Indazole–Quinolone Hybrids as Anti-Virulence Agents against Pseudomonas aeruginosa
Abstract
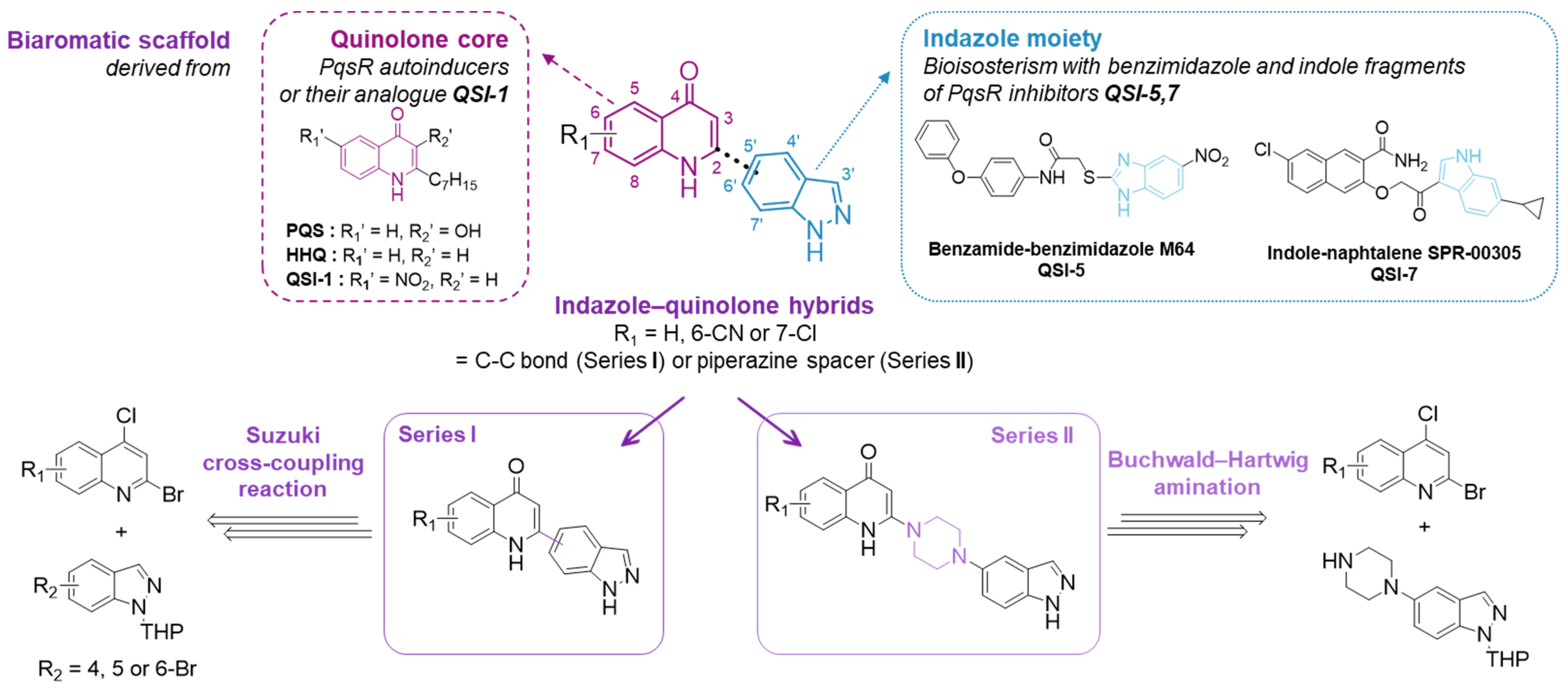
1. Introduction
2. Results and Discussion
2.1. Chemistry
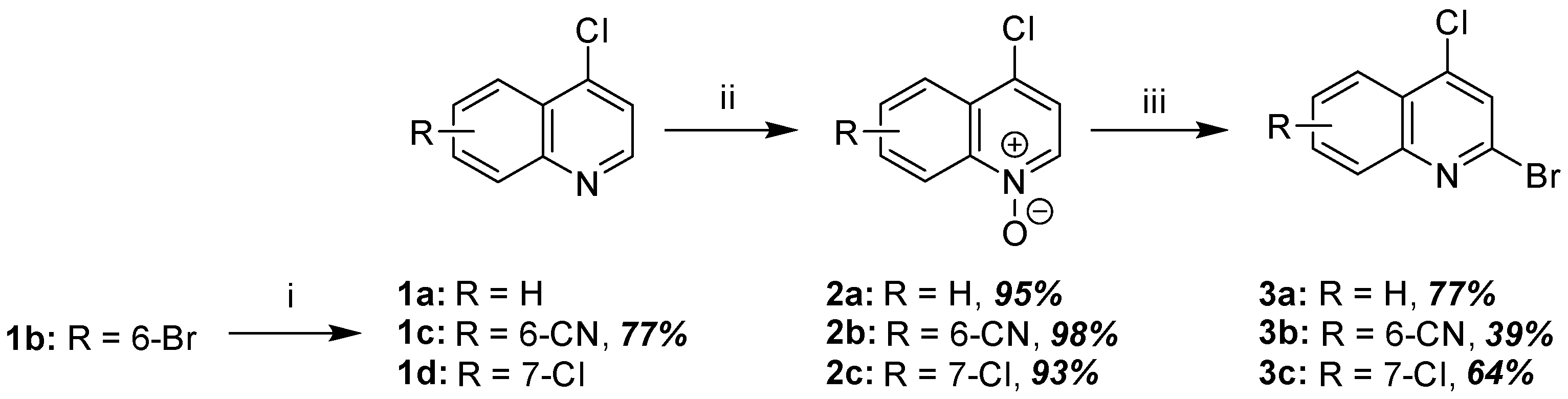
2.1.1. Synthesis of 2-Bromo-4-Chloroquinoline Precursors
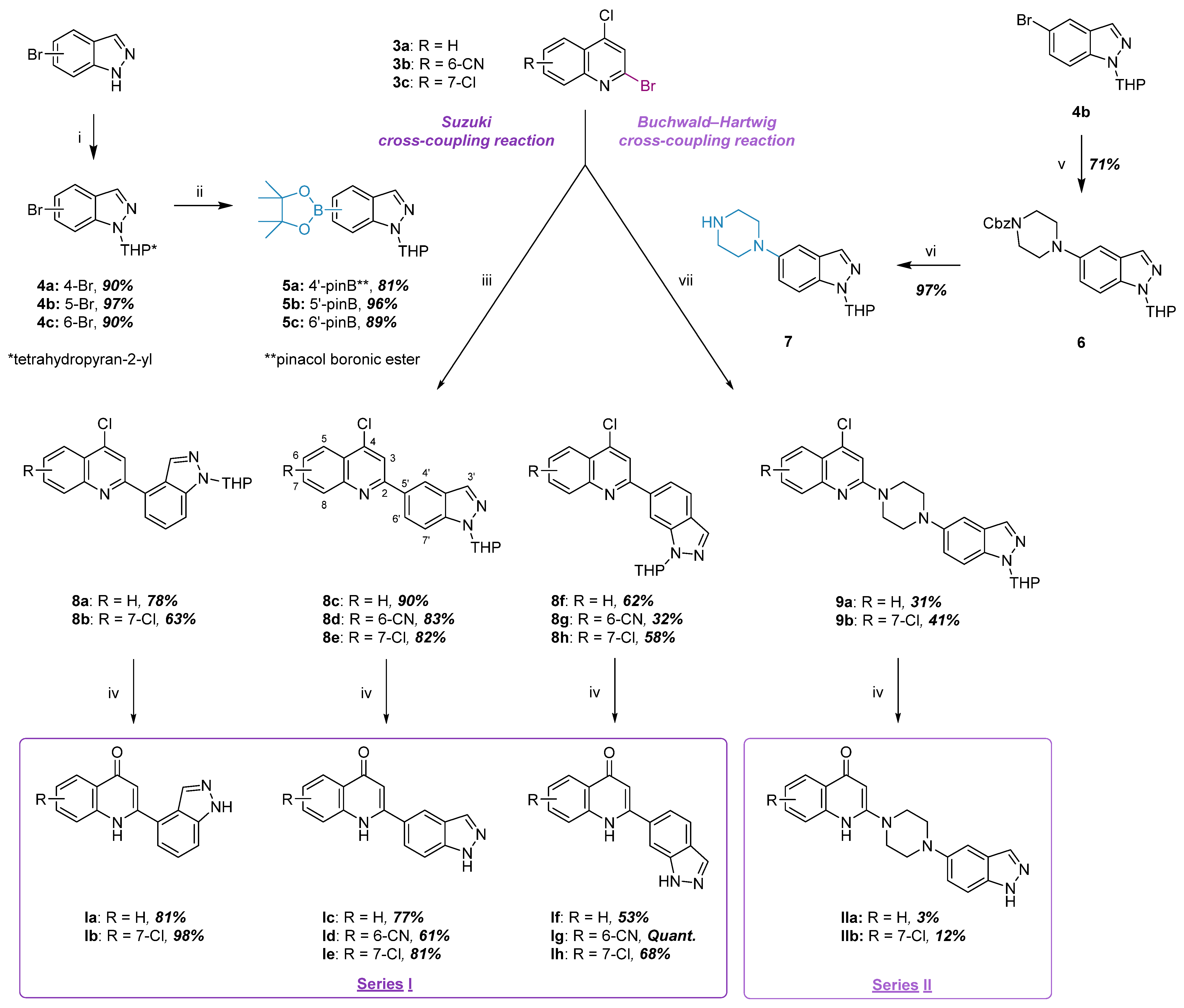
2.1.2. Synthesis of Indazole–Quinolone Hybrids
2.2. Physicochemical and Biological Prerequisite Evaluation
2.2.1. In Silico-Predicted Physicochemical Properties
2.2.2. Pseudomonal Minimum Inhibitory Concentrations (MICs)
2.2.3. Eukaryotic Cytotoxicity
2.3. In Vitro Anti-Virulence Evaluation
2.3.1. Anti-Biofilm Activity
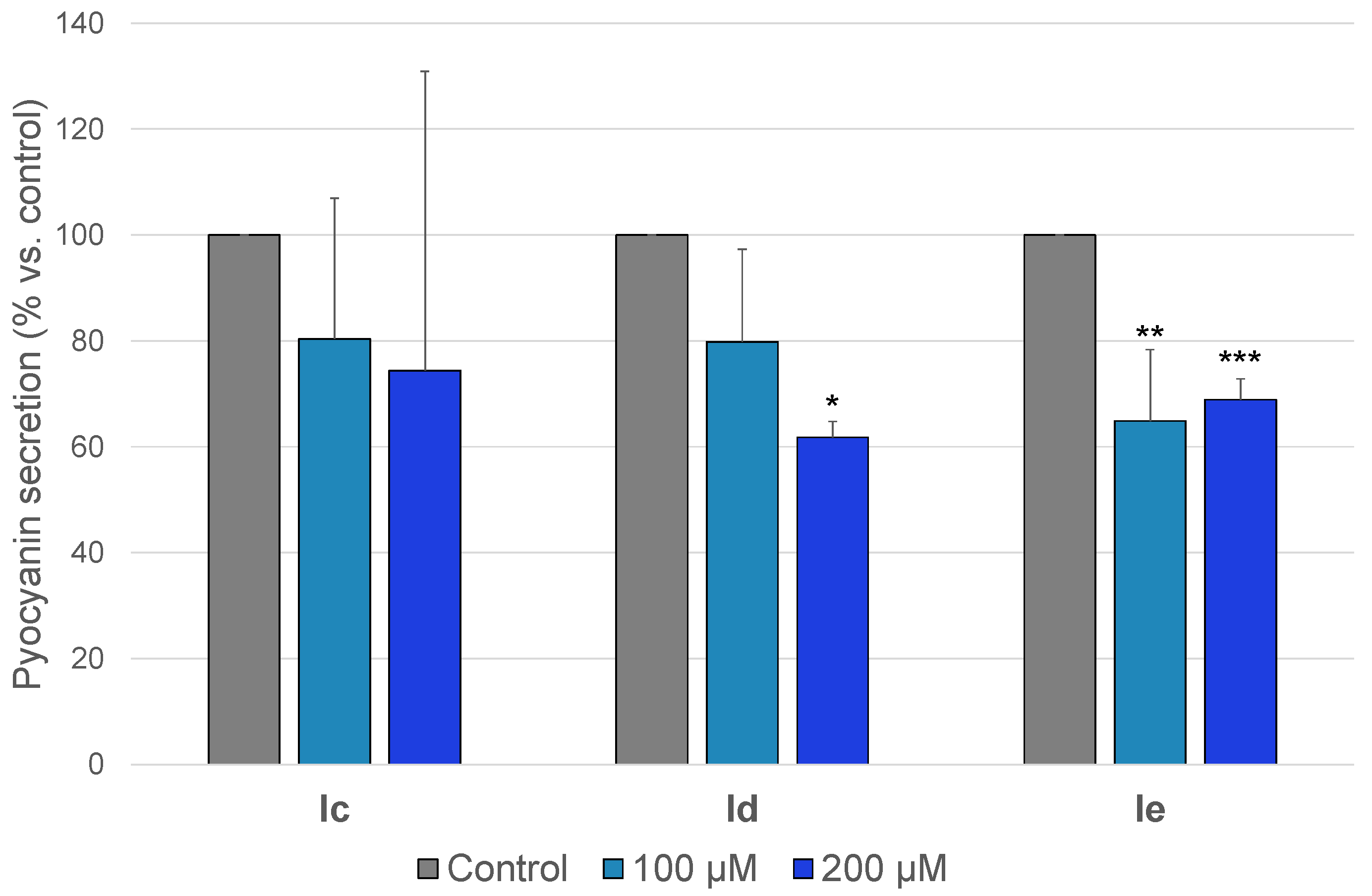
2.3.2. Anti-Pyocyanin Activity
2.4. Anti-QS Studies
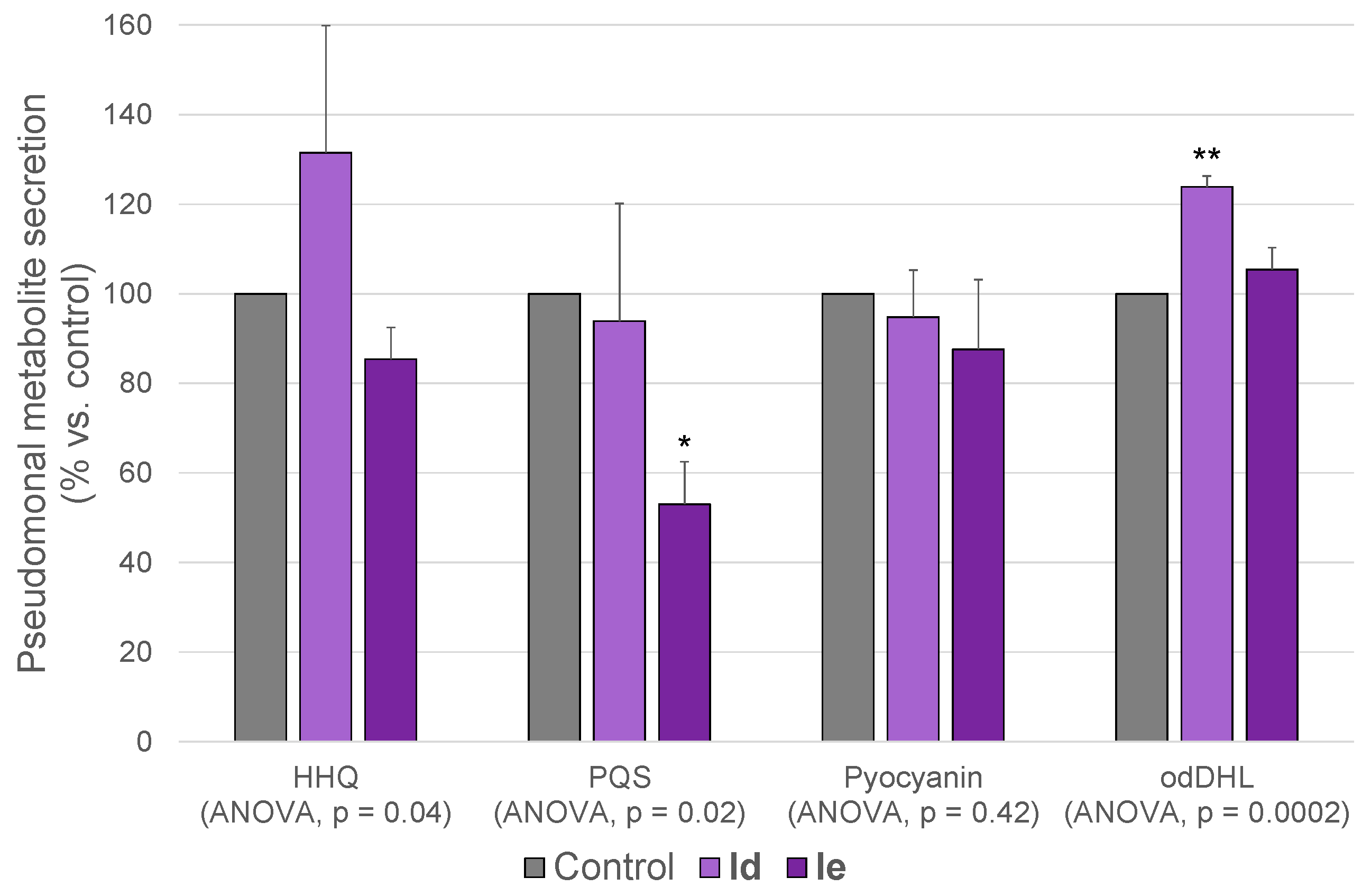
2.4.1. Metabolomics
2.4.2. Molecular Modelling
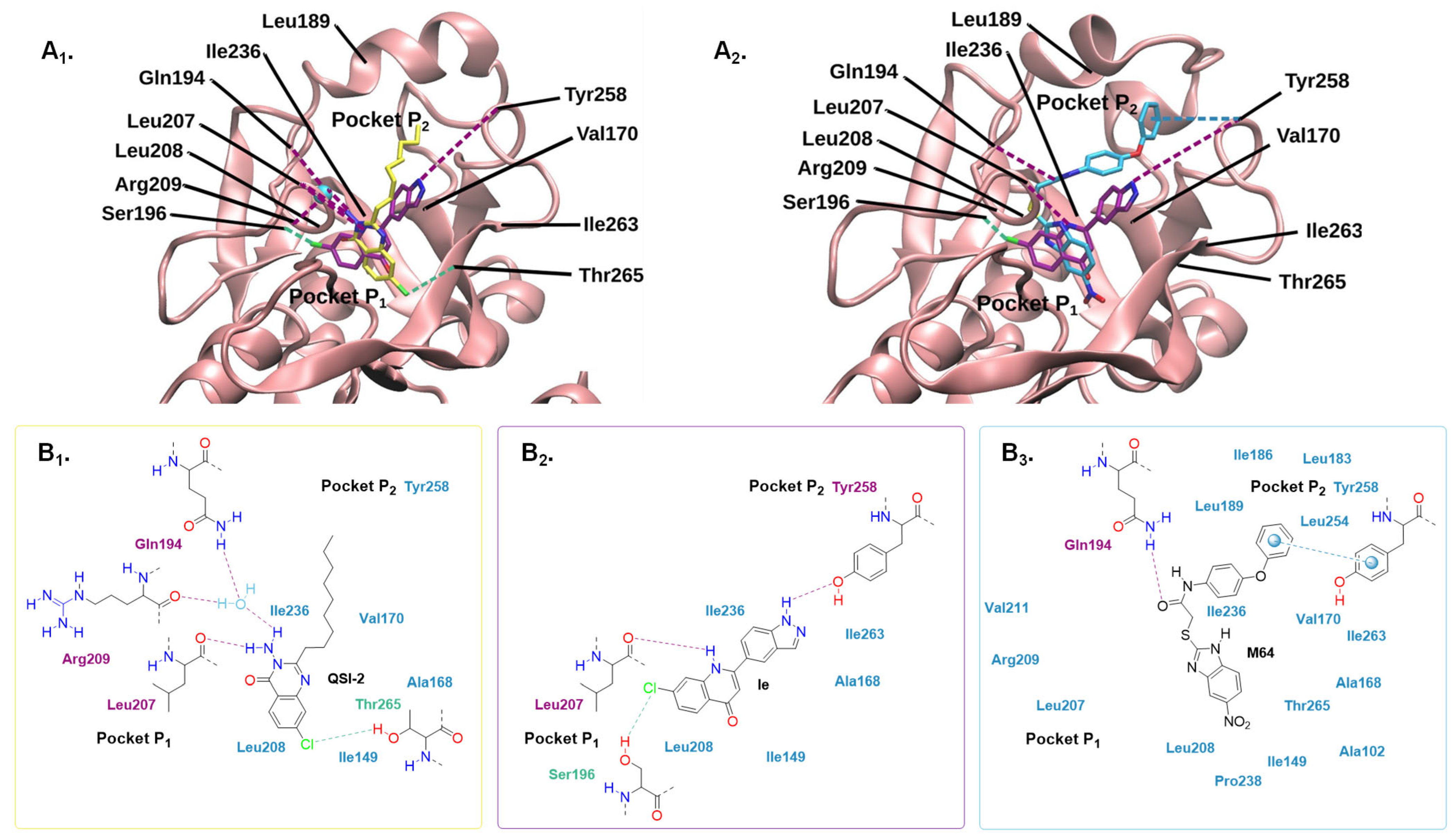
3. Materials and Methods
3.1. Chemistry
3.1.1. Synthesis of Indazole–Quinoline Hybrids 8a-h and 9a,b
- General procedure for the synthesis of compounds 8a–h
- 4-chloro-2-(1-(tetrahydro-2H-pyran-2-yl)-1H-indazol-4-yl)quinoline (8a)
- 4,7-dichloro-2-(1-(tetrahydro-2H-pyran-2-yl)-1H-indazol-4-yl)quinoline (8b)
- 4-chloro-2-(1-(tetrahydro-2H-pyran-2-yl)-1H-indazol-5-yl)quinoline (8c)
- 4-chloro-2-(1-(tetrahydro-2H-pyran-2-yl)-1H-indazol-5-yl)quinoline-6-carbonitrile (8d)
- 4,7-dichloro-2-(1-(tetrahydro-2H-pyran-2-yl)-1H-indazol-5-yl)quinoline (8e)
- 4-chloro-2-(1-(tetrahydro-2H-pyran-2-yl)-1H-indazol-6-yl)quinoline (8f)
