Abstract
Therapeutic phages against pathogenic bacteria should kill the bacteria efficiently before the latter evolve resistance against the phages. While many factors contribute to phage efficiency in killing bacteria, such as phage attachment to host, delivery of phage genome into the host, phage mechanisms against host defense, phage biosynthesis rate, and phage life cycle, this paper focuses only on the optimization of phage mRNA for efficient translation. Phage mRNA may not be adapted to its host translation machinery for three reasons: (1) mutation disrupting adaptation, (2) a recent host switch leaving no time for adaptation, and (3) multiple hosts with different translation machineries so that adaptation to one host implies suboptimal adaptation to another host. It is therefore important to optimize phage mRNAs in therapeutic phages. Theoretical and practical principles based on many experiments were developed and applied to phages engineered against a drug-resistant Mycobacterium abscessus that infected a young cystic fibrosis patient. I provide a detailed genomic evaluation of the three therapeutic phages with respect to translation initiation, elongation, and termination, by making use of both experimental results and highly expressed genes in the host. For optimizing phage genes against M. abscessus, the start codon should be AUG. The DtoStart distance from base-pairing between the Shine-Dalgarno (SD) sequence and the anti-SD sequence should be 14–16. The stop codon should be UAA. If UAG or UGA is used as a stop codon, they should be followed by nucleotide U. Start codon, SD, or stop codon should not be embedded in a secondary structure that may obscure the signals and interfere with their decoding. The optimization framework should be generally applicable to developing therapeutic phages against bacterial pathogens.
1. Introduction
Phage therapy against bacterial infection has a long history, with success stories in both mice [1] and humans [2]. It gained momentum in recent years due to the alarming increase of bacterial resistance to antimicrobial drugs [3,4,5]. Bacterial resistance to penicillin became known soon after its discovery in 1928 and its routine medical applications in 1940 [6,7]. However, it is the recent emergence and prevalence of multidrug-resistant bacteria [8] that highlights the urgency of research in therapeutic phages as an attractive alternative to antibiotics [9,10]. What is particularly promising is that phage therapy works not only through phages lysing the bacterial pathogen, but also enhancing the human immune response against the pathogen by disabling one or more immune evasion mechanisms of the pathogen [11]. Thus, the bacterial pathogen is attacked by phages from within and by human immune responses from without.
1.1. General Criteria for Optimizing Therapeutic Phages against Pathogenic Bacteria
A desirable therapeutic phage must have (1) a high bactericidal efficiency [12], (2) limited side effects [13], (3) a low chance of drug resistance [14], (4) a low cost of development [15], and (5) a low human immune response so that the therapeutic phage will not be cleared before it has a chance to kill the pathogenic bacteria [16,17,18,19]. However, among these desirable properties, the bactericidal efficiency is obviously the most important.
There are many other processes contributing to the bactericidal efficiency of phages, such as phage attachment to host, delivery of phage DNA into the host, susceptibility of phage DNA to host deoxyribonucleases, phage life cycle, viral genome replication, viral transcription, etc. All these factors contribute to generation time. However, the main objective of this paper is to develop principles for phage mRNA optimization for increased translation efficiency and to illustrate the application of these principles to improve the design of three phages against a bacterial pathogen, Mycobacterium abscessus. I should emphasize that this optimization of phage mRNAs is relevant only after the phage has already been shown to lyse the bacterial pathogen.
1.2. Principles of Optimizing Phage mRNA
A translation machinery is made of ribosomes, translation initiation factors, tRNAs, tRNA-charging enzymes, and release factors, as well as the energy that drives the translation machinery ([20], pp. 522–635). Optimization of mRNA for efficient translation depends mainly on two factors: selection and mutation [21,22,23]. Natural selection has operated for millions of years to optimize the translation machinery in bacteria. An organism that cannot mass-produce its proteins efficiently would be eliminated by its more efficient competitors. Well-adapted phages typically have their mRNA mimicking the highly expressed genes in their bacterial hosts to take advantage of the host translation machinery [24,25,26,27]. However, rapidly replicating bacteria such as Escherichia coli with generation time of only 20–30 min [28] are expected to approach translation optimum closer than slow-replicating bacteria such as M. abscessus with generation time of 4–5 h [29]. Mutations in bacterial genomes disrupt optimizing selection on their encoded mRNAs. It also disrupts adaptation of phage mRNA to their host environment [24,25], so that phages [24,25] or human viruses [30] with a high mutation rate are associated with less adaptation than those with relatively a low mutation rate.
Efficient translation requires efficient initiation, elongation, and termination, as well as mRNA stability, and limits the rate of biosynthesis of both bacteria [21,31,32,33] and phage [24,25,27,34]. Translation initiation is often the limiting step [35,36,37,38,39]. Efficient initiation generally requires (1) AUG as a start codon, (2) a well-positioned base-pairing between the Shine-Dalgarno (SD) sequence and the anti-SD (aSD) of the free 3’ end of the small ribosomal rRNA [40,41,42,43,44,45], and (3) no strong secondary structure that would embed the start codon or SD and obscure them from being decoded by initiation tRNA and aSD, respectively [46].
With efficient translation initiation, translation elongation becomes rate-limiting [31,33]. Codon-anticodon adaptation is invariably observed in rapidly replicating organisms [21,38,47,48]. Highly expressed genes such as ribosomal protein genes exhibit strong preferences of codons decoded by the most abundant tRNAs [31,47,48,49,50]. The same pattern was also found in phages [24,25,26,27]. Codon optimization is associated with an increased growth rate in E. coli. Genes with optimal codon usage tend to generate more proteins. Experimental replacement of minor codons by major codons, or vice versa, typically leads to increased or decreased translation rates.
Translation terminations in bacterial species are mediated by one or two release factors RF1 and RF2 encoded by prfA and prfB, with RF1 decoding UAA and UAG and RF2 decoding UAA and UGA. All three stop codons can be misread by tRNAs in bacterial species, and UGA appears to be the leakiest of the three, with a readthrough frequency of at least 10−3 to 10−2 in Salmonella typhimurium [51] and E. coli [52,53]. Stop codons UAA and UAG can also be leaky in bacteria [54,55]. Natural UAG readthrough frequency is mostly within the range of 1.1 × 10−4 to 7 × 10−3, depending on the nature of the downstream nucleotides [55,56,57,58]. The readthrough rate is the lowest for UAA, at frequencies [59] from 9 × 10−4 to less than 1 × 10−5 [55]. Overall, the available experimental data suggest that readthrough error rate in bacteria species is in the order of UGA > UAG > UAA [53,60,61,62,63,64,65]. The hypothesis that such small error rates should be biologically important may seem weak. However, I present empirical results to show that it is because highly expressed genes exhibit strong preferences of UAA against UGA.
Termination efficiency also depends on the nucleotide immediately downstream of the stop codon [66,67,68], leading to the proposal of the tetranucleotide stop signal including the +4 site [23,66,68,69,70,71]. The best documented case involves translation of prfB (encoding RF2) in E. coli in which an inframe UGA stop codon is followed by nucleotide C [72,73,74]. When RF2 is abundant, the inframe UGA is decoded correctly to terminate translation, generating a short nonfunctional peptide. When RF2 is rare, UGA is not decoded. A +1 frameshift leads to translation of GAC at a different coding frame, generating a functional RF2.
Phage proteins ideally should be produced in exact stoichiometric ratios if phage protein composition is fully known [75]. While this paper does not address stoichiometry directly, an understanding of the translation machinery and translation efficiency should facilitate stoichiometric optimization. For example, if protein A is 30 times as many as protein B in the final assembled virion, but only 3 times as many as protein B in the host bacterial cell, then increasing translation efficiency for protein A would bring us closer to the 10:1 stoichiometric ratio.
The optimization principles above have been derived mostly from model species, such as E. coli as a representative of the gram-negative bacteria and Bacillus subtilis as a representative of the gram-positive bacteria. For a specific bacterial pathogen, one must examine features of its translation machinery by contrasting its highly expressed and lowly expressed genes.
1.3. A Case Study: Optimizing Phages against M. abscessus
A recent successful application of phage therapy targets antibiotic-resistant M. abscessus [2]. The patient was 15 years old, suffered from cystic fibrosis, and underwent a lung transplantation, but was subsequently infected by an antibiotics-resistant bacterial pathogen. Genomic sequencing identified the pathogen as M. abscessus subsp. massiliense strain GD01, which is a common nontuberculous lung pathogen [76]. The three Mycobacterium phages targeting M. abscessus were Muddy, ZoeJ, and BPs from screening a large number of potential phage candidates [2]. The original study [2] was a compassionate therapeutic application for a terminally ill patient, and therefore could not afford much time for phage mRNA optimization.
Three significant findings emerged from our detailed genomic analysis of the pathogenic host and the three phages. Firstly, the translation machinery of M. abscessus strongly favors AUG as a start codon and UAA as a stop codon. They are strongly preferred by highly expressed genes in M. abscessus. Thus, therapeutic phages should be engineered to have AUG and UAA as start and stop codons, respectively. Secondly, secondary structure stability is much reduced near start and stop codons, especially in highly expressed genes. This is consistent with the hypothesis that a strong secondary structure embedding initiation and termination signals may interfere with the decoding of these signals. Therapeutic phage mRNA should be engineered to reduce secondary structure near important translation signals such as start codon, SD sequence, and stop codon. Thirdly, protein-coding genes in phage Muddy would benefit more from codon optimization than the other two phages. This is because M. abscessus, like M. tuberculosis [59,77], has a GC-rich genome, with its tRNA anticodons strongly in favor of G-ending and C-ending codons. Phage Muddy is relatively AT-rich, so its codon usage deviates more from the optimum than the other two GC-rich phages. Lastly, if stop codons UGA and UAG must be allowed in phage mRNA, they should be engineered to have nucleotide U at the site immediately following the stop codon.
