, 490–498. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mozzarelli, A.M.; Simanshu, D.K.; Castel, P. Functional and structural insights into RAS effector proteins. Mol. Cell 2024, 84, 2807–2821, Erratum in Mol. Cell 2024, 84, 3163–3164. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tang, R.; Shuldiner, E.G.; Kelly, M.; Murray, C.W.; Hebert, J.D.; Andrejka, L.; Tsai, M.K.; Hughes, N.W.; Parker, M.I.; Cai, H.; et al. Multiplexed screens identify RAS paralogues HRAS and NRAS as suppressors of KRAS-driven lung cancer growth. Nat. Cell Biol. 2023, 25, 159–169. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Alawieh, D.; Cysique-Foinlan, L.; Willekens, C.; Renneville, A. RAS mutations in myeloid malignancies: Revisiting old questions with novel insights and therapeutic perspectives. Blood Cancer J. 2024, 14, 72. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, S.; Wu, Z.; Li, T.; Li, Y.; Wang, W.; Hao, Q.; Xie, X.; Wan, D.; Jiang, Z.; Wang, C.; et al. Mutational spectrum and prognosis in NRAS-mutated acute myeloid leukemia. Sci. Rep. 2020, 10, 12152. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gelb, B.D.; Tartaglia, M. Noonan syndrome and related disorders: Dysregulated RAS-mitogen activated protein kinase signal transduction. Hum. Mol. Genet. 2006, 15 (Suppl. S2), R220–R226. [Google Scholar] [CrossRef] [PubMed]
- Aoki, Y.; Niihori, T.; Banjo, T.; Okamoto, N.; Mizuno, S.; Kurosawa, K.; Ogata, T.; Takada, F.; Yano, M.; Ando, T.; et al. Gain-of-function mutations in RIT1 cause Noonan syndrome, a RAS/MAPK pathway syndrome. Am. J. Hum. Genet. 2013, 93, 173–180. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jongmans, M.C.; van der Burgt, I.; Hoogerbrugge, P.M.; Noordam, K.; Yntema, H.G.; Nillesen, W.M.; Kuiper, R.P.; Ligtenberg, M.J.; van Kessel, A.G.; van Krieken, J.H.; et al. Cancer risk in patients with Noonan syndrome carrying a PTPN11 mutation. Eur. J. Hum. Genet. 2011, 19, 870–874. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Escherich, C.S.; Li, Z.; Barnett, K.R.; Li, Y.; Walker, M.; Yoshimura, S.; Yang, W.; Huang, X.; Yu, J.; Stock, W.; et al. Differentiation-dependent EBF1 activity determines CD22 transcription and leukemia sensitivity to inotuzumab ozogamicin. Blood 2025, 146, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Gökbuget, N.; Kneba, M.; Raff, T.; Trautmann, H.; Bartram, C.-R.; Arnold, R.; Fietkau, R.; Freund, M.; Ganser, A.; Ludwig, W.-D.; et al. Adult patients with acute lymphoblastic leukemia and molecular failure display a poor prognosis and are candidates for stem cell transplantation and targeted therapies. Blood 2012, 120, 1868–1876. [Google Scholar] [CrossRef]
- Su, J.; Song, Y.; Zhu, Z.; Huang, X.; Fan, J.; Qiao, J.; Mao, F. Cell-cell communication: New insights and clinical implications. Signal Transduct. Target. Ther. 2024, 9, 196. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tóthová, Z.; Šemeláková, M.; Solárová, Z.; Tomc, J.; Debeljak, N.; Solár, P. The Role of PI3K/AKT and MAPK Signaling Pathways in Erythropoietin Signalization. Int. J. Mol. Sci. 2021, 22, 7682. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef]
- van der Ploeg, P.; Uittenboogaard, A.; Thijs, A.M.J.; Westgeest, H.M.; Boere, I.A.; Lambrechts, S.; van de Stolpe, A.; Bekkers, R.L.M.; Piek, J.M.J. The effectiveness of monotherapy with PI3K/AKT/mTOR pathway inhibitors in ovarian cancer: A meta-analysis. Gynecol. Oncol. 2021, 163, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Browne, I.M.; Okines, A.F.C. Resistance to Targeted Inhibitors of the PI3K/AKT/mTOR Pathway in Advanced Oestrogen-Receptor-Positive Breast Cancer. Cancers 2024, 16, 2259. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lill, C.B.; Fitter, S.; Zannettino, A.C.W.; Vandyke, K.; Noll, J.E. Molecular and cellular mechanisms of chemoresistance in paediatric pre-B cell acute lymphoblastic leukaemia. Cancer Metastasis Rev. 2024, 43, 1385–1399. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pourhassan, H.; Murphy, L.; Aldoss, I. Glucocorticoid Therapy in Acute Lymphoblastic Leukemia: Navigating Short-Term and Long-Term Effects and Optimal Regimen Selection. Curr. Hematol. Malig. Rep. 2024, 19, 175–185. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jabbarzadeh Kaboli, P.; Salimian, F.; Aghapour, S.; Xiang, S.; Zhao, Q.; Li, M.; Wu, X.; Du, F.; Zhao, Y.; Shen, J.; et al. Akt-targeted therapy as a promising strategy to overcome drug resistance in breast cancer—A comprehensive review from chemotherapy to immunotherapy. Pharmacol. Res. 2020, 156, 104806. [Google Scholar] [CrossRef] [PubMed]
- Tanno, S.; Tanno, S.; Mitsuuchi, Y.; Altomare, D.A.; Xiao, G.H.; Testa, J.R. AKT activation up-regulates insulin-like growth factor I receptor expression and promotes invasiveness of human pancreatic cancer cells. Cancer Res. 2001, 61, 589–593. [Google Scholar]
- Chandarlapaty, S.; Sawai, A.; Scaltriti, M.; Rodrik-Outmezguine, V.; Grbovic-Huezo, O.; Serra, V.; Majumder, P.K.; Baselga, J.; Rosen, N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 2011, 19, 58–71. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tamburini, J.; Chapuis, N.; Bardet, V.; Park, S.; Sujobert, P.; Willems, L.; Ifrah, N.; Dreyfus, F.; Mayeux, P.; Lacombe, C.; et al. Mammalian target of rapamycin (mTOR) inhibition activates phosphatidylinositol 3-kinase/Akt by up-regulating insulin-like growth factor-1 receptor signaling in acute myeloid leukemia: Rationale for therapeutic inhibition of both pathways. Blood 2008, 111, 379–382. [Google Scholar] [CrossRef]
- Bertacchini, J.; Heidari, N.; Mediani, L.; Capitani, S.; Shahjahani, M.; Ahmadzadeh, A.; Saki, N. Targeting PI3K/AKT/mTOR network for treatment of leukemia. Cell. Mol. Life Sci. 2015, 72, 2337–2347. [Google Scholar] [CrossRef]
- Garrett, J.T.; Olivares, M.G.; Rinehart, C.; Granja-Ingram, N.D.; Sánchez, V.; Chakrabarty, A.; Dave, B.; Cook, R.S.; Pao, W.; McKinely, E.; et al. Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. Proc. Natl. Acad. Sci. USA 2011, 108, 5021–5026. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bahar, M.E.; Kim, H.J.; Kim, D.R. Targeting the RAS/RAF/MAPK pathway for cancer therapy: From mechanism to clinical studies. Signal Transduct. Target. Ther. 2023, 8, 455. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tandon, A.; Kuriappan, J.A.; Dubey, V. Translocation Tales: Unraveling the MYC Deregulation in Burkitt Lymphoma for Innovative Therapeutic Strategies. Lymphatics 2023, 1, 97–117. [Google Scholar] [CrossRef]
- Mlakar, V.; Morel, E.; Mlakar, S.J.; Ansari, M.; Gumy-Pause, F. A review of the biological and clinical implications of RAS-MAPK pathway alterations in neuroblastoma. J. Exp. Clin. Cancer Res. 2021, 40, 189. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, Y.; Dong, Q.; Cui, Y. Synergistic inhibition of MEK and reciprocal feedback networks for targeted intervention in malignancy. Cancer Biol. Med. 2019, 16, 415–434. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Houde, N.; Beuret, L.; Bonaud, A.; Fortier-Beaulieu, S.P.; Truchon-Landry, K.; Aoidi, R.; Pic, É.; Alouche, N.; Rondeau, V.; Schlecht-Louf, G.; et al. Fine-tuning of MEK signaling is pivotal for limiting B and T cell activation. Cell Rep. 2022, 38, 110223. [Google Scholar] [CrossRef] [PubMed]
- Kun, E.; Tsang, Y.T.M.; Ng, C.W.; Gershenson, D.M.; Wong, K.K. MEK inhibitor resistance mechanisms and recent developments in combination trials. Cancer Treat. Rev. 2021, 92, 102137. [Google Scholar] [CrossRef] [PubMed]