- 4-chloro-2-(1-(tetrahydro-2H-pyran-2-yl)-1H-indazol-6-yl)quinoline-6-carbonitrile (8g)
- 4,7-dichloro-2-(1-(tetrahydro-2H-pyran-2-yl)-1H-indazol-6-yl)quinoline (8h)
- General procedure for synthesis of compounds 9a,b
- 4-chloro-2-(4-(1-(tetrahydro-2H-pyran-2-yl)-1H-indazol-5-yl)piperazin-1-yl)quinoline (9a)
- 4,7-dichloro-2-(4-(1-(tetrahydro-2H-pyran-2-yl)-1H-indazol-5-yl)piperazin-1-yl)quinoline (9b)
3.1.2. Synthesis of Indazole–Quinolone Hybrids Ia-h and IIa,b
- 2-(1H-indazol-4-yl)-4-quinolone (Ia)
- 7-chloro-2-(1H-indazol-4-yl)-4-quinolone (Ib)
- 2-(1H-indazol-5-yl)-4-quinolone (Ic)
- 2-(1H-indazol-5-yl)-4-oxo-1,4-dihydroquinoline-6-carbonitrile (Id)
- 7-chloro-2-(1H-indazol-5-yl)-4-quinolone (Ie)
- 2-(1H-indazol-6-yl)-4-quinolone (If)
- 2-(1H-indazol-6-yl)-4-oxo-1,4-dihydroquinoline-6-carbonitrile (Ig)
- 7-chloro-2-(1H-indazol-6-yl)-4-quinolone (Ih)
- 2-(4-(1H-indazol-5-yl)piperazin-1-yl)-4-quinolone (IIa)
- 7-chloro-2-(4-(1H-indazol-5-yl)piperazin-1-yl)-4-quinolone (IIb)
3.2. Biological Assays
3.2.1. MIC Determination
3.2.2. Cytotoxicity Assay
3.2.3. Biofilm Formation Assay
3.2.4. Pyocyanin Quantification Assay
3.2.5. Metabolomics
3.3. Molecular Modelling
3.4. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Mulcahy, L.R.; Isabella, V.M.; Lewis, K. Pseudomonas aeruginosa Biofilms in Disease. Microb. Ecol. 2014, 68, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef] [PubMed]