2. Results
I present mRNA optimization specific to the M. abscessus strain GD01, which is the pathogen against which the three-phage cocktail was used. Results from GD01A and GD01B are nearly identical, so only results from GD01A were presented unless otherwise indicated.
2.1. Translation Initiation
I present results to show that the translation machinery of M. abscessus prefers AUG as a start codon, a much-reduced secondary structure stability in sequences near the start codon and SD sequence, and SD/aSD base-pairing with a DtoStart within the range of 13–18. Phage mRNAs should be so modified to achieve efficient translation.
2.1.1. Start Codon Usage
AUG is typically the preferred start codon in [27,35,78], which is also true in M. abscessus (Table 1.), together with E. coli and B. subtilis. The AUG usage as a start codon is presented for HEGs (highly expressed protein-coding genes, including ribosomal protein genes and the 11 highly expressed genes listed in Materials and Methods) and REST (all annotated protein-coding genes excluding HEG). HEGs consistently use AUG as a start codon more frequently than REST (Table 1), consistent with previous studies [27,34,79,80] that AUG is the start codon favored by the bacterial translation machinery. This effect is highly significant (p = 0.00986) when tested with a three-way log-linear model (SPECIES*START*EXPRESSION, where START has AUG and non-AUG categories and EXPRESSION has HEG and REST categories).

Table 1.
Rapidly replicating bacterial species tend to prefer AUG as a start codon more strongly than slowly replicating species.
Generalizing the early hypothesis that highly expressed genes are more optimized than lowly expressed genes [50], one may also hypothesize that rapidly replicating bacterial species should be under stronger selection pressure to optimize their translation machinery than slowly replicating organisms. E. coli replicates once every 20–30 min [28], B. subtilis once every 30–70 min [81], but M. abscessus once every 4–5 h [29]. If AUG is universally a better start codon than other alternatives, then one would predict that AUG should be used most frequently by E. coli, followed by B. subtilis, and finally by M. abscessus. This prediction is empirically substantiated (Table 1). Both HEG and REST genes use AUG as a start codon most frequently in E. coli, but least frequently in M. abscessus. This species difference is also highly significant (p < 0.00001) when tested with the three-way log-linear model. The start codon preference is AUG > GUG > UUG > Other (i.e., CUG+AUA+AUC+AUU, Table 2), which is consistent with other bacterial and archaeal species [78].

Table 2.
Start codon usages in M. abscessus and the three phages in counts and percentages in parentheses.
Many publications have documented GUG and UUG as suboptimal to AUG for translation initiation, although most of studies involve the translation machinery in E. coli. A recent study [80] tested all 64 codons for their initiation efficiencies as start codons. Among the three canonical start codons (AUG, GUG, and UUG), translation initiation efficiency is consistently in the order of AUG > GUG > UUG [79,80]. The hypothesis proposed to explain this pattern states that a nucleotide mismatch weakens the pairing affinity between a start codon and the decoding tRNA anticodon, leading to reduced translation initiation efficiency [78]. This is the same hypothesis proposed previously for AUA as a suboptimal start codon [79]. The prediction from this hypothesis is that, if temperature is increased leading to further weakening of the base-pairing affinity, then start codons such as AUA or UUG with a nucleotide mismatch would exhibit reduced efficiency of translation initiation. Indeed, when temperature was increased from 21 °C to 37 °C, initiation efficiency decreased dramatically for mRNA with AUA as a start codon, but increased for mRNA with AUG as a start codon [79]. The hypothesis also explains the finding of an earlier study in which ACG as a start codon exhibits the same temperature sensitivity as AUA [82]. The frequency of start codon usage, in the order of AUG > GUG > UUG, has been documented in diverse bacterial and archaeal species [78], suggesting perhaps a universal preference of AUG as a start codon in prokaryotic translation machinery.
The three phages do not use rare start codons CUG, AUA, AUC, or AUU (Table 2), which is similar to HEGs in the bacterial host. Phage BPs exhibits the highest AUG usage, higher than HEG genes of the bacterial host (87.30% vs. 78.57%). However, phages Muddy and ZoeJ still have many suboptimal GUG and UUG as start codons. Three of the four UUG start codons in Muddy genes (Table 2) are in hypothetical genes. The remaining UUG is in a gene encoding ATPase, which is typically needed only during genome packaging [83]. Since it is not a structural protein and not needed in large quantities, there might be little selection for its translation efficiency, so a UUG start codon is tolerated. All genes encoding structural proteins in Muddy, such as capsids, minor tail proteins, and minor tail subunits, as well as tail assembly chaperones, use AUG as start codon. Thus, changing UUG to AUG in ATPase in Muddy may not result in a more efficient phage, although more phage ATPase than necessary may be produced.
The four UUG start codons in ZoeJ genes (Table 2) also include three in hypothetical genes. The remaining one is in a gene encoding lysin A, which the phage uses, together with holins, to lyse the bacterial cell wall in the late phase of the lytic cycle [84]. Phage lysins have been used by themselves as antibacterial agents [84,85]. A tentative hypothesis is that both ATPase and lysins are not mass-produced proteins such as capsids and tail proteins, so no strong selection operates on them to optimize their translation.
2.1.2. Secondary Structure Flanking the Start Codon
The minimum requirement for translation initiation is an accessible start codon that is not embedded in a strong secondary structure [22,35,46,79,86]. Previous experimental data based on engineered E. coli genes have shown a strong negative association between secondary structure stability near the start codon and translation efficiency in E. coli [31,32,33]. I measure the secondary structure stability by the minimum folding energy (MFE) computed over sliding windows of 40, 50, and 60 nucleotides. The patterns are similar from the three different window sizes and only results from the window size of 40 nt are shown below.
MFE equal to 0 means no secondary structure, and a stronger secondary structure corresponds to a more negative MFE value. Secondary structure stability decreases conspicuously in sequences flanking the translation initiation sequence in both M. abscessus (Figure 1A) and the three phages (Figure 1B). The weakest secondary structure corresponds to the SD sequence upstream of the start codon (Figure 1.). This is consistent with the interpretation [27,34] that a strong secondary structure embedding the SD sequence or the start codon is selected against because it prevents the translation initiation signal (SD and start codon) from being decoded by the aSD sequence and the initiation tRNA, respectively.
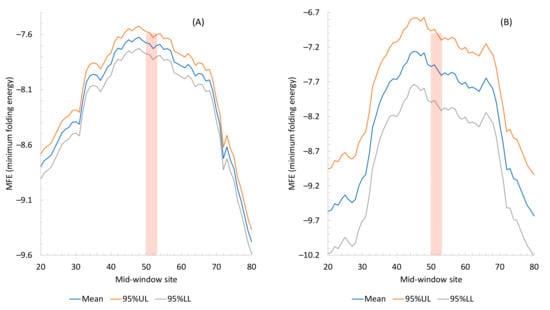
Figure 1.
Secondary structure stability, measured by MFE (minimum free energy), decreases near translation initiation signal. The 95% upper and lower limits (95% UL and 95% LL) are also plotted. The mid-window site (horizontal axis) indicates the middle of the sliding window of 40 nt. The start codon, indicated by the vertical bar, occupies the sites 51, 52, and 53. (A) From all 4707 protein-coding genes in M. abscessus. (B) From all 226 protein-coding genes in the three phages (63 in BPs, 71 in Muddy, and 92 in ZoeJ), with 95% UL and LL wider than in (A) because of a smaller number of genes than that in M. abscessus.
The three phages are consistent in exhibiting a pattern of reduced secondary structures in sequences flanking the start codon (Figure 2). However, there are highly significant differences among the three phage species (ANOVA on the three phage species blocked by the sliding windows, p < 2 × 10−16). The secondary structure is significantly weaker (p < 2 × 10−16) in Muddy genes (Figure 2B) than in BPs genes (Figure 2A) but stronger in ZoeJ genes (Figure 2C) than in BPs genes (p < 2 × 10−16). If I may extrapolate from experimental results in E. coli [31,32,33] where reduced secondary structure is associated with increased translation efficiency, then genes in Phage Muddy are expected to be translated more efficiently than those in the other two phages.
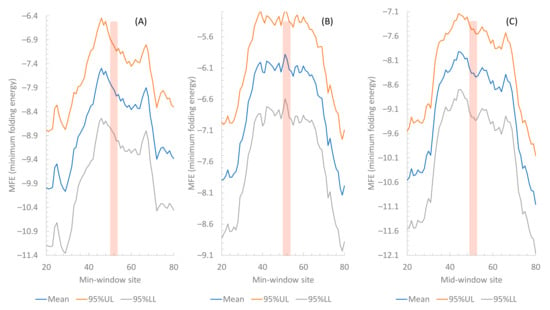
Figure 2.
Comparison of MFE (mean and 95% upper and lower limits) along a sliding window of 40 nt among three phage species. The vertical bar indicates the position of the start codon. (A) Phage BPs (n = 63). (B) Phage Muddy (n = 71). (C) Phage ZoeJ (n = 92).
There could be two nonexclusive explanations for the difference in MFE among the three phage species. The first involves differential selection. Phage Muddy is naturally lytic and the other two phages are lysogenic before being genetically engineered to disable lysogeny [2]. Lytic phages are more frequently under optimizing selection than lysogenic phages because most genes in a lysogenic phage do not express during the lysogenic phase and are consequently at the mercy of mutations [27]. Given that adaptation represents a balance between selection and mutation, the prolonged accumulation of mutation during the lysogenic phase is likely to contribute to a suboptimal state. Therefore, genes in phages BPs and ZoeJ should have stronger (suboptimal) secondary structures near the translation initiation signals than the genes in Phage Muddy. The second explanation is differential mutation bias. Phage Muddy is more AT-rich than the other two phages (genomic AT% is 41.2% for Muddy, but 33.4% for BPs and 31.5% for ZoeJ). Secondary structure from an AT-rich mRNA is expected to be, on average, weaker than that from a GC-rich mRNA.
