Protein SUMOylation and Its Functional Role in Nuclear Receptor Control
Abstract
1. Introduction
2. Post-Translational Protein Modifications
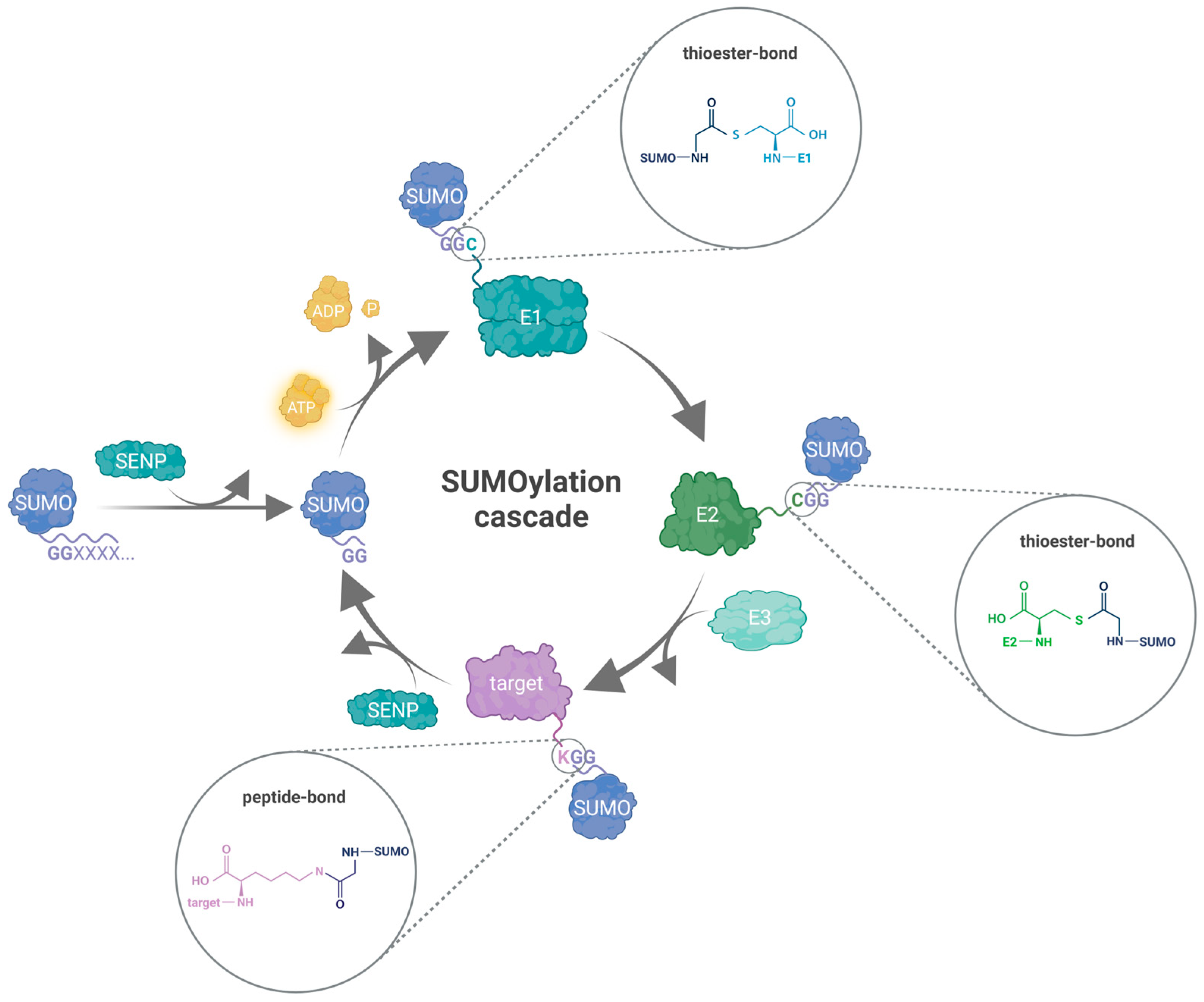
3. The Protein SUMOylation Cascade
3.1. SUMO-E1 Activating Enzyme
3.2. SUMO-E2 Conjugating Enzyme
3.3. SUMO-E3 Proteins
3.4. SUMO Proteases
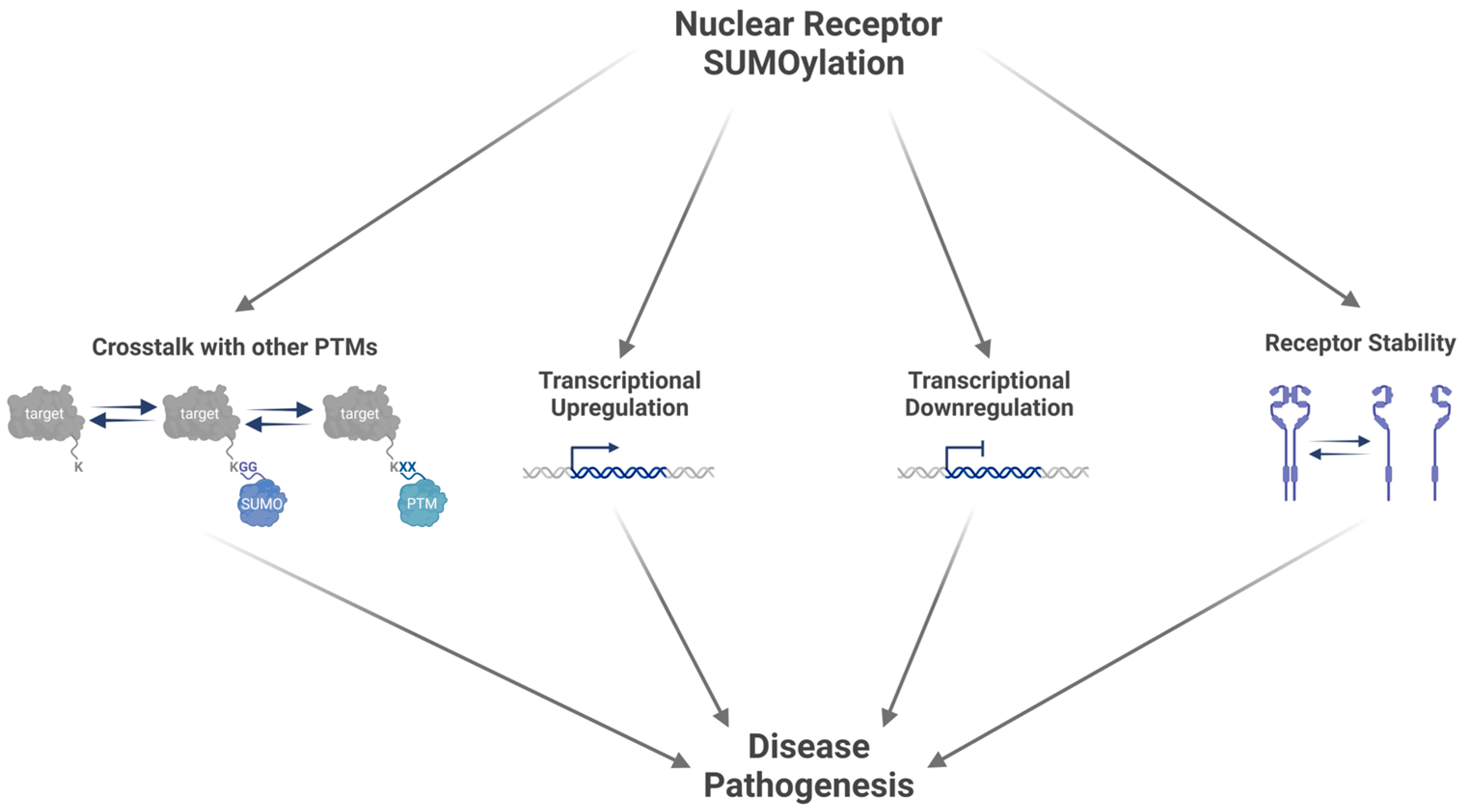
4. Receptor Protein SUMOylation
4.1. SUMOylation of Nuclear Receptors Influences the Regulation of Transcriptional Activity
| Receptor | UniProt | SUMO Isoform | SUMOylation Site | Transcriptional Effect | References |
|---|---|---|---|---|---|
| estrogen receptor 1 (ESR1, NR3A1) | P03372 | SUMO1 | K266 K268 K299 K302 K303 | transcription activity repression | [64] |
| androgen receptor (AR, NR3C4) | P10275 | SUMO1 | K386 K520 | transcription activity repression | [67,68,69] |
| liver X receptor beta (LXR-β, NR1H2) | F1D8P7 | SUMO2 SUMO3 | K409 K447 | promoter trans-repression | [70,71] |
| farnesoid X receptor (FXR, NR1H4) | B6ZGS9 | SUMO1 | K132 K289 | transcription activity trans- repression of inflammation genes; ligand-activated transcription inhibition | [72,73] |
| testicular receptor 2 (TR2, NR2C1) | P13056 | SUMO1 | K250 | transcription activity repression | [74,75] |
| mineralcorticoid receptor (MR, NR3C2) | P08235 | SUMO1 | K89 K399 K428 K494 | transcription activity repression | [76,77,78] |
| thyroid hormone receptor beta (THR-β, NR1A2) | P10828 | SUMO1 SUMO3 | K50 K146 K443 | ligand activated transcription induction | [79] |
| peroxisome proliferator-activated receptor gamma (PPAR-γ, NR1C3) | P37231 | SUMO1 | K107 K395 | transcription activity activation | [80,81] |
| steroidogenic factor 1 (SF1, NR5A1) | Q13285 | SUMO1 SUMO2 | K119 K194 | synergistic SOX9 transcription activity repression | [82,83,84] |
| liver receptor homolog 1 (LRH-1, NR5A2) | Q8WY08 | SUMO1 | K270 | transcription activity trans- repression of inflammatory genes | [70,85,86] |
| glucocorticoid receptor (GR, NR3C1) | P04150 | SUMO1 | K293 | transcription activity repression | [87,88] |
| progesterone receptor (PR, NR3C3) | P06401 | SUMO1 | K7 K388 K531 | inhibits hormone-dependent transcription activation | [89,90,91] |
| pregnane X receptor (PXR, NR1I2) | H0Y8E2 | SUMO3 | K108 K128 K129 | transcription activity trans- repression of inflammatory genes | [92,93,94,95] |
| retinoic acid receptor alpha (RAR-α, NR1B1) | P10276 | SUMO2 | K166 K171 K399 | receptor localization inhibition; transcription activity repression | [96] |
| nuclear receptor ROR alpha (ROR-α, NR1F1) | P35398 | SUMO1 SUMO2 | K240 | transcription activity activation | [97] |
| retinoid X receptor alpha (RXR-α, NR2B1) | P19793 | SUMO1 | K108 | transcription activity repression | [98] |
| thyroid hormone receptor alpha (THR-α, NR1A1) | P10827 | SUMO1 SUMO3 | K283 K389 | ligand-activated transcription induction | [79] |
| peroxisome proliferator- activated receptor alpha (PPAR-α, NR1C1) | Q07869 | SUMO1 | K185 | transcription activity trans- repression | [99] |
| nuclear receptor subfamily 4 group A member 2 (NUR1, NR4A2) | P43354 | SUMO2 SUMO3 | K558 K577 | transcription activity trans- repression of inflammatory genes | [100,101] |
| vitamin D3 receptor (VDR, NR1I1) | P11473 | SUMO2 | K91 | transcription activity repression | [102,103] |
| small heterodimer partner (SHP, NR0B2) | Q15466 | SUMO2 | K68 | transcription activity repression | [104] |
| nuclear receptor subfamily 4 group A member 1 (NUR77, NR4A1) | H3BSB9 | SUMO1 | K102 K558 K577 | transcription activity repression | [100] |
4.2. SUMOylation Influences Nuclear Receptor Stability
4.3. Crosstalk with Other Post-Translational Modifications Modifies Nuclear Receptor Activity
5. Implications of Receptor SUMOylation in Disease Pathogenesis
6. Summary and Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ramazi, S.; Allahverdi, A.; Zahiri, J. Evaluation of post-translational modifications in histone proteins: A review on histone modification defects in developmental and neurological disorders. J. Biosci. 2020, 45, 135. [Google Scholar] [CrossRef]
- Han, Z.-J.; Feng, Y.-H.; Gu, B.-H.; Li, Y.-M.; Chen, H. The post-translational modification, SUMOylation, and cancer (Review). Int. J. Oncol. 2018, 52, 1081–1094. [Google Scholar] [CrossRef]
- Doerig, C.; Rayner, J.C.; Scherf, A.; Tobin, A.B. Post-translational protein modifications in malaria parasites. Nat. Rev. Microbiol. 2015, 13, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Hickey, C.M.; Wilson, N.R.; Hochstrasser, M. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 2012, 13, 755–766. [Google Scholar] [CrossRef]
- Sehat, B.; Tofigh, A.; Lin, Y.; Trocmé, E.; Liljedahl, U.; Lagergren, J.; Larsson, O. SUMOylation mediates the nuclear translocation and signaling of the IGF-1 receptor. Sci. Signal. 2010, 3, ra10. [Google Scholar] [CrossRef] [PubMed]