- Chadha, J.; Harjai, K.; Chhibber, S. Revisiting the Virulence Hallmarks of Pseudomonas aeruginosa: A Chronicle through the Perspective of Quorum Sensing. Environ. Microbiol. 2022, 24, 2630–2656. [Google Scholar] [CrossRef] [PubMed]
- Duplantier, M.; Lohou, E.; Sonnet, P. Quorum Sensing Inhibitors to Quench, P. Aeruginosa Pathogenicity. Pharmaceuticals 2021, 14, 1262. [Google Scholar] [CrossRef] [PubMed]
- Brindhadevi, K.; LewisOscar, F.; Mylonakis, E.; Shanmugam, S.; Verma, T.N.; Pugazhendhi, A. Biofilm and Quorum Sensing Mediated Pathogenicity in Pseudomonas aeruginosa. Process Biochem. 2020, 96, 49–57. [Google Scholar] [CrossRef]
- Rather, M.A.; Saha, D.; Bhuyan, S.; Jha, A.N.; Mandal, M. Quorum Quenching: A Drug Discovery Approach against Pseudomonas aeruginosa. Microbiol. Res. 2022, 264, 127173. [Google Scholar] [CrossRef] [PubMed]
- García-Reyes, S.; Soberón-Chávez, G.; Cocotl-Yanez, M. The Third Quorum-Sensing System of Pseudomonas aeruginosa: Pseudomonas Quinolone Signal and the Enigmatic PqsE Protein. J. Med. Microbiol. 2020, 69, 25–34. [Google Scholar] [CrossRef]
- Lin, J.; Cheng, J.; Wang, Y.; Shen, X. The Pseudomonas Quinolone Signal (PQS): Not Just for Quorum Sensing Anymore. Front. Cell. Infect. Microbiol. 2018, 8, 230. [Google Scholar] [CrossRef]
- Kamal, A.A.M.; Petrera, L.; Eberhard, J.; Hartmann, R.W. Structure–Functionality Relationship and Pharmacological Profiles of Pseudomonas aeruginosa Alkylquinolone Quorum Sensing Modulators. Org. Biomol. Chem. 2017, 15, 4620–4630. [Google Scholar] [CrossRef]
