Abstract
The cycle number (nc) of a recycling receptor is defined as the average number of round trips (cell surface–endosome–cell surface) the receptor can make before it is degraded. This characteristic parameter of recycling receptors can be easily determined from the receptor’s half-life (t½, the time in which 50% of the receptor is degraded) and cycling time (Tc, the time a receptor needs to complete a round trip). Relationship analyses revealed that nc increases linearly with increasing t½ and decreases exponentially with increasing Tc. For commonly observed t½ and Tc values, it was calculated that recycling receptors have nc values of <300. In addition, it was found that recycling receptors in cancer cells have generally smaller nc values (<100), whereas recycling receptors in normal cells have larger nc values (>100). Based on this latter finding, the cycle number nc may be a useful criterion for distinguishing between cancer and normal cells.
1. Introduction
Recycling receptors are cell surface proteins that are used by cells for the endocytosis of extracellular macromolecules [,]. In general, after binding its ligand, the receptor clusters with other receptors in clathrin-coated pits. The receptor–ligand complex is internalized in coated vesicles which fuse with the endosome. Usually, within the endosome, the low luminal pH of this compartment leads to the dissociation of the ligand from the receptor. While the ligand is transported to the lysosome, where it is degraded, the receptor returns to the cell surface to bind another ligand and initiates another cycle of endocytosis (a scheme of the recycling process of surface receptors is shown in Figure 1).
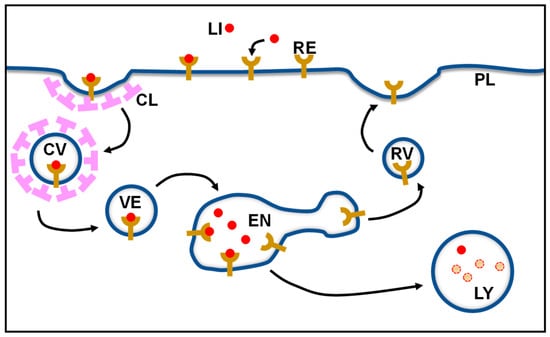
Figure 1.
Schematic representation of the surface receptor recycling process. CL, clathrin; CV, clathrin-coated vesicle; EN, endosome; LI, ligand; LY, lysosome; PL, plasma membrane; RE, receptor; RV, recycling vesicle.
Although the recycling of receptors has been studied in great detail in past decades, one parameter has not been paid much attention: the cycle number, i.e., the number of round trips a receptor undertakes before it is degraded. This may be due to the fact that the cycle number cannot be determined directly. Very few estimations of receptor cycle numbers have been published in the literature, ranging from 300 to 1000 cycles [,]. However, as shown herein, these values are, to some extent, hugely overestimated. Obviously, the cycle number depends on the half-life (the period of time required for half of the receptor molecules to be degraded) and on the cycling time (the time needed for the receptor to complete one round trip) of the receptor. The longer the half-life and the shorter the cycling time, the greater the cycle number is. This work analyzed the relationship between the cycle number, half-life, and cycling time of recycling receptors. In addition, the cycle numbers of different recycling receptors in normal cells and cancer cells were computed and compared.
2. Calculation of the Average Cycle Number of Recycling Receptors
The cycling time of a receptor can be easily calculated from the total number of functional receptors divided by the rate of ligand uptake [,]. It has been shown that this value is similar to the cycling time obtained from the sum of the individual rate constants [,]. The half-life of a receptor can be readily determined via radioactive metabolic labelling experiments [].
The cycle number of a recycling receptor was calculated using a previously developed equation []. In brief, the average cycle number (nc) of a receptor before it is degraded can be computed from the number of receptor molecules remaining after each cycle (Nc) divided by the number of receptor molecules (N0) at the time t0.
The number of receptor molecules remaining after each cycle (Nc) is provided by:
where t½ is the half-life, Tc is the cycling time, and n is the number of cycles of the receptor. Together, Equations (1) and (2) provide:
which can be simplified to:
Equation (4) equals to:
The solution for Equation (5) is:
3. Relationship between Cycle Number, Half-Life, and Cycling Time
To understand how the cycle number nc is linked with the half-life t½ and the cycling time Tc of a receptor, nc was determined as a function of t½ and Tc for given Tc and t½ values, respectively, using Equation (6). As depicted in Figure 2A, nc is linearly dependent on t½ for given values of Tc. With an increase in t½, nc also increases. This is plausible as a receptor with a longer half-life can undertake more round trips. Although it appears that with increasing Tc values the increase in nc diminishes (the slopes of the linear regressions for different Tc values decrease), the fold increase in nc over time is the same for all Tc values. As illustrated in Figure 2B, nc decreases exponentially with an increasing Tc for given values of t½. Thus, with an increasing Tc, nc becomes smaller and smaller and less dependent on Tc. This makes sense because with an increase in the cycling time (Tc), the number of round trips (i.e., the cycle number (nc)) should decrease. Furthermore, when the cycling time approaches the half-life (t½), the cycle number should be increasingly determined by only the half-life of the receptor. The relationship among the three variables, nc, t½, and Tc, is shown in Figure 3 in the form of a 3D surface plot. From the 3D graph, it can be seen clearly that with increasing t½ values and decreasing Tc values, nc values increase greatly.