- Treuter, E.; Venteclef, N. Transcriptional control of metabolic and inflammatory pathways by nuclear receptor SUMOylation. Biochim. Biophys. Acta 2011, 1812, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Flotho, A.; Melchior, F. Sumoylation: A regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013, 82, 357–385. [Google Scholar] [CrossRef]
- Kroonen, J.S.; Vertegaal, A.C.O. Targeting SUMO Signaling to Wrestle Cancer. Trends Cancer 2021, 7, 496–510. [Google Scholar] [CrossRef]
- Reiter, K.H.; Ramachandran, A.; Xia, X.; Boucher, L.E.; Bosch, J.; Matunis, M.J. Characterization and Structural Insights into Selective E1-E2 Interactions in the Human and Plasmodium falciparum SUMO Conjugation Systems. J. Biol. Chem. 2016, 291, 3860–3870. [Google Scholar] [CrossRef]
- Chang, H.-M.; Yeh, E.T.H. SUMO: From Bench to Bedside. Physiol. Rev. 2020, 100, 1599–1619. [Google Scholar] [CrossRef]
- Pichler, A.; Fatouros, C.; Lee, H.; Eisenhardt, N. SUMO conjugation—A mechanistic view. Biomol. Concepts 2017, 8, 13–36. [Google Scholar] [CrossRef]
- Choi, J.-H.; Park, J.-Y.; Park, S.P.; Lee, H.; Han, S.; Park, K.H.; Suh, Y.H. Regulation of mGluR7 trafficking by SUMOylation in neurons. Neuropharmacology 2016, 102, 229–235. [Google Scholar] [CrossRef]
- Hendriks, I.A.; Lyon, D.; Young, C.; Jensen, L.J.; Vertegaal, A.C.O.; Nielsen, M.L. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat. Struct. Mol. Biol. 2017, 24, 325–336. [Google Scholar] [CrossRef]
- Liu, W.; Zeng, M.; Fu, N. Functions of nuclear receptors SUMOylation. Clin. Chim. Acta 2021, 516, 27–33. [Google Scholar] [CrossRef]
- Adorisio, S.; Fierabracci, A.; Muscari, I.; Liberati, A.M.; Ayroldi, E.; Migliorati, G.; Thuy, T.T.; Riccardi, C.; Delfino, D.V. SUMO proteins: Guardians of immune system. J. Autoimmun. 2017, 84, 21–28. [Google Scholar] [CrossRef]
- Bawa-Khalfe, T.; Yeh, E.T.H. SUMO Losing Balance: SUMO Proteases Disrupt SUMO Homeostasis to Facilitate Cancer Development and Progression. Genes Cancer 2010, 1, 748–752. [Google Scholar] [CrossRef]
- Yau, T.-Y.; Molina, O.; Courey, A.J. SUMOylation in development and neurodegeneration. Development 2020, 147, dev175703. [Google Scholar] [CrossRef]
- Zamaraev, A.V.; Kopeina, G.S.; Prokhorova, E.A.; Zhivotovsky, B.; Lavrik, I.N. Post-translational Modification of Caspases: The Other Side of Apoptosis Regulation. Trends Cell Biol. 2017, 27, 322–339. [Google Scholar] [CrossRef]
- Wang, S.; Osgood, A.O.; Chatterjee, A. Uncovering post-translational modification-associated protein-protein interactions. Curr. Opin. Struct. Biol. 2022, 74, 102352. [Google Scholar] [CrossRef]
- Seet, B.T.; Dikic, I.; Zhou, M.-M.; Pawson, T. Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 2006, 7, 473–483. [Google Scholar] [CrossRef]
- Patel, D.J.; Wang, Z. Readout of epigenetic modifications. Annu. Rev. Biochem. 2013, 82, 81–118. [Google Scholar] [CrossRef]
- Venne, A.S.; Kollipara, L.; Zahedi, R.P. The next level of complexity: Crosstalk of posttranslational modifications. Proteomics 2014, 14, 513–524. [Google Scholar] [CrossRef]
- Issar, N.; Roux, E.; Mattei, D.; Scherf, A. Identification of a novel post-translational modification in Plasmodium falciparum: Protein sumoylation in different cellular compartments. Cell. Microbiol. 2008, 10, 1999–2011. [Google Scholar] [CrossRef]
- Schlott, A.C.; Holder, A.A.; Tate, E.W. N-Myristoylation as a Drug Target in Malaria: Exploring the Role of N-Myristoyltransferase Substrates in the Inhibitor Mode of Action. ACS Infect. Dis. 2018, 4, 449–457. [Google Scholar] [CrossRef]
- Hochstrasser, M. Origin and function of ubiquitin-like proteins. Nature 2009, 458, 422–429. [Google Scholar] [CrossRef]
- Goldstein, G.; Scheid, M.; Hammerling, U.; Schlesinger, D.H.; Niall, H.D.; Boyse, E.A. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc. Natl. Acad. Sci. USA 1975, 72, 11–15. [Google Scholar] [CrossRef]
- Nakamura, N. Ubiquitin System. Int. J. Mol. Sci. 2018, 19, 1080. [Google Scholar] [CrossRef]
- Wilkinson, K.A.; Henley, J.M. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 2010, 428, 133–145. [Google Scholar] [CrossRef]
- Su, H.-L.; Li, S.S.-L. Molecular features of human ubiquitin-like SUMO genes and their encoded proteins. Gene 2002, 296, 65–73. [Google Scholar] [CrossRef]
- Liang, Y.-C.; Lee, C.-C.; Yao, Y.-L.; Lai, C.-C.; Schmitz, M.L.; Yang, W.-M. SUMO5, a Novel Poly-SUMO Isoform, Regulates PML Nuclear Bodies. Sci. Rep. 2016, 6, 26509. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, H.; Hinchey, J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000, 275, 6252–6258. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Li, M.; Zhang, Y.; Yang, P.; Eckenrode, S.; Hopkins, D.; Zheng, W.; Purohit, S.; Podolsky, R.H.; Muir, A.; et al. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat. Genet. 2004, 36, 837–841. [Google Scholar] [CrossRef]