If a reduction in secondary structure stability in sequences flanking the start codon is functionally important, then one would expect highly expressed genes (HEGs) to exhibit a stronger pattern than an average gene in M. abscessus. This prediction is strongly supported (Figure 3). These results suggest that genes in the three phages can be modified to reduce secondary structure to improve their translation initiation efficiency.
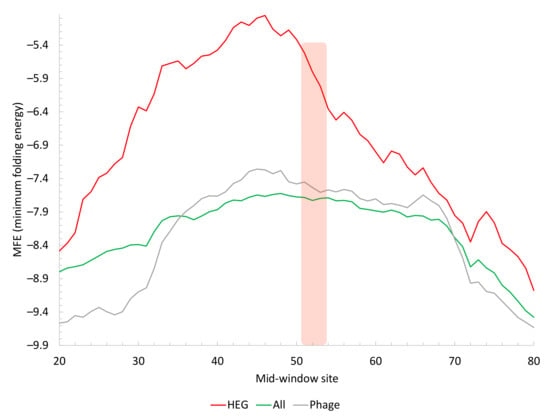
Figure 3.
Change in secondary structure stability, measured by MFE, in sequences flanking the start codon. Highly expressed genes (HEG, n = 73) in M. abscessus have weak secondary structures (less negative MFE) relative to all genes (All, n = 4707) in M. abscessus or genes in the three phages (Phage, n = 226). Mean MFE values along a sliding window of 40 nt were plotted. The vertical bar indicates the position of the start codon.
I have previously reported that ATPase in Muddy and Lysin A in GPs have UUG as a start codon in their mRNAs, and hypothesized that these proteins may be lowly expressed relative to structure proteins and consequently have experienced less selection for increasing their translation efficiency. If ATPase and lysins are indeed lowly expressed, then the selection for reducing secondary structure in sequences flanking their start signals should also be weak. I contrasted MFE along a sliding window for ATPase and lysin genes from the three phages against structural protein genes (Figure 4). Secondary structure, as indicated by MFE, is indeed weaker for structural protein genes than for ATPase and lysin genes. Thus, both the use of UUG as a stop codon and the relatively strong secondary structure near the translation initiation signal in ATPase and lysin genes appear consistent with the hypothesis that ATPase and lysins are not mass-produced and their genes have not experienced strong selection for increasing translation efficiency.
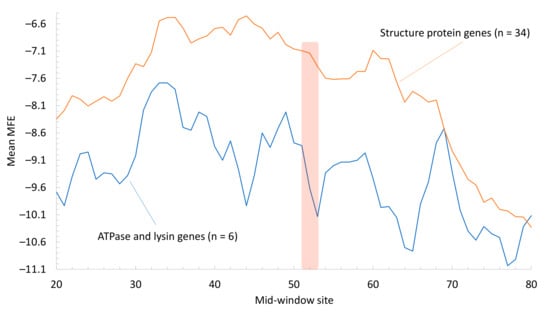
Figure 4.
Contrast of average MFE between genes encoding structure proteins (representing mass-produced phage proteins, including capsid proteins, head-to-tail adaptors, head-to-tail stoppers, major tail subunits, minor tail proteins, tail assembly chaperones, tail terminators, minor tail subunits, and scaffolding proteins, n = 34 from all three phages) and those encoding ATPase and lysins (representing non-mass-produced phage proteins, n = 6 from all three phages). MFE is calculated along a sliding window of 40 nt. The vertical bar indicates the position of the start codon.
2.1.3. Shine-Dalgarno Paring
In addition to a good start codon and the lack of a strong secondary structure embedding the start codon, an efficiently translated bacterial or phage mRNA must have a well-positioned SD sequence. Highly expressed bacterial genes or phage genes are more likely to have a well-positioned Shine-Dalgarno sequence than lowly expressed genes [22,27].
SD/aSD base-pairing is illustrated with two ribosomal protein genes rpsT and rpsI in M. abscessus (Figure 5A) to highlight a common misconception of an optimal distance between SD and the start codon. The distance (L1) between the SD sequence for rpsT (AAGGA, Figure 5A) and the start codon is greater than that (L2) for rpsI (SD = AGGUG, Figure 5A). As both ribosomal proteins are highly expressed, one may wonder if the differences between L1 and L2 matter in translation initiation. A model of SD/aSD base-pairing [27,34] to juxtapose the start codon and the initiation tRNA anticodon (Figure 5B) shows SD1 and SD2 by base-pairing with different aSD sequences at the 3’ end of the small subunit (ssu) rRNA; both serve to align the start codon against the anticodon of the tRNA. In spite of the differences between L1 and L2, the distance DtoStart remains the same, being 15 for both rpsT and rpsI. If DtoStart changes, then the aligned start codon and the initiation tRNA anticodon would be shifted out of alignment (Figure 5B).
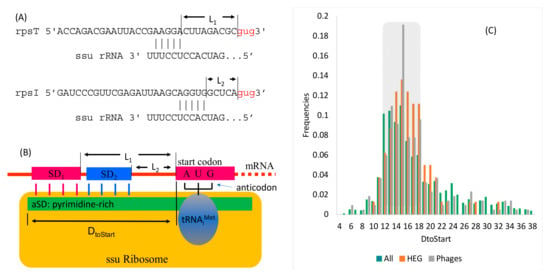
Figure 5.
Determining the optimal position of base-pairing between the Shine-Dalgarno (SD) sequence and the anti-SD (aSD) sequence at the 3’ tail of the small subunit ribosomal RNA (ssu rRNA). (A) SD/aSD base-pairing for two highly expressed ribosomal protein genes (rpsT and rpsI). (B) A model of SD/aSD base-pairing for positioning the start codon against the anticodon of the initiation tRNA, illustrating the distance (DtoStart) between the 3’ end of the ssu rRNA and the start codon. (C) Frequency distribution of DtoStart for all protein-coding genes in M. abscessus (All), highly expressed genes in M. abscessus (HEG), and protein-coding genes in the three phages (Phages). The gray bar indicates the favored DtoStart range (12–18).
The model in Figure 5B predicts that DtoStart must be constrained within a narrow range. This prediction is substantiated by the frequency distributions of DtoStart (Figure 5C) which suggests that a good DtoStart should be within the range between 13 and 18. This constrained DtoStart range is also visible in the phage genes (Figure 5C). Therefore, genes encoding mass-produced proteins in therapeutic phages targeting M. abscessus should be engineered to have their DtoStart within the range of 13 and 18.
I previously hypothesized that ATPase in Muddy and Lysin A in BPs are not subject to strong selection for translation efficiency. They both have UUG as the start codon, and their secondary structures near their start signals are more stable than more highly expressed structure protein genes (Figure 4). ATPase mRNA in Muddy does not have effective SD/aSD base-pairing; the maximum number of consecutive base-pairs is three. Lysin A mRNA has an SD/aSD base-pairing with DtoStart = 18, which is at the upper limit of the preferred DtoStart range of 13–18 (Figure 5C).
It has long been noted that both base-pairing strength and the position of SD/aSD affect translation initiation efficiency [82]. When SD/aSD involves only four consecutive base-pairs, DtoStart is widely distributed in M. abscessus genes (Figure 6A). However, the distribution of DtoStart becomes narrower and peakier when the SD/aSD base-pair increases to seven, when the preferred DtoStart = 17 (Figure 6A). The figure does not include genes with eight or more consecutive base-pairs because there are too few of such genes to form a meaningful distribution.
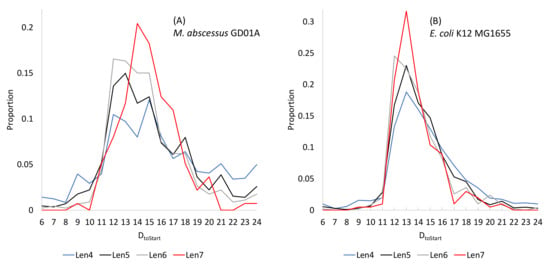
Figure 6.
The relationships between DtoStart and SD/aSD base-pairing length in (A) M abscessus and (B) E. coli. Len4 to Len7 represent four to seven consecutive base-pairings between SD and aSD. The distribution becomes increasingly peaky from four consecutive base-pairs to seven consecutive base-pairs.
The same relationship between DtoStart and SD/aSD is visible in E. coli (Figure 6B), although the distribution is overall much narrower in E. coli genes than in M. abscessus genes. E. coli replicates much more rapidly than M. abscessus, which may indicate stronger selection for more efficient translation in E. coli than in M. abscessus. The most frequent DtoStart is 13 in E. coli.
2.2. Translation Elongation and Codon-Anticodon Adaptation
Codon-anticodon adaptation depends on both tRNA-mediated selection and mutation. Different tRNAs differ in availability. Highly expressed genes preferentially use codons corresponding to the most abundant tRNA to increase translation efficiency [21,38,47,48]. This pattern is particularly strong in highly expressed genes in rapidly replicating unicellular organisms such as bacterial species [21,48,50,87] or yeast species [21,48]. Protein production increases with codon optimization, especially when translation initiation is efficient [31,33].