- Colombo, I.; Garg, S.; Danesh, A.; Bruce, J.; Shaw, P.; Tan, Q.; Quevedo, R.; Braunstein, M.; Oza, A.M.; Pugh, T.; et al. Heterogeneous alteration of the ERBB3-MYC axis associated with MEK inhibitor resistance in a KRAS-mutated low-grade serous ovarian cancer patient. Cold Spring Harb. Mol. Case Stud. 2019, 5, a004341. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kawasaki, Y.; Sakimura, A.; Park, C.M.; Tomaru, R.; Tanaka, T.; Ozawa, T.; Zhou, Y.; Narita, K.; Kishi, H.; Muraguchi, A.; et al. Feedback control of ErbB2 via ERK-mediated phosphorylation of a conserved threonine in the juxtamembrane domain. Sci. Rep. 2016, 6, 31502. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, K.; Zheng, M.; Lu, R.; Du, J.; Zhao, Q.; Li, Z.; Li, Y.; Zhang, S. The role of CDC25C in cell cycle regulation and clinical cancer therapy: A systematic review. Cancer Cell Int. 2020, 20, 213. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lax, I.; Wong, A.; Lamothe, B.; Lee, A.; Frost, A.; Hawes, J.; Schlessinger, J. The docking protein FRS2alpha controls a MAP kinase-mediated negative feedback mechanism for signaling by FGF receptors. Mol. Cell 2002, 10, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Li, Y. Receptor tyrosine kinases: Biological functions and anticancer targeted therapy. MedComm 2023, 4, e446. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kiyatkin, A.; van Alderwerelt van Rosenburgh, I.K.; Klein, D.E.; Lemmon, M.A. Kinetics of receptor tyrosine kinase activation define ERK signaling dynamics. Sci. Signal. 2020, 13, eaaz5267. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Akhavan, D.; Pourzia, A.L.; Nourian, A.A.; Williams, K.J.; Nathanson, D.; Babic, I.; Villa, G.R.; Tanaka, K.; Nael, A.; Yang, H.; et al. De-repression of PDGFRβ transcription promotes acquired resistance to EGFR tyrosine kinase inhibitors in glioblastoma patients. Cancer Discov. 2013, 3, 534–547. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Benedettini, E.; Sholl, L.M.; Peyton, M.; Reilly, J.; Ware, C.; Davis, L.; Vena, N.; Bailey, D.; Yeap, B.Y.; Fiorentino, M.; et al. Met activation in non-small cell lung cancer is associated with de novo resistance to EGFR inhibitors and the development of brain metastasis. Am. J. Pathol. 2010, 177, 415–423. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Meyer, A.S.; Miller, M.A.; Gertler, F.B.; Lauffenburger, D.A. The receptor AXL diversifies EGFR signaling and limits the response to EGFR-targeted inhibitors in triple-negative breast cancer cells. Sci. Signal. 2013, 6, ra66. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Boewe, A.S.; Wrublewsky, S.; Hoppstädter, J.; Götz, C.; Kiemer, A.K.; Menger, M.D.; Laschke, M.W.; Ampofo, E. C-Myc/H19/miR-29b axis downregulates nerve/glial (NG)2 expression in glioblastoma multiforme. Mol. Ther. Nucleic Acids 2024, 35, 102120. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Osthus, R.C.; Shim, H.; Kim, S.; Li, Q.; Reddy, R.; Mukherjee, M.; Xu, Y.; Wonsey, D.; Lee, L.A.; Dang, C.V. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J. Biol. Chem. 2000, 275, 21797–21800. [Google Scholar] [CrossRef] [PubMed]
- Frank, S.B.; Berger, P.L.; Ljungman, M.; Miranti, C.K. Human prostate luminal cell differentiation requires NOTCH3 induction by p38-MAPK and MYC. J. Cell Sci. 2017, 130, 1952–1964. [Google Scholar] [CrossRef] [PubMed]
- Wang, V.; Davis, D.A.; Veeranna, R.P.; Haque, M.; Yarchoan, R. Characterization of the activation of protein tyrosine phosphatase, receptor-type, Z polypeptide 1 (PTPRZ1) by hypoxia inducible factor-2 alpha. PLoS ONE 2010, 5, e9641. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yu, J.; Li, J.; Zhang, S.; Xu, X.; Zheng, M.; Jiang, G.; Li, F. IGF-1 induces hypoxia-inducible factor 1α-mediated GLUT3 expression through PI3K/Akt/mTOR dependent pathways in PC12 cells. Brain Res. 2012, 1430, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Gao, Q.; Yin, C.; Zou, M.; Lu, K.; Liu, W.; Zhu, Y.; Zhang, M.; Cheng, R. The CD146-HIF-1α axis regulates epithelial cell migration and alveolar maturation in a mouse model of bronchopulmonary dysplasia. Lab. Investig. 2022, 102, 794–804. [Google Scholar] [CrossRef]
- Sun, L.; Liu, Y.; Lin, S.; Shang, J.; Liu, J.; Li, J.; Yuan, S.; Zhang, L. Early growth response gene-1 and hypoxia-inducible factor-1α affect tumor metastasis via regulation of tissue factor. Acta Oncol. 2013, 52, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Evens, A.M.; Bhalla, S.; Singh, A.; James, C.; Yang, S.; Prachand, S.; Dokic, D.; Gascoyne, R.D.; Winter, J.N.; et al. Hypoxia Inducible Factor 1á (HIF-1á) Regulates CD20 Expression in Lymphoma Cells: Possible Implications for Rituximab Based Therapy in Diffuse Large B Cell Lymphoma (DLBCL). Blood 2009, 114, 1698. [Google Scholar] [CrossRef]
- Marshall, A.D.; Lagutina, I.; Grosveld, G.C. PAX3-FOXO1 induces cannabinoid receptor 1 to enhance cell invasion and metastasis. Cancer Res. 2011, 71, 7471–7480. [Google Scholar] [CrossRef] [PubMed]
- Shao, F.; Liu, Z.; Wei, Q.; Yu, D.; Zhao, M.; Zhang, X.; Gao, X.; Fan, Z.; Wang, S. FOXO1 orchestrates the intestinal homeostasis via neuronal signaling in group 3 innate lymphoid cells. J. Exp. Med. 2023, 220, e20230133. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gopinath, S.D.; Webb, A.E.; Brunet, A.; Rando, T.A. FOXO3 promotes quiescence in adult muscle stem cells during the process of self-renewal. Stem Cell Rep. 2014, 2, 414–426. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pyrzynska, B.; Dwojak, M.; Zerrouqi, A.; Morlino, G.; Zapala, P.; Miazek, N.; Zagozdzon, A.; Bojarczuk, K.; Bobrowicz, M.; Siernicka, M.; et al. FOXO1 promotes resistance of non-Hodgkin lymphomas to anti-CD20-based therapy. Oncoimmunology 2018, 7, e1423183. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gu, Z.; Churchman, M.L.; Roberts, K.G.; Moore, I.; Zhou, X.; Nakitandwe, J.; Hagiwara, K.; Pelletier, S.; Gingras, S.; Berns, H.; et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat. Genet. 2019, 51, 296–307. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dai, Y.T.; Zhang, F.; Fang, H.; Li, J.F.; Lu, G.; Jiang, L.; Chen, B.; Mao, D.D.; Liu, Y.F.; Wang, J.; et al. Transcriptome-wide subtyping of pediatric and adult T cell acute lymphoblastic leukemia in an international study of 707 cases. Proc. Natl. Acad. Sci. USA 2022, 119, e2120787119. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Beder, T.; Hansen, B.T.; Hartmann, A.M.; Zimmermann, J.; Amelunxen, E.; Wolgast, N.; Walter, W.; Zaliova, M.; Antić, Ž.; Chouvarine, P.; et al. The Gene Expression Classifier ALLCatchR Identifies B-cell Precursor ALL Subtypes and Underlying Developmental Trajectories Across Age. Hemasphere 2023, 7, e939. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Uhlen, M.; Karlsson, M.J.; Zhong, W.; Tebani, A.; Pou, C.; Mikes, J.; Lakshmikanth, T.; Forsström, B.; Edfors, F.; Odeberg, J.; et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 2019, 366, eaax9198. [Google Scholar] [CrossRef] [PubMed]
- Bausch-Fluck, D.; Goldmann, U.; Müller, S.; van Oostrum, M.; Müller, M.; Schubert, O.T.; Wollscheid, B. The in silico human surfaceome. Proc. Natl. Acad. Sci. USA 2018, 115, E10988–E10997. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gene Ontology Consortium. Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Singh, A.; Miranda Bedate, A.; von Richthofen, H.J.; Vijver, S.V.; van der Vlist, M.; Kuhn, R.; Yermanos, A.; Kuball, J.J.; Kesmir, C.; Pascoal Ramos, M.I.; et al. A novel bioinformatics pipeline for the identification of immune inhibitory receptors as potential therapeutic targets. Elife 2024, 13, RP92870. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Szklarczyk, D.; Nastou, K.; Koutrouli, M.; Kirsch, R.; Mehryary, F.; Hachilif, R.; Hu, D.; Peluso, M.E.; Huang, Q.; Fang, T.; et al. The STRING database in 2025, protein networks with directionality of regulation. Nucleic Acids Res. 2025, 53, D730–D737. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Leo, I.R.; Aswad, L.; Stahl, M.; Kunold, E.; Post, F.; Erkers, T.; Struyf, N.; Mermelekas, G.; Joshi, R.N.; Gracia-Villacampa, E.; et al. Integrative multi-omics and drug response profiling of childhood acute lymphoblastic leukemia cell lines. Nat. Commun. 2022, 13, 1691. [Google Scholar] [CrossRef]
- Sofen, H.; Bissonnette, R.; Yosipovitch, G.; Silverberg, J.I.; Tyring, S.; Loo, W.J.; Zook, M.; Lee, M.; Zou, L.; Jiang, G.L.; et al. Efficacy and safety of vixarelimab, a human monoclonal oncostatin M receptor β antibody, in moderate-to-severe prurigo nodularis: A randomised, double-blind, placebo-controlled, phase 2a study. EClinicalMedicine 2023, 57, 101826. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Geethadevi, A.; Nair, A.; Parashar, D.; Ku, Z.; Xiong, W.; Deng, H.; Li, Y.; George, J.; McAllister, D.M.; Sun, Y.; et al. Oncostatin M Receptor-Targeted Antibodies Suppress STAT3 Signaling and Inhibit Ovarian Cancer Growth. Cancer Res. 2021, 81, 5336–5352. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lee, B.Y.; Hogg, E.K.J.; Below, C.R.; Kononov, A.; Blanco-Gomez, A.; Heider, F.; Xu, J.; Hutton, C.; Zhang, X.; Scheidt, T.; et al. Heterocellular OSM-OSMR signalling reprograms fibroblasts to promote pancreatic cancer growth and metastasis. Nat. Commun. 2021, 12, 7336. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hoffmann, R.M.; Crescioli, S.; Mele, S.; Sachouli, E.; Cheung, A.; Chui, C.K.; Andriollo, P.; Jackson, P.J.M.; Lacy, K.E.; Spicer, J.F.; et al. A Novel Antibody-Drug Conjugate (ADC) Delivering a DNA Mono-Alkylating Payload to Chondroitin Sulfate Proteoglycan (CSPG4)-Expressing Melanoma. Cancers 2020, 12, 1029. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mungra, N.; Biteghe, F.A.N.; Malindi, Z.; Huysamen, A.M.; Karaan, M.; Hardcastle, N.S.; Bunjun, R.; Chetty, S.; Naran, K.; Lang, D.; et al. CSPG4 as a target for the specific killing of triple-negative breast cancer cells by a recombinant SNAP-tag-based antibody-auristatin F drug conjugate. J. Cancer Res. Clin. Oncol. 2023, 149, 12203–12225. [Google Scholar] [CrossRef]