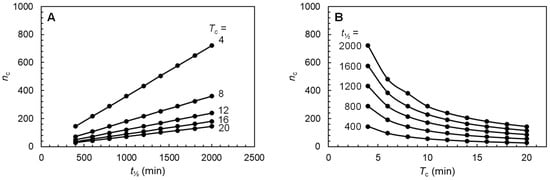
Figure 2.
Plots of cycle number nc as a function of half-life t½ for given cycling time Tc (A) and of Tc for given half-life t½ (B). The range of values for t½ and Tc are within the ranges of half-lives and cycling times normally observed for recycling receptors. The cycle number was calculated using Equation (6).
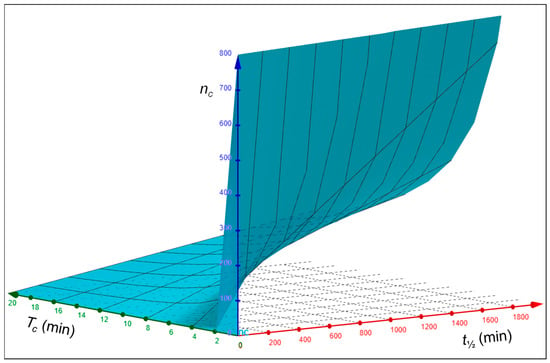
Figure 3.
Three-dimensional surface plot of the relationship between the half-lives t½ cycling times Tc, and cycle numbers nc of recycling receptors. The 3D graph was created with the GeoGebra 3D Calculator [].
Values of nc for t½ and Tc values in the ranges normally found for recycling receptors (400–2000 min and 4–20 min, respectively) are presented in Table 1. From the data, it is clear that nc values are usually smaller than 300 and do not assume values of 300–1000, as previously estimated [,]. Only for t½ values ≥1000 min in combination with Tc values ≤8 min does nc assume values of >300 (see grey highlighted numbers in Table 1). Table 1 is also useful for rough estimations of nc values.

Table 1.
Values of cycle numbers (nc) of recycling receptors for given half-lives (t½) and cycling times (Tc). The cycle numbers were calculated using Equation (6).
4. Cycle Numbers of Classical Recycling Receptors
Using Equation (6), the cycle numbers nc of the recycling receptors for asialoglycoprotein, low-density lipoprotein, mannose, and transferrin were calculated (Table 2). It was found that the nc values for these classical recycling receptors ranged between 38 and 240. These nc values are smaller than the previously suggested receptor cycle numbers of 300–1000 [,]. Moreover, the calculated nc values correspond well with measured cycle numbers. For example, it was found that the low-density lipoprotein receptor can undergo up to 150 cycles in fibroblasts [], which is in close agreement with the calculated nc value of 144 (Table 2).
The nc values determined for recycling receptors in this study, which are almost an order of magnitude smaller, are more compatible with the physiological stress a receptor experiences during the recycling process. Recycling requires that a receptor is not denatured when passing repeatedly through the acidic environment of the endosome. In the acidic compartment, a receptor must undergo substantial conformational changes to release its ligand [] but must not become irreversibly damaged. Thus, round trips of more than 300 may harm a receptor in such a manner that it will lose its function.

Table 2.
Calculated nc values for recycling receptors. The nc values were computed using Equation (6) and published t½ and Tc values. The sources of the t½ and Tc values are indicated.
Table 2.
Calculated nc values for recycling receptors. The nc values were computed using Equation (6) and published t½ and Tc values. The sources of the t½ and Tc values are indicated.