- Kunz, K.; Piller, T.; Müller, S. SUMO-specific proteases and isopeptidases of the SENP family at a glance. J. Cell Sci. 2018, 131, jcs211904. [Google Scholar] [CrossRef]
- Sampson, D.A.; Wang, M.; Matunis, M.J. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 2001, 276, 21664–21669. [Google Scholar] [CrossRef] [PubMed]
- Yau, T.-Y.; Sander, W.; Eidson, C.; Courey, A.J. SUMO Interacting Motifs: Structure and Function. Cells 2021, 10, 2825. [Google Scholar] [CrossRef]
- Gong, L.; Li, B.; Millas, S.; Yeh, E.T. Molecular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex. FEBS Lett. 1999, 448, 185–189. [Google Scholar] [CrossRef]
- Kagey, M.H.; Melhuish, T.A.; Wotton, D. The Polycomb Protein Pc2 Is a SUMO E3. Cell 2003, 113, 127–137. [Google Scholar] [CrossRef]
- Olsen, S.K.; Capili, A.D.; Lu, X.; Tan, D.S.; Lima, C.D. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature 2010, 463, 906–912. [Google Scholar] [CrossRef]
- Desterro, J.M.; Rodriguez, M.S.; Kemp, G.D.; Hay, R.T. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 1999, 274, 10618–10624. [Google Scholar] [CrossRef]
- Lois, L.M.; Lima, C.D. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005, 24, 439–451. [Google Scholar] [CrossRef]
- Varejão, N.; Lascorz, J.; Li, Y.; Reverter, D. Molecular mechanisms in SUMO conjugation. Biochem. Soc. Trans. 2020, 48, 123–135. [Google Scholar] [CrossRef]
- Gong, L.; Kamitani, T.; Fujise, K.; Caskey, L.S.; Yeh, E.T. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 1997, 272, 28198–28201. [Google Scholar] [CrossRef]
- Yang, S.-H.; Galanis, A.; Witty, J.; Sharrocks, A.D. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006, 25, 5083–5093. [Google Scholar] [CrossRef]
- Mohideen, F.; Capili, A.D.; Bilimoria, P.M.; Yamada, T.; Bonni, A.; Lima, C.D. A molecular basis for phosphorylation-dependent SUMO conjugation by the E2 UBC9. Nat. Struct. Mol. Biol. 2009, 16, 945–952. [Google Scholar] [CrossRef]
- Lamoliatte, F.; Caron, D.; Durette, C.; Mahrouche, L.; Maroui, M.A.; Caron-Lizotte, O.; Bonneil, E.; Chelbi-Alix, M.K.; Thibault, P. Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nat. Commun. 2014, 5, 5409. [Google Scholar] [CrossRef]
- Lascorz, J.; Codina-Fabra, J.; Reverter, D.; Torres-Rosell, J. SUMO-SIM interactions: From structure to biological functions. Semin. Cell Dev. Biol. 2022, 132, 193–202. [Google Scholar] [CrossRef]
- Hay, R.T. SUMO: A history of modification. Mol. Cell 2005, 18, 1–12. [Google Scholar] [CrossRef]
- Johnson, E.S. Protein modification by SUMO. Annu. Rev. Biochem. 2004, 73, 355–382. [Google Scholar] [CrossRef]
- Streich, F.C.; Lima, C.D. Structural and functional insights to ubiquitin-like protein conjugation. Annu. Rev. Biophys. 2014, 43, 357–379. [Google Scholar] [CrossRef]
- Yeh, E.T.; Gong, L.; Kamitani, T. Ubiquitin-like proteins: New wines in new bottles. Gene 2000, 248, 1–14. [Google Scholar] [CrossRef]
- Shin, E.J.; Shin, H.M.; Nam, E.; Kim, W.S.; Kim, J.-H.; Oh, B.-H.; Yun, Y. DeSUMOylating isopeptidase: A second class of SUMO protease. EMBO Rep. 2012, 13, 339–346. [Google Scholar] [CrossRef]
- Schulz, S.; Chachami, G.; Kozaczkiewicz, L.; Winter, U.; Stankovic-Valentin, N.; Haas, P.; Hofmann, K.; Urlaub, H.; Ovaa, H.; Wittbrodt, J.; et al. Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 2012, 13, 930–938. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, D.; Dasso, M. Modification in reverse: The SUMO proteases. Trends Biochem. Sci. 2007, 32, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef]
- Ullrich, A.; Schlessinger, J. Signal transduction by receptors with tyrosine kinase activity. Cell 1990, 61, 203–212. [Google Scholar] [CrossRef]
- Mangelsdorf, D.J.; Thummel, C.; Beato, M.; Herrlich, P.; Schütz, G.; Umesono, K.; Blumberg, B.; Kastner, P.; Mark, M.; Chambon, P.; et al. The nuclear receptor superfamily: The second decade. Cell 1995, 83, 835–839. [Google Scholar] [CrossRef]
- Cuatrecasas, P. Membrane receptors. Annu. Rev. Biochem. 1974, 43, 169–214. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 4th ed.; Garland Science Taylor & Francis Group: New York, NY, USA, 2002. [Google Scholar]
- Seeler, J.-S.; Dejean, A. SUMO and the robustness of cancer. Nat. Rev. Cancer 2017, 17, 184–197. [Google Scholar] [CrossRef]
- Cubeñas-Potts, C.; Matunis, M.J. SUMO: A multifaceted modifier of chromatin structure and function. Dev. Cell 2013, 24, 1–12. [Google Scholar] [CrossRef]
- Ao, X.; Li, S.; Xu, Z.; Yang, Y.; Chen, M.; Jiang, X.; Wu, H. Sumoylation of TCF21 downregulates the transcriptional activity of estrogen receptor-alpha. Oncotarget 2016, 7, 26220–26234. [Google Scholar] [CrossRef]