Mutation can disrupt codon-anticodon adaptation. For example, A-biased mutation in HIV-1 disrupts the codon adaptation of HIV-1 genes to the human tRNA pool, which favors C-ending and G-ending codons in some codon families [30]. I used nucleotide frequencies of sequences between genes in M. abscessus as a proxy of mutation spectrum (Table 3). The GC% of 59.71% indicates GC-biased mutation in M. abscessus, as previously observed in M. tuberculosis [59]. If there is no tRNA-mediated selection, then the nucleotide frequencies at the third codon site should have similar GC%. However, the GC% at the third codon site is much greater (Table 3), especially so in highly expressed genes (HEGs). This suggests strong selection in favor of C-ending and G-ending codons, consistent with the observation that M. abscessus tRNAs feature C and G at the wobble site favoring C- and G-ending codons. This preference of C- and G-ending codons is particularly strong in highly expressed genes (HEG-CS3, Table 3).

Table 3.
Nucleotide frequencies from sequences between genes (Between) and from the third codon site of all coding sequences (CS3) and of highly expressed genes (HEG-CS3). Column “NNuc” is the total nucleotide count, and the last four columns show nucleotide proportions.
The tRNA-mediated selection is strong against codon AUA in the Ile codon family in M. abscessus species. Among the same 48 tRNA genes shared by three M. abscessus genomes (GD01A, GD01B and GZ002), only a single tRNAIle/GAU exists for decoding Ile codons, with a GAU anticodon decoding AUC and AUU codons. The translation of AUA would require a noncanonical G/A wobble between two purines. Highly expressed genes (HEGs) in GD01A have only 2 AUA codons out of 755 Ile codons, indicative of strong selection against AUA codons. The other Ile codons in HEGs include 689 AUC and 64 AUU codons. Anticodon GAU forms perfect base-pairing with codon AUC but wobble with codon AUU, which explains the preference of AUC over AUU.
Codon adaptation mediated by differential tRNA availability was traditionally measured by the codon adaptation index (CAI) [88] and its improved version [89]. Calculation of CAI requires a codon usage reference of known highly expressed genes, e.g., ribosomal protein genes. One main shortcoming of CAI is that it does not take background mutation bias into consideration. This motivated the development of a more general index of translation elongation (ITE) of which CAI is a special case when there is no background mutation bias [31]. ITE is implemented in DAMBE [90] with compiled codon usage for many commonly used species.
Among the three groups of genes in the M. abscessus genome, HEGs naturally have the highest mean ITE, followed by REST and pseudogenes (Table 4). It is not surprising that pseudogenes should have the poorest codon adaptation measured by ITE because they are not subject to tRNA-mediated natural selection optimizing their codon usage. In contrast, functional genes, especially highly expressed ones, are subject to selection to increase translation elongation efficiency. The difference in ITE between HEGs and REST is greater in the reference genome (Mean ± SE = 0.770736 ± 0.006869 for HEGs and 0.639903 ± 0.000789 for REST) than in GD01.

Table 4.
Comparison of ITE (index of translation elongation) among different groups of genes in M. abscessus GD01A.
The three phages differ significantly in ITE (Table 4) based on an ANOVA test (F = 19.747, DFbetweenGroup = 2, DFwithinGroup = 223, p < 0.00001). Muddy genes have the lowest mean ITE (Table 4). However, even Muddy genes have ITEs significantly higher than M. abscessus pseudogenes based on a two-sample t-test (t = 3.9933, DF = 99, p = 0.0001, two-tailed test). This suggests positive selection for codon optimization. Two factors may contribute to the low ITE in Muddy genes. The first factor involves overlapped genes or frameshifted sites that are in one reading frame for one gene but in another reading frame for another gene. Muddy has 513 of such functionally constrained sites, but BPs and ZoeJ have only 217 and 255 of such sites, respectively. Since optimizing codons for one gene at such sites would alter protein sequence in another gene, and since altering protein sequence is typically deleterious, natural selection would not favor such codon optimization that would result in deleterious amino acid replacement. Therefore, Muddy genes are less amenable to codon optimization, and consequently have lower ITEs than the genes in the other two phages. The second contributing factor invokes differential mutation bias among the three phages and is based on the following observations: All major codons in M. abscessus (i.e., codons preferred in highly expressed genes and typically decoded by the most abundant tRNA) are NNC or NNG codons. This suggests that GC-biased mutation is in the same direction as tRNA-mediated selection. However, the Muddy genome is more AT-rich than the other two phage genomes (41.2% for Muddy, but 33.4% for BPs and 31.5% for ZoeJ), suggesting a less GC-biased mutation in Muddy than the other two phages. Thus, less GC-biased mutations in Muddy would shift codons away from the optimal NNC or NNG codons. In short, highly expressed Muddy genes would benefit more from codon optimization than genes in the other two phage species.
There are other factors affecting codon adaptation. A phage just recently switched to a new bacterial host will not have time to evolve adaptation in the new host. Moreover, a phage parasitizing multiple hosts with different tRNA pools would experience fluctuating selection, i.e., adaptation to the tRNA pool in one host may leads to suboptimal codon usage in another host. All these factors could contribute to suboptimal codon usage in phage genes.
One complication in optimizing phage codon usage is the potential change of the tRNA pool in the host cell between early and late phase of the infection cycle. In HIV-1, early genes have similar codon usage as host genes, which is expected because these early genes are translated with a normal tRNA pool shared with host gene translation. However, late genes in HIV-1 exhibit a different codon usage bias. Experiments measuring tRNA abundance suggests that late HIV-1 genes adapt to the late tRNA pool that is different from the tRNA pool in the normal cell [30]. Interestingly, a parallel case has also been documented in phage lambda [91].
2.3. Termination Signals
Our results highlight three features involving stop signals in M. abscessus. First, UAA is the optimal stop codon although UGA is the most frequently used stop codon. UAA prefers a +4G but UGA and UAGs prefer a +4U. The secondary structure stability is greatly reduced in sequences flanking a stop codon.
2.3.1. Identification of Optimal Stop Signals
UGA stop codon is used most frequently in M. abscessus genes (Table 5), similar to M. tuberculosis [59]. The latter, as well as other GC-rich bacterial species, expresses much more RF2 than RF1 and uses UGA as a stop codon much more frequently than UAG or UAA codons. However, highly expressed genes in these GC-rich genomes invariably prefer UAA stop codons [23,59]. This is also true in M. abscessus (Table 5)with a higher proportion of genes using UAA in HEGs than in REST genes (21.9% for HEG vs. 15.6% for REST, Table 5). UAA in bacterial species, especially in E. coli, has been shown to have the lowest error rate in translation termination with almost any nucleotide at the +4 site [53,60,61,62,63,64,65]. Without contrasting between the HEG and REST, one might be misled to think that UGA, being the most frequently used, is the preferred stop codon by the translation machinery in M. abscessus.

Table 5.
Stop codon usages (counts and percentages in parentheses) of genes in M. abscessus GD01A and its three phages (Muddy, BPs and ZoeJ).
Among the three phages, Muddy has the highest proportion of genes using the UAA stop codon, comparable to the proportion in HEGs. The lower proportion of genes using UAA in BPSs and ZoeJ may reflect the fact that they are lysogenic phages in which most genes are not under selection during the lysogenic phase. However, it could also result from stronger GC-biased mutation in these two phages than in Muddy. Given that highly expressed genes prefer UAA stop codons [59], which is consistent with the results in Table 5, one should change stop codons in highly expressed phage genes to UAA.
2.3.2. Differential Nucleotide Preference at the +4 Site
The +4 site is the nucleotide site immediately downstream of the stop codon. The association between stop codon and +4 nucleotide for all 4707 genes in M. abscessus (ALL subtable in Table 6) partially reflects mutation bias, i.e., a preponderance of +4C (43.8%, 40.4%, and 36.5% for UAA, UAG, and UGA, respectively. Table 6). This pattern is surprising because +4C is known to reduce termination efficiency in diverse organisms, from bacteria [56,58,67,92] to yeast [93,94,95] and other eukaryotes [96,97,98]. A +4C increases readthrough error particularly strongly for stop codon UGA [63,92,99].

Table 6.
Associations between stop codons and the +4 nucleotide in numbers (percentages) of genes.
One explanation for this preponderance of +4C is the GC-richness of the M. abscessus genome, i.e., GC-biased mutation creates +4C even if it is suboptimal. If +4C reduces translation termination efficiency and accuracy in M. abscessus as in other bacteria, then I should expect +4C to be avoided in HEGs [98]. This expectation is supported by empirical data for HEGs (HEG subtable in Table 5), with UAA associating with +4G, and UAG and UGA codons associating with +4U (HEG subtable in Table 6). This is consistent with previous studies that +4U decreases stop codon misreading and is preferred by highly expressed genes in diverse bacterial species [67,71,92,93,100]. Therefore, stop codon UAA in phage mRNA intended to replicate in M. abscessus should be engineered to have +4G and stop codons UAG and UGA should have +4U.
2.3.3. Reduction in Secondary Structure Stability in Sequences Flanking the Stop Codon
The tetranucleotide stop signal, just like the translation initiation signal represented by the SD sequence and the start codon, would not be available for decoding if it is embedded in a strong secondary structure. I have previously shown reduced secondary structure stability in sequences flanking the initiation signal. This is also true for sequences flanking the stop codon, not only for genes in M. abscessus (Figure 7A), but also in the three phage species (Figure 7B).
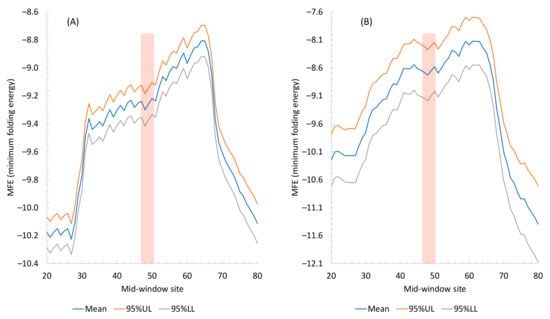
Figure 7.