- Breij, E.C.; de Goeij, B.E.; Verploegen, S.; Schuurhuis, D.H.; Amirkhosravi, A.; Francis, J.; Miller, V.B.; Houtkamp, M.; Bleeker, W.K.; Satijn, D.; et al. An antibody-drug conjugate that targets tissue factor exhibits potent therapeutic activity against a broad range of solid tumors. Cancer Res. 2014, 74, 1214–1226. [Google Scholar] [CrossRef] [PubMed]
- Kirchhoff, D.; Stelte-Ludwig, B.; Lerchen, H.G.; Wengner, A.M.; Ahsen, O.V.; Buchmann, P.; Märsch, S.; Mahlert, C.; Greven, S.; Dietz, L.; et al. IL3RA-Targeting Antibody-Drug Conjugate BAY-943 with a Kinesin Spindle Protein Inhibitor Payload Shows Efficacy in Preclinical Models of Hematologic Malignancies. Cancers 2020, 12, 3464. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ogitani, Y.; Aida, T.; Hagihara, K.; Yamaguchi, J.; Ishii, C.; Harada, N.; Soma, M.; Okamoto, H.; Oitate, M.; Arakawa, S.; et al. DS-8201a, A Novel HER2-Targeting ADC with a Novel DNA Topoisomerase I Inhibitor, Demonstrates a Promising Antitumor Efficacy with Differentiation from T-DM1. Clin. Cancer Res. 2016, 22, 5097–5108. [Google Scholar] [CrossRef] [PubMed]
- Lewis Phillips, G.D.; Li, G.; Dugger, D.L.; Crocker, L.M.; Parsons, K.L.; Mai, E.; Blättler, W.A.; Lambert, J.M.; Chari, R.V.; Lutz, R.J.; et al. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res. 2008, 68, 9280–9290. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Guo, Y.; Wang, Z.; Wu, M.; Peng, W.; Sun, L.; Sun, J.; Li, M.; Zhu, J. A Dose Escalation Phase Ia Study of Anti-CD20 Antibody Drug Conjugate, MRG001 in Relapsed/Refractory Advanced Non-Hodgkin Lymphom. Blood 2021, 138 (Suppl. S1), 2490. [Google Scholar] [CrossRef]
- Demetri, G.D.; Luke, J.J.; Hollebecque, A.; Powderly, J.D., 2nd; Spira, A.I.; Subbiah, V.; Naumovski, L.; Chen, C.; Fang, H.; Lai, D.W.; et al. First-in-Human Phase I Study of ABBV-085, an Antibody-Drug Conjugate Targeting LRRC15, in Sarcomas and Other Advanced Solid Tumors. Clin. Cancer Res. 2021, 27, 3556–3566. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.D.; Atallah, E.; Rizzieri, D.; Walter, R.B.; Chung, K.Y.; Spira, A.; Stock, W.; Tallman, M.S.; Cruz, H.G.; Boni, J.; et al. Camidanlumab tesirine, an antibody-drug conjugate, in relapsed/refractory CD25-positive acute myeloid leukemia or acute lymphoblastic leukemia: A phase I study. Leuk. Res. 2020, 95, 106385. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Wang, Z.; Li, M.; Yang, N.; Lu, H.; Zhang, Z.; Dong, Y.; Chen, Y.; Zhu, Z.; Tong, A.; et al. A novel SLC3A2-targeting antibody-drug conjugate exerts potent antitumor efficacy in head and neck squamous cell cancer. Transl. Oncol. 2024, 45, 101981. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shi, J.; Yan, J.; Huang, Y.; Guo, J.; Kong, Y.; Meng, X.; Liu, S.-H. Abstract 739, AMT-253, a first-in-class MUC18-targeting antibody-drug conjugate, for the treatment of MUC18-positive solid tumors. Cancer Res. 2024, 84 (Suppl. S6), 739. [Google Scholar] [CrossRef]
- Jonasch, E.; Call, J.; Kambhampati, S.; Caimi, P.; Curti, B.; Diefenbach, C.; Heath, E.; Park, S.; Kornblum, N.; Li, W.; et al. Phase 1/2 study of PRO1160, a CD70-directed antibody-drug conjugate, in patients with advanced solid tumors and hematologic malignancies. Oncologist 2023, 28, S14–S15. [Google Scholar] [CrossRef]
- Rosen, L.S.; Wesolowski, R.; Baffa, R.; Liao, K.H.; Hua, S.Y.; Gibson, B.L.; Pirie-Shepherd, S.; Tolcher, A.W. A phase I, dose-escalation study of PF-06650808, an anti-Notch3 antibody-drug conjugate, in patients with breast cancer and other advanced solid tumors. Investig. New Drugs 2020, 38, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, J.; Grandits, M.; Palhares, L.C.G.F.; Mele, S.; Nakamura, M.; López-Abente, J.; Crescioli, S.; Laddach, R.; Romero-Clavijo, P.; Cheung, A.; et al. Anti-cancer pro-inflammatory effects of an IgE antibody targeting the melanoma-associated antigen chondroitin sulfate proteoglycan 4. Nat. Commun. 2023, 14, 2192. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hoffmeister, L.M.; Orhan, E.; Walter, C.; Niktoreh, N.; Hanenberg, H.; von Neuhoff, N.; Reinhardt, D.; Schneider, M. Impact of KMT2A Rearrangement and CSPG4 Expression in Pediatric Acute Myeloid Leukemia. Cancers 2021, 13, 4817. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Smith, F.O.; Rauch, C.; Williams, D.E.; March, C.J.; Arthur, D.; Hilden, J.; Lampkin, B.C.; Buckley, J.D.; Buckley, C.V.; Woods, W.G.; et al. The human homologue of rat NG2, a chondroitin sulfate proteoglycan, is not expressed on the cell surface of normal hematopoietic cells but is expressed by acute myeloid leukemia blasts from poor-prognosis patients with abnormalities of chromosome band 11q23. Blood 1996, 87, 1123–1133. [Google Scholar] [CrossRef]
- Chen, X.; Habib, S.; Alexandru, M.; Chauhan, J.; Evan, T.; Troka, J.M.; Rahimi, A.; Esapa, B.; Tull, T.J.; Ng, W.Z.; et al. Chondroitin Sulfate Proteoglycan 4 (CSPG4) as an Emerging Target for Immunotherapy to Treat Melanoma. Cancers 2024, 16, 3260. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, K.; Yong, J.; Zauner, R.; Wally, V.; Whitelock, J.; Sajinovic, M.; Kopecki, Z.; Liang, K.; Scott, K.F.; Mellick, A.S. Chondroitin Sulfate Proteoglycan 4 as a Marker for Aggressive Squamous Cell Carcinoma. Cancers 2022, 14, 5564. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kurokawa, T.; Imai, K. Chondroitin sulfate proteoglycan 4, An attractive target for antibody-based immunotherapy. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2024, 100, 293–308. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ilieva, K.M.; Cheung, A.; Mele, S.; Chiaruttini, G.; Crescioli, S.; Griffin, M.; Nakamura, M.; Spicer, J.F.; Tsoka, S.; Lacy, K.E.; et al. Chondroitin Sulfate Proteoglycan 4 and Its Potential As an Antibody Immunotherapy Target across Different Tumor Types. Front. Immunol. 2018, 8, 1911. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Saegusa, S.; Fukaya, M.; Kakegawa, W.; Tanaka, M.; Katsumata, O.; Sugawara, T.; Hara, Y.; Itakura, M.; Okubo, T.; Sato, T.; et al. Mice lacking EFA6C/Psd2, a guanine nucleotide exchange factor for Arf6, exhibit lower Purkinje cell synaptic density but normal cerebellar motor functions. PLoS ONE 2019, 14, e0216960. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sun, D.; Guo, Y.; Tang, P.; Li, H.; Chen, L. Arf6 as a therapeutic target: Structure, mechanism, and inhibitors. Acta Pharm. Sin. B. 2023, 13, 4089–4104. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vega-Benedetti, A.F.; Loi, E.; Moi, L.; Blois, S.; Fadda, A.; Antonelli, M.; Arcella, A.; Badiali, M.; Giangaspero, F.; Morra, I.; et al. Clustered protocadherins methylation alterations in cancer. Clin. Epigenetics 2019, 11, 100. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dallosso, A.R.; Øster, B.; Greenhough, A.; Thorsen, K.; Curry, T.J.; Owen, C.; Hancock, A.L.; Szemes, M.; Paraskeva, C.; Frank, M.; et al. Long-range epigenetic silencing of chromosome 5q31 protocadherins is involved in early and late stages of colorectal tumorigenesis through modulation of oncogenic pathways. Oncogene 2012, 31, 4409–4419. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bernardo-Castineira, C.; Valdés, N.; Celada, L.; Martinez, A.S.J.; Sáenz-de-Santa-María, I.; Bayón, G.F.; Fernández, A.F.; Sierra, M.I.; Fraga, M.F.; Astudillo, A.; et al. Epigenetic Deregulation of Protocadherin PCDHGC3 in Pheochromocytomas/Paragangliomas Associated With SDHB Mutations. J. Clin. Endocrinol. Metab. 2019, 104, 5673–5692. [Google Scholar] [CrossRef] [PubMed]
- Gabbert, L.; Dilling, C.; Meybohm, P.; Burek, M. Deletion of Protocadherin Gamma C3 Induces Phenotypic and Functional Changes in Brain Microvascular Endothelial Cells In Vitro. Front. Pharmacol. 2020, 11, 590144. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Feldheim, J.; Wend, D.; Lauer, M.J.; Monoranu, C.M.; Glas, M.; Kleinschnitz, C.; Ernestus, R.I.; Braunger, B.M.; Meybohm, P.; Hagemann, C.; et al. Protocadherin Gamma C3 (PCDHGC3) Is Strongly Expressed in Glioblastoma and Its High Expression Is Associated with Longer Progression-Free Survival of Patients. Int. J. Mol. Sci. 2022, 23, 8101. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Busche, S.; Ge, B.; Vidal, R.; Spinella, J.F.; Saillour, V.; Richer, C.; Healy, J.; Chen, S.H.; Droit, A.; Sinnett, D.; et al. Integration of high-resolution methylome and transcriptome analyses to dissect epigenomic changes in childhood acute lymphoblastic leukemia. Cancer Res. 2013, 73, 4323–4336. [Google Scholar] [CrossRef] [PubMed]