- Kuo, H.-Y.; Chang, C.-C.; Jeng, J.-C.; Hu, H.-M.; Lin, D.-Y.; Maul, G.G.; Kwok, R.P.S.; Shih, H.-M. SUMO modification negatively modulates the transcriptional activity of CREB-binding protein via the recruitment of Daxx. Proc. Natl. Acad. Sci. USA 2005, 102, 16973–16978. [Google Scholar] [CrossRef] [PubMed]
- Tiefenbach, J.; Novac, N.; Ducasse, M.; Eck, M.; Melchior, F.; Heinzel, T. SUMOylation of the corepressor N-CoR modulates its capacity to repress transcription. Mol. Biol. Cell 2006, 17, 1643–1651. [Google Scholar] [CrossRef] [PubMed]
- Sentis, S.; Le Romancer, M.; Bianchin, C.; Rostan, M.-C.; Corbo, L. Sumoylation of the estrogen receptor alpha hinge region regulates its transcriptional activity. Mol. Endocrinol. 2005, 19, 2671–2684. [Google Scholar] [CrossRef] [PubMed]
- Callewaert, L.; Verrijdt, G.; Haelens, A.; Claessens, F. Differential effect of small ubiquitin-like modifier (SUMO)-ylation of the androgen receptor in the control of cooperativity on selective versus canonical response elements. Mol. Endocrinol. 2004, 18, 1438–1449. [Google Scholar] [CrossRef]
- Ouyang, J.; Valin, A.; Gill, G. Regulation of transcription factor activity by SUMO modification. Methods Mol. Biol. 2009, 497, 141–152. [Google Scholar] [CrossRef]
- Poukka, H.; Karvonen, U.; Janne, O.A.; Palvimo, J.J. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc. Natl. Acad. Sci. USA 2000, 97, 14145–14150. [Google Scholar] [CrossRef]
- Kotaja, N.; Karvonen, U.; Jänne, O.A.; Palvimo, J.J. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 2002, 22, 5222–5234. [Google Scholar] [CrossRef]
- Nishida, T.; Yasuda, H. PIAS1 and PIASxalpha function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J. Biol. Chem. 2002, 277, 41311–41317. [Google Scholar] [CrossRef]
- Venteclef, N.; Jakobsson, T.; Ehrlund, A.; Damdimopoulos, A.; Mikkonen, L.; Ellis, E.; Nilsson, L.-M.; Parini, P.; Jänne, O.A.; Gustafsson, J.-A.; et al. GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 and LXRbeta in the hepatic acute phase response. Genes Dev. 2010, 24, 381–395. [Google Scholar] [CrossRef]
- Ghisletti, S.; Huang, W.; Ogawa, S.; Pascual, G.; Lin, M.-E.; Willson, T.M.; Rosenfeld, M.G.; Glass, C.K. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARgamma. Mol. Cell 2007, 25, 57–70. [Google Scholar] [CrossRef]
- Vavassori, P.; Mencarelli, A.; Renga, B.; Distrutti, E.; Fiorucci, S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J. Immunol. 2009, 183, 6251–6261. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniyan, N.; Luo, Y.; Sun, A.-Q.; Suchy, F.J. SUMOylation of the farnesoid X receptor (FXR) regulates the expression of FXR target genes. J. Biol. Chem. 2013, 288, 13850–13862. [Google Scholar] [CrossRef]
- Gupta, P.; Ho, P.-C.; Huq, M.M.; Ha, S.G.; Park, S.W.; Khan, A.A.; Tsai, N.-P.; Wei, L.-N. Retinoic acid-stimulated sequential phosphorylation, PML recruitment, and SUMOylation of nuclear receptor TR2 to suppress Oct4 expression. Proc. Natl. Acad. Sci. USA 2008, 105, 11424–11429. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Hu, X.; Gupta, P.; Lin, Y.-P.; Ha, S.G.; Wei, L.-N. SUMOylation of Tr2 orphan receptor involves Pml and fine-tunes Oct4 expression in stem cells. Nat. Struct. Mol. Biol. 2007, 14, 68–75. [Google Scholar] [CrossRef]
- Tallec, L.P.-L.; Kirsh, O.; Lecomte, M.-C.; Viengchareun, S.; Zennaro, M.-C.; Dejean, A.; Lombès, M. Protein inhibitor of activated signal transducer and activator of transcription 1 interacts with the N-terminal domain of mineralocorticoid receptor and represses its transcriptional activity: Implication of small ubiquitin-related modifier 1 modification. Mol. Endocrinol. 2003, 17, 2529–2542. [Google Scholar] [CrossRef]
- Tirard, M.; Almeida, O.F.X.; Hutzler, P.; Melchior, F.; Michaelidis, T.M. Sumoylation and proteasomal activity determine the transactivation properties of the mineralocorticoid receptor. Mol. Cell. Endocrinol. 2007, 268, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Yokota, K.; Shibata, H.; Kurihara, I.; Kobayashi, S.; Suda, N.; Murai-Takeda, A.; Saito, I.; Kitagawa, H.; Kato, S.; Saruta, T.; et al. Coactivation of the N-terminal transactivation of mineralocorticoid receptor by Ubc9. J. Biol. Chem. 2007, 282, 1998–2010. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Kogai, T.; Schultz, J.J.; Mody, K.; Brent, G.A. Thyroid hormone receptor isoform-specific modification by small ubiquitin-like modifier (SUMO) modulates thyroid hormone-dependent gene regulation. J. Biol. Chem. 2012, 287, 36499–36508. [Google Scholar] [CrossRef]
- Choi, S.-S.; Park, J.; Choi, J.H. Revisiting PPARγ as a target for the treatment of metabolic disorders. BMB Rep. 2014, 47, 599–608. [Google Scholar] [CrossRef]