Secondary structure stability, measured by MFE (minimum free energy), decreases near the stop signal. The 95% upper and lower limits (95% UL and 95% LL) are also plotted. The midwindow site (horizontal axis) indicates the middle of the sliding window of 40 nt. The stop codon, indicated by the vertical bar, occupies the sites 48, 49, and 50. (A) From all 4912 protein-coding genes in M. abscessus. (B) From all 226 protein-coding genes in the three phages (63 in BPs, 71 in Muddy, and 92 in ZoeJ), with 95% UL and LL wider than in (A) due to a smaller number of genes than that in M. abscessus.
If the reduction in secondary structure stability near the stop codon is important for translation termination, then one should expect the highly expressed genes in M. abscessus to exhibit even a stronger pattern than the average of all M. abscessus genes shown in Figure 7A. This is indeed the case (Figure 8). The change in MFE near the stop codon is the most dramatic in HEGs in M. abscessus (Figure 8). The phage genes have weaker secondary structure than an average M. abscessus gene. However, the weak secondary structure in HEGs (Figure 8) suggests that one should aim to further reduce the secondary structure stability of phage mRNAs.
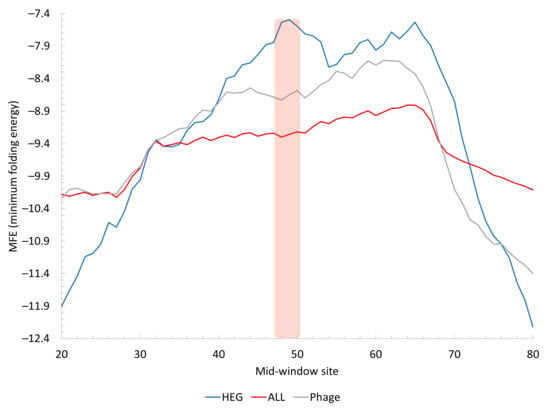
Figure 8.
Secondary structure stability, measured by MFE (minimum folding energy) in sequences flanking the stop codon. Highly expressed genes (HEG, n = 67) in M. abscessus have weak secondary structure (less negative MFE) relative to all genes (ALL, n = 4912) in M. abscessus or genes in the three phages (Phage, n = 226). Mean MFE values along a sliding window of 40 nt were plotted. The vertical bar indicates the position of the stop codon.
Different optimizations can conceivably conflict with each other. For example, the importance of having a weak secondary structure in sequences flanking the start and the stop codons would conflict with the optimization of codons immediately downstream of the start codon or immediately upstream of the stop codon because replacing a minor codon by a major codon could increase local secondary structure. Integrated software is needed to facilitate simultaneous optimization of the three subprocesses of translation.
3. Discussion
Will translation optimization necessarily increase phage replication and consequently bactericidal efficiency? If translation is not a limiting factor in phage replication, then we should not observe translation adaptation of phage genes to host translation machinery. The ubiquitous observation of adaptation of phage genes to host translation machinery suggests that optimization of translation efficiency is beneficial to the phage proliferation and that the selection that optimizes translation has overwhelmed the disruptive effect of mutation. Ideally one should produce viral proteins in the right proportion. It is not clear what the optimal proportion of different viral proteins is, but the stoichiometric ratio, if known, should make a good approximation. If a virion needs 100 copies of protein A and 10 copies of protein B, then one should engineer the phage to make proteins A and B in such proportions. However, phage infection and replication depend on many factors so translation efficiency may not be a limiting factor. This may well be true even within the translation framework. For example, optimization of codon usage in genes with low translation initiation efficiency contributes little to protein production [31,32,33].
There are many concerns in phage therapy [101]. It is more difficult to elucidate the potential side effects of therapeutic phages than antibiotics. The latter are mostly small molecules with limited numbers of functional groups. In contrast, phage proteins are macromolecules with far more functional groups that can potentially interact with many cellular components and immune systems. While phages have not been reported to become human pathogens, they are obviously capable of modulating microbiomes in our digestive system [102,103,104] and causing diseases [105,106]. Some phages were intended to kill intracellular pathogens phagocytosed into human cells [107,108], i.e., they can enter eukaryotic cells. Free phages can travel across multiple human tissues [102,109,110]. T7 phages administered through mouse tail veins can be recovered in uterus and fetal tissues [111]. The potential interaction of such phages with human cells is unpredictable. Experimental studies are clearly needed to investigate potential side effects of engineered phages.
It is often assumed that phages are highly host-specific. However, viruses frequently switch hosts in evolution. Bacterial hosts such as E. coli can evolve resistance to phage T4. In this particular context, it is beneficial for phage T4 to find new hosts, i.e., nonresistant strains of E. coli or other bacterial species. It turns out that the host specificity in T4 (and close relatives of T4) depends heavily on the C-terminus of the distal tail fiber which binds to the host receptor [2]. Mutations at this distal tail fiber can allow phage T4 to enter bacterial species that belong to different families, e.g., Yersinia pseudotuberculosis or even different orders, e.g., vibrio species [112]. Host switching also occurs in mammalian viruses, e.g., the feline panleukopenia virus switching to dogs to become canine parvovirus [113,114] (p. 793), the avian influenza virus traveling from birds to mammals, coronaviruses passed from bats or camels to humans.
One may ask what platform can be used to optimize phage mRNA in a phage genome, and create an infectious phage with the optimized genome. There are two possible approaches. The first makes use of the method that created the first bacterial cell with a chemically synthesized genome [115]. One can incorporate an optimized phage genome into a bacterial genome, and then induce the lytic phage to generate new phages. This should be suitable for DNA phages. The second would follow the reverse genetics approach [116,117,118,119] which was used to generate a full-length coronavirus genome and make it infectious. The approach potentially can be used to generate a full-length phage genome and make it infectious. It is suitable for RNA phages.
One of the problems in the development of phage therapies is that naturally occurring phages may not be patentable because many of them have been in use over the past century. The proposed mRNA optimization would help biopharmaceutical companies to patent therapeutic phage. However, phage optimization can be applied in nontherapeutic scenarios. For example, phages have been used to decontaminate food items [101,120]. Such phages would also benefit from such phage mRNA optimizations.
4. Materials and Methods
The pathogenic M. abscessus subsp. massiliense strain GD01 has two variants with complete genomic sequences (NZ_CP035923 for GD01A and NZ_CP035924 for GD01B). These were downloaded from GenBank, together with the reference genome of M. abscessus (NZ_CP034181, strain GZ002), which is better-annotated. For example, genes tig and tsf have their gene names in the reference genome but only a locus_tag in the genomes for GD01A and GD01B. The reference genome allows us to match locus_tags to gene names. I also downloaded the three therapeutic phage genomes (BPs: EU568876; Muddy: NC_022054; ZoeJ: NC_024147).
The M. abscessus reference genome includes 4942 protein-coding genes (CDSs), of which 30 are annotated as pseudogenes. The GD01A and GD01B genomes have 4770 and 4759 CDSs, respectively, and 63 and 61 pseudogenes, respectively. These pseudogenes were excluded from data analysis involving translation initiation, elongation, and termination because some of them have lost the 5’ or 3’ end of their functional counterparts, and some have frameshifting mutations.
Software DAMBE (version 7.3.32) [90] was used to extract coding sequences (CDSs) and rRNA genes from the GenBank files. I classified the bacterial host genes (GD01A strain) into three groups: (1) known highly expressed genes (HEG, n = 73), (2) the rest of the genes that may include both highly expressed and lowly expressed genes (REST, n = 4634), and 3) pseudogenes (PSEUDO, n = 63). The HEG group consists of all ribosomal protein genes as well as 11 genes (eno, dnaK, tsf, atpD, fusA, atpA, tuf, ilvC, tig, glnA, and groL) previously characterized as highly expressed in four bacterial species [121]: E. coli, Haemophilus influenzae, Vibrio cholerae, and B. subtilis. Two genes (eno and groL) have two paralogs in GD01A.
To test the prediction that secondary structure should be reduced near important translation signals such as start codon, SD sequence, and stop codon, I measured secondary structure stability by the minimum folding energy (MFE) implemented in DAMBE [90] that uses the Vienna RNA fold library [122] for secondary structure characterization. For quantifying MFE in sequences flanking the start codon, 50 nt (nucleotides) immediately upstream of the start codon and 50 nt downstream of the start codon were extracted, with the start codon occupying sites 51–53 in the resulting 100 nt sequence. A sliding window of 40 nt was used to characterize the change of MFE along the sequence. Alternative sliding windows of 50 nt and 60 nt were also used. This was similarly done for quantifying MFE in sequences flanking the stop codon; 50 nt at the 3′ end of the coding sequence (including the stop codon) as well as 50 nt immediately downstream of the stop codon were extracted, with the stop codon occupying sites 48–50. The same sliding window approach was used to measure the change of MFE along the sequence.
To characterize the optimal pairing configuration between SD and aSD, I measured the DtoStart distance, which is the number of nucleotides between the 3′ end of 16S rRNA and the start codon given an SD/aSD pairing. Previous studies have shown DtoStart to be constrained to a narrow range for highly expressed bacterial genes [90].
All sets of sequences extracted from the GD01A genome (NZ_CP035923) and used in this manuscript are included in the Supplementary Files. The file name convention is explained below. There are six gene groups: MaHEG (n = 73, where Ma is abbreviation for M. abscessus), MaREST (n = 4635), MaPSEUDO (n = 63), PhageBPs (n = 63), PhageMuddy (n = 71), and PhageZoeJ (n = 92). Each gene group features three types of sequences: (1) CDSs, (2) Start_50_50 sequences (50 nt of 5’ UTR immediately upstream of the start codon plus 50 nt of the CDS, including the start codon occupying sites 51–53), (3) Stop_50_50 sequences (50 nt of 3′ end of CDS followed by 50 nt of 3′ UTR, with the stop codon occupying sites 48–50), and (4) Up30 sequences (30 nt immediately upstream of the start codon) for characterizing strength and position of SD sequence. There should be 6 × 4 = 24 sequence files. However, because pseudogenes often miss 5′ ends or 3′ ends, only CDS sequences, but not the last three types of sequences, were compiled for pseudogenes, so the total number of files is 21. The file name follows the format of GeneGroup_SequenceType.FAS. For example, the file containing CDSs of the highly expressed genes in M. abscessus is named MaHEG_CDS.FAS. The first and the last genes may not have 50 nt upstream or downstream of the coding sequence. For example, the first CDS in the phage BPs genome starts at site 43. This gene is not included in PhageBPs_Start_50_50.FAS file.