- Chatterton, Z.; Morenos, L.; Mechinaud, F.; Ashley, D.M.; Craig, J.M.; Sexton-Oates, A.; Halemba, M.S.; Parkinson-Bates, M.; Ng, J.; Morrison, D.; et al. Epigenetic deregulation in pediatric acute lymphoblastic leukemia. Epigenetics 2014, 9, 459–467. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cui, T.; Chen, Y.; Yang, L.; Knösel, T.; Zöller, K.; Huber, O.; Petersen, I. DSC3 expression is regulated by p53, and methylation of DSC3 DNA is a prognostic marker in human colorectal cancer. Br. J. Cancer 2011, 104, 1013–1019. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cui, T.; Chen, Y.; Yang, L.; Knösel, T.; Huber, O.; Pacyna-Gengelbach, M.; Petersen, I. The p53 target gene desmocollin 3 acts as a novel tumor suppressor through inhibiting EGFR/ERK pathway in human lung cancer. Carcinogenesis 2012, 33, 2326–2333. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.; Yang, L.; Ma, Y.; Petersen, I.; Chen, Y. Desmocollin 3 has a tumor suppressive activity through inhibition of AKT pathway in colorectal cancer. Exp. Cell Res. 2019, 378, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhang, Y.; Ren, X.; Li, D.; Fu, H.; Liu, C.; Zhou, W.; Liu, Q.; Liu, Q.; Wu, M. Leucine-rich repeat containing 4 act as an autophagy inhibitor that restores sensitivity of glioblastoma to temozolomide. Oncogene 2020, 39, 4551–4566. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, M.; Huang, C.; Gan, K.; Huang, H.; Chen, Q.; Ouyang, J.; Tang, Y.; Li, X.; Yang, Y.; Zhou, H.; et al. LRRC4, a putative tumor suppressor gene, requires a functional leucine-rich repeat cassette domain to inhibit proliferation of glioma cells in vitro by modulating the extracellular signal-regulated kinase/protein kinase B/nuclear factor-kappaB pathway. Mol. Biol. Cell 2006, 17, 3534–3542. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, M.; Huang, C.; Li, X.; Li, X.; Gan, K.; Chen, Q.; Tang, Y.; Tang, K.; Shen, S.; Li, G. LRRC4 inhibits glioblastoma cell proliferation, migration, and angiogenesis by downregulating pleiotropic cytokine expression and responses. J. Cell. Physiol. 2008, 214, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Ren, X.; Fu, H.; Li, D.; Chen, X.; Zu, X.; Liu, Q.; Wu, M. LRRC4 mediates the formation of circular RNA CD44 to inhibitGBM cell proliferation. Mol. Ther. Nucleic Acids 2021, 26, 473–487. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lv, Z.; Wang, T.; Cao, X.; Sun, M.; Qu, Y. The role of receptor-type protein tyrosine phosphatases in cancer. Precis. Med. Sci. 2023, 12, 48–57. [Google Scholar] [CrossRef]
- Zheng, C.; Liu, T.; Wang, A.Q.; Chen, X.A.; Zhang, R.Z.; Wang, X.C.; Lv, C.Y.; Pan, R.L.; Wang, O.C.; Lu, X.C. Protein tyrosine phosphatase receptor type kappa (PTPRK) revisited: Evolving insights into structure, function, and pathology. J. Transl. Med. 2025, 23, 534. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Young, K.A.; Wojdyla, K.; Lai, T.; Mulholland, K.E.; Aldaz Casanova, S.; Antrobus, R.; Andrews, S.R.; Biggins, L.; Mahler-Araujo, B.; Barton, P.R.; et al. The receptor protein tyrosine phosphatase PTPRK promotes intestinal repair and catalysis-independent tumour suppression. J. Cell Sci. 2024, 137, jcs261914. [Google Scholar] [CrossRef]
- Stevenson, W.S.; Best, O.G.; Przybylla, A.; Chen, Q.; Singh, N.; Koleth, M.; Pierce, S.; Kennedy, T.; Tong, W.; Kuang, S.Q.; et al. DNA methylation of membrane-bound tyrosine phosphatase genes in acute lymphoblastic leukaemia. Leukemia 2014, 28, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Novellino, L.; De Filippo, A.; Deho, P.; Perrone, F.; Pilotti, S.; Parmiani, G.; Castelli, C. PTPRK negatively regulates transcriptional activity of wild type and mutated oncogenic beta-catenin and affects membrane distribution of beta-catenin/E-cadherin complexes in cancer cells. Cell Signal. 2008, 20, 872–883. [Google Scholar] [CrossRef] [PubMed]
- Priebe, V.; Drakul, A.; Zeyen, S.; Bornhauser, B.; Santoro, R.; Bourquin, J.P. P331, The Oncogenic Transcription Factor Tcf3, :Hlf Marks a Distinct Enhancer-Promoter Interaction Network in T(17;19) Positive Childhood Acute Lymphoblastic Leukemia. Hemasphere 2023, 7, e88138e3. [Google Scholar] [CrossRef] [PubMed Central]
- Navarrete-Meneses, M.D.P.; Pérez-Vera, P. Alteraciones epigenéticas en leucemia linfoblástica aguda [Epigenetic alterations in acute lymphoblastic leukemia]. Bol. Med. Hosp. Infant. Mex. 2017, 74, 243–264. (In Spanish) [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Lin, Y.C.; Chen, C.C. Lysophosphatidic Acid Receptor Antagonists and Cancer: The Current Trends, Clinical Implications, and Trials. Cells 2021, 10, 1629. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Geraldo, L.H.M.; Spohr, T.C.L.S.; Amaral, R.F.D.; Fonseca, A.C.C.D.; Garcia, C.; Mendes, F.A.; Freitas, C.; dosSantos, M.F.; Lima, F.R.S. Role of lysophosphatidic acid and its receptors in health and disease: Novel therapeutic strategies. Signal Transduct. Target. Ther. 2021, 6, 45. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bokaii Hosseini, Z.; Rajabi, F.; Morovatshoar, R.; Ashrafpour, M.; Behboodi, P.; Zareie, D.; Natami, M. Downregulation of LPAR1 Promotes Invasive Behavior in Papillary Thyroid Carcinoma Cells. Cancer Inform. 2024, 23, 11769351241277012. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hu, X.; Haney, N.; Kropp, D.; Kabore, A.F.; Johnston, J.B.; Gibson, S.B. Lysophosphatidic acid (LPA) protects primary chronic lymphocytic leukemia cells from apoptosis through LPA receptor activation of the anti-apoptotic protein AKT/PKB. J. Biol. Chem. 2005, 280, 9498–9508. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, G.H.; Cheon, J.; Kim, D.; Jun, H.S. Lysophosphatidic Acid Promotes Epithelial-Mesenchymal Transition in Kidney Epithelial Cells via the LPAR1/MAPK-AKT/KLF5 Signaling Pathway in Diabetic Nephropathy. Int. J. Mol. Sci. 2022, 23, 10497. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, X.; Cao, D.; Zheng, X.; Wang, G.; Liu, M. Tissue factor as a new target for tumor therapy-killing two birds with one stone: A narrative review. Ann. Transl. Med. 2022, 10, 1250. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ren, Z.; Xue, Y.; Liu, L.; Zhang, X.; Pei, J.; Zhang, Y.; Wang, Y.; Yu, K. Tissue factor overexpression in triple-negative breast cancer promotes immune evasion by impeding T-cell infiltration and effector function. Cancer Lett. 2023, 565, 216221. [Google Scholar] [CrossRef] [PubMed]
- Koizume, S.; Miyagi, Y. Tissue factor in cancer-associated thromboembolism: Possible mechanisms and clinical applications. Br. J. Cancer 2022, 127, 2099–2107. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mechtcheriakova, D.; Wlachos, A.; Holzmüller, H.; Binder, B.R.; Hofer, E. Vascular endothelial cell growth factor-induced tissue factor expression in endothelial cells is mediated by EGR-1. Blood 1999, 93, 3811–3823. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.Z.; Wei, M.N.; Huang, J.R.; Zhang, Z.J.; Zhang, W.J.; Jiang, Q.W.; Yang, Y.; Wang, H.Y.; Jin, H.L.; Wang, K.; et al. Targeting TF-AKT/ERK-EGFR Pathway Suppresses the Growth of Hepatocellular Carcinoma. Front. Oncol. 2019, 9, 150. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hu, Z. Tissue factor as a new target for CAR-NK cell immunotherapy of triple-negative breast cancer. Sci. Rep. 2020, 10, 2815. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Guo, X.; Yang, Y.; Tang, J.; Xiang, J. Ephs in cancer progression: Complexity and context-dependent nature in signaling, angiogenesis and immunity. Cell Commun. Signal. 2024, 22, 299. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, X.; Zhang, C.; Deng, M.; Jiang, Y.; He, Z.; Qian, H. EFNB1 levels determine distinct drug response patterns guiding precision therapy for B-cell neoplasms. iScience 2023, 27, 108667. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zheng, Y.; Shi, J. EFNB1 drives glioma progression and shapes the immune microenvironment: A potential prognostic biomarker. Discov. Oncol. 2025, 16, 249. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chatzikalil, E.; Stergiou, I.E.; Papadakos, S.P.; Konstantinidis, I.; Theocharis, S. The Clinical Relevance of the EPH/Ephrin Signaling Pathway in Pediatric Solid and Hematologic Malignancies. Int. J. Mol. Sci. 2024, 25, 3834. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mekkaoui, L.; Rassart, C.; Rozen, L.; Janssens, A.; Bron, D.; Ferster, A.; Cantinieaux, B. Use of CD 123 Expression on Blasts from AML, ALL and RAEB As Minimal Residual Disease Marker. Blood 2015, 126, 5402. [Google Scholar] [CrossRef]
- Liu, J.; Tan, X.; Ma, Y.Y.; Liu, Y.; Gao, L.; Gao, L.; Kong, P.; Peng, X.G.; Zhang, X.; Zhang, C. Study on the Prognostic Value of Aberrant Antigen in Patients With Acute B Lymphocytic Leukemia. Clin. Lymphoma Myeloma Leuk. 2019, 19, e349–e358. [Google Scholar] [CrossRef] [PubMed]