- Katafuchi, T.; Holland, W.L.; Kollipara, R.K.; Kittler, R.; Mangelsdorf, D.J.; Kliewer, S.A. PPARγ-K107 SUMOylation regulates insulin sensitivity but not adiposity in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 12102–12111. [Google Scholar] [CrossRef]
- Chen, W.-Y.; Lee, W.-C.; Hsu, N.-C.; Huang, F.; Chung, B.-C. SUMO modification of repression domains modulates function of nuclear receptor 5A1 (steroidogenic factor-1). J. Biol. Chem. 2004, 279, 38730–38735. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, T.; Mizusaki, H.; Mukai, T.; Ogawa, H.; Baba, D.; Shirakawa, M.; Hatakeyama, S.; Nakayama, K.I.; Yamamoto, H.; Kikuchi, A.; et al. Small ubiquitin-like modifier 1 (SUMO-1) modification of the synergy control motif of Ad4 binding protein/steroidogenic factor 1 (Ad4BP/SF-1) regulates synergistic transcription between Ad4BP/SF-1 and Sox9. Mol. Endocrinol. 2004, 18, 2451–2462. [Google Scholar] [CrossRef] [PubMed]
- Suda, N.; Shibata, H.; Kurihara, I.; Ikeda, Y.; Kobayashi, S.; Yokota, K.; Murai-Takeda, A.; Nakagawa, K.; Oya, M.; Murai, M.; et al. Coactivation of SF-1-mediated transcription of steroidogenic enzymes by Ubc9 and PIAS1. Endocrinology 2011, 152, 2266–2277. [Google Scholar] [CrossRef]
- Chalkiadaki, A.; Talianidis, I. SUMO-dependent compartmentalization in promyelocytic leukemia protein nuclear bodies prevents the access of LRH-1 to chromatin. Mol. Cell. Biol. 2005, 25, 5095–5105. [Google Scholar] [CrossRef]
- Ogawa, H.; Komatsu, T.; Hiraoka, Y.; Morohashi, K. Transcriptional Suppression by Transient Recruitment of ARIP4 to Sumoylated nuclear receptor Ad4BP/SF-1. Mol. Biol. Cell 2009, 20, 4235–4245. [Google Scholar] [CrossRef][Green Version]
- Tian, S.; Poukka, H.; Palvimo, J.J.; Jänne, O.A. Small ubiquitin-related modifier-1 (SUMO-1) modification of the glucocorticoid receptor. Biochem. J. 2002, 367 Pt 3, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Impens, F.; Radoshevich, L.; Cossart, P.; Ribet, D. Mapping of SUMO sites and analysis of SUMOylation changes induced by external stimuli. Proc. Natl. Acad. Sci. USA 2014, 111, 12432–12437. [Google Scholar] [CrossRef]
- Man, J.-H.; Li, H.-Y.; Zhang, P.-J.; Zhou, T.; He, K.; Pan, X.; Liang, B.; Li, A.-L.; Zhao, J.; Gong, W.-L.; et al. PIAS3 induction of PRB sumoylation represses PRB transactivation by destabilizing its retention in the nucleus. Nucleic Acids Res. 2006, 34, 5552–5566. [Google Scholar] [CrossRef][Green Version]
- Daniel, A.R.; Faivre, E.J.; Lange, C.A. Phosphorylation-dependent antagonism of sumoylation derepresses progesterone receptor action in breast cancer cells. Mol. Endocrinol. 2007, 21, 2890–2906. [Google Scholar] [CrossRef]
- Abdel-Hafiz, H.; Dudevoir, M.L.; Horwitz, K.B. Mechanisms underlying the control of progesterone receptor transcriptional activity by SUMOylation. J. Biol. Chem. 2009, 284, 9099–9108. [Google Scholar] [CrossRef]
- Hu, G.; Xu, C.; Staudinger, J.L. Pregnane X receptor is SUMOylated to repress the inflammatory response. J. Pharmacol. Exp. Ther. 2010, 335, 342–350. [Google Scholar] [CrossRef]
- Rogers, R.S.; Parker, A.; Vainer, P.D.; Elliott, E.; Sudbeck, D.; Parimi, K.; Peddada, V.P.; Howe, P.G.; D’Ambrosio, N.; Ruddy, G.; et al. The Interface between Cell Signaling Pathways and Pregnane X Receptor. Cells 2021, 10, 3262. [Google Scholar] [CrossRef]
- Priyanka; Kotiya, D.; Rana, M.; Subbarao, N.; Puri, N.; Tyagi, R.K. Transcription regulation of nuclear receptor PXR: Role of SUMO-1 modification and NDSM in receptor function. Mol. Cell. Endocrinol. 2016, 420, 194–207. [Google Scholar] [CrossRef]
- Cui, W.; Sun, M.; Galeva, N.; Williams, T.D.; Azuma, Y.; Staudinger, J.L. SUMOylation and Ubiquitylation Circuitry Controls Pregnane X Receptor Biology in Hepatocytes. Drug Metab. Dispos. Biol. Fate Chem. 2015, 43, 1316–1325. [Google Scholar] [CrossRef]
- Zhu, L.; Santos, N.C.; Kim, K.H. Small ubiquitin-like modifier-2 modification of retinoic acid receptor-alpha regulates its subcellular localization and transcriptional activity. Endocrinology 2009, 150, 5586–5595. [Google Scholar] [CrossRef]
- Hwang, E.J.; Lee, J.M.; Jeong, J.; Park, J.H.; Yang, Y.; Lim, J.-S.; Kim, J.H.; Baek, S.H.; Kim, K.I. SUMOylation of RORalpha potentiates transcriptional activation function. Biochem. Biophys. Res. Commun. 2009, 378, 513–517. [Google Scholar] [CrossRef]
- Choi, S.J.; Chung, S.S.; Rho, E.J.; Lee, H.W.; Lee, M.H.; Choi, H.-S.; Seol, J.H.; Baek, S.H.; Bang, O.S.; Chung, C.H. Negative modulation of RXRalpha transcriptional activity by small ubiquitin-related modifier (SUMO) modification and its reversal by SUMO-specific protease SUSP1. J. Biol. Chem. 2006, 281, 30669–30677. [Google Scholar] [CrossRef]