5. Conclusions
Natural phage mRNAs may not be optimized to their host translation machinery for three reasons: (1) a phage has recently switched to a new host with no time for adaptation, (2) a high mutation rate is disrupting adaptation, and (3) a phage is parasitizing multiple hosts with different translation machineries so that adaptation to one host would lead to suboptimal adaptation to another host. For these reasons, phage mRNAs must be optimized for efficient translation in a specific bacterial pathogen. I have detailed the optimization principles for the three subprocesses of translation: initiation, elongation, and termination. The optimization principles can be further enhanced by taking stoichiometric ratios of different phage proteins. However, stoichiometric ratios of phage component proteins are unknown for most phages.
Supplementary Materials
Twenty-one sequence files in FASTA format are available online at https://www.mdpi.com/article/10.3390/ddc2010012/s1. The naming convention and content of these files are specified in Materials and Methods.
Funding
This research was funded by Discovery Grant from Natural Science and Engineering Research Council (NSERC) of Canada, grant number RGPIN/2018-03878.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
I thank P. Aris, D. Gray, A. Kruglikov, Y. Wei, and J. Xu for discussion. Jeremy Y. Xia contributed significantly to data compilation and analysis. I thank the reviewers for their helpful comments.
Conflicts of Interest
The author declares no conflict of interest.
References
- Capparelli, R.; Parlato, M.; Borriello, G.; Salvatore, P.; Iannelli, D. Experimental phage therapy against Staphylococcus aureus in mice. Antimicrob. Agents Chemother. 2007, 51, 2765–2773. [Google Scholar] [CrossRef] [PubMed]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Brives, C.; Pourraz, J. Phage therapy as a potential solution in the fight against AMR: Obstacles and possible futures. Palgrave Commun. 2020, 6, 100. [Google Scholar] [CrossRef]
- Gurney, J.; Pradier, L.; Griffin, J.S.; Gougat-Barbera, C.; Chan, B.K.; Turner, P.E.; Kaltz, O.; Hochberg, M.E. Phage steering of antibiotic-resistance evolution in the bacterial pathogen, Pseudomonas aeruginosa. Evol. Med. Public Health 2020, 2020, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Abraham, E.P.; Chain, E. An enzyme from bacteria able to destroy penicillin. Rev. Infect. Dis. 1940, 10, 677–678. [Google Scholar] [CrossRef]
- Abraham, E.P.; Chain, E.; Fletcher, C.M.; Florey, H.W.; Gardner, A.D.; Heatley, N.G.; Jennings, M.A. Further observations on penicillin. Lancet 1941, 238, 177–189. [Google Scholar] [CrossRef]
- Jernigan, J.A.; Hatfield, K.M.; Wolford, H.; Nelson, R.E.; Olubajo, B.; Reddy, S.C.; McCarthy, N.; Paul, P.; McDonald, L.C.; Kallen, A.; et al. Multidrug-Resistant Bacterial Infections in U.S. Hospitalized Patients, 2012–2017. N. Engl. J. Med. 2020, 382, 1309–1319. [Google Scholar] [CrossRef]
- Meile, S.; Du, J.; Dunne, M.; Kilcher, S.; Loessner, M.J. Engineering therapeutic phages for enhanced antibacterial efficacy. Curr. Opin. Virol. 2022, 52, 182–191. [Google Scholar] [CrossRef]
- Ferry, T.; Kolenda, C.; Briot, T.; Souche, A.; Lustig, S.; Josse, J.; Batailler, C.; Pirot, F.; Medina, M.; Leboucher, G.; et al. Past and Future of Phage Therapy and Phage-Derived Proteins in Patients with Bone and Joint Infection. Viruses 2021, 13, 2414. [Google Scholar] [CrossRef] [PubMed]
- Johansen, M.D.; Alcaraz, M.; Dedrick, R.M.; Roquet-Banères, F.; Hamela, C.; Hatfull, G.F.; Kremer, L. Mycobacteriophage–antibiotic therapy promotes enhanced clearance of drug-resistant Mycobacterium abscessus. Dis. Model. Mech. 2021, 14, dmm049159. [Google Scholar] [CrossRef]
- Knezevic, P.; Aleksic Sabo, V. Combining bacteriophages with other antibacterial agents to combat bacteria. In Phage Therapy: A Practical Approach; Górski, A., Międzybrodzki, R., Borysowski, J., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 257–293. [Google Scholar]
- Gabard, J.; Jault, P. How to Achieve a Good Phage Therapy Clinical Trial? In Phage Therapy: A Practical Approach; Górski, A., Międzybrodzki, R., Borysowski, J., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 147–168. [Google Scholar]
- Tzipilevich, E.; Pollak-Fiyaksel, O.; Shraiteh, B.; Ben-Yehuda, S. Bacteria elicit a phage tolerance response subsequent to infection of their neighbors. EMBO J. 2022, 41, e109247. [Google Scholar] [CrossRef] [PubMed]
- García, R.; Latz, S.; Romero, J.; Higuera, G.; García, K.; Bastías, R. Bacteriophage Production Models: An Overview. Front. Microbiol. 2019, 10, 1187. [Google Scholar] [CrossRef]
- Krut, O.; Bekeredjian-Ding, I. Contribution of the Immune Response to Phage Therapy. J. Immunol. 2018, 200, 3037–3044. [Google Scholar] [CrossRef] [PubMed]
- Loh, B.; Gondil, V.S.; Manohar, P.; Khan, F.M.; Yang, H.; Leptihn, S. Encapsulation and Delivery of Therapeutic Phages. Appl. Environ. Microbiol. 2020, 87, e01979-20. [Google Scholar] [CrossRef] [PubMed]
- Cunha, L.D.; Silva, A.L.N.; Ribeiro, J.M.; Mascarenhas, D.P.A.; Quirino, G.F.S.; Santos, L.L.; Flavell, R.A.; Zamboni, D.S. AIM2 Engages Active but Unprocessed Caspase-1 to Induce Noncanonical Activation of the NLRP3 Inflammasome. Cell Rep. 2017, 20, 794–805. [Google Scholar] [CrossRef]
- Hornung, V.; Ablasser, A.; Charrel-Dennis, M.; Bauernfeind, F.; Horvath, G.; Caffrey, D.R.; Latz, E.; Fitzgerald, K.A. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 2009, 458, 514–518. [Google Scholar] [CrossRef]
- Weaver, R.F. Molecular Biology, 5th ed.; McGraw-Hill: Boston, MA, USA, 2012. [Google Scholar]
- Xia, X. How optimized is the translational machinery in Escherichia coli, Salmonella typhimurium and Saccharomyces cerevisiae? Genetics 1998, 149, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. Bioinformatics and translation initiation. In Bioinformatics and the Cell: Modern Computational Approaches in Genomics, Proteomics and Transcriptomics; Springer: Cham, Switzerland, 2018; pp. 173–195. [Google Scholar]
- Xia, X. Bioinformatics and translation termination in bacteria. In Bioinformatics and the Cell: Modern Computational Approaches in Genomics, Proteomics and Transcriptomics; Springer: Cham, Switzerland, 2018; pp. 239–254. [Google Scholar]
- Chithambaram, S.; Prabhakaran, R.; Xia, X. Differential Codon Adaptation between dsDNA and ssDNA Phages in Escherichia coli. Mol. Biol. Evol. 2014, 31, 1606–1617. [Google Scholar] [CrossRef] [PubMed]
- Chithambaram, S.; Prabhakaran, R.; Xia, X. The Effect of Mutation and Selection on Codon Adaptation in Escherichia coli Bacteriophage. Genetics 2014, 197, 301–315. [Google Scholar] [CrossRef]
- Prabhakaran, R.; Chithambaram, S.; Xia, X. Aeromonas phages encode tRNAs for their overused codons. Int. J. Comput. Biol. Drug Des. 2014, 7, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Prabhakaran, R.; Chithambaram, S.; Xia, X. Escherichia coli and Staphylococcus phages: Effect of translation initiation efficiency on differential codon adaptation mediated by virulent and temperate lifestyles. J. Gen. Virol. 2015, 96, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Sezonov, G.; Joseleau-Petit, D.; D’Ari, R. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef]
- Cortes, M.A.; Nessar, R.; Singh, A.K. Laboratory maintenance of Mycobacterium abscessus. Curr. Protoc. Microbiol. 2010, 18, 10D.1.1–10D.1.12. [Google Scholar] [CrossRef]
- Van Weringh, A.; Ragonnet-Cronin, M.; Pranckeviciene, E.; Pavon-Eternod, M.; Kleiman, L.; Xia, X. HIV-1 Modulates the tRNA Pool to Improve Translation Efficiency. Mol. Biol. Evol. 2011, 28, 1827–1834. [Google Scholar] [CrossRef]
- Xia, X. A Major Controversy in Codon-Anticodon Adaptation Resolved by a New Codon Usage Index. Genetics 2015, 199, 573–579. [Google Scholar] [CrossRef]
- Kudla, G.; Murray, A.W.; Tollervey, D.; Plotkin, J.B. Coding-Sequence Determinants of Gene Expression in Escherichia coli. Science 2009, 324, 255–258. [Google Scholar] [CrossRef]
- Tuller, T.; Waldman, Y.Y.; Kupiec, M.; Ruppin, E. Translation efficiency is determined by both codon bias and folding energy. Proc. Natl. Acad. Sci. USA 2010, 107, 3645–3650. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. Optimizing Phage Translation Initiation. OBM Genet. 2019, 3, 16. [Google Scholar] [CrossRef]