- Al-Mudallal, S.S.; Shaker, H.; Dede, H. Assessment of Expression and Prognostic Significance of Interleukin 3 Receptor Alpha Subunit (CD123) in Childhood Acute Lymphoblastic Leukemia. Int. J. Med. Res. Prof. 2016, 2, 247–254. [Google Scholar]
- Pelosi, E.; Castelli, G.; Testa, U. CD123 a Therapeutic Target for Acute Myeloid Leukemia and Blastic Plasmocytoid Dendritic Neoplasm. Int. J. Mol. Sci. 2023, 24, 2718. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hercus, T.R.; Kan, W.L.T.; Broughton, S.E.; Tvorogov, D.; Ramshaw, H.S.; Sandow, J.J.; Nero, T.L.; Dhagat, U.; Thompson, E.J.; Shing, K.S.C.T.; et al. Role of the β Common (βc) Family of Cytokines in Health and Disease. Cold Spring Harb. Perspect. Biol. 2018, 10, a028514. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hara, T.; Miyajima, A. Function and signal transduction mediated by the interleukin 3 receptor system in hematopoiesis. Stem Cells 1996, 14, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Songyang, Z.; Baltimore, D.; Cantley, L.C.; Kaplan, D.R.; Franke, T.F. Interleukin 3-dependent survival by the Akt protein kinase. Proc. Natl. Acad. Sci. USA 1997, 94, 11345–11350. [Google Scholar] [CrossRef] [PubMed]
- Kovtun, Y.; Jones, G.E.; Adams, S.; Harvey, L.; Audette, C.A.; Wilhelm, A.; Bai, C.; Rui, L.; Laleau, R.; Liu, F.; et al. A CD123-targeting antibody-drug conjugate, IMGN632, designed to eradicate AML while sparing normal bone marrow cells. Blood Adv. 2018, 2, 848–858. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ogata, H.; Ford, D.; Kouttab, N.; King, T.C.; Vita, N.; Minty, A.; Stoeckler, J.; Morgan, D.; Girasole, C.; Morgan, J.W.; et al. Regulation of interleukin-13 receptor constituents on mature human B lymphocytes. J. Biol. Chem. 1998, 273, 9864–9871. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Wang, J.; Cheng, K.; Lu, Q.; Zhao, R.; Wang, S.; Zhang, Q.; Ge, L.; Pan, J.; Song, G.; et al. IL13Rα1 prevents a castration resistant phenotype of prostate cancer by targeting hexokinase 2 for ubiquitin-mediated degradation. Cancer Biol. Med. 2021, 19, 1008–1028. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, S.; Olde Heuvel, F.; Rehman, R.; Aousji, O.; Froehlich, A.; Li, Z.; Jark, R.; Zhang, W.; Conquest, A.; Woelfle, S.; et al. Interleukin-13 and its receptor are synaptic proteins involved in plasticity and neuroprotection. Nat. Commun. 2023, 14, 200. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Prokopchuk, O.; Liu, Y.; Henne-Bruns, D.; Kornmann, M. Interleukin-4 enhances proliferation of human pancreatic cancer cells: Evidence for autocrine and paracrine actions. Br. J. Cancer 2005, 92, 921–928. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shi, J.; Song, X.; Traub, B.; Luxenhofer, M.; Kornmann, M. Involvement of IL-4, IL-13 and Their Receptors in Pancreatic Cancer. Int. J. Mol. Sci. 2021, 22, 2998. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kollewe, A.; Schwarz, Y.; Oleinikov, K.; Raza, A.; Haupt, A.; Wartenberg, P.; Wyatt, A.; Boehm, U.; Ectors, F.; Bildl, W.; et al. Subunit composition, molecular environment, and activation of native TRPC channels encoded by their interactomes. Neuron 2022, 110, 4162–4175.e7. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Kim, J.; Park, C.H.; Jeong, B.; So, I. Direct modulation of TRPC ion channels by Gα proteins. Front. Physiol. 2024, 15, 1362987. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Song, H.B.; Jun, H.O.; Kim, J.H.; Fruttiger, M.; Kim, J.H. Suppression of transient receptor potential canonical channel 4 inhibits vascular endothelial growth factor-induced retinal neovascularization. Cell Calcium 2015, 57, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.; Prontera, P.; Sarchielli, P.; Tonelli, A.; Bassi, M.T.; Cupini, L.M.; Caproni, S.; Siliquini, S.; Donti, E.; Calabresi, P. A novel ATP1A2 gene mutation in familial hemiplegic migraine and epilepsy. Cephalalgia 2014, 34, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, N.; Carmine-Simmen, K.; Nair, R.; Wang, C.; Moghadas-Jafari, S.; Blaser, H.; Tran-Thanh, D.; Wang, D.; Wang, P.; Wang, J.; et al. Amplification of a calcium channel subunit CACNG4 increases breast cancer metastasis. EBioMedicine 2020, 52, 102646. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Halatsch, M.E.; Löw, S.; Mursch, K.; Hielscher, T.; Schmidt, U.; Unterberg, A.; Vougioukas, V.I.; Feuerhake, F. Candidate genes for sensitivity and resistance of human glioblastoma multiforme cell lines to erlotinib. Lab. Investig. J. Neurosurg. 2009, 111, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.L.; Gu, F.M.; Wang, Z.; Jiang, J.H.; Yao, L.Q.; Tan, C.J.; Huang, X.Y.; Ke, A.W.; Dai, Z.; Fan, J.; et al. Activation of PI3K/AKT and MAPK pathway through a PDGFRβ-dependent feedback loop is involved in rapamycin resistance in hepatocellular carcinoma. PLoS ONE 2012, 7, e33379. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, H.; Bajraszewski, N.; Wu, E.; Wang, H.; Moseman, A.P.; Dabora, S.L.; Griffin, J.D.; Kwiatkowski, D.J. PDGFRs are critical for PI3K/Akt activation and negatively regulated by mTOR. J. Clin. Investig. 2007, 117, 730–738. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Miyazaki, B.; Ueno, T.; Sugiyama, M.; Kojima, S.; Arakawa, A.; Tao, K.; Tanimura, K.; Shiraishi, K.; Yagishita, S.; Kohsaka, S.; et al. Promoter swapping of truncated PDGFRB drives Ph-like acute lymphoblastic leukemia. npj Precis. Oncol. 2023, 7, 132. [Google Scholar] [CrossRef]
- Wang, X.; Yue, M.; Cheung, J.P.Y.; Cheung, P.W.H.; Fan, Y.; Wu, M.; Wang, X.; Zhao, S.; Khanshour, A.M.; Rios, J.J.; et al. Impaired glycine neurotransmission causes adolescent idiopathic scoliosis. J. Clin. Investig. 2024, 134, e168783. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Raiteri, L.; Raiteri, M. Functional ‘glial’ GLYT1 glycine transporters expressed in neurons. J. Neurochem. 2010, 114, 647–653. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Feng, T.; Xie, Y.; Swamiappan, S.; Zhou, Y.; Zhou, Y.; Zhou, H.; Peng, X. Recent Advances in the Development and Application of Cell-Loaded Collagen Scaffolds. Int. J. Mol. Sci. 2025, 26, 4009. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Trono, P.; Ottavi, F.; Rosano’, L. Novel insights into the role of Discoidin domain receptor 2 (DDR2) in cancer progression: A new avenue of therapeutic intervention. Matrix Biol. 2024, 125, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.M.; Lee, F.Y.F.; Jones, R.T.; Kimball, A.K.; Saravia, E.; Graziano, R.F.; Coleman, B.; Menard, K.; Yan, J.; Michaud, E.; et al. Targeting DDR2 enhances tumor response to anti-PD-1 immunotherapy. Sci. Adv. 2019, 5, eaav2437. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ren, T.; Zhang, W.; Liu, X.; Zhao, H.; Zhang, J.; Zhang, J.; Li, X.; Zhang, Y.; Bu, X.; Shi, M.; et al. Discoidin domain receptor 2 (DDR2) promotes breast cancer cell metastasis and the mechanism implicates epithelial mesenchymal transition programme under hypoxia. J. Pathol. 2014, 234, 526–537. [Google Scholar] [CrossRef]
- Vessella, T.; Xiang, S.; Xiao, C.; Stilwell, M.; Fok, J.; Shohet, J.; Rozen, E.; Zhou, H.S.; Wen, Q. DDR2 signaling and mechanosensing orchestrate neuroblastoma cell fate through different transcriptome mechanisms. FEBS Open Bio. 2024, 14, 867–882. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hinz, N.; Baranowsky, A.; Horn, M.; Kriegs, M.; Sibbertsen, F.; Smit, D.J.; Clezardin, P.; Lange, T.; Schinke, T.; Jücker, M. Knockdown of AKT3 Activates HER2 and DDR Kinases in Bone-Seeking Breast Cancer Cells, Promotes Metastasis In Vivo and Attenuates the TGFβ/CTGF Axis. Cells 2021, 10, 430. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fujikawa, A.; Sugawara, H.; Tanaka, T.; Matsumoto, M.; Kuboyama, K.; Suzuki, R.; Tanga, N.; Ogata, A.; Masumura, M.; Noda, M. Targeting PTPRZ inhibits stem cell-like properties and tumorigenicity in glioblastoma cells. Sci. Rep. 2017, 7, 5609. [Google Scholar] [CrossRef]
- Rush, L.J.; Raval, A.; Funchain, P.; Johnson, A.J.; Smith, L.; Lucas, D.M.; Bembea, M.; Liu, T.H.; Heerema, N.A.; Rassenti, L.; et al. Epigenetic profiling in chronic lymphocytic leukemia reveals novel methylation targets. Cancer Res. 2004, 64, 2424–2433. [Google Scholar] [CrossRef] [PubMed]
- Motiwala, T.; Majumder, S.; Kutay, H.; Smith, D.S.; Neuberg, D.S.; Lucas, D.M.; Byrd, J.C.; Grever, M.; Jacob, S.T. Methylation and silencing of protein tyrosine phosphatase receptor type O in chronic lymphocytic leukemia. Clin. Cancer Res. 2007, 13, 3174–3181. [Google Scholar] [CrossRef] [PubMed] [PubMed Central][Green Version]
