Abstract
Fragment libraries have a major significance in drug discovery due to their role in de novo design and enumerating large and ultra-large compound libraries. Although several fragment libraries are commercially available, most are derived from synthetic compounds. The number of fragment libraries derived from natural products is still being determined. Still, they represent a rich source of building blocks to generate pseudo-natural products and bioactive synthetic compounds inspired by natural products. In this work, we generated and analyzed a fragment library of natural products from Colombia, a highly diverse geographical region where fragment libraries are yet to be reported. We also generated and reported fragment libraries of three novel natural product libraries and, as a reference, the most updated version of FDA-approved drugs. In line with the principles of open science, the fragment libraries developed in this study are freely available.
1. Introduction
Natural products and their analogsanalogues have major contributions to drug discovery. Out of all the drugs approved for clinical use by 2020, approximately 25% are natural products or derived from them [1]. Natural products have a diversity of privileged scaffolds [2,3] and molecular fragments [4,5]. An approach to maximizing the use of natural products for drug discovery using computational techniques is through the assembly and organization of natural products in compound databases [6]. Examples of large natural product databases are the Collection of Open NatUral ProdUcts (COCONUT) [7], SuperNatural 3.0 [8], and the Universal Natural Product Database [9].
Natural product’s’ unique and complex structures make them attractive to generate fragment libraries. Such libraries can be used as building blocks for de novo design and building bioactive pseud-natural products [10]. In previous studies, we generated, analyzed, and made publicly fragmented libraries from natural products. In the first one, we obtained fragments libraries from natural products, drug-like compounds tested for biological activity, and synthetic accessible compounds [4]. In that analysis, we noted that the compounds and the molecular fragments derived from natural products were the most diverse and complex regarding their chemical structure. In an independent project [6], we generated, analyzed, and made publicly available a fragment library that was obtained from natural products and food chemicals, emphasizing that compounds and fragments derived from natural products and food chemicals had the largest diversity.
Colombia is a country with a large biodiversity of plants, with more than 28,900 species reported, distributed across all its bioregions and ecosystems [11]. Natural products from Colombia Coffee Region (NPDBEjeCol) is a recently constructed database that contains a unique and attractive set of compounds for drug discovery [12]. NPDBEjeCol is a collection of over 200 natural products isolated and characterized in Colombia derived from plants, specifically from the Coffee Region. As shown in this study, the distinctive structural features of compounds in the NPDBEjeCol database make them very attractive sources for generating fragment libraries.
This study aimed to generate and characterize a fragment library from NPDBEjeCol and other Latin America data sets of natural products of interest for drug discovery. Latin America is a geographical region with high biodiversity. Following the principles of open science, fragment libraries are made freely available and can be used in de novo design, including the design of pseudo-natural products.
2. Results and Discussion
The results and discussion are described in the following five sub-sections. The methods used to obtain the results are detailed in Section 3, which are based on the compound databases and computational protocols published in references [12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27].
2.1. Unique and Overlapping Compounds and Fragments
Figure 1 illustrates the number of unique and overlapping compounds and fragments between the databases studied in this work. Figure 1 shows that all five data sets have three compounds in common, and there are 5166 unique compounds among all data sets comprising NPDBEjeCol (157), BIOFACQUIM (529), NUBBEDB (1944) PeruNPDB (188), and 2348 compounds from FDA-approved drugs. The largest overlap between NPDBEjeCol and the data sets analyzed in this study was with NUBBEDB, sharing 34 molecules.
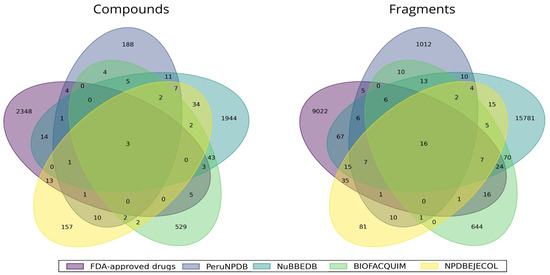
Figure 1.
Unique and overlapping compounds and fragments from NPDBEjeCol (yellow), BIOFACQUIM (green), NuBBEDB (blue), PeruNPDB (dark blue), and FDA-approved drugs (purple).
Concerning the molecular fragments generated, there were 26,540 unique fragment structures among all databases, comprising 81 from NPDBEjeCol, 644 from BIOFACQUIM, 15,781 from NUBBEDB, 1012 from PeruNPDB, and 9022 fragments from FDA-approved drugs. Interestingly, NPDBEjeCol did not share any fragments with BIOFACQUIM but shared 35 fragments with FDA drugs and 15 with NUBBEDB. NPDBEjeCol yielded 200 fragments in total, out of which 16 are shared by all five fragment libraries.