| Receptor | Cell Type | t½(min) | Tc (min) | nc |
| Asialoglycoprotein | HepG2 cells | 720 [] | 15.9 [] | 65 |
| Rat hepatocytes | 1200 [] | 7.2 [] | 240 | |
| Low-density lipoprotein | Human fibroblasts | 1200 [] | 12 [] | 144 |
| Mannose | Macrophages | 1980 [] | 15 [] | 190 |
| Transferrin | HeLa | 1140 [] | 21 [] | 78 |
| HepG2 cells | 420 [] | 15.8 [] | 38 | |
| K562 cells | 480 [] | 12.5 [] | 55 | |
| Trypanosoma brucei | 426 [] | 10.7 [] | 57 |
5. Cycle Numbers of Recycling Receptors Distinguish between Cancer and Normal Cells
It is interesting to note that receptors in cancer cells have smaller cycle numbers than receptors in noncancerous cells freshly prepared from tissues (Table 2). For instance, recycling receptors in human hepatocarcinoma HepG2 cells [], human erythroleukaemia K562 cells [], and human immortalized cancerous HeLa cells [] have cycle numbers between 38 and 79, whereas recycling receptors in human fibroblasts (freshly prepared from the foreskin of a newborn boy []) and rat hepatocytes (freshly isolated from a rat liver []) have cycle numbers of 144 and 237, respectively. In addition, the mannose receptor in cultured macrophages has an nc value of >100 (Table 2). The difference in the cycle numbers of recycling receptors in cancer cells and normal cells is probably associated with differences in metabolic fluxes and nutritional needs between these cells []. As fast-proliferating cells, cancer cells have an upregulated metabolic activity and, accordingly, a higher protein turnover than normal cells []. This is reflected in the shorter half-life of receptors in cancer cells compared with normal cells (Table 2). This suggestion is further supported by the finding that the transferrin receptor of the fast-proliferating protozoan parasite Trypanosoma brucei also has a short half-life and a small cycle number (Table 2).
The cycle number may be affected by ligand-induced signaling of the receptor. This, however, depends on the receptor-mediated endocytic pathway []. For receptors that are endocytosed only after they have bound a ligand, the cycle number may be lower when there is a lack of a ligand as in this case, the receptor would remain at the cell surface for a longer time. On the other hand, for receptors that are continuously internalized even in the absence of a ligand, the cycle number will be unaffected by ligand binding. Receptors for low-density lipoprotein, transferrin, and asialoglycoprotein belong to this latter group [,,].
There is evidence that cancer cells show increased rates of clathrin-mediated endocytosis []. As clathrin-mediated endocytosis plays an important role in the process of ligand uptake by recycling receptors, it could be assumed that these receptors would be recycled faster in cancer cells than in normal cells and therefore would have smaller cycling times (Tc values). However, this is not the case. It rather seems that the recycling receptors in cancer cells tend to have longer cycling times, although no statistically significant difference between the Tc values for recycling receptors in cancer and normal cells was observed (unpaired t-test: p = 0.1422). This finding may indicate that the clathrin-mediated endocytosis rate in cancer cells is actually not different from the rate in normal cells. As the cycling times of the different recycling receptors differ only by a factor of 3, it seems that the cycle number nc is mainly determined by the half-lives of the receptors.
6. Conclusions
This study has shown that the average number of round trips (cycle number) of a recycling receptor can be easily determined using the receptor’s half-life and cycling number. The cycle numbers of classical recycling receptors range between 40 and 240. In cancer cells, the cycle numbers of receptors are <100, while in normal cells, they are >100. Thus, the cycle number of recycling receptors may be used as a characteristic to differentiate cancer cells from normal cells.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The author declares no conflict of interest.
References
- Brown, M.S.; Anderson, R.G.W.; Goldstein, J.L. Recycling receptors: The round-trip itinerary of migrant membrane proteins. Cell 1983, 32, 663–667. [Google Scholar] [CrossRef] [PubMed]
- Stahl, P.; Schwartz, A.L. Receptor-mediated endocytosis. J. Clin. Investig. 1986, 77, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Warren, R.; Doyle, D. Turnover of the surface proteins and the receptor for serum asialoglycoproteins in primary cultures of rat hepatocytes. J. Biol. Chem. 1981, 256, 1346–1355. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, A.L.; Fridovich, S.E.; Lodish, H.F. Kinetics of internalization and recycling of the asialoglycoprotein receptor in a hepatoma cell line. J. Biol. Chem. 1982, 257, 4230–4237. [Google Scholar] [CrossRef]
- Chiechanover, A.; Schwartz, A.L.; Dautry-Varsat, A.; Lodish, H.F. Kinetics of internalization and recycling of transferrin and the transferrin receptor in a human hepatoma cell line. Effect of lysosomotropic agents. J. Biol. Chem. 1983, 258, 9681–9689. [Google Scholar] [CrossRef]
- Harford, J.; Ashwell, G. Assessment of receptor recycling in mammalian hepatocystes: Perspectives based on current techniques. Methods Enzymol. 1985, 109, 232–246. [Google Scholar]
- Kabiri, M.; Steverding, D. Studies on the recycling of the transferrin receptor in Trypanosoma brucei using an inducible gene expression system. Eur. J. Biochem. 2000, 267, 3309–3314. [Google Scholar] [CrossRef]
- GeoGebra. 3D Calculator. Available online: https://www.geogebra.org/3d (accessed on 5 March 2023).