- Ilangovan, A.; Fletcher, M.; Rampioni, G.; Pustelny, C.; Rumbaugh, K.; Heeb, S.; Cámara, M.; Truman, A.; Chhabra, S.R.; Emsley, J.; et al. Structural Basis for Native Agonist and Synthetic Inhibitor Recognition by the Pseudomonas aeruginosa Quorum Sensing Regulator PqsR (MvfR). PLoS Pathog. 2013, 9, e1003508. [Google Scholar] [CrossRef]
- Li, Y.-B.; Liu, J.; Huang, Z.-X.; Yu, J.-H.; Xu, X.-F.; Sun, P.-H.; Lin, J.; Chen, W.-M. Design, Synthesis and Biological Evaluation of 2-Substituted 3-Hydroxy-6-Methyl-4H-Pyran-4-One Derivatives as Pseudomonas aeruginosa Biofilm Inhibitors. Eur. J. Med. Chem. 2018, 158, 753–766. [Google Scholar] [CrossRef]
- Liu, J.; Hou, J.-S.; Li, Y.-B.; Miao, Z.-Y.; Sun, P.-H.; Lin, J.; Chen, W.-M. Novel 2-Substituted 3-Hydroxy-1, 6-Dimethylpyridin-4 (1 H)-Ones as Dual-Acting Biofilm Inhibitors of Pseudomonas aeruginosa. J. Med. Chem. 2020, 63, 10921–10945. [Google Scholar] [CrossRef]
- Soukarieh, F.; Liu, R.; Romero, M.; Roberston, S.N.; Richardson, W.; Lucanto, S.; Oton, E.V.; Qudus, N.R.; Mashabi, A.; Grossman, S.; et al. Hit Identification of New Potent PqsR Antagonists as Inhibitors of Quorum Sensing in Planktonic and Biofilm Grown Pseudomonas aeruginosa. Front. Chem. 2020, 8, 204. [Google Scholar] [CrossRef]
- Maura, D.; Rahme, L.G. Pharmacological Inhibition of the Pseudomonas aeruginosa MvfR Quorum-Sensing System Interferes with Biofilm Formation and Potentiates Antibiotic-Mediated Biofilm Disruption. Antimicrob. Agents Chemother. 2017, 61, e01362-17. [Google Scholar] [CrossRef]