2.2. Fragment Analysis
Figure 2 shows the chemical structures of the ten most frequent and unique fragments in all databases, and the frequency and percentage of each fragment in the corresponding data setdataset. The chemical structures were drawn using Marvinsketch. For NPDBEjeCol, the fragment with the largest abundance (1.01%) is for a hydroxylated linear chain and a double bond (Figure 2A). Three fragments have linear chains with at least one double bond with more than seven carbons. In these fragments are two structures with a tetrahydropyran ring, two with a phenol ring, and one with benzene. Also, carbonyl groups are evident, forming aldehydes and ketones. For BIOFACQUIM (Figure 2B), the ten most frequent fragments show 0.3% abundance; most of the structures have at least one oxygen atom, except one that is only a linear chain of six carbons. Also, eight of ten fragments present cycles, three have only heterocycles, one only carbocycle, and four a mix carbocycles and a heterocycle, of which there are benzene, dihydropyran, tetrahydrofuran rings and, predominantly, delta lactone. The most frequent fragments from NUBBEDB (Figure 2C) are diverse with fragment structures that include benzene, delta lactone, and pyrrolidone rings. The other frequent fragments are linear chains, all of which contain the presence of carbonyls (aldehydes). The fragments from the PeruNPDB (Figure 2D) reveal the presence of one or more rings, with the most abundant fragment featuring two fused rings (1,4-hidroxybenzene and dihydrofuran). All fragments are oxygenated with hydroxyl, carbonyl, and carboxyl groups. The ring most common is tetrahydropyran. The FDA fragments (Figure 2E) have a nitrogen function (amines and amides), distinguishing them as the only set with nitrogen structures. Additionally, three structures present sulfur with thiazolidine rings, another characteristic not found in the other common fragment sets. Six out of ten fragments also contain carbonyl, hydroxyl, and carboxyl groups.
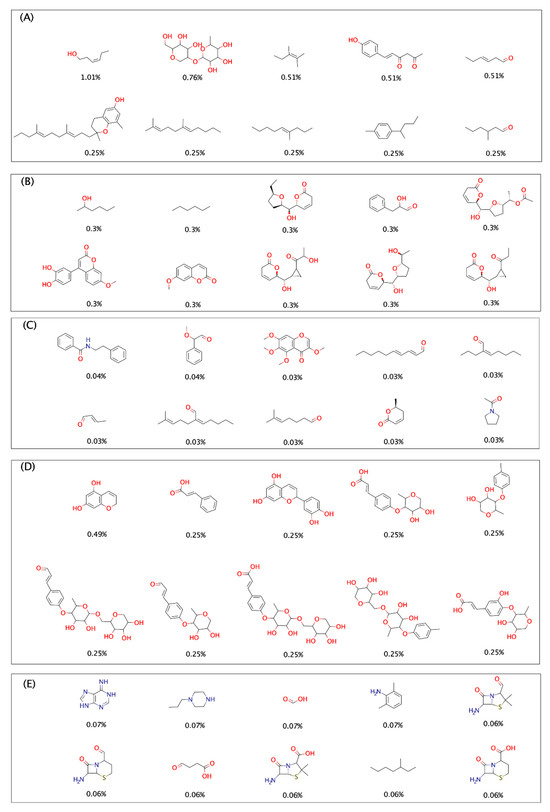
Figure 2.
The ten most frequent and unique fragments from (A) NPDBEjeCol, (B) BIOFACQUIM, (C) NuBBEDB, (D) PeruNPDB, and (E) FDA-approved drugs. The percentage of each fragment within its database is indicated below the chemical structures.
2.3. Structural Content, Composition, and Complexity of the Compound and Fragment Libraries
Table 1 summarizes the structural composition of all compounds in NPDBEjeCol and reference databases. Specifically, Table 1 reports the mean distribution values of various atom types, rings, heterocycles, and atom types, such as the fraction of sp3 carbon atoms, spiro, and bridgehead atoms as indicators of structural complexity. Using the same set of constitutional descriptors, Table 2 summarizes the profile of the fragment libraries obtained from NPDBEjeCol and the other four compound databases. The mean values of the distribution are reported.

Table 1.
Structural composition of compound libraries: NPDBEjeCol and reference databases a.

Table 2.
Structural composition of the fragment libraries from NPDBEejeCol and reference databases a.
The median values of the molecular weight and carbon atoms indicate that compounds in NPDBEjeCol (234 and 14, respectively) are, in general, smaller than the other natural product databases analyzed in this study as well as the FDA-approved drugs. The median values for the other reference databases are between 358 and 386 of molecular weight and 19–22 carbon atoms. Similarly, natural products from NPDBEjeCol are smaller than natural products from COCONUT (for example, a median of 26 carbon atoms as recently reported).5 Analogous conclusions can be derived, considering the number of heavy atoms. NPDBEjeCol has an average of 17 heavy atoms, while the reference databases are between 26 and 28 atoms. The count of oxygen and nitrogen atoms also shows significant differences. For NPDBEjeCol, there are, on average, three oxygen atoms, while for the other data sets, there are between four and six. Regarding nitrogen atoms, there are closeness values between NPDBEjeCol and BIOFACQUIM, but generally, all databases except FDA-approved drugs have less than one nitrogen atom on average. FDA-approved drugs have, on average, two nitrogen atoms. For sp3 carbon fractions and chiral carbons, the values are close for all databases. Concerning the presence of rings, reference databases on average have three rings, while in NPDBEjeCol, the value is lower (two rings). This same trend is observed for NPDBEjeCol and other databases in ring types. For spiro atoms, the value of NPDBEjeCol is very close to FDA-approved drugs and further away from the other three databases. For bridgehead atoms, NPDBEjeCol has the highest value, while PeruNPDB has the lowest, all values being less than one.