- Pourcet, B.; Pineda-Torra, I.; Derudas, B.; Staels, B.; Glineur, C. SUMOylation of human peroxisome proliferator-activated receptor alpha inhibits its trans-activity through the recruitment of the nuclear corepressor NCoR. J. Biol. Chem. 2010, 285, 5983–5992. [Google Scholar] [CrossRef] [PubMed]
- Zárraga-Granados, G.; Muciño-Hernández, G.; Sánchez-Carbente, M.R.; Villamizar-Gálvez, W.; Peñas-Rincón, A.; Arredondo, C.; Andrés, M.E.; Wood, C.; Covarrubias, L.; Castro-Obregón, S. The nuclear receptor NR4A1 is regulated by SUMO modification to induce autophagic cell death. PLoS ONE 2020, 15, e0222072. [Google Scholar] [CrossRef] [PubMed]
- García-Yagüe, Á.J.; Cuadrado, A. Mechanisms of NURR1 Regulation: Consequences for Its Biological Activity and Involvement in Pathology. Int. J. Mol. Sci. 2023, 24, 12280. [Google Scholar] [CrossRef] [PubMed]
- Jena, S.; Lee, W.-P.; Doherty, D.; Thompson, P.D. PIAS4 represses vitamin D receptor-mediated signaling and acts as an E3-SUMO ligase towards vitamin D receptor. J. Steroid Biochem. Mol. Biol. 2012, 132, 24–31. [Google Scholar] [CrossRef]
- Zenata, O.; Vrzal, R. Fine tuning of vitamin D receptor (VDR) activity by post-transcriptional and post-translational modifications. Oncotarget 2017, 8, 35390–35402. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Kwon, S.; Byun, S.; Xiao, Z.; Park, S.; Wu, S.-Y.; Chiang, C.-M.; Kemper, B.; Kemper, J.K. Critical role of RanBP2-mediated SUMOylation of Small Heterodimer Partner in maintaining bile acid homeostasis. Nat. Commun. 2016, 7, 12179. [Google Scholar] [CrossRef]
- Kaikkonen, S.; Jääskeläinen, T.; Karvonen, U.; Rytinki, M.M.; Makkonen, H.; Gioeli, D.; Paschal, B.M.; Palvimo, J.J. SUMO-specific protease 1 (SENP1) reverses the hormone-augmented SUMOylation of androgen receptor and modulates gene responses in prostate cancer cells. Mol. Endocrinol. 2009, 23, 292–307. [Google Scholar] [CrossRef]
- Wen, S.; Niu, Y.; Huang, H. Posttranslational regulation of androgen dependent and independent androgen receptor activities in prostate cancer. Asian J. Urol. 2020, 7, 203–218. [Google Scholar] [CrossRef]
- Traboulsi, T.; El Ezzy, M.; Dumeaux, V.; Audemard, E.; Mader, S. Role of SUMOylation in differential ERα transcriptional repression by tamoxifen and fulvestrant in breast cancer cells. Oncogene 2019, 38, 1019–1037. [Google Scholar] [CrossRef]
- Seeler, J.-S.; Dejean, A. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 2003, 4, 690–699. [Google Scholar] [CrossRef]
- Neyret-Kahn, H.; Benhamed, M.; Ye, T.; Le Gras, S.; Cossec, J.-C.; Lapaquette, P.; Bischof, O.; Ouspenskaia, M.; Dasso, M.; Seeler, J.; et al. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res. 2013, 23, 1563–1579. [Google Scholar] [CrossRef]
- Chen, P.; Li, B.; Ou-Yang, L. Role of estrogen receptors in health and disease. Front. Endocrinol. 2022, 13, 839005. [Google Scholar] [CrossRef] [PubMed]
- Filardo, E.J.; Thomas, P. Minireview: G protein-coupled estrogen receptor-1, GPER-1: Its mechanism of action and role in female reproductive cancer, renal and vascular physiology. Endocrinology 2012, 153, 2953–2962. [Google Scholar] [CrossRef] [PubMed]
- Kuiper, G.G.; Enmark, E.; Pelto-Huikko, M.; Nilsson, S.; Gustafsson, J.A. Cloning of a novel receptor expressed in rat prostate and ovary. Proc. Natl. Acad. Sci. USA 1996, 93, 5925–5930. [Google Scholar] [CrossRef]
- Fuentes, N.; Silveyra, P. Estrogen receptor signaling mechanisms. Adv. Protein Chem. Struct. Biol. 2019, 116, 135–170. [Google Scholar] [CrossRef]
- Yu, E.J.; Kim, S.-H.; Kim, M.J.; Seo, W.-Y.; Song, K.-A.; Kang, M.-S.; Yang, C.K.; Stallcup, M.R.; Kim, J.H. SUMOylation of ZFP282 potentiates its positive effect on estrogen signaling in breast tumorigenesis. Oncogene 2013, 32, 4160–4168. [Google Scholar] [CrossRef]
- Qin, Y.; Yuan, H.; Chen, X.; Yang, X.; Xing, Z.; Shen, Y.; Dong, W.; An, S.; Qi, Y.; Wu, H. SUMOylation Wrestles with the Occurrence and Development of Breast Cancer. Front. Oncol. 2021, 11, 659661. [Google Scholar] [CrossRef]
- Stanisić, V.; Lonard, D.M.; O’Malley, B.W. Modulation of steroid hormone receptor activity. Prog. Brain Res. 2010, 181, 153–176. [Google Scholar] [CrossRef]
- Gelmann, E.P. Molecular biology of the androgen receptor. J. Clin. Oncol. 2002, 20, 3001–3015. [Google Scholar] [CrossRef]
- Chang, C.; Saltzman, A.; Yeh, S.; Young, W.; Keller, E.; Lee, H.J.; Wang, C.; Mizokami, A. Androgen receptor: An overview. Crit. Rev. Eukaryot. Gene Expr. 1995, 5, 97–125. [Google Scholar] [CrossRef]
- Aurilio, G.; Cimadamore, A.; Mazzucchelli, R.; Lopez-Beltran, A.; Verri, E.; Scarpelli, M.; Massari, F.; Cheng, L.; Santoni, M.; Montironi, R. Androgen Receptor Signaling Pathway in Prostate Cancer: From Genetics to Clinical Applications. Cells 2020, 9, 2653. [Google Scholar] [CrossRef]