- Gualerzi, C.O.; Pon, C.L. Initiation of mRNA translation in bacteria: Structural and dynamic aspects. Cell. Mol. Life Sci. CMLS 2015, 72, 4341–4367. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Kurland, C.G. Ram ribosomes are defective proofreaders. Mol. Gen. Genet. 1983, 191, 378–381. [Google Scholar] [CrossRef] [PubMed]
- Bulmer, M. The effect of context on synonymous codon usage in genes with low codon usage bias. Nucleic Acids Res. 1990, 18, 2869–2873. [Google Scholar] [CrossRef]
- Bulmer, M. The selection-mutation-drift theory of synonymous codon usage. Genetics 1991, 129, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Liljenstrom, H.; von Heijne, G. Translation rate modification by preferential codon usage: Intragenic position effects. J. Theor. Biol. 1987, 124, 43–55. [Google Scholar] [CrossRef]
- Shine, J.; Dalgarno, L. The 3’-terminal sequence of Escherichia coli 16S ribosomal RNA: Complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 1974, 71, 1342–1346. [Google Scholar] [CrossRef]
- Shine, J.; Dalgarno, L. Identical 3’-terminal octanucleotide sequence in 18S ribosomal ribonucleic acid from different eukaryotes. A proposed role for this sequence in the recognition of terminator codons. Biochem. J. 1974, 141, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Shine, J.; Dalgarno, L. Determinant of cistron specificity in bacterial ribosomes. Nature 1975, 254, 34–38. [Google Scholar] [CrossRef]
- Hui, A.; de Boer, H.A. Specialized ribosome system: Preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl. Acad. Sci. USA 1987, 84, 4762–4766. [Google Scholar] [CrossRef] [PubMed]
- Steitz, J.A.; Jakes, K. How ribosomes select initiator regions in mRNA: Base pair formation between the 3’ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 1975, 72, 4734–4738. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, T.; Weissmann, C. Inhibition of Qbeta RNA 70S ribosome initiation complex formation by an oligonucleotide complementary to the 3’ terminal region of E. coli 16S ribosomal RNA. Nature 1978, 275, 770–772. [Google Scholar] [CrossRef]
- Nakamoto, T. A unified view of the initiation of protein synthesis. Biochem. Biophys. Res. Commun. 2006, 341, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Ikemura, T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 1981, 146, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. Bioinformatics and translation elongation. In Bioinformatics and the Cell: Modern Computational Approaches in Genomics, Proteomics and Transcriptomics; Springer: Cham, Switzerland, 2018; pp. 197–238. [Google Scholar]
- Ikemura, T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J. Mol. Biol. 1982, 158, 573–597. [Google Scholar] [PubMed]
- Gouy, M.; Gautier, C. Codon usage in bacteria: Correlation with gene expressivity. Nucleic Acids Res. 1982, 10, 7055–7064. [Google Scholar] [CrossRef] [PubMed]
- Roth, J.R. UGA nonsense mutations in Salmonella typhimurium. J. Bacteriol. 1970, 102, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.F.; Fan, D.P.; Brenner, S. A strong suppressor specific for UGA. Nature 1967, 214, 452–453. [Google Scholar] [CrossRef]
- Strigini, P.; Brickman, E. Analysis of specific misreading in Escherichia coli. J. Mol. Biol. 1973, 75, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Jones, D.S.; Khorana, H.G. A further study of misreading of codons induced by streptomycin and neomycin using ribopolynucleotides containing two nucleotides in alternating sequence as templates. J. Mol. Biol. 1966, 18, 48–57. [Google Scholar] [CrossRef]
- Ryden, S.M.; Isaksson, L.A. A temperature-sensitive mutant of Escherichia coli that shows enhanced misreading of UAG/A and increased efficiency for some tRNA nonsense suppressors. Mol. Gen. Genet. 1984, 193, 38–45. [Google Scholar] [CrossRef]
- Bossi, L. Context effects: Translation of UAG codon by suppressor tRNA is affected by the sequence following UAG in the message. J. Mol. Biol. 1983, 164, 73–87. [Google Scholar] [CrossRef]
- Bossi, L.; Roth, J.R. The influence of codon context on genetic code translation. Nature 1980, 286, 123. [Google Scholar] [CrossRef]
- Miller, J.H.; Albertini, A.M. Effects of surrounding sequence on the suppression of nonsense codons. J. Mol. Biol. 1983, 164, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Wang, J.; Xia, X. Coevolution between Stop Codon Usage and Release Factors in Bacterial Species. Mol. Biol. Evol. 2016, 33, 2357–2367. [Google Scholar] [CrossRef]
- Geller, A.I.; Rich, A. A UGA termination suppression tRNATrp active in rabbit reticulocytes. Nature 1980, 283, 41–46. [Google Scholar] [CrossRef]
- Jorgensen, F.; Adamski, F.M.; Tate, W.P.; Kurland, C.G. Release factor-dependent false stops are infrequent in Escherichia coli. J. Mol. Biol. 1993, 230, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Parker, J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 1989, 53, 273–298. [Google Scholar] [CrossRef]
- Tate, W.P.; Mansell, J.B.; Mannering, S.A.; Irvine, J.H.; Major, L.L.; Wilson, D.N. UGA: A dual signal for ‘stop’ and for recoding in protein synthesis. Biochemistry 1999, 64, 1342–1353. [Google Scholar]
- Cesar Sanchez, J.; Padron, G.; Santana, H.; Herrera, L. Elimination of an HuIFN alpha 2b readthrough species, produced in Escherichia coli, by replacing its natural translational stop signal. J. Biotechnol. 1998, 63, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.Y.; Hui, J.O.; Haniu, M.; Tsai, L.B. Analysis of translational termination of recombinant human methionyl-neurotrophin 3 in Escherichia coli. Biochem. Biophys. Res. Commun. 1995, 211, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Tate, W.P.; Brown, C.M. Translational termination: “Stop” for protein synthesis or “pause” for regulation of gene expression. Biochemistry 1992, 31, 2443–2450. [Google Scholar] [CrossRef]
- Tate, W.P.; Mannering, S.A. Three, four or more: The translational stop signal at length. Mol. Microbiol. 1996, 21, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Xia, X. The Role of +4U as an Extended Translation Termination Signal in Bacteria. Genetics 2017, 205, 539–549. [Google Scholar] [CrossRef]
- Konecki, D.S.; Aune, K.C.; Tate, W.; Caskey, C.T. Characterization of reticulocyte release factor. J. Biol. Chem. 1977, 252, 4514–4520. [Google Scholar] [CrossRef] [PubMed]
- McCaughan, K.K.; Brown, C.M.; Dalphin, M.E.; Berry, M.J.; Tate, W.P. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. USA 1995, 92, 5431–5435. [Google Scholar] [CrossRef]
- Brown, C.M.; Stockwell, P.A.; Trotman, C.N.; Tate, W.P. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 1990, 18, 6339–6345. [Google Scholar] [CrossRef] [PubMed]
- Craigen, W.J.; Caskey, C.T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature 1986, 322, 273–275. [Google Scholar] [CrossRef]
- Craigen, W.J.; Cook, R.G.; Tate, W.P.; Caskey, C.T. Bacterial peptide chain release factors: Conserved primary structure and possible frameshift regulation of release factor 2. Proc. Natl. Acad. Sci. USA 1985, 82, 3616–3620. [Google Scholar] [CrossRef]
- Curran, J.F.; Yarus, M. Use of tRNA suppressors to probe regulation of Escherichia coli release factor 2. J. Mol. Biol. 1988, 203, 75–83. [Google Scholar] [CrossRef]
- Arisaka, F. Stoichiometry of protein interactions in bacteriophage tail assembly. In Stoichiometry and Research—The Importance of Quantity in Biomedicine; Innocenti, A., Ed.; IntechOpen: London, UK, 2012; pp. 243–258. [Google Scholar]
- Griffith, D.E.; Daley, C.L. Treatment of Mycobacterium abscessus Pulmonary Disease. Chest 2022, 161, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Andersson, G.E.; Sharp, P.M. Codon usage in the Mycobacterium tuberculosis complex. Microbiology 1996, 142, 915–925. [Google Scholar] [CrossRef]
- Belinky, F.; Rogozin, I.B.; Koonin, E.V. Selection on start codons in prokaryotes and potential compensatory nucleotide substitutions. Sci. Rep. 2017, 7, 12422. [Google Scholar] [CrossRef] [PubMed]
- Hartz, D.; McPheeters, D.S.; Gold, L. Influence of mRNA determinants on translation initiation in Escherichia coli. J. Mol. Biol. 1991, 218, 83–97. [Google Scholar] [CrossRef]
- Hecht, A.; Glasgow, J.; Jaschke, P.R.; Bawazer, L.A.; Munson, M.S.; Cochran, J.R.; Endy, D.; Salit, M. Measurements of translation initiation from all 64 codons in E. coli. Nucleic Acids Res. 2017, 45, 3615–3626. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, M.E.; Hauser, P.M.; Sharpe, R.G.; Errington, J. Bacillus subtilis cell cycle as studied by fluorescence microscopy: Constancy of cell length at initiation of DNA replication and evidence for active nucleoid partitioning. J. Bacteriol. 1998, 180, 547–555. [Google Scholar] [CrossRef]