The fragment libraries generated from NPDBEjeCol had the smallest size as compared to the size of fragment libraries generated from other compound libraries (as measured by the number of carbon atoms, number of heavy atoms, and molecular weight) (Table 2). For carbon atoms, NPDBEjeCol contains 10, while other databases change from 16 to 25 atoms, with 25 in NuBBEDB. This trend is similar to molecular weight, with NPDBEjeCol displaying the smallest fragments and NuBBEDB the largest. The difference between NPDBEjeCol and the reference data sets is significant for oxygen and heavy atoms. Specifically, NPDBEjeCol contains two oxygen atoms, while the other databases range from four to nine. For heavy atoms, NPDBEjeCol reports 12, compared in the other database between 22 and 34 atoms. In sp3 carbon fractions, the values are closely aligned across all databases, underscoring the identical values for NPDBEjeCol and NuBBEDB, the highest values for five sets of molecules.
Structural Complexity
Structural complexity is a property that has been associated with the drug likeness of compound data sets [28]. Comparisons of the structural complexity of approved drugs and compounds under pre and clinical development have suggested that compounds in clinical use have, in general, an increased fraction of sp3 carbons that has been taken as a rough measure of structural complexity. Also, complexity has been considered as an indicator of selectivity or deceased promiscuity regarding biological interactions between small-molecule libraries [29,30]. Yet, structural complexity, although intuitive in principle, can be quantified in different ways systematically and consistently and today is an area of research, both computationally and experimentally [31,32]. One of the simplest but consistent ways to quantify complexity that has been adopted in cross-comparisons is the fraction of sp3 carbons [30,33]. Considering this metric as an approximate but well-established metric to quantify the structural complexity of compound data sets [33], we concluded that compounds in NPDBEjeCol were the most complex of all natural product data sets (median value of 0.63)—including natural products from COCONUT, which have a reported value of 0.51 [5]—and were more complex than the set of approved drugs (Table 1). The second most complex natural product database was PeruNPDB, and all four natural product data sets had an overall larger median value of the fraction of sp3 carbons than approved drugs, as previously noted for other natural product collections [29,34]. Similarly, compounds in NPDBEjeCol had the largest median values of bridgehead atoms (0.41) of all databases. In natural products, this knowledge is relevant because the increased structural complexity has been associated with the overall increased target selectivity or less promiscuity as compared to synthetic organic compounds with a smaller proportion of sp3 carbons [29,34].
Like the values obtained for entire chemical structures, the fragment libraries obtained from NPDBEjeCol and NuBBEDB had the largest fraction of sp3 carbons (median values of 0.63) (Table 2), followed by fragment libraries from PeruNPDB and BIOFACQUIM, and finally from approved drugs. Notably, in general, the complexity of fragment libraries from NPDBEjeCol and NuBBEDB was higher than the complexity of fragments generated from COCONUT, which had a reported value of 0.56 [5].
2.4. Structural Diversity
The fingerprint-based structural diversity of the compound and fragment libraries was measured with cumulative distribution functions of the pairwise similarity values that were calculated with the Tanimoto Coefficient and Morgan2, Morgan3, and MACCS keys fingerprints (see Section 3, Section 2.3). The results are summarized in Figure 3 and Figure 4. Figure 3 indicates that the chemical structures of the NPDBEjeCol database are the most diverse of the natural product databases regarding the three molecular fingerprints, and they are as diverse as the FDA-approved drug data set. The next most diverse natural product set is PeruNPDB, followed by NuBBEDB and BIOFACQUIM, which are the least diverse.
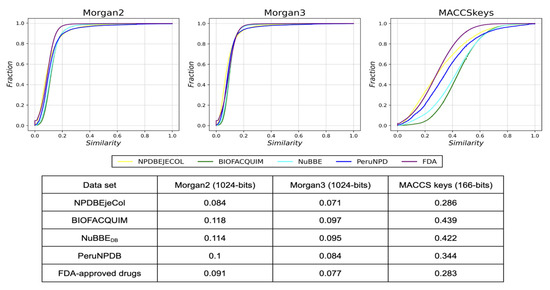
Figure 3.
Cumulative distribution functions of the pairwise Tanimoto similarity using MACCS keys (166-bits), Morgan2, and Morgan3 fingerprints of the compound libraries. NPDBEjeCol (yellow), BIOFACQUIM (green), NuBBEDB (blue), PeruNPD (dark blue), and FDA-approved drugs (purple). This table summarizes the median value of the distributions.
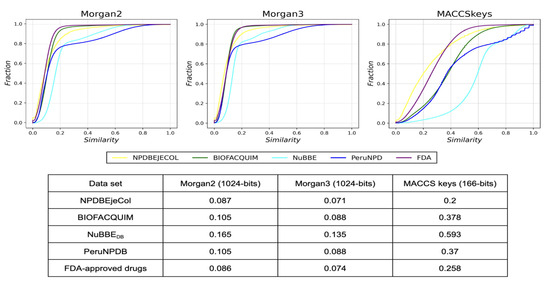
Figure 4.