- Starkey, M.; Lepine, F.; Maura, D.; Bandyopadhaya, A.; Lesic, B.; He, J.; Kitao, T.; Righi, V.; Milot, S.; Tzika, A.; et al. Identification of Anti-Virulence Compounds That Disrupt Quorum-Sensing Regulated Acute and Persistent Pathogenicity. PLoS Pathog. 2014, 10, e1004321. [Google Scholar] [CrossRef]
- Singh, V.K.; Almpani, M.; Maura, D.; Kitao, T.; Ferrari, L.; Fontana, S.; Bergamini, G.; Calcaterra, E.; Pignaffo, C.; Negri, M.; et al. Tackling Recalcitrant Pseudomonas aeruginosa Infections in Critical Illness via Anti-Virulence Monotherapy. Nat. Commun. 2022, 13, 5103. [Google Scholar] [CrossRef]
- Zahler, R. Aryloxyacetylindoles and Analogs as Antibiotic Tolerance Inhibitors. Patent WO 2016/112088 A1, 14 July 2016. [Google Scholar]
- Maura, D.; Drees, S.L.; Bandyopadhaya, A.; Kitao, T.; Negri, M.; Starkey, M.; Lesic, B.; Milot, S.; Déziel, E.; Zahler, R.; et al. Polypharmacology Approaches against the Pseudomonas aeruginosa MvfR Regulon and Their Application in Blocking Virulence and Antibiotic Tolerance. ACS Chem. Biol. 2017, 12, 1435–1443. [Google Scholar] [CrossRef]
- Wang, D.; Jia, H.; Wang, W.; Wang, Z. A Practical and Mild Chlorination of Fused Heterocyclic N-Oxides. Tetrahedron Lett. 2014, 55, 7130–7132. [Google Scholar] [CrossRef]
- Crestey, F.; Lohou, E.; Stiebing, S.; Collot, V.; Rault, S. Protected Indazole Boronic Acid Pinacolyl Esters: Facile Syntheses and Studies of Reactivities in Suzuki–Miyaura Cross-Coupling and Hydroxydeboronation Reactions. Synlett 2009, 4, 615–619. [Google Scholar] [CrossRef]
