Abstract
Introduction: Prostate cancer (PCa) is one of the deadliest and most common causes of malignancy and death in men worldwide, with a higher prevalence and mortality in developing countries specifically. Factors such as age, family history, race and certain genetic mutations are some of the factors contributing to the occurrence of PCa in men. Recent advances in technology and algorithms gave rise to the computer-aided diagnosis (CAD) of PCa. With the availability of medical image datasets and emerging trends in state-of-the-art machine and deep learning techniques, there has been a growth in recent related publications. Materials and Methods: In this study, we present a systematic review of PCa diagnosis with medical images using machine learning and deep learning techniques. We conducted a thorough review of the relevant studies indexed in four databases (IEEE, PubMed, Springer and ScienceDirect) using the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. With well-defined search terms, a total of 608 articles were identified, and 77 met the final inclusion criteria. The key elements in the included papers are presented and conclusions are drawn from them. Results: The findings show that the United States has the most research in PCa diagnosis with machine learning, Magnetic Resonance Images are the most used datasets and transfer learning is the most used method of diagnosing PCa in recent times. In addition, some available PCa datasets and some key considerations for the choice of loss function in the deep learning models are presented. The limitations and lessons learnt are discussed, and some key recommendations are made. Conclusion: The discoveries and the conclusions of this work are organized so as to enable researchers in the same domain to use this work and make crucial implementation decisions.
1. Introduction
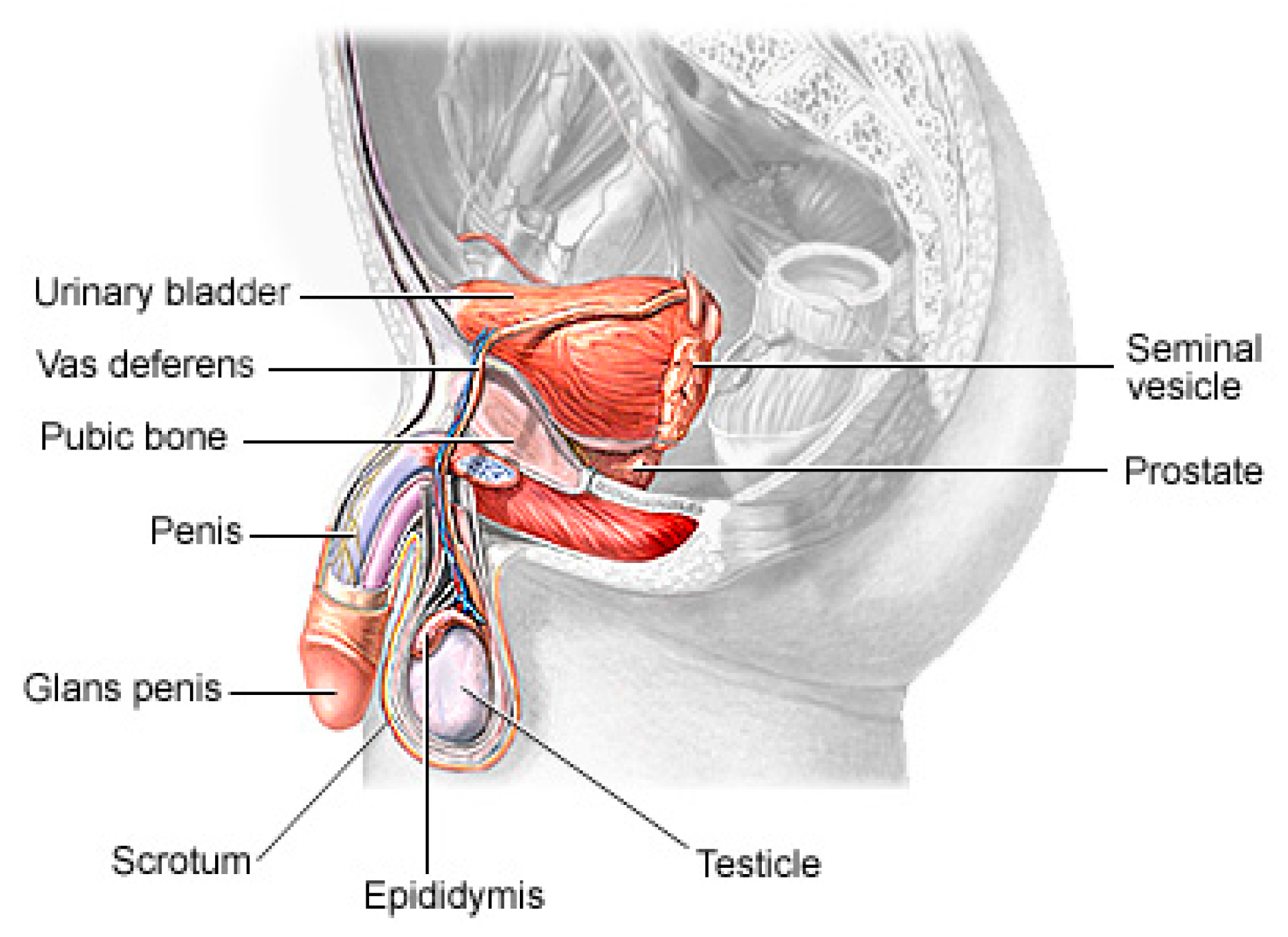
Prostate cancer (PCa) is the second most lethal and prevalent non-cutaneous tumor in males globally [1]. Published statistics from the American Cancer Society (ACS) show that it is the most common cancer in American men after skin cancer, with 288 and 300 new cases in 2023, resulting in about 34,700 deaths. By 2030, it is anticipated that there will be 11 million cancer deaths, which would be a record high [2]. Worldwide, this type of cancer affects many males, with developing and underdeveloped countries having a higher prevalence and higher mortality rates [3]. PCa is a type of cancer that develops in the prostate gland, a small walnut-shaped gland located below the bladder in men [4]. The male reproductive system contains the prostate, which is a small gland that is located under the