Cumulative distribution functions of the pairwise Tanimoto similarity using MACCS key (166-bits), Morgan2, and Morgan3 fingerprints of the fragment libraries. Fragments from NPDBEjeCol (yellow), BIOFACQUIM (green), NuBBEDB (blue), PeruNPD (dark blue), and FDA-approved drugs (purple). This table summarizes the median value of the distributions.
The fragment library obtained from NPDBEjeCol was also the most diverse as measured by all three fingerprints (Figure 4), with comparable and high fingerprint-based diversity as the approved drugs from the FDA. BIOFACQUIM and PeruNPDB had similar diversity, followed by NuBBEDB, which was the least diverse of the fragment libraries analyzed.
2.5. Visual Representation of the Chemical Space and Chemical Multiverse
A visual representation of the chemical space of the compounds and fragment libraries was conducted with t-SNE and PCA. As detailed in the Section 3, we used multiple descriptors for a more comprehensive analysis of the chemical space, i.e., chemical multiverse. The visual representations were based on the t-SNE and PCA of similarity matrices of the pairwise comparisons of the similarity computed with the Tanimoto coefficient and three fingerprints. Two similarity matrices were generated, one with 5724 dimensions (Figure 5 and Figure 6) (compounds) and the second with 27,395 dimensions (Figure 7 and Figure 8) (fragments). The four figures show a comparison in the chemical space of NPDBEjeCol as compared to the chemical space of the other four natural product databases and the set of approved drugs.
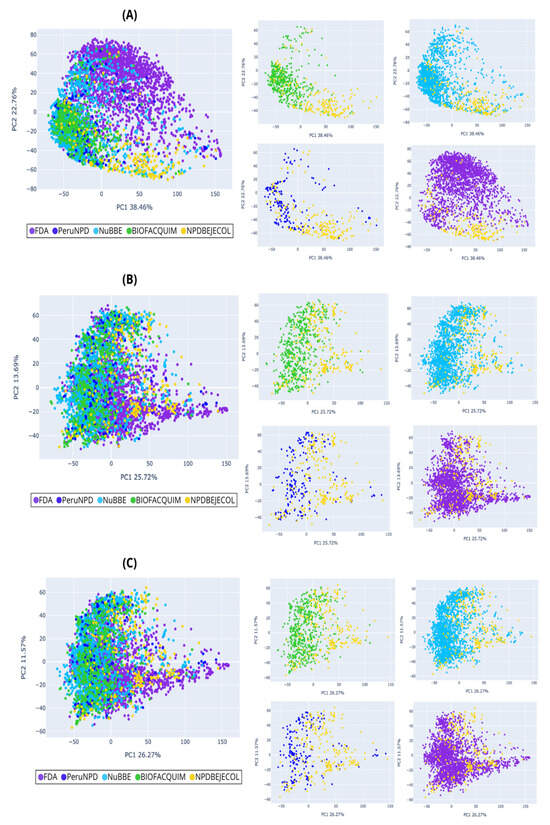
Figure 5.
Chemical multiverse visualization of the compound libraries: NPDBEjeCol and reference databases using principal component analysis based on (A) MACCS keys, (B) Morgan2, and (C) Morgan3 fingerprints. Compounds from NPDBEjeCol (yellow), BIOFACQUIM (green), NuBBEDB (blue), PeruNPDB (dark blue), and FDA-approved drugs (purple).
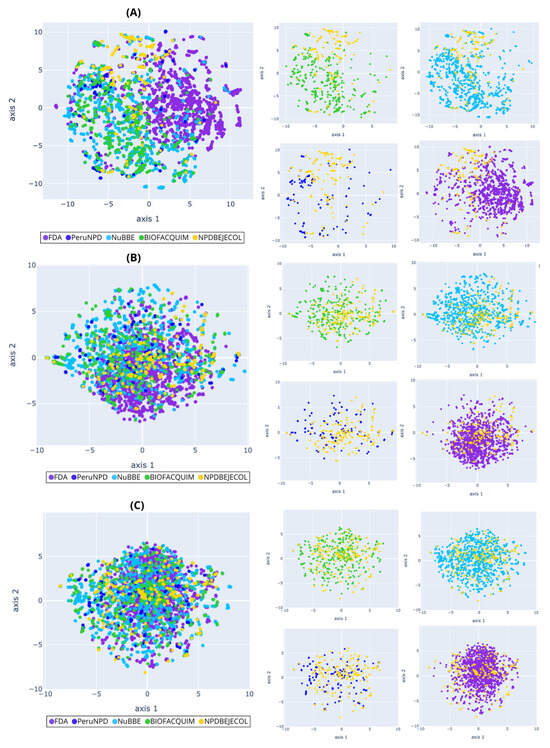
Figure 6.