- Goldstein, J.L.; Brown, M.S.; Anderson, R.G.W.; Russell, D.W.; Schneider, W.J. Receptor-mediated endocytosis: Concepts emerging from the LDL receptor system. Ann. Rev. Cell Biol. 1985, 1, 1–39. [Google Scholar] [CrossRef]
- DiPaola, M.; Maxfield, F.R. Conformational changes in the receptors for epidermal growth factor and asialoglycoproteins induced by the mildly acidic pH found in endocytic vesicles. J. Biol. Chem. 1984, 259, 9163–9171. [Google Scholar] [CrossRef]
- Bischoff, J.; Lodish, H.F. Two asialoglycoprotein receptor polypeptides in human hepatoma cells. J. Biol. Chem. 1987, 262, 11825–11832. [Google Scholar] [CrossRef]
- Basu, S.K.; Goldstein, J.L.; Anderson, R.G.W.; Brown, M.S. Monensin interrupts the recycling of low density lipoprotein receptors in human fibroblasts. Cell 1981, 24, 493–502. [Google Scholar] [CrossRef]
- Lennartz, M.R.; Coles, F.S.; Stahl, P.D. Biosynthesis and processing of the mannose receptor in human macrophages. J. Biol. Chem. 1989, 264, 2385–2390. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, E.A.; Mikoryak, C.A.; Draper, R.K. Turnover of the transferrin receptor is not influenced by removing most of the extracellular domain. J. Biol. Chem. 1991, 266, 21125–21130. [Google Scholar] [CrossRef]
- Bleil, J.D.; Bretscher, M.S. Transferrin receptor and its recycling in HeLa cell. EMBO J. 1982, 1, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Volz, B.; Orberger, G.; Porwoll, S.; Hauri, H.-P.; Tauber, R. Selective reentry of recycling cell surface glycoproteins to the biosynthetic pathway in human hepatocarcinoma HepG2 cells. J. Cell Biol. 1995, 130, 537–551. [Google Scholar] [CrossRef]
- Weissman, A.M.; Klausner, R.D.; Rao, K.; Harford, J.B. Exposure of K562 cells to anti-receptor monoclonal antibody OKT9 results in rapid redistribution and enhanced degradation of the transferrin receptor. J. Cell Biol. 1986, 102, 951–958. [Google Scholar] [CrossRef]
- Knowles, B.B.; Howe, C.C.; Aden, D.P. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 1980, 209, 497–499. [Google Scholar] [CrossRef]
- Klein, E.; Ben-Bassat, H.; Neumann, H.; Ralph, P.; Zeuthen, J.; Polliack, A.; Vánky, F. Properties of the K562 cell line, derived from a patient with chronic myeloid leukemia. Int. J. Cancer 1976, 18, 421–431. [Google Scholar] [CrossRef]
- Lucey, B.P.; Nelson-Rees, W.A.; Hutchins, G.M. Henrietta Lacks, HeLa cells, and cell culture contamination. Arch. Pathol. Lab. Med. 2009, 133, 1463–1467. [Google Scholar] [CrossRef]
- Brown, M.S.; Goldstein, J.L. Regulation of the activity of the low density lipoprotein receptor in human fibroblasts. Cell 1975, 6, 307–316. [Google Scholar] [CrossRef]
- Neagu, M.; Constantin, C.; Popescu, J.D.; Zipeto, D.; Tzanakakis, G.; Nikitovic, D.; Fenga, C.; Stratakis, C.A.; Spandidos, D.A.; Tsatsakis, A.M. Inflammation and metabolism in cancer cell—Mitochondria key player. Front. Oncol. 2019, 9, 348. [Google Scholar] [CrossRef]
- Xiao, Z.; Dai, Z.; Locasale, J.W. Metabolic landscape of the tumor microenvironment at single cell resolution. Nat. Commun. 2019, 10, 3763. [Google Scholar] [CrossRef]
- Hopkins, C.R.; Trowbridge, I.S. Internalization and processing of transferrin and the transferrin receptor in human carcinoma A 431 cells. J. Cell Biol. 1983, 97, 508–521. [Google Scholar] [CrossRef]
- Berg, T.; Blomhoff, R.; Naess, L.; Tolleshaug, H.; Drevon, C.A. Monensin inhibits receptor-mediated endocytosis of asialoglycoproteins in hepatocytes. Exp. Cell Res. 1983, 148, 319–330. [Google Scholar] [CrossRef]
- Khan, I.; Steeg, P.S. Endocytosis: A pivotal pathway for regulating metastasis. Br. J. Cancer 2021, 124, 66–75. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).