- Papadimitriou, E.; Kanellopoulou, V.K. Protein Tyrosine Phosphatase Receptor Zeta 1 as a Potential Target in Cancer Therapy and Diagnosis. Int. J. Mol. Sci. 2023, 24, 8093. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Muto, M.; Suzuki, H.; Suzuki, Y. New Insights and Future Perspectives of APRIL in IgA Nephropathy. Int. J. Mol. Sci. 2024, 25, 10340. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Albini, A.; Di Paola, L.; Mei, G.; Baci, D.; Fusco, N.; Corso, G.; Noonan, D. Inflammation and cancer cell survival: TRAF2 as a key player. Cell Death Dis. 2025, 16, 292. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cascalho, M.; Platt, J.L. TNFRSF13B Diversification Fueled by B Cell Responses to Environmental Challenges—A Hypothesis. Front. Immunol. 2021, 12, 634544. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mamara, A.; Germenis, A.E.; Kompoti, M.; Palassopoulou, M.; Mandala, E.; Banti, A.; Giannakoulas, N.; Speletas, M. TACI expression and signaling in chronic lymphocytic leukemia. J. Immunol. Res. 2015, 2015, 478753. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tai, Y.T.; Lin, L.; Xing, L.; Cho, S.-F.; Yu, T.; Acharya, C.; Wen, K.; Hsieh, P.A.; Dulos, J.; van Elsas, A.; et al. APRIL signaling via TACI mediates immunosuppression by T regulatory cells in multiple myeloma: Therapeutic implications. Leukemia 2019, 33, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Creative-Bioloabs: Anti-TNFRSF13B (Tabalumab)-MC-Vc-PAB-MMAE.; CAT#ADC-W-1907. Available online: https://www.creative-biolabs.com/adc/tabalumab-mc-vc-pab-mmae-adc-2188.htm (accessed on 4 June 2025).
- Bagherie-Lachidan, M.; Reginensi, A.; Pan, Q.; Zaveri, H.P.; Scott, D.A.; Blencowe, B.J.; Helmbacher, F.; McNeill, H. Stromal Fat4 acts non-autonomously with Dchs1/2 to restrict the nephron progenitor pool. Development 2015, 142, 2564–2573. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.K.; Keck, J.M.; Eide, C.A.; Bottomly, D.; Traer, E.; Tyner, J.W.; McWeeney, S.K.; Tognon, C.E.; Druker, B.J. ERBB2/HER2 mutations are transforming and therapeutically targetable in leukemia. Leukemia 2020, 34, 2798–2804. [Google Scholar] [CrossRef]
- Giacoletto, C.J.; Valente, L.J.; Brown, L.; Patterson, S.; Gokhale, R.; Mockus, S.M.; Grody, W.W.; Deng, H.W.; Rotter, J.I.; Schiller, M.R. New Gain-of-Function Mutations Prioritize Mechanisms of HER2 Activation. medRxiv 2025. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Irwin, M.E.; Nelson, L.D.; Santiago-O’Farrill, J.M.; Knouse, P.D.; Miller, C.P.; Palla, S.L.; Siwak, D.R.; Mills, G.B.; Estrov, Z.; Li, S.; et al. Small molecule ErbB inhibitors decrease proliferative signaling and promote apoptosis in philadelphia chromosome-positive acute lymphoblastic leukemia. PLoS ONE 2013, 8, e70608. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Grant, S.; Qiao, L.; Dent, P. Roles of ERBB family receptor tyrosine kinases, and downstream signaling pathways, in the control of cell growth and survival. Front. Biosci. 2002, 7, d376–d389. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Wang, J.; Shen, S.; Ni, X.; Gong, Z.; Zheng, B.; Sun, W.; Suo, T.; Liu, H.; Ni, X.; et al. ERBB2 S310F mutation independently activates PI3K/AKT and MAPK pathways through homodimers to contribute gallbladder carcinoma growth. Med. Oncol. 2022, 39, 64. [Google Scholar] [CrossRef] [PubMed]
- Vaudry, D.; Falluel-Morel, A.; Bourgault, S.; Basille, M.; Burel, D.; Wurtz, O.; Fournier, A.; Chow, B.K.; Hashimoto, H.; Galas, L.; et al. Pituitary adenylate cyclase-activating polypeptide and its receptors: 20 years after the discovery. Pharmacol. Rev. 2009, 61, 283–357. [Google Scholar] [CrossRef] [PubMed]
- Asano, S.; Yamasaka, M.; Ozasa, K.; Sakamoto, K.; Hayata-Takano, A.; Nakazawa, T.; Hashimoto, H.; Waschek, J.A.; Ago, Y. Vasoactive intestinal peptide-VIPR2 signaling regulates tumor cell migration. Front. Oncol. 2022, 12, 852358. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Asano, S.; Ono, A.; Baba, K.; Uehara, T.; Sakamoto, K.; Hayata-Takano, A.; Nakazawa, T.; Yanamoto, S.; Tanimoto, K.; Hashimoto, H.; et al. Blockade of vasoactive intestinal peptide receptor 2 (VIPR2) signaling suppresses cyclin D1-dependent cell-cycle progression in MCF-7 cells. J. Pharmacol. Sci. 2024, 154, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Eon Kuek, L.; Leffler, M.; Mackay, G.A.; Hulett, M.D. The MS4A family: Counting past 1, 2 and 3. Immunol. Cell Biol. 2016, 94, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Pavlasova, G.; Mraz, M. The regulation and function of CD20, an “enigma” of B-cell biology and targeted therapy. Haematologica 2020, 105, 1494–1506. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mahadevia, H.; Ananthamurugan, M.; Shah, K.; Desai, A.; Shrestha, A. A Review of Anti-CD20 Antibodies in the Management of B-Cell Lymphomas. Lymphatics 2024, 2, 10–24. [Google Scholar] [CrossRef]
- Dworzak, M.N.; Schumich, A.; Printz, D.; Pötschger, U.; Husak, Z.; Attarbaschi, A.; Basso, G.; Gaipa, G.; Ratei, R.; Mann, G.; et al. CD20 up-regulation in pediatric B-cell precursor acute lymphoblastic leukemia during induction treatment: Setting the stage for anti-CD20 directed immunotherapy. Blood 2008, 112, 3982–3988. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bonavida, B. ‘Rituximab-induced inhibition of antiapoptotic cell survival pathways: Implications in chemo/immunoresistance, rituximab unresponsiveness, prognostic and novel therapeutic interventions’. Oncogene 2007, 26, 3629–3636. [Google Scholar] [CrossRef]
- Sanchez, R.; Ribera, J.; Morgades, M.; Ayala, R.; Onecha, E.; Ruiz-Heredia, Y.; Juárez-Rufián, A.; de Nicolás, R.; Sánchez-Pina, J.; Vives, S.; et al. A novel targeted RNA-Seq panel identifies a subset of adult patients with acute lymphoblastic leukemia with BCR-ABL1-like characteristics. Blood Cancer J. 2020, 10, 43. [Google Scholar] [CrossRef] [PubMed]
- Russell, L.J.; Capasso, M.; Vater, I.; Akasaka, T.; Bernard, O.A.; Calasanz, M.J.; Chandrasekaran, T.; Chapiro, E.; Gesk, S.; Griffiths, M.; et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood 2009, 114, 2688–2698. [Google Scholar] [CrossRef] [PubMed]
- Chuang, S.; Chu, A.; Hurtado, R.; Tirado, C.A. Integrative Insights into Philadelphia-like B-Cell Acute Lymphoblastic Leukemia: A Genetic and Molecular Landscape. Diagnostics 2025, 15, 385. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tasian, S.K.; Doral, M.Y.; Borowitz, M.J.; Wood, B.L.; Chen, I.M.; Harvey, R.C.; Gastier-Foster, J.M.; Willman, C.L.; Hunger, S.P.; Mullighan, C.G.; et al. Aberrant STAT5 and PI3K/mTOR pathway signaling occurs in human CRLF2-rearranged B-precursor acute lymphoblastic leukemia. Blood 2012, 120, 833–842. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Toiyama, Y.; Mizoguchi, A.; Kimura, K.; Hiro, J.; Inoue, Y.; Tutumi, T.; Miki, C.; Kusunoki, M. TTYH2, a human homologue of the Drosophila melanogaster gene tweety, is up-regulated in colon carcinoma and involved in cell proliferation and cell aggregation. World J. Gastroenterol. 2007, 13, 2717–2721. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Leung, H.H.; Mansour, C.; Rousseau, M.; Nakhla, A.; Kiselyov, K.; Venkatachalam, K.; Wong, C.O. Drosophila tweety facilitates autophagy to regulate mitochondrial homeostasis and bioenergetics in Glia. Glia 2024, 72, 433–451. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kang, H.; Chen, I.M.; Wilson, C.S.; Bedrick, E.J.; Harvey, R.C.; Atlas, S.R.; Devidas, M.; Mullighan, C.G.; Wang, X.; Murphy, M.; et al. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood 2010, 115, 1394–1405. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bakr, M.N.; Takahashi, H.; Kikuchi, Y. CHRNA1 and its correlated-myogenesis/cell cycle genes are prognosis-related markers of metastatic melanoma. Biochem. Biophys. Rep. 2023, 33, 101425. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kim, J.Y.; Kim, H.; Chung, W.S.; Park, H. Selective regulation of corticostriatal synapses by astrocytic phagocytosis. Nat. Commun. 2025, 16, 2504. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Charlet, J.; Tomari, A.; Dallosso, A.R.; Szemes, M.; Kaselova, M.; Curry, T.J.; Almutairi, B.; Etchevers, H.C.; McConville, C.; Malik, K.T.; et al. Genome-wide DNA methylation analysis identifies MEGF10 as a novel epigenetically repressed candidate tumor suppressor gene in neuroblastoma. Mol. Carcinog. 2017, 56, 1290–1301. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, X.; Liu, Y.; Hu, Y.; Su, F.; Wang, Z.; Chen, Y.; Zhuang, Z. Leucine rich repeat containing 15 promotes triple-negative breast cancer proliferation and invasion via the ITGB1/FAK/PI3K signalling pathway. Sci. Rep. 2025, 15, 14535. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cui, J.; Dean, D.; Wei, R.; Hornicek, F.J.; Ulmert, D.; Duan, Z. Expression and clinical implications of leucine-rich repeat containing 15 (LRRC15) in osteosarcoma. J. Orthop. Res. 2020, 38, 2362–2372. [Google Scholar] [CrossRef] [PubMed]