Chemical multiverse visualization of the compound libraries: NPDBEjeCol and reference databases using t-SNE based on (A) MACCS keys, (B) Morgan2, and (C) Morgan3 fingerprints. Compounds from NPDBEjeCol (yellow), BIOFACQUIM (green), NuBBEDB (blue), PeruNPDB (dark blue), and FDA-approved drugs (purple).
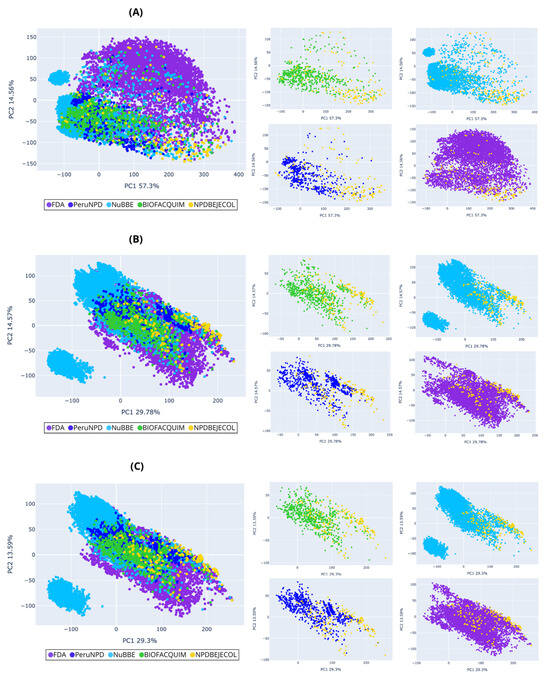
Figure 7.
Chemical multiverse visualization of the fragment libraries: NPDBEjeCol and reference databases using principal component analysis based on (A) MACCS keys, (B) Morgan2, and (C) Morgan3 fingerprints. Compounds from NPDBEjeCol (yellow), BIOFACQUIM (green), NuBBEDB (blue), PeruNPDB (dark blue), and FDA-approved drugs (purple).
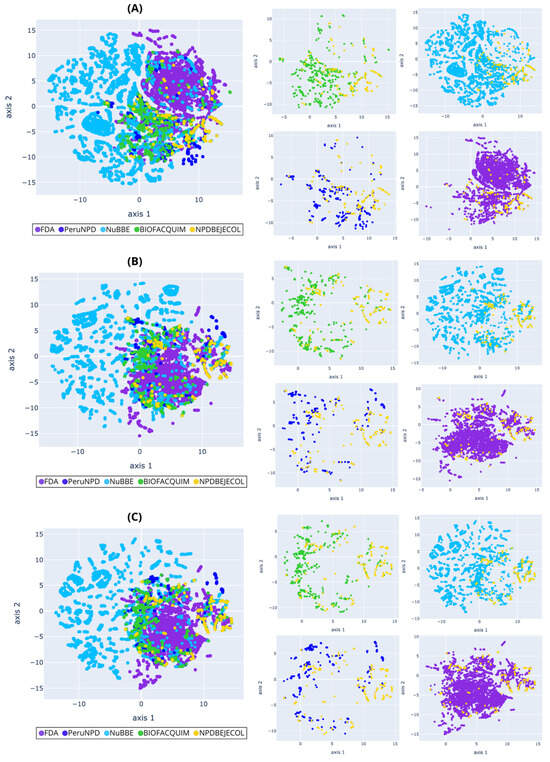
Figure 8.
Chemical multiverse visualization of the fragment libraries: NPDBEjeCol and reference databases using t-SNE based on (A) MACCS keys, (B) Morgan2, and (C) Morgan3 fingerprints. Compounds from NPDBEjeCol (yellow), BIOFACQUIM (green), NuBBEDB (blue), PeruNPDB (dark blue), and FDA-approved drugs (purple).
The PCA for compounds (Figure 5) shows that the chemical space based on MACCS keys recover more information from the variables due to principal component 1 recovering 38.46% of the variance, principal component 2 recovering 22.76% of the variance, and the accumulated variance recovered by the two principal components represented in Figure 5A was 61.22%, followed by the PCA based on Morgan2 (39.41%, Figure 5B) and Morgan3 (37.84%, Figure 5C). Similarly, the PCA for fragments showed that chemical space based on MACCS keys recover more information from the variables because principal component 1 recovered 57.3% and principal component 2 recovered 14.56%, and the accumulated variance recovered by two principal components represented in Figure 7A was 71.86%, followed by the PCA based on Morgan2 (44.35%, Figure 7B) and Morgan3 (42.89%, Figure 7C). Compounds from natural products and FDA-approved drugs share similar chemical space using Morgan2 and Morgan3 fingerprints. The PCA based on MACCS keys for compounds, the FDA-approved drugs covering different regions of the chemical space, and the all-natural product compounds (NPDBEjeCol, BIOFACQUIM, NuBBEDB, and PeruNPD) converge in the same chemical space. The PCA based on MACCS keys for fragments split the chemical space of the FDA-approved drugs into two regions. The less-dense region converges with the chemical space of fragments derived from natural products (NPDBEjeCol, BIOFACQUIM, NuBBEDB and PeruNPD).