- Wu, F.; Mo, Y.-Y. Ubiquitin-like protein modifications in prostate and breast cancer. Front. Biosci. 2007, 12, 700–711. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tang, Z.-R.; Zhang, R.; Lian, Z.-X.; Deng, S.-L.; Yu, K. Estrogen-Receptor Expression and Function in Female Reproductive Disease. Cells 2019, 8, 1123. [Google Scholar] [CrossRef] [PubMed]
- Faltas, C.L.; LeBron, K.A.; Holz, M.K. Unconventional Estrogen Signaling in Health and Disease. Endocrinology 2020, 161, bqaa030. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Ashikaga, E.; Rubin, P.P.; Heimann, M.J.; Hildick, K.L.; Bishop, P.; Girach, F.; Josa-Prado, F.; Tang, L.T.H.; Carmichael, R.E.; et al. Receptor trafficking and the regulation of synaptic plasticity by SUMO. Neuromol. Med. 2013, 15, 692–706. [Google Scholar] [CrossRef]
- Zhou, H.J.; Xu, Z.; Wang, Z.; Zhang, H.; Zhuang, Z.W.; Simons, M.; Min, W. SUMOylation of VEGFR2 regulates its intracellular trafficking and pathological angiogenesis. Nat. Commun. 2018, 9, 3303. [Google Scholar] [CrossRef]
- van Beekum, O.; Fleskens, V.; Kalkhoven, E. Posttranslational modifications of PPAR-gamma: Fine-tuning the metabolic master regulator. Obesity 2009, 17, 213–219. [Google Scholar] [CrossRef]
- Mal, S.; Dwivedi, A.R.; Kumar, V.; Kumar, N.; Kumar, B.; Kumar, V. Role of Peroxisome Proliferator-Activated Receptor Gamma (PPARγ) in Different Disease States: Recent Updates. Curr. Med. Chem. 2021, 28, 3193–3215. [Google Scholar] [CrossRef]
- Wadosky, K.M.; Willis, M.S. The story so far: Post-translational regulation of peroxisome proliferator-activated receptors by ubiquitination and SUMOylation. Am. J. Physiol. Heart Circ. Physiol. 2012, 302, H515–H526. [Google Scholar] [CrossRef]
- Floyd, Z.E.; Stephens, J.M. Control of peroxisome proliferator-activated receptor gamma2 stability and activity by SUMOylation. Obes. Res. 2004, 12, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Miyazono, K.; Kamiya, Y.; Miyazawa, K. SUMO amplifies TGF-beta signalling. Nat. Cell Biol. 2008, 10, 635–637. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Huang, R.; Yuan, L. Crosstalk of intracellular post-translational modifications in cancer. Arch. Biochem. Biophys. 2019, 676, 108138. [Google Scholar] [CrossRef]
- Yang, X.-J.; Seto, E. Lysine acetylation: Codified crosstalk with other posttranslational modifications. Mol. Cell 2008, 31, 449–461. [Google Scholar] [CrossRef]
- Woodsmith, J.; Stelzl, U. Studying post-translational modifications with protein interaction networks. Curr. Opin. Struct. Biol. 2014, 24, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Tomasi, M.L.; Ramani, K. SUMOylation and phosphorylation cross-talk in hepatocellular carcinoma. Transl. Gastroenterol. Hepatol. 2018, 3, 20. [Google Scholar] [CrossRef] [PubMed]
- Davies, L.; Karthikeyan, N.; Lynch, J.T.; Sial, E.-A.; Gkourtsa, A.; Demonacos, C.; Krstic-Demonacos, M. Cross talk of signaling pathways in the regulation of the glucocorticoid receptor function. Mol. Endocrinol. 2008, 22, 1331–1344. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Xia, Y.; He, J.; Wang, J.; Li, J.; Ye, M.; Jin, X. The SUMOylation and ubiquitination crosstalk in cancer. J. Cancer Res. Clin. Oncol. 2023, 149, 16123–16146. [Google Scholar] [CrossRef] [PubMed]
- Watts, F.Z. Starting and stopping SUMOylation. What regulates the regulator? Chromosoma 2013, 122, 451–463. [Google Scholar] [CrossRef]
- Zhang, P.-J.; Zhao, J.; Li, H.-Y.; Man, J.-H.; He, K.; Zhou, T.; Pan, X.; Li, A.-L.; Gong, W.-L.; Jin, B.-F.; et al. CUE domain containing 2 regulates degradation of progesterone receptor by ubiquitin-proteasome. EMBO J. 2007, 26, 1831–1842. [Google Scholar] [CrossRef]
- Kumar, R.; Sabapathy, K. RNF4-A Paradigm for SUMOylation-Mediated Ubiquitination. Proteomics 2019, 19, e1900185. [Google Scholar] [CrossRef]
- Celen, A.B.; Sahin, U. Sumoylation on its 25th anniversary: Mechanisms, pathology, and emerging concepts. FEBS J. 2020, 287, 3110–3140. [Google Scholar] [CrossRef]
- Sriramachandran, A.M.; Dohmen, R.J. SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta 2014, 1843, 75–85. [Google Scholar] [CrossRef]
- Chang, Y.-C.; Oram, M.K.; Bielinsky, A.-K. SUMO-Targeted Ubiquitin Ligases and Their Functions in Maintaining Genome Stability. Int. J. Mol. Sci. 2021, 22, 5391. [Google Scholar] [CrossRef]
- Kim, S.O.; Ono, K.; Tobias, P.S.; Han, J. Orphan Nuclear Receptor Nur77 Is Involved in Caspase-independent Macrophage Cell Death. J. Exp. Med. 2003, 197, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xie, F.; Zhang, J.; Dijke, P.T.; Zhou, F. SUMO-triggered ubiquitination of NR4A1 controls macrophage cell death. Cell Death Differ. 2017, 24, 1530–1539. [Google Scholar] [CrossRef]
- Hu, M.; Luo, Q.; Alitongbieke, G.; Chong, S.; Xu, C.; Xie, L.; Chen, X.; Zhang, D.; Zhou, Y.; Wang, Z.; et al. Celastrol-Induced Nur77 Interaction with TRAF2 Alleviates Inflammation by Promoting Mitochondrial Ubiquitination and Autophagy. Mol. Cell 2017, 66, 141–153.e6. [Google Scholar] [CrossRef] [PubMed]