- Mohajeri, A.; Shahamirian, M. Pi-Electron Delocalization in Aza Derivatives of Naphthalene and Indole. Comput. Theor. Chem. 2011, 976, 19–29. [Google Scholar] [CrossRef]
- Lohou, E.; Collot, V.; Stiebing, S.; Rault, S. Direct Access to 3-Aminoindazoles by Buchwald–Hartwig C–N Coupling Reaction. Synthesis 2011, 16, 2651–2663. [Google Scholar] [CrossRef]
- Ferreira, I.C.F.R.; Queiroz, M.-J.R.P.; Kirsch, G. Synthesis of Diarylamines in the Benzo [b] Thiophene Series Bearing Electron Donating or Withdrawing Groups by Buchwald–Hartwig C–N Coupling. Tetrahedron 2003, 59, 975–981. [Google Scholar] [CrossRef]
- O’Neill, P.; Biagini, G.; Ward, S.A.; Berry, N.G.; Nixon, G.; Amewu, R.K.; Pidathala, C.; Hong, W.D.; Gibbons, P.; Leung, S.C.; et al. Antimalarial Compounds. Patent WO 2012/069856 A1, 31 May 2012. [Google Scholar]
- O’Shea, R.; Moser, H.E. Physicochemical Properties of Antibacterial Compounds: Implications for Drug Discovery. J. Med. Chem. 2008, 51, 2871–2878. [Google Scholar] [CrossRef]
- Turkina, M.V.; Vikström, E. Bacteria-Host Crosstalk: Sensing of the Quorum in the Context of Pseudomonas aeruginosa Infections. J. Innate Immun. 2019, 11, 263–279. [Google Scholar] [CrossRef]
- Schrödinger Release. Epik, 2023-3; Schrödinger, LLC.: New York, NY, USA, 2023.
- Schrödinger Release. QikProp, 2023-3; Schrödinger, LLC.: New York, NY, USA, 2023.
- Entryway. Available online: https://www.entry-way.org/ (accessed on 8 March 2023).