- Storey, C.M.; Cheng, M.; Altai, M.; Park, J.E.; Tran, J.; Lueong, S.S.; Thorek, D.; Mao, L.; Zedan, W.; Yuen, C.; et al. Key Regulatory Elements of the TGFβ-LRRC15 Axis Predict Disease Progression and Immunotherapy Resistance Across Cancer Types. bioRxiv 2024. [Google Scholar] [CrossRef]
- Purcell, J.W.; Tanlimco, S.G.; Hickson, J.; Fox, M.; Sho, M.; Durkin, L.; Uziel, T.; Powers, R.; Foster, K.; McGonigal, T.; et al. LRRC15 Is a Novel Mesenchymal Protein and Stromal Target for Antibody-Drug Conjugates. Cancer Res. 2018, 78, 4059–4072. [Google Scholar] [CrossRef] [PubMed]
- Dalpati, N.; Rai, S.K.; Sharma, P.; Sarangi, P.P. Integrins and integrin-driven secretory pathways as multi-dimensional regulators of tumor-associated macrophage recruitment and reprogramming in tumor microenvironment. Matrix Biol. 2025, 135, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Ji, W.; Cheng, X.; Lv, L.; Yu, X.; Wang, N.; Li, M.; Hu, T.; Shi, Z. Revealing a Novel Methylated Integrin Alpha-8 Related to Extracellular Matrix and Anoikis Resistance Using Proteomic Analysis in the Immune Microenvironment of Lung Adenocarcinoma. Mol. Biotechnol. 2025, 67, 1137–1155. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.; Koh, Y.; Park, H.; Kim, D.Y.; Kim, D.C.; Byun, J.M.; Lee, H.J.; Yoon, S.S. Highly Expressed Integrin-α8 Induces Epithelial to Mesenchymal Transition-Like Features in Multiple Myeloma with Early Relapse. Mol. Cells 2016, 39, 898–908. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yurchenko, M.Y.; Kovalevska, L.M.; Shlapatska, L.M.; Berdova, G.G.; Clark, E.A.; Sidorenko, S.P. CD150 regulates JNK1/2 activation in normal and Hodgkin’s lymphoma B cells. Immunol. Cell Biol. 2010, 88, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Mikhalap, S.V.; Shlapatska, L.M.; Yurchenko, O.V.; Yurchenko, M.Y.; Berdova, G.G.; Nichols, K.E.; Clark, E.A.; Sidorenko, S.P. The adaptor protein SH2D1A regulates signaling through CD150 (SLAM) in B cells. Blood 2004, 104, 4063–4070. [Google Scholar] [CrossRef] [PubMed]
- Gordiienko, I.; Shlapatska, L.; Kholodniuk, V.; Sklyarenko, L.; Gluzman, D.F.; Clark, E.A.; Sidorenko, S.P. The interplay of CD150 and CD180 receptor pathways contribute to the pathobiology of chronic lymphocytic leukemia B cells by selective inhibition of Akt and MAPK signaling. PLoS ONE 2017, 12, e0185940. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- von Wenserski, L.; Schultheiß, C.; Bolz, S.; Schliffke, S.; Simnica, D.; Willscher, E.; Gerull, H.; Wolters-Eisfeld, G.; Riecken, K.; Fehse, B.; et al. SLAMF receptors negatively regulate B cell receptor signaling in chronic lymphocytic leukemia via recruitment of prohibitin-2. Leukemia 2021, 35, 1073–1086. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chinen, T.; Kannan, A.K.; Levine, A.G.; Fan, X.; Klein, U.; Zheng, Y.; Gasteiger, G.; Feng, Y.; Fontenot, J.D.; Rudensky, A.Y. An essential role for the IL-2 receptor in Treg cell function. Nat. Immunol. 2016, 17, 1322–1333. [Google Scholar] [CrossRef] [PubMed]
- Peerlings, D.; Mimpen, M.; Damoiseaux, J. The IL-2—IL-2 receptor pathway: Key to understanding multiple sclerosis. J. Transl. Autoimmun. 2021, 4, 100123. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nguyen, C.H.; Schlerka, A.; Grandits, A.M.; Koller, E.; van der Kouwe, E.; Vassiliou, G.S.; Staber, P.B.; Heller, G.; Wieser, R. IL2RA Promotes Aggressiveness and Stem Cell-Related Properties of Acute Myeloid Leukemia. Cancer Res. 2020, 80, 4527–4539. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.; Brennan, S.; Milne, T.A.; Chen, W.Y.; Li, Y.; Hurtz, C.; Kweon, S.M.; Zickl, L.; Shojaee, S.; Neuberg, D.; et al. Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia. Cancer Discov. 2012, 2, 1004–1023. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Robinson, M.E.; Sun, R.; Kume, K.; Ma, N.; Cosgun, K.N.; Chan, L.N.; Leveille, E.; Geng, H.; Vykunta, V.S.; et al. Dynamic phosphatase-recruitment controls B-cell selection and oncogenic signaling. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Carrà, G.; Cartellà, A.; Maffeo, B.; Circosta, P.; Cignetti, A.; Pautasso, M.; Familiari, U.; Volante, M.; Dragani, M.E.E.; Cambrin, G.R.; et al. Interleukin-2 Receptor Alpha Chain, Also Called CD25, Is a Potential Target in Acute Lymphoblastic Leukemia. Blood 2020, 136 (Suppl. S1), 11–12. [Google Scholar] [CrossRef]
- Sidahmed-Adrar, N.; Ottavi, J.F.; Benzoubir, N.; Ait Saadi, T.; Bou Saleh, M.; Mauduit, P.; Guettier, C.; Desterke, C.; Le Naour, F. Tspan15 Is a New Stemness-Related Marker in Hepatocellular Carcinoma. Proteomics 2019, 19, e1900025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhang, Z.; Li, L.; Qin, Y.R.; Liu, H.; Jiang, C.; Zeng, T.T.; Li, M.Q.; Xie, D.; Li, Y.; et al. TSPAN15 interacts with BTRC to promote oesophageal squamous cell carcinoma metastasis via activating NF-κB signaling. Nat. Commun. 2018, 9, 1423. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, H.; Song, Q.; Shang, K.; Li, Y.; Jiang, L.; Yang, L. Tspan protein family: Focusing on the occurrence, progression, and treatment of cancer. Cell Death Discov. 2024, 10, 187. [Google Scholar] [CrossRef]
- Sims, N.A.; Lévesque, J.P. Oncostatin M: Dual Regulator of the Skeletal and Hematopoietic Systems. Curr. Osteoporos. Rep. 2024, 22, 80–95. [Google Scholar] [CrossRef]
- Lantieri, F.; Bachetti, T. OSM/OSMR and Interleukin 6 Family Cytokines in Physiological and Pathological Condition. Int. J. Mol. Sci. 2022, 23, 11096. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Polak, K.L.; Tamagno, I.; Parameswaran, N.; Smigiel, J.; Chan, E.R.; Yuan, X.; Rios, B.; Jackson, M.W. Oncostatin-M and OSM-Receptor Feed-Forward Activation of MAPK Induces Separable Stem-like and Mesenchymal Programs. Mol. Cancer Res. 2023, 21, 975–990. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhou, M.H.; Yang, Q.M. NUP214 fusion genes in acute leukemia (Review). Oncol. Lett. 2014, 8, 959–962. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dai, W.; Xu, Y.; Mo, S.; Li, Q.; Yu, J.; Wang, R.; Ma, Y.; Ni, Y.; Xiang, W.; Han, L.; et al. GLUT3 induced by AMPK/CREB1 axis is key for withstanding energy stress and augments the efficacy of current colorectal cancer therapies. Signal Transduct. Target. Ther. 2020, 5, 177. [Google Scholar] [CrossRef]
- Yan, B.; Li, X.; Peng, M.; Zuo, Y.; Wang, Y.; Liu, P.; Ren, W.; Jin, X. The YTHDC1/GLUT3/RNF183 axis forms a positive feedback loop that modulates glucose metabolism and bladder cancer progression. Exp. Mol. Med. 2023, 55, 1145–1158. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.; Sim, H.I.; Yun, B.; Park, Y.; Jin, H.S. Revisiting T-cell adhesion molecules as potential targets for cancer immunotherapy: CD226 and CD2. Exp. Mol. Med. 2024, 56, 2113–2126. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, Z.; Qi, G.; Miller, J.S.; Zheng, S.G. CD226, An Emerging Role in Immunologic Diseases. Front. Cell Dev. Biol. 2020, 8, 564. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sakano, Y.; Sakano, K.; Hurrell, B.P.; Helou, D.G.; Shafiei-Jahani, P.; Kazemi, M.H.; Li, X.; Shen, S.; Hilser, J.R.; Hartiala, J.A.; et al. Blocking CD226 regulates type 2 innate lymphoid cell effector function and alleviates airway hyperreactivity. J. Allergy Clin. Immunol. 2024, 153, 1406–1422.e6. [Google Scholar] [CrossRef]
- Liu, P.; Zhang, C.; Liao, Y.; Liu, J.; Huang, J.; Xia, M.; Chen, M.; Tan, H.; He, W.; Xu, M.; et al. High expression of PTPRM predicts poor prognosis and promotes tumor growth and lymph node metastasis in cervical cancer. Cell Death Dis. 2020, 11, 687. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, Q.; Zhang, N.; Du, X.; Xu, G.; Yan, X. CD146, from a melanoma cell adhesion molecule to a signaling receptor. Signal Transduct. Target. Ther. 2020, 5, 148. [Google Scholar] [CrossRef]