- Hietakangas, V.; Anckar, J.; Blomster, H.A.; Fujimoto, M.; Palvimo, J.J.; Nakai, A.; Sistonen, L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. USA 2006, 103, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, T.; Koga, H.; Shimotohno, K. Transcriptional activity of peroxisome proliferator-activated receptor gamma is modulated by SUMO-1 modification. J. Biol. Chem. 2004, 279, 29551–29557. [Google Scholar] [CrossRef]
- Yamashita, D.; Yamaguchi, T.; Shimizu, M.; Nakata, N.; Hirose, F.; Osumi, T. The transactivating function of peroxisome proliferator-activated receptor gamma is negatively regulated by SUMO conjugation in the amino-terminal domain. Genes Cells Devoted Mol. Cell. Mech. 2004, 9, 1017–1029. [Google Scholar] [CrossRef]
- Rangwala, S.M.; Rhoades, B.; Shapiro, J.S.; Rich, A.S.; Kim, J.K.; Shulman, G.I.; Kaestner, K.H.; Lazar, M.A. Genetic modulation of PPARgamma phosphorylation regulates insulin sensitivity. Dev. Cell 2003, 5, 657–663. [Google Scholar] [CrossRef]
- Sahin, U.; de Thé, H.; Lallemand-Breitenbach, V. Sumoylation in Physiology, Pathology and Therapy. Cells 2022, 11, 814. [Google Scholar] [CrossRef]
- Andreou, A.M.; Tavernarakis, N. SUMOylation and cell signalling. Biotechnol. J. 2009, 4, 1740–1752. [Google Scholar] [CrossRef]
- Talamillo, A.; Ajuria, L.; Grillo, M.; Barroso-Gomila, O.; Mayor, U.; Barrio, R. SUMOylation in the control of cholesterol homeostasis. Open Biol. 2020, 10, 200054. [Google Scholar] [CrossRef]
- Zeng, M.; Liu, W.; Hu, Y.; Fu, N. Sumoylation in liver disease. Clin. Chim. Acta 2020, 510, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Mikkonen, L.; Hirvonen, J.; Jänne, O.A. SUMO-1 regulates body weight and adipogenesis via PPARγ in male and female mice. Endocrinology 2013, 154, 698–708. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Zhang, W.; Yin, L.; Shi, Z.; Luan, J.; Chen, L.; Liu, L. The Potential Roles of Post-Translational Modifications of PPARγ in Treating Diabetes. Biomolecules 2022, 12, 1832. [Google Scholar] [CrossRef]
- Yuan, W.; Ma, C.; Zhou, Y.; Wang, M.; Zeng, G.; Huang, Q. Negative regulation of eNOS-NO signaling by over-SUMOylation of PPARγ contributes to insulin resistance and dysfunction of vascular endothelium in rats. Vasc. Pharmacol. 2019, 122–123, 106597. [Google Scholar] [CrossRef]
- Gervois, P.; Fruchart, J.-C.; Staels, B. Drug Insight: Mechanisms of action and therapeutic applications for agonists of peroxisome proliferator-activated receptors. Nat. Clin. Pract. Endocrinol. Metab. 2007, 3, 145–156. [Google Scholar] [CrossRef]
- Karamouzis, M.V.; Konstantinopoulos, P.A.; Badra, F.A.; Papavassiliou, A.G. SUMO and estrogen receptors in breast cancer. Breast Cancer Res. Treat. 2008, 107, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Sarfstein, R.; Pasmanik-Chor, M.; Yeheskel, A.; Edry, L.; Shomron, N.; Warman, N.; Wertheimer, E.; Maor, S.; Shochat, L.; Werner, H. Insulin-like growth factor-I receptor (IGF-IR) translocates to nucleus and autoregulates IGF-IR gene expression in breast cancer cells. J. Biol. Chem. 2012, 287, 2766–2776. [Google Scholar] [CrossRef]
- Sehat, B.; Andersson, S.; Vasilcanu, R.; Girnita, L.; Larsson, O. Role of ubiquitination in IGF-1 receptor signaling and degradation. PLoS ONE 2007, 2, e340. [Google Scholar] [CrossRef]
- Bettermann, K.; Benesch, M.; Weis, S.; Haybaeck, J. SUMOylation in carcinogenesis. Cancer Lett. 2012, 316, 113–125. [Google Scholar] [CrossRef]
- Lin, Y.; Liu, H.; Waraky, A.; Haglund, F.; Agarwal, P.; Jernberg-Wiklund, H.; Warsito, D.; Larsson, O. SUMO-modified insulin-like growth factor 1 receptor (IGF-1R) increases cell cycle progression and cell proliferation. J. Cell. Physiol. 2017, 232, 2722–2730. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wild, N.; Kaiser, C.S.; Wunderlich, G.; Liebau, E.; Wrenger, C. Protein SUMOylation and Its Functional Role in Nuclear Receptor Control. Receptors 2024, 3, 408-424. https://doi.org/10.3390/receptors3030020
Wild N, Kaiser CS, Wunderlich G, Liebau E, Wrenger C. Protein SUMOylation and Its Functional Role in Nuclear Receptor Control. Receptors. 2024; 3(3):408-424. https://doi.org/10.3390/receptors3030020
Chicago/Turabian StyleWild, Nele, Charlotte Sophia Kaiser, Gerhard Wunderlich, Eva Liebau, and Carsten Wrenger. 2024. "Protein SUMOylation and Its Functional Role in Nuclear Receptor Control" Receptors 3, no. 3: 408-424. https://doi.org/10.3390/receptors3030020
APA StyleWild, N., Kaiser, C. S., Wunderlich, G., Liebau, E., & Wrenger, C. (2024). Protein SUMOylation and Its Functional Role in Nuclear Receptor Control. Receptors, 3(3), 408-424. https://doi.org/10.3390/receptors3030020