- Richter, M.F.; Drown, B.S.; Riley, A.P.; Garcia, A.; Shirai, T.; Svec, R.L.; Hergenrother, P.J. Predictive Compound Accumulation Rules Yield a Broad-Spectrum Antibiotic. Nature 2017, 545, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.F.; Hergenrother, P.J. The Challenge of Converting Gram-Positive-Only Compounds into Broad-Spectrum Antibiotics: Challenges in Developing Broad-Spectrum Antibiotics. Ann. N. Y. Acad. Sci. 2019, 1435, 18–38. [Google Scholar] [CrossRef] [PubMed]
- Prajapati, J.D.; Kleinekathöfer, U.; Winterhalter, M. How to Enter a Bacterium: Bacterial Porins and the Permeation of Antibiotics. Chem. Rev. 2021, 121, 5158–5192. [Google Scholar] [CrossRef]
- Sharma, P.C.; Jain, A.; Jain, S.; Pahwa, R.; Yar, M.S. Ciprofloxacin: Review on Developments in Synthetic, Analytical, and Medicinal Aspects. J. Enzym. Inhib. Med. Chem. 2010, 25, 577–589. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Kuehn, M.J. Outer-Membrane Vesicles from Gram-Negative Bacteria: Biogenesis and Functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Schertzer, J.W.; Yong, X. Molecular Conformation Affects the Interaction of the Pseudomonas Quinolone Signal with the Bacterial Outer Membrane. J. Biol. Chem. 2019, 294, 1089–1094. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing [EUCAST]. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 13.0. Available online: https://www.eucast.org (accessed on 27 April 2023).
- Grace, A.; Sahu, R.; Owen, D.R.; Dennis, V.A. Pseudomonas aeruginosa Reference Strains PAO1 and PA14: A Genomic, Phenotypic, and Therapeutic Review. Front. Microbiol. 2022, 13, 1023523. [Google Scholar] [CrossRef] [PubMed]
- Haney, E.; Trimble, M.; Cheng, J.; Vallé, Q.; Hancock, R. Critical Assessment of Methods to Quantify Biofilm Growth and Evaluate Antibiofilm Activity of Host Defence Peptides. Biomolecules 2018, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Savitskii, M.V.; Moskaleva, N.E.; Brito, A.; Zigangirova, N.A.; Soloveva, A.V.; Sheremet, A.B.; Bondareva, N.E.; Lubenec, N.L.; Kuznetsov, R.M.; Samoylov, V.M.; et al. Pharmacokinetics, Quorum-Sensing Signal Molecules and Tryptophan-Related Metabolomics of the Novel Anti-Virulence Drug Fluorothiazinon in a Pseudomonas aeruginosa-Induced Pneumonia Murine Model. J. Pharm. Biomed. Anal. 2023, 236, 115739. [Google Scholar] [CrossRef] [PubMed]
- Kitao, T.; Lepine, F.; Babloudi, S.; Walte, F.; Steinbacher, S.; Maskos, K.; Blaesse, M.; Negri, M.; Pucci, M.; Zahler, B.; et al. Molecular Insights into Function and Competitive Inhibition of Pseudomonas aeruginosa Multiple Virulence Factor Regulator. mBio 2018, 9, e02158-17. [Google Scholar] [CrossRef] [PubMed]
- Shandil, S.; Yu, T.T.; Sabir, S.; Black, D.S.; Kumar, N. Synthesis of Novel Quinazolinone Analogues for Quorum Sensing Inhibition. Antibiotics 2023, 12, 1227. [Google Scholar] [CrossRef] [PubMed]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A Program to Generate Schematic Diagrams of Protein-Ligand Interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand-Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Lohou, E.; Sopkova-de Oliveira Santos, J.; Schumann-Bard, P.; Boulouard, M.; Stiebing, S.; Rault, S.; Collot, V. New Hypotheses for the Binding Mode of 4- and 7-Substituted Indazoles in the Active Site of Neuronal Nitric Oxide Synthase. Bioorg. Med. Chem. 2012, 20, 5296–5304. [Google Scholar] [CrossRef]
- Savage, S.; McClory, A.; Zhang, H.; Cravillion, T.; Lim, N.-K.; Masui, C.; Robinson, S.J.; Han, C.; Ochs, C.; Rege, P.D.; et al. Synthesis of Selective Estrogen Receptor Degrader GDC-0810 via Stereocontrolled Assembly of a Tetrasubstituted All-Carbon Olefin. J. Org. Chem. 2018, 83, 11571–11576. [Google Scholar] [CrossRef] [PubMed]
- M07-A10; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard-Tenth Edition. CLSI: Wayne, PA, USA, 2015; ISBN 978-1-56238-987-1.
- O’Toole, G.A. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2011, e2437. [Google Scholar] [CrossRef]
- Bortolotti, D.; Trapella, C.; Bragonzi, A.; Marchetti, P.; Zanirato, V.; Alogna, A.; Gentili, V.; Cervellati, C.; Valacchi, G.; Sicolo, M.; et al. Conjugation of LasR Quorum-Sensing Inhibitors with Ciprofloxacin Decreases the Antibiotic Tolerance of P. aeruginosa Clinical Strains. J. Chem. 2019, 2019, 8143739. [Google Scholar] [CrossRef]
- Geddis, S.M.; Coroama, T.; Forrest, S.; Hodgkinson, J.T.; Welch, M.; Spring, D.R. Synthesis and Biological Evaluation of 1,2-Disubstituted 4-Quinolone Analogues of Pseudonocardia Sp. Natural Products. Beilstein J. Org. Chem. 2018, 14, 2680–2688. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Sanner, M.F. Python: A Programming Language for Software Integration and Development. J. Mol. Graph. Model. 1999, 17, 57–61. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting Modern Challenges in Visualization and Analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 19 October 2023).