Fragments derived from NuBBEDB have a unique fragment subset different from fragments generated from NPDBEjeCol, BIOFACQUIM, PeruNPD, and FDA-approved drugs, and they are shown in PCA and t-SNE based on MACCS keys, Morgan2, and Morgan3 (Figure 7 and Figure 8). Overall, the visualizations of the multiverse showed the large diversity of compounds in NPDBEjeCol. This large diversity is particularly notable in the large area of the chemical space as compared to drugs approved for clinical use. These results agree with the large fingerprint-based diversity analyzed in Section 3.4.
3. Materials and Methods
Table 3 summarizes the compound databases analyzed in this work, including the number of compounds before and after standardization, described in Section 2.1. One such database is NPDBEjeCol [12], a compilation of natural products isolated and characterized in Colombia. BIOFACQUIM is a collection of natural products isolated and characterized in Mexico [13]. The Nuclei of Bioassays, Ecophysiology, and Biosynthesis of Natural Products Database (NuBBEDB) is a collection of compounds from Brazil [14], and PeruNPDB is a database of natural products from Peru [15]. FDA-approved drugs were retrieved from DrugBank [16].

Table 3.
Compound libraries are studied in this work.
3.1. Data Set Standardization
Data set compounds encoded as SMILES strings [17] were standardized using open-source chemoinformatic toolkits RDKit version 2024.09.1 (ETH Zurich, Zurich, Switzerland) [18] and MolVS version 0.1.1 (Massachusetts Institute of Technology (MIT), Cambridge, MA, USA) [19], using a protocol described by Sánchez-Cruz et al. [20]. Briefly, chemical compounds were selected if they had chemical elements such as H, B, C, N, O, F, Si, P, S, Cl, Se, Br, and I. Stereochemistry information was retained. Compounds with multiple components were split, and the largest component was retained. The remaining compounds were neutralized and reionized to generate the corresponding canonical tautomer. Finally, duplicated compounds were deleted.
3.2. Fragment Analysis
In this study, molecular fragments were obtained with the validated REtrosyntheric Combinatorial Analysis Procedure (RECAP) [21], a function implemented in RDKit. The RECAP breaks eleven possible bonds such as amide, ester, urea, olefin, amine, aromatic nitrogen-aliphatic carbon, lactam nitrogen-aliphatic carbon, aromatic carbon-aromatic carbon, quaternary nitrogen, and sulphonamide. Compounds with molecular weight above 1000 Da were excluded from the fragmentation.
3.3. Structural Diversity and Complexity
Compounds and molecular fragments were analyzed regarding structural diversity, complexity, atoms, and ring content. The structural diversity was evaluated with the pairwise similarity values calculated with the Tanimoto coefficient [22], for both Morgan fingerprints with radius 2 (Morgan2, 1024-bits) and radius 3 (Morgan3, 1024-bits) [23], and Molecular ACCes System (MACCS) keys (166-bits) [24]. The structural differences between compound and fragment data sets were evaluated by comparing the profiles of 14 descriptors: The number of atoms of carbon, nitrogen, oxygen, and heavy atoms, the number of rings and heterocycles (both aliphatic and aromatic), spiro atoms, bridgehead atoms, the fraction of sp3 carbons, and chiral carbons.
3.4. Chemical Space and Chemical Multiverse
Chemical space has been defined as an M-dimensional cartesian space, and each dimension represents the descriptors or features encoding a molecule. The length of descriptor sets defines the number of dimensions of each chemical space [25]. The chemical multiverse is a natural extension of the concept of chemical space, and it has been conceptualized as a group of alternative chemical spaces of a set of compounds, each defined by a distinct set of molecular descriptors [26]. To generate a visual representation of the chemical space, we used two dimensionality reduction techniques, namely, t-distributed stochastic neighbor embedding (t-SNE) [27] and principal component analysis (PCA) based on the similarity matrices computed with the Tanimoto coefficient and MACCS keys, Morgan2, and Morgan3 fingerprints.
4. Conclusions
We generated a fragment library of the natural product collection from Colombia, NPDBEjeCol with 200 fragments, and three other major databases from Latin America: BIOFACQUIM, NuBBEDB, and PeruNPDB. In total, there were 26,540 unique fragment structures among all five databases. In agreement with open science and its symbiosis with artificial intelligence and other computational applications [35], all fragment libraries are freely available at https://github.com/DIFACQUIM/Fragment-Library-of-Colombian-Natural-Products.git (accessed on 1 October 2024). Analysis of the most frequent and unique fragments revealed that a hydroxylated linear chain and double bond were unique fragments with the largest abundance in NPDBEjeCol. Also, the fragments in the PeruNPDB were unique, with a large distinct proportion of oxygen atoms. Analysis of the structural complexity as measured by the fraction of sp3 carbons revealed that compounds from NPDBEjeCol were the most complex, with similar conclusions for the fragment libraries obtained from this novel data set. The overall high structural complexity of compounds in NPDBEjeCol and its associated fragment library make this collection highly attractive for drug discovery, particularly for designing selective compounds. From the analysis of structural diversity, we concluded that chemical structures and fragments derived from NPDBEjeCol are the most diverse of the natural product databases regarding the three molecular fingerprints, and they are as diverse as the set of approved drugs. Comparative visualization of the chemical multiverse of natural products in NPDBEjeCol and its related fragment library agreed with its large diversity. The fragment libraries from NPDBEjeCol and other natural product collections, plus the fragment library from the FDA drugs, add up to the fragment libraries of larger natural product databases such as COCONUT and can be used to design pseudo-natural products.