- Joshkon, A.; Heim, X.; Dubrou, C.; Bachelier, R.; Traboulsi, W.; Stalin, J.; Fayyad-Kazan, H.; Badran, B.; Foucault-Bertaud, A.; Leroyer, A.S.; et al. Role of CD146 (MCAM) in Physiological and Pathological Angiogenesis-Contribution of New Antibodies for Therapy. Biomedicines 2020, 8, 633. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Malamud, M.; Brown, G.D. The Dectin-1 and Dectin-2 clusters: C-type lectin receptors with fundamental roles in immunity. EMBO Rep. 2024, 25, 5239–5264. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Freund, P.; Porpaczy, E.A.; Le, T.; Gruber, M.; Pausz, C.; Staber, P.; Jäger, U.; Vanura, K. Cannabinoid Receptors Are Overexpressed in CLL but of Limited Potential for Therapeutic Exploitation. PLoS ONE 2016, 11, e0156693. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lopez-Cardona, A.P.; Pérez-Cerezales, S.; Fernández-González, R.; Laguna-Barraza, R.; Pericuesta, E.; Agirregoitia, N.; Gutiérrez-Adán, A.; Agirregoitia, E. CB1 cannabinoid receptor drives oocyte maturation and embryo development via PI3K/Akt and MAPK pathways. FASEB J. 2017, 31, 3372–3382. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, C.; Ochsenbein, A.F. Immune checkpoints regulate acute myeloid leukemia stem cells. Leukemia 2025, 39, 1277–1293. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sauer, T.; Parikh, K.; Sharma, S.; Omer, B.; Sedloev, D.; Chen, Q.; Angenendt, L.; Schliemann, C.; Schmitt, M.; Müller-Tidow, C.; et al. CD70-specific CAR T cells have potent activity against acute myeloid leukemia without HSC toxicity. Blood 2021, 138, 318–330. [Google Scholar] [CrossRef]
- Marques-Piubelli, M.L.; Kumar, B.; Basar, R.; Panowski, S.; Srinivasan, S.; Norwood, K.; Prashad, S.; Szenes, V.; Balakumaran, A.; Arandhya, A.; et al. Increased expression of CD70 in relapsed acute myeloid leukemia after hypomethylating agents. Virchows Arch. 2024, 485, 937–941. [Google Scholar] [CrossRef]
- Sun, Q.; Zabrocki, P.; Seyfried, F.; de Haard, H.; Kronnie, G.; Debatin, K.M.; Hinrich Meyer, L. P317, Anti-Leukemia Activity of Cd70-Directed Immunotherapy in B Cell Precursor Acute Lymphoblastic Leukemia. Hemasphere 2023, 7, e6339407. [Google Scholar] [CrossRef] [PubMed Central]
- Sawada, H.; Daugherty, A. BEST3-Mediated MEKK2/3 Activation: A Novel Therapeutic Target in Aortopathies. Circulation 2023, 148, 607–609. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lee, W.K.; Chakraborty, P.K.; Roussa, E.; Wolff, N.A.; Thévenod, F. ERK1/2-dependent bestrophin-3 expression prevents ER-stress-induced cell death in renal epithelial cells by reducing CHOP. Biochim. Biophys. Acta 2012, 1823, 1864–1876. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Shahine, A.; Cheng, T.Y.; Chen, Y.L.; Ng, S.W.; Balaji, G.R.; Farquhar, R.; Gras, S.; Hardman, C.S.; Altman, J.D.; et al. CD1 lipidomes reveal lipid-binding motifs and size-based antigen-display mechanisms. Cell 2023, 186, 4583–4596.e13. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bagchi, S.; Li, S.; Wang, C.R. CD1b-autoreactive T cells recognize phospholipid antigens and contribute to antitumor immunity against a CD1b+ T cell lymphoma. Oncoimmunology 2016, 5, e1213932. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gonzalez, R.; Reinberg, D. The Notch pathway: A guardian of cell fate during neurogenesis. Curr. Opin. Cell Biol. 2025, 95, 102543. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Lanzas, R.; Barclay, J.; Hardas, A.; Kalampalika, F.; Jiménez-Pompa, A.; Gallipoli, P.; Ganuza, M. A CADASIL NOTCH3 mutation leads to clonal hematopoiesis and expansion of Dnmt3a-R878H hematopoietic clones. Leukemia 2025, 39, 460–472. [Google Scholar] [CrossRef] [PubMed]
- Giuli, M.V.; Diluvio, G.; Giuliani, E.; Franciosa, G.; Di Magno, L.; Pignataro, M.G.; Tottone, L.; Nicoletti, C.; Besharat, Z.M.; Peruzzi, G.; et al. Notch3 contributes to T-cell leukemia growth via regulation of the unfolded protein response. Oncogenesis 2020, 9, 93. [Google Scholar] [CrossRef]
- Lu, J.; Xue, X.; Wang, H.; Hao, Y.; Yang, Q. Notch1 activation and inhibition in T-cell acute lymphoblastic leukemia subtypes. Exp. Hematol. 2025, 148, 104771. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhang, Y.; Xin, S.; Jin, L.; Cao, Q.; Wang, H.; Wang, K.; Liu, X.; Tang, C.; Li, W.; et al. NOTCH3 promotes docetaxel resistance of prostate cancer cells through regulating TUBB3 and MAPK signaling pathway. Cancer Sci. 2024, 115, 412–426. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, C.; Ge, H.; Shen, C.; Hu, D.; Zhao, X.; Qin, R.; Wang, Y. NOTCH3 promotes malignant progression of bladder cancer by directly regulating SPP1 and activating PI3K/AKT pathway. Cell Death Dis. 2024, 15, 840. [Google Scholar] [CrossRef]
- Gong, X.; Wang, X.; Jin, J.; Gong, Z. The novel functions of chemokines in lung cancer progression. Front. Immunol. 2025, 16, 1607225. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Maciocia, P.M.; Wawrzyniecka, P.A.; Maciocia, N.C.; Burley, A.; Karpanasamy, T.; Devereaux, S.; Hoekx, M.; O’Connor, D.; Leon, T.; Rapoz-D’Silva, T.; et al. Anti-CCR9 chimeric antigen receptor T cells for T-cell acute lymphoblastic leukemia. Blood 2022, 140, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Qiao, X.; Fang, Y.; Guo, R.; Bai, P.; Liu, S.; Li, T.; Jiang, Y.; Wei, S.; Na, Z.; et al. Epigenetics-targeted drugs: Current paradigms and future challenges. Signal Transduct. Target. Ther. 2024, 9, 332. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Johnson-Holiday, C.; Singh, R.; Johnson, E.L.; Grizzle, W.E.; Lillard, J.W., Jr.; Singh, S. CCR9-CCL25 interactions promote cisplatin resistance in breast cancer cell through Akt activation in a PI3K-dependent and FAK-independent fashion. World J. Surg. Oncol. 2011, 9, 46. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Johnson, E.L.; Singh, R.; Johnson-Holiday, C.M.; Grizzle, W.E.; Partridge, E.E.; Lillard, J.W., Jr.; Singh, S. CCR9 interactions support ovarian cancer cell survival and resistance to cisplatin-induced apoptosis in a PI3K-dependent and FAK-independent fashion. J. Ovarian Res. 2010, 3, 15. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tu, Z.; Xiao, R.; Xiong, J.; Tembo, K.M.; Deng, X.; Xiong, M.; Liu, P.; Wang, M.; Zhang, Q. CCR9 in cancer: Oncogenic role and therapeutic targeting. J. Hematol. Oncol. 2016, 9, 10. [Google Scholar] [CrossRef]
- Fang, J.; Guo, L.; Zhang, Y.; Guo, Q.; Wang, M.; Wang, X. The target atlas for antibody-drug conjugates across solid cancers. Cancer Gene Ther. 2024, 31, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Tumey, L.N. An Overview of the Current ADC Discovery Landscape. Methods Mol. Biol. 2020, 2078, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Buongervino, S.; Lane, M.V.; Garrigan, E.; Zhelev, D.V.; Dimitrov, D.S.; Bosse, K.R. Antibody-Drug Conjugate Efficacy in Neuroblastoma: Role of Payload, Resistance Mechanisms, Target Density, and Antibody Internalization. Mol. Cancer Ther. 2021, 20, 2228–2239. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lee, S.H.R.; Li, Z.; Tai, S.T.; Oh, B.L.Z.; Yeoh, A.E.J. Genetic Alterations in Childhood Acute Lymphoblastic Leukemia: Interactions with Clinical Features and Treatment Response. Cancers 2021, 13, 4068. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Harrer, D.C.; Schenkel, C.; Berking, C.; Herr, W.; Abken, H.; Dörrie, J.; Schaft, N. Decitabine-Mediated Upregulation of CSPG4 in Ovarian Carcinoma Cells Enables Targeting by CSPG4-Specific CAR-T Cells. Cancers 2022, 14, 5033. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yoshimura, S.; Li, Z.; Gocho, Y.; Yang, W.; Crews, K.R.; Lee, S.H.R.; Roberts, K.G.; Mullighan, C.G.; Relling, M.V.; Yu, J.; et al. Impact of Age on Pharmacogenomics and Treatment Outcomes of B-Cell Acute Lymphoblastic Leukemia. J. Clin. Oncol. 2024, 42, 3478–3490. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Apavaloaei, A.; Zhao, Q.; Hesnard, L.; Cahuzac, M.; Durette, C.; Larouche, J.D.; Hardy, M.P.; Vincent, K.; Brochu, S.; Laverdure, J.P.; et al. Tumor antigens preferentially derive from unmutated genomic sequences in melanoma and non-small cell lung cancer. Nat. Cancer 2025, 6, 1419–1437. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]

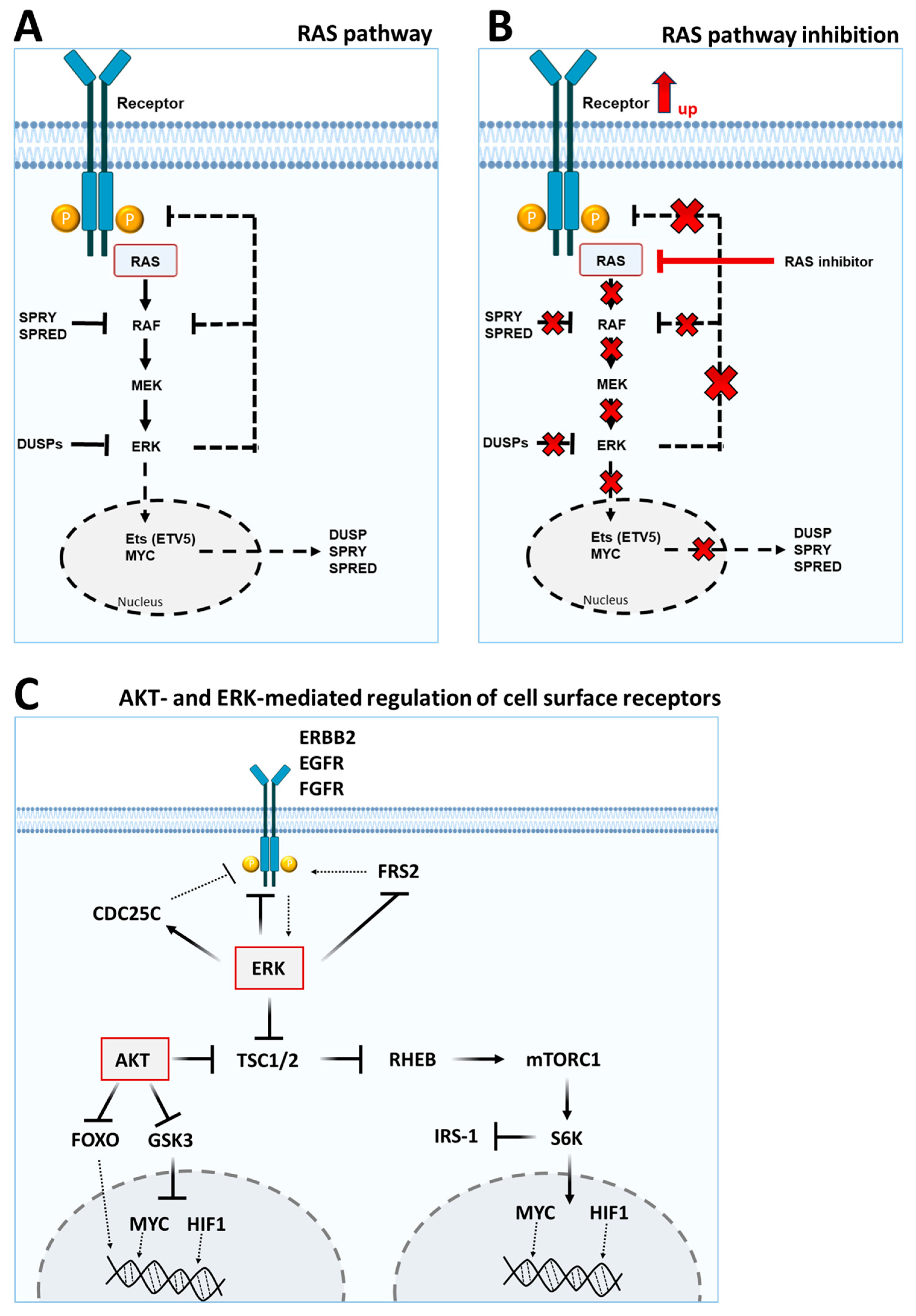

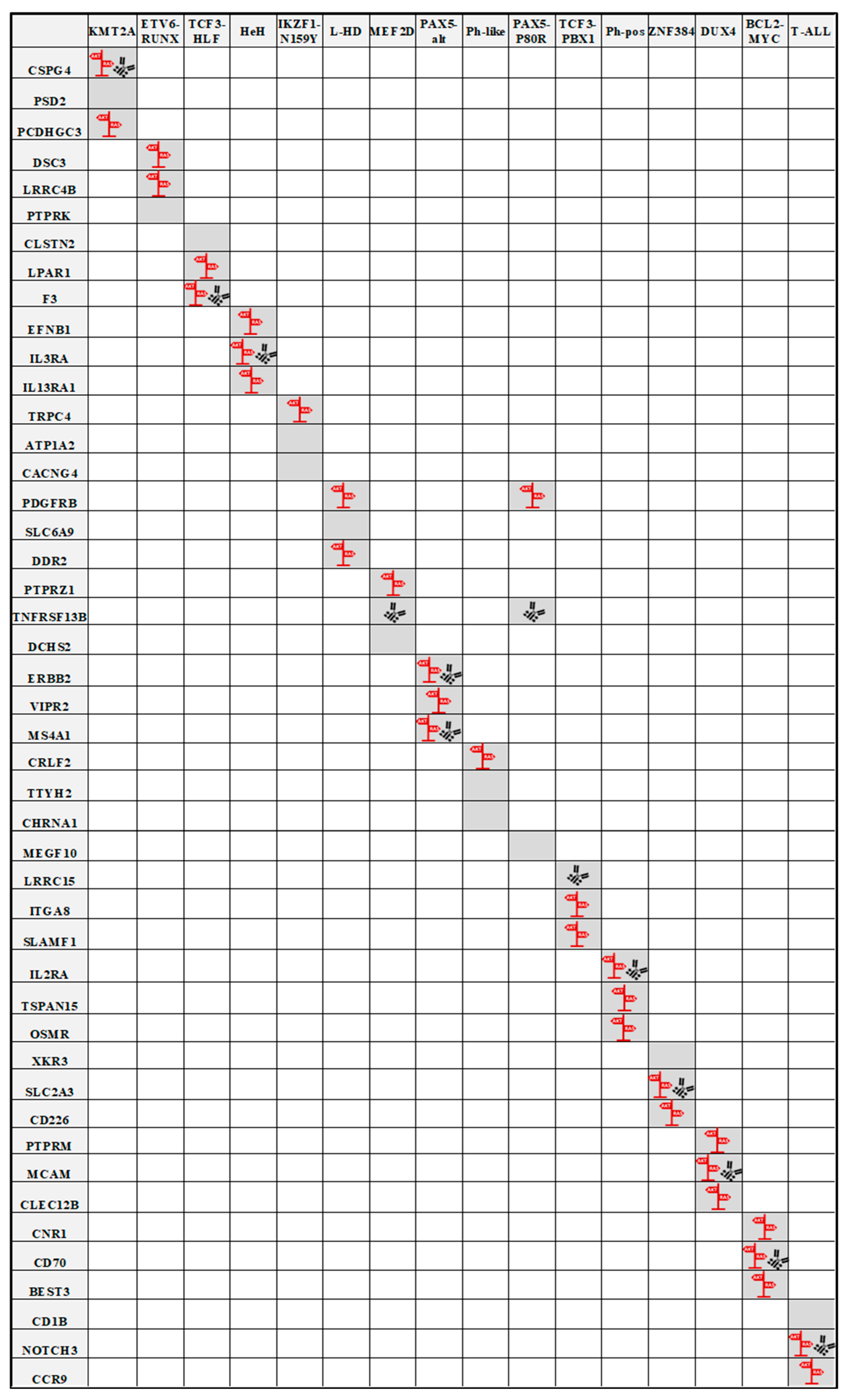
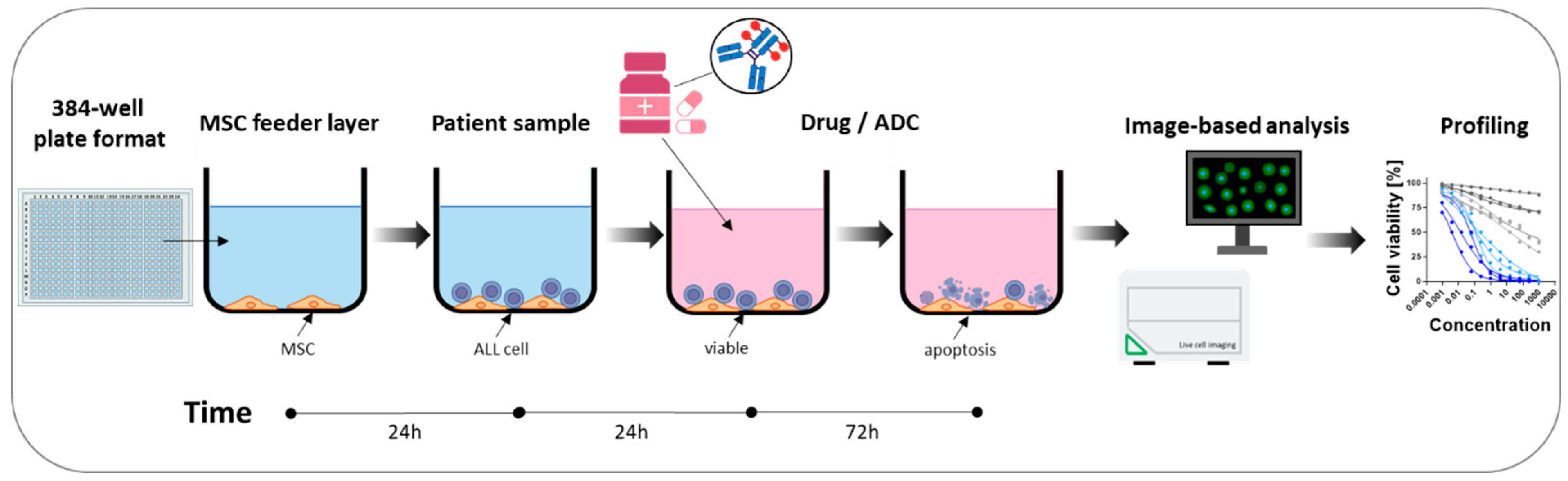
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ehm, P.A.H.; Rehbach, C. Relevance of AKT and RAS Signaling Pathways for Antibody–Drug Conjugate Immunotherapies in Acute Lymphoblastic Leukemia. Lymphatics 2025, 3, 33. https://doi.org/10.3390/lymphatics3040033
Ehm PAH, Rehbach C. Relevance of AKT and RAS Signaling Pathways for Antibody–Drug Conjugate Immunotherapies in Acute Lymphoblastic Leukemia. Lymphatics. 2025; 3(4):33. https://doi.org/10.3390/lymphatics3040033
Chicago/Turabian StyleEhm, Patrick A. H., and Christoph Rehbach. 2025. "Relevance of AKT and RAS Signaling Pathways for Antibody–Drug Conjugate Immunotherapies in Acute Lymphoblastic Leukemia" Lymphatics 3, no. 4: 33. https://doi.org/10.3390/lymphatics3040033
APA StyleEhm, P. A. H., & Rehbach, C. (2025). Relevance of AKT and RAS Signaling Pathways for Antibody–Drug Conjugate Immunotherapies in Acute Lymphoblastic Leukemia. Lymphatics, 3(4), 33. https://doi.org/10.3390/lymphatics3040033