- Signorell, A.; Aho, K.; Alfons, A.; Anderegg, N.; Aragon, T.; Arachchige, C.; Arppe, A.; Baddeley, A.; Barton, K.; Bolker, B.; et al. DescTools: Tools for Descriptive Statistics. R Package Version 0.99.47. Available online: https://cran.r-project.org/package=DescTools (accessed on 19 October 2023).

| PqsR Inhibitors | Anti-QS Properties * | Anti-Virulence Properties ** | Ref. | |||
|---|---|---|---|---|---|---|
| % Inhibition or IC50 | Anti-Biofilm Activity | Anti-Pyocyanin Activity | ||||
| % Inhibition or IC50 | % Inhibition or IC50 | |||||
| Alkylquinolone autoinducer analogue QSI | QSI-1 ‡ |  | 0.051/1.32 µM a/b | ND *** | 1.9 µM j | [11] |
| QSI-2 ‡ | 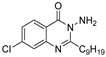 | 5 µM c | Observable using fluorescence microscopy | 50% at 50 µM k | [12] | |
| QSI-3 † | 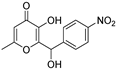 | 45% at 40 µM d | 20.31 µM h | 60% at 40 µM k | [13,14] | |
| QSI-4 † |  | 50% at 20 µM d | 6.57 µM h | 60% at 20 µM k | [14] | |
| Alkylquinolone autoinducer non-analogue QSI | QSI-5 †,✦ (M64) | 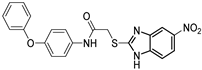 | 0.32/1.22 µM c/e | 50% at 10 µM h,i | 0.3 µM j | [15,16,17] |
| QSI-6 †,✦ (D88) |  | 1.31 µM f | 50% at 10 µM i | 0.53 µM j | [18] | |
| QSI-7 ‡,✦ (SPR00305) |  | 0.05–0.25 µM g | ND | 0.05–0.25 µM j | [19] | |
| Tested Compound | MW (g/mol) | Activity Against Bacterial Growth a | Eukaryotic Cytotoxicity (% Inhibition at 100 µM) b | Anti-Virulence Evaluation c | ||||
|---|---|---|---|---|---|---|---|---|
| MIC (µg/mL) | MIC (µM) | Anti-Biofilm Activity (% Inhibition) | Anti-Pyocyanin Activity (% Inhibition) | |||||
| 25 µM | 100 µM | 100 µM | 200 µM | |||||
| Ciprofloxacin | 331.34 | 0.06 | 0.18 | ND * | ND | ND | ND | ND |
| QSI-3 | 277.23 | >128 | >462 | ND | −16 | −20 | ND | ND |
| Ia | 261.28 | >128 | >490 | 31 | ø ** | ø | ND | ND |
| Ib | 295.73 | >128 | >433 | ND | 32 | 39 | ND | ND |
| Ic | 261.28 | >128 | >490 | 42 | ø | ø | ø | ø |
| Id | 286.29 | >128 | >447 | Not cytotoxic | 17 | ø | ø | 38 |
| Ie | 295.73 | >128 | >433 | 33 | 35 | 34 | 35 | 31 |
| If | 261.28 | >128 | >490 | Not cytotoxic | 15 | ø | ND | ND |
| Ig | 286.29 | >128 | >447 | ND | ø | 22 | ND | ND |
| Ih | 295.73 | >128 | >433 | Not cytotoxic | 29 | 30 | ND | ND |
| Metabolite | Retention Time (min) | Base Peak in the Mass Spectrum (ESI−) | |
|---|---|---|---|
| Ion | m/z | ||
| HHQ | 15.32 | [M + Cl]− | 278.1317 |
| PQS | 15.34 | [M − H]− | 258.1500 |
| Pyocyanin | 10.39 | [M + FA * − H]− | 255.0775 |
| odDHL | 15.6 | [M − H]− | 296.1867 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanot, M.; Duplantier, M.; Dalle, C.; Ren, Y.; Da Nascimento, S.; Becker, J.-P.; Taudon, N.; Lohou, E.; Sonnet, P. Indazole–Quinolone Hybrids as Anti-Virulence Agents against Pseudomonas aeruginosa. Drugs Drug Candidates 2024, 3, 512-536. https://doi.org/10.3390/ddc3030030
Hanot M, Duplantier M, Dalle C, Ren Y, Da Nascimento S, Becker J-P, Taudon N, Lohou E, Sonnet P. Indazole–Quinolone Hybrids as Anti-Virulence Agents against Pseudomonas aeruginosa. Drugs and Drug Candidates. 2024; 3(3):512-536. https://doi.org/10.3390/ddc3030030
Chicago/Turabian StyleHanot, Marie, Marine Duplantier, Céline Dalle, Yani Ren, Sophie Da Nascimento, Jean-Paul Becker, Nicolas Taudon, Elodie Lohou, and Pascal Sonnet. 2024. "Indazole–Quinolone Hybrids as Anti-Virulence Agents against Pseudomonas aeruginosa" Drugs and Drug Candidates 3, no. 3: 512-536. https://doi.org/10.3390/ddc3030030
APA StyleHanot, M., Duplantier, M., Dalle, C., Ren, Y., Da Nascimento, S., Becker, J.-P., Taudon, N., Lohou, E., & Sonnet, P. (2024). Indazole–Quinolone Hybrids as Anti-Virulence Agents against Pseudomonas aeruginosa. Drugs and Drug Candidates, 3(3), 512-536. https://doi.org/10.3390/ddc3030030