Author Contributions
A.L.C.-H. and J.R.R.-P.: investigation, methodology, data curation, writing—original draft, review and editing. H.F.C.-H., H.A.V.-S. and M.Á.C.-F.: investigation, methodology, supervision, funding acquisition. M.Á.C.-F.: writing—review and editing. J.L.M.-F.: conceptualization, supervision, writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
A.L.C.-H. is thankful to CONAHCyT, Mexico, for the Ph.D. scholarship number 847870. We also thank Marilia Valli, Adriano D. Andricopulo, and Vanderlan da S. Bolzani for making NuBBEDB accessible. Authors thank the Technological University of Pereira (UTP) through the Vicerrectoría de investigaciones, Innovación y Extensión for the development of the funded project: “Desarrollo de una biblioteca de productos naturales aislados y caracterizados de especies vegetales estudiadas en la región del Eje Cafetero, Colombia” code E3-23-1. We also thank the computational resources for using the Miztli supercomputer at UNAM under the project LANCAD-UNAM-DGTIC-335. We are also grateful to DGAPA, UNAM, Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT), Grant No. IG200124.
Data Availability Statement
The data presented in this study are openly available in Github at https://github.com/DIFACQUIM/Fragment-Library-of-Colombian-Natural-Products.git (accessed on 1 October 2024).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; International Natural Product Sciences Taskforce; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Grigalunas, M.; Brakmann, S.; Waldmann, H.J. Chemical Evolution of Natural Product Structure. Am. Chem. Soc. 2022, 144, 3314–3329. [Google Scholar] [CrossRef] [PubMed]
- Chávez-Hernández, A.L.; Sánchez-Cruz, N.; Medina-Franco, J.L. A Fragment Library of Natural Products and its Comparative Chemoinformatic Characterization. Mol. Inform. 2020, 39, e2000050. [Google Scholar] [CrossRef] [PubMed]
- Chávez-Hernández, A.L.; Sánchez-Cruz, N.; Medina-Franco, J.L. Fragment Library of Natural Products and Compound Databases for Drug Discovery. Biomolecules 2020, 10, 1518. [Google Scholar] [CrossRef]
- Chen, Y.; Kirchmair, J. Cheminformatics in Natural Product-based Drug Discovery. Mol. Inform. 2020, 39, e2000171. [Google Scholar] [CrossRef]
- Sorokina, M.; Merseburger, P.; Rajan, K.; Yirik, M.A.; Steinbeck, C.J. COCONUT online: Collection of Open Natural Products database. J. Cheminformatics 2021, 13, 2. [Google Scholar] [CrossRef]
- Gallo, K.; Kemmler, E.; Goede, A.; Becker, F.; Dunkel, M.; Preissner, R.; Banerjee, P. SuperNatural 3.0—a database of natural products and natural product-based derivatives. Nucleic Acids Res. 2023, 51, D654–D659. [Google Scholar] [CrossRef]
- Gu, J.; Gui, Y.; Chen, L.; Yuan, G.; Lu, H.-Z.; Xu, X. Use of Natural Products as Chemical Library for Drug Discovery and Network Pharmacology. PLoS ONE 2013, 8, e62839. [Google Scholar] [CrossRef]
- Karageorgis, G.; Foley, D.J.; Laraia, L.; Brakmann, S.; Waldmann, H. Pseudo Natural Products-Chemical Evolution of Natural Product Structure. Angew. Chem. Int. Ed Engl. 2021, 60, 15705–15723. [Google Scholar] [CrossRef]
- Catálogo de Plantas y Líquenes de Colombia. Available online: https://ipt.biodiversidad.co/sib/resource?r=catalogo_plantas_liquenes (accessed on 22 April 2024).
- Rodríguez-Pérez, J.R.; Valencia-Sanchez, H.A.; Mosquera-Martinez, O.M.; Gómez-García, A.; Medina-Franco, J.L.; Cortes-Hernandez, H.F. Fragment Library of Colombian Natural Products: Generation and Comparative Chemoinformatic Analysis. ChemRxiv 2024. [Google Scholar] [CrossRef]
- Pilón-Jiménez, B.A.; Saldívar-González, F.I.; Díaz-Eufracio, B.I.; Medina-Franco, J.L. BIOFACQUIM: A Mexican Compound Database of Natural Products. Biomolecules 2019, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Barazorda-Ccahuana, H.L.; Ranilla, L.G.; Candia-Puma, M.A.; Cárcamo-Rodriguez, E.G.; Centeno-Lopez, A.E.; Davila-Del-Carpio, G.; Medina-Franco, J.L.; Chávez-Fumagalli, M.A. PeruNPDB: The Peruvian Natural Products Database for in silico drug screening. Sci. Rep. 2023, 13, 7577. [Google Scholar] [CrossRef] [PubMed]
- Pilon, A.C.; Valli, M.; Dametto, A.C.; Pinto, M.E.F.; Freire, R.T.; Castro-Gamboa, I.; Andricopulo, A.D.; Bolzani, V.S. NuBBEDB: An updated database to uncover chemical and biological information from Brazilian biodiversity. Sci. Rep. 2017, 7, 7215. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef]
- Weininger, D.; Weininger, A.; Weininger, J.L. SMILES. 2. Algorithm for generation of unique SMILES notation. J. Chem. Inf. Comput. Sci. 1989, 29, 97–101. [Google Scholar] [CrossRef]
- RDKit. Available online: https://www.rdkit.org (accessed on 8 January 2022).