- Shinedling, S.; Gayle, M.; Pribnow, D.; Gold, L. Mutations affecting translation of the bacteriophage T4 rIIB gene cloned in Escherichia coli. Mol. Gen. Genet. 1987, 207, 224–232. [Google Scholar] [CrossRef]
- Sun, S.; Kondabagil, K.; Gentz, P.M.; Rossmann, M.G.; Rao, V.B. The Structure of the ATPase that Powers DNA Packaging into Bacteriophage T4 Procapsids. Mol. Cell 2007, 25, 943–949. [Google Scholar] [CrossRef]
- Jofre, J.; Muniesa, M. Minimal Methods of Processing|Potential Use of Phages and Lysins. In Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Tortorello, M.L., Eds.; Academic Press: Oxford, UK, 2014; pp. 752–758. [Google Scholar]
- Vázquez, R.; García, E.; García, P. Phage Lysins for Fighting Bacterial Respiratory Infections: A New Generation of Antimicrobials. Front. Immunol. 2018, 9, 2252. [Google Scholar] [CrossRef] [PubMed]
- De Smit, M.H.; van Duin, J. Control of prokaryotic translational initiation by mRNA secondary structure. Prog. Nucleic Acid Res. Mol. Biol. 1990, 38, 1–35. [Google Scholar] [PubMed]
- Ikemura, T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: A proposal for a synonymous codon choice that is optimal for the E coli translational system. J. Mol. Biol. 1981, 151, 389–409. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M.; Li, W.H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. An Improved Implementation of Codon Adaptation Index. Evol. Bioinform. 2007, 3, 53–58. [Google Scholar] [CrossRef]
- Xia, X. DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2018, 35 Pt 4, 1550–1552. [Google Scholar] [CrossRef]
- Goz, E.; Mioduser, O.; Diament, A.; Tuller, T. Evidence of translation efficiency adaptation of the coding regions of the bacteriophage lambda. DNA Res. 2017, 24, 333–342. [Google Scholar] [CrossRef]
- Poole, E.S.; Brown, C.M.; Tate, W.P. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. Embo. J. 1995, 14, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Beznoskova, P.; Gunisova, S.; Valasek, L.S. Rules of UGA-N decoding by near-cognate tRNAs and analysis of readthrough on short uORFs in yeast. RNA 2016, 22, 456–466. [Google Scholar] [CrossRef]
- Jungreis, I.; Chan, C.S.; Waterhouse, R.M.; Fields, G.; Lin, M.F.; Kellis, M. Evolutionary Dynamics of Abundant Stop Codon Readthrough. Mol. Biol. Evol. 2016, 33, 3108–3132. [Google Scholar] [CrossRef] [PubMed]
- Jungreis, I.; Lin, M.F.; Spokony, R.; Chan, C.S.; Negre, N.; Victorsen, A.; White, K.P.; Kellis, M. Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res. 2011, 21, 2096–2113. [Google Scholar] [CrossRef] [PubMed]
- Manuvakhova, M.; Keeling, K.; Bedwell, D.M. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA 2000, 6, 1044–1055. [Google Scholar] [CrossRef] [PubMed]
- Dabrowski, M.; Bukowy-Bieryllo, Z.; Zietkiewicz, E. Translational readthrough potential of natural termination codons in eucaryotes--The impact of RNA sequence. RNA Biol. 2015, 12, 950–958. [Google Scholar] [CrossRef]
- Xia, X. Detailed Dissection and Critical Evaluation of the Pfizer/BioNTech and Moderna mRNA Vaccines. Vaccines 2021, 9, 734. [Google Scholar] [CrossRef]
- Brown, C.M.; Tate, W.P. Direct recognition of mRNA stop signals by Escherichia coli polypeptide chain release factor two. J. Biol. Chem. 1994, 269, 33164–33170. [Google Scholar] [CrossRef]
- Brown, C.M.; Stockwell, P.A.; Trotman, C.N.; Tate, W.P. The signal for the termination of protein synthesis in procaryotes. Nucleic Acids Res. 1990, 18, 2079–2086. [Google Scholar] [CrossRef]
- Pirnay, J.-P.; De Vos, D.; Verbeken, G.; Merabishvili, M.; Chanishvili, N.; Vaneechoutte, M.; Zizi, M.; Laire, G.; Lavigne, R.; Huys, I.; et al. The Phage Therapy Paradigm: Prêt-à-Porter or Sur-mesure? Pharm. Res. 2011, 28, 934–937. [Google Scholar] [CrossRef] [PubMed]
- Barr, J.J. A bacteriophages journey through the human body. Immunol. Rev. 2017, 279, 106–122. [Google Scholar] [CrossRef] [PubMed]
- De Sordi, L.; Khanna, V.; Debarbieux, L. The Gut Microbiota Facilitates Drifts in the Genetic Diversity and Infectivity of Bacterial Viruses. Cell Host Microbe 2017, 22, 801–808.e803. [Google Scholar] [CrossRef] [PubMed]
- Shkoporov, A.N.; Hill, C. Bacteriophages of the Human Gut: The "Known Unknown" of the Microbiome. Cell Host Microbe 2019, 25, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Manrique, P.; Dills, M.; Young, M.J. The Human Gut Phage Community and Its Implications for Health and Disease. Viruses 2017, 9, 141. [Google Scholar] [CrossRef]
- Norman, J.M.; Handley, S.A.; Baldridge, M.T.; Droit, L.; Liu, C.Y.; Keller, B.C.; Kambal, A.; Monaco, C.L.; Zhao, G.; Fleshner, P.; et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015, 160, 447–460. [Google Scholar] [CrossRef]
- Kaur, S.; Harjai, K.; Chhibber, S. Bacteriophage-aided intracellular killing of engulfed methicillin-resistant Staphylococcus aureus (MRSA) by murine macrophages. Appl. Microbiol. Biotechnol. 2014, 98, 4653–4661. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sun, L.; Wei, R.; Gao, Q.; He, T.; Xu, C.; Liu, X.; Wang, R. Intracellular Staphylococcus aureus Control by Virulent Bacteriophages within MAC-T Bovine Mammary Epithelial Cells. Antimicrob. Agents Chemother. 2017, 61, e01990-16. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.; Baker, K.; Padman, B.S.; Patwa, R.; Dunstan, R.A.; Weston, T.A.; Schlosser, K.; Bailey, B.; Lithgow, T.; Lazarou, M.; et al. Bacteriophage Transcytosis Provides a Mechanism To Cross Epithelial Cell Layers. mBio 2017, 8, e01874-17. [Google Scholar] [CrossRef] [PubMed]
- Tetz, G.; Tetz, V. Bacteriophages as New Human Viral Pathogens. Microorganisms 2018, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.S.; Chauhan, D.P.; Carrier, E. In utero detection of T7 phage after systemic administration to pregnant mice. Biotechniques 2004, 37, 81–83. [Google Scholar] [CrossRef]
- Tétart, F.; Repoila, F.; Monod, C.; Krisch, H.M. Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesin. J. Mol. Biol. 1996, 258, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Voorhees, I.E.H.; Lee, H.; Allison, A.B.; Lopez-Astacio, R.; Goodman, L.B.; Oyesola, O.O.; Omobowale, O.; Fagbohun, O.; Dubovi, E.J.; Hafenstein, S.L.; et al. Limited Intrahost Diversity and Background Evolution Accompany 40 Years of Canine Parvovirus Host Adaptation and Spread. J. Virol. 2019, 94, 108760. [Google Scholar] [CrossRef] [PubMed]
- Flint, S.J.; Enquist, L.W.; Racaniello, V.R.; Skalka, A.M. Principles of Virology: Molecular Biology, Pathogenesis, and Control of Animal Viruses, 2nd ed.; ASM Press: Washington, DC, USA, 2004; p. 918. [Google Scholar]
- Gibson, D.G.; Glass, J.I.; Lartigue, C.; Noskov, V.N.; Chuang, R.Y.; Algire, M.A.; Benders, G.A.; Montague, M.G.; Ma, L.; Moodie, M.M.; et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 2010, 329, 52–56. [Google Scholar] [CrossRef]
- Becker, M.M.; Graham, R.L.; Donaldson, E.F.; Rockx, B.; Sims, A.C.; Sheahan, T.; Pickles, R.J.; Corti, D.; Johnston, R.E.; Baric, R.S.; et al. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl. Acad. Sci. USA 2008, 105, 19944–19949. [Google Scholar] [CrossRef] [PubMed]
- Yount, B.; Curtis, K.M.; Fritz, E.A.; Hensley, L.E.; Jahrling, P.B.; Prentice, E.; Denison, M.R.; Geisbert, T.W.; Baric, R.S. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 2003, 100, 12995–13000. [Google Scholar] [CrossRef] [PubMed]
- Scobey, T.; Yount, B.L.; Sims, A.C.; Donaldson, E.F.; Agnihothram, S.S.; Menachery, V.D.; Graham, R.L.; Swanstrom, J.; Bove, P.F.; Kim, J.D.; et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 2013, 110, 16157–16162. [Google Scholar] [CrossRef]
- Menachery, V.D.; Yount, B.L., Jr.; Debbink, K.; Agnihothram, S.; Gralinski, L.E.; Plante, J.A.; Graham, R.L.; Scobey, T.; Ge, X.Y.; Donaldson, E.F.; et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015, 21, 1508–1513. [Google Scholar] [CrossRef] [PubMed]
- Thiel, K. Old dogma, new tricks—21st Century phage therapy. Nat. Biotechnol. 2004, 22, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Karlin, S.; Mrazek, J.; Campbell, A.; Kaiser, D. Characterizations of highly expressed genes of four fast-growing bacteria. J. Bacteriol. 2001, 183, 5025–5040. [Google Scholar] [CrossRef] [PubMed]
- Hofacker, I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003, 31, 3429–3431. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).