- MolVS. Available online: https://molvs.readthedocs.io/en/latest/ (accessed on 8 January 2022).
- Sánchez-Cruz, N.; Pilón-Jiménez, B.A.; Medina-Franco, J.L. Functional group and diversity analysis of BIOFACQUIM: A Mexican natural product database. F1000Research 2019, 8, 2071. [Google Scholar] [CrossRef]
- Lewell, X.Q.; Judd, D.B.; Watson, S.P.; Hann, M.M. RECAP—retrosynthetic combinatorial analysis procedure: A powerful new technique for identifying privileged molecular fragments with useful applications in combinatorial chemistry. J. Chem. Inf. Comput. Sci. 1998, 38, 511–522. [Google Scholar] [CrossRef]
- Jaccard, P. étude Comparative de la distribuition florale dans une portion des Alpes et des Jura. Bull. Soc. Vaud. Sci. Nat. 1901, 37, 547–579. [Google Scholar]
- Rogers, D.; Hahn, M.J. Extended-Connectivity Fingerprints. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef]
- Durant, J.L.; Leland, B.A.; Henry, D.R.; Nourse, J.G. Reoptimization of MDL keys for use in drug discovery. J. Chem. Inf. Comput. Sci. 2002, 42, 1273–1280. [Google Scholar] [CrossRef] [PubMed]
- Virshup, A.M.; Contreras-García, J.; Wipf, P.; Yang, W.; Beratan, D.N. Stochastic Voyages into Uncharted Chemical Space Produce a Representative Library of All Possible Drug-Like Compounds. J. Am. Chem. Soc. 2013, 135, 7296–7303. [Google Scholar] [CrossRef] [PubMed]
- Medina-Franco, J.L.; Chávez-Hernández, A.L.; López-López, E.; Saldívar-González, F.I. Chemical multiverse: An expanded view of chemical space. Mol. Inform. 2022, 41, e2200116. [Google Scholar] [CrossRef] [PubMed]
- Van der Maaten, L.; Hinton, G.J. Visualizing High-Dimensional Data Using t-SNE. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Wei, W.; Cherukupalli, S.; Jing, L.; Liu, X.; Zhan, P. Fsp3: A new parameter for drug-likeness. Drug Discov. Today 2020, 25, 1839–1845. [Google Scholar] [CrossRef]
- Clemons, P.A.; Bodycombe, N.E.; Carrinski, H.A.; Wilson, J.A.; Shamji, A.F.; Wagner, B.K.; Koehler, A.N.; Schreiber, S.L. Small molecules of different origins have distinct distributions of structural complexity that correlate with protein-binding profiles. Proc. Natl. Acad. Sci. USA 2010, 107, 18787–18792. [Google Scholar] [CrossRef]
- Lovering, F. Escape from Flatland 2: Complexity and promiscuity. Med. Chem. Commun. 2013, 4, 515–519. [Google Scholar] [CrossRef]
- Oprea, T.I.; Bologa, C.J. Molecular Complexity: You Know It When You See It. Med. Chem. 2023, 66, 12710–12714. [Google Scholar] [CrossRef]
- Jirasek, M.; Sharma, A.; Bame, J.R.; Mehr, S.H.M.; Bell, N.; Marshall, S.M.; Mathis, C.; MacLeod, A.; Cooper, G.J.T.; Swart, M.; et al. Investigating and Quantifying Molecular Complexity Using Assembly Theory and Spectroscopy. ACS Cent. Sci. 2024; in press. [Google Scholar] [CrossRef]
- Lovering, F.; Bikker, J.; Humblet, C.J. Escape from flatland: Increasing saturation as an approach to improving clinical success. Med. Chem. 2009, 52, 6752–6756. [Google Scholar] [CrossRef]
- Lachance, H.; Wetzel, S.; Kumar, K.; Waldmann, H.J. Charting, navigating, and populating natural product chemical space for drug discovery. Med. Chem. 2012, 55, 5989–6001. [Google Scholar] [CrossRef] [PubMed]
- Brinkhaus, H.O.; Rajan, K.; Schaub, J.; Zielesny, A.; Steinbeck, C. Open data and algorithms for open science in AI-driven molecular informatics. Curr. Opin. Struct. Biol. 2023, 79, 102542. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).