bladder and in front of the rectum. It surrounds the urethra, which is the tube that carries urine from the bladder out of the body. The primary function of the prostate (Figure 1) is to produce and secrete a fluid that makes up a part of semen, which is the fluid that carries sperm during ejaculation [5]. The development of PCa in an individual can be caused by a variety of circumstances including age (older men are more likely to develop prostate cancer), family history (having a close relative who has prostate cancer increases the risk), race (African American males are more likely to develop prostate cancer) and specific genetic mutations [6,7].
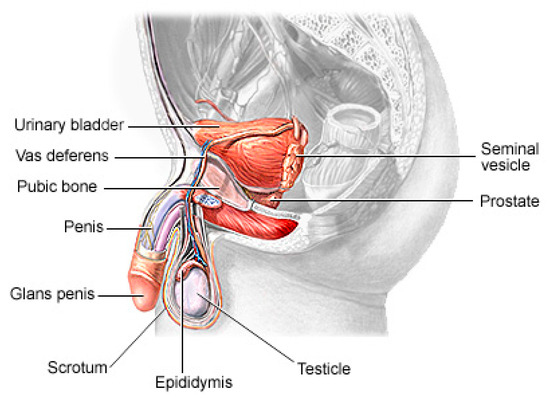
Figure 1.
The physiology of a human prostate.
The recent advances in sophisticated computers and algorithms in recent decades have paved the way for improved PCa diagnosis and treatment [8]. Computer-aided diagnosis (CAD) refers to the use of computer algorithms and technology to assist healthcare professionals in the prognosis and diagnosis of patients [9]. CAD systems are designed to serve as Decision Support (or Expert) Systems, which analyze medical data, such as images or test results, and provide experts with additional information or suggestions to aid in the interpretation and diagnosis of various medical conditions. They are commonly used in medical imaging fields to detect anomalies or assist in the interpretation and analysis of medical images such as X-rays, Computed Tomography (CT) scans, Magnetic Resonance Imaging (MRI) scans and mammograms [10]. These systems use pattern recognition, machine learning algorithms and deep learning algorithms to identify specific features or patterns that may indicate the presence or absence of a disease or condition [11]. It can also help radiologists by highlighting regions of interest (ROI) or by providing quantitative measurements for further analysis. Soft computing techniques play a major role in decision making across several sectors of the field of medical image analysis [12,13]. Deep learning, a branch of artificial intelligence, has shown promising performance in the identification of patterns and the classification of medical images [14,15].
Several studies have investigated some CAD solutions to identify PCa by analyzing medical images as a decision support tool for an effective and efficient diagnosis process, easing these tasks as well as reducing human errors and effort. Also, there is an avalanche of review and survey papers published in this area that summarize and organizes recent works and aid in the understanding of the state-of-the-art in this field, discussing the trends and recommending future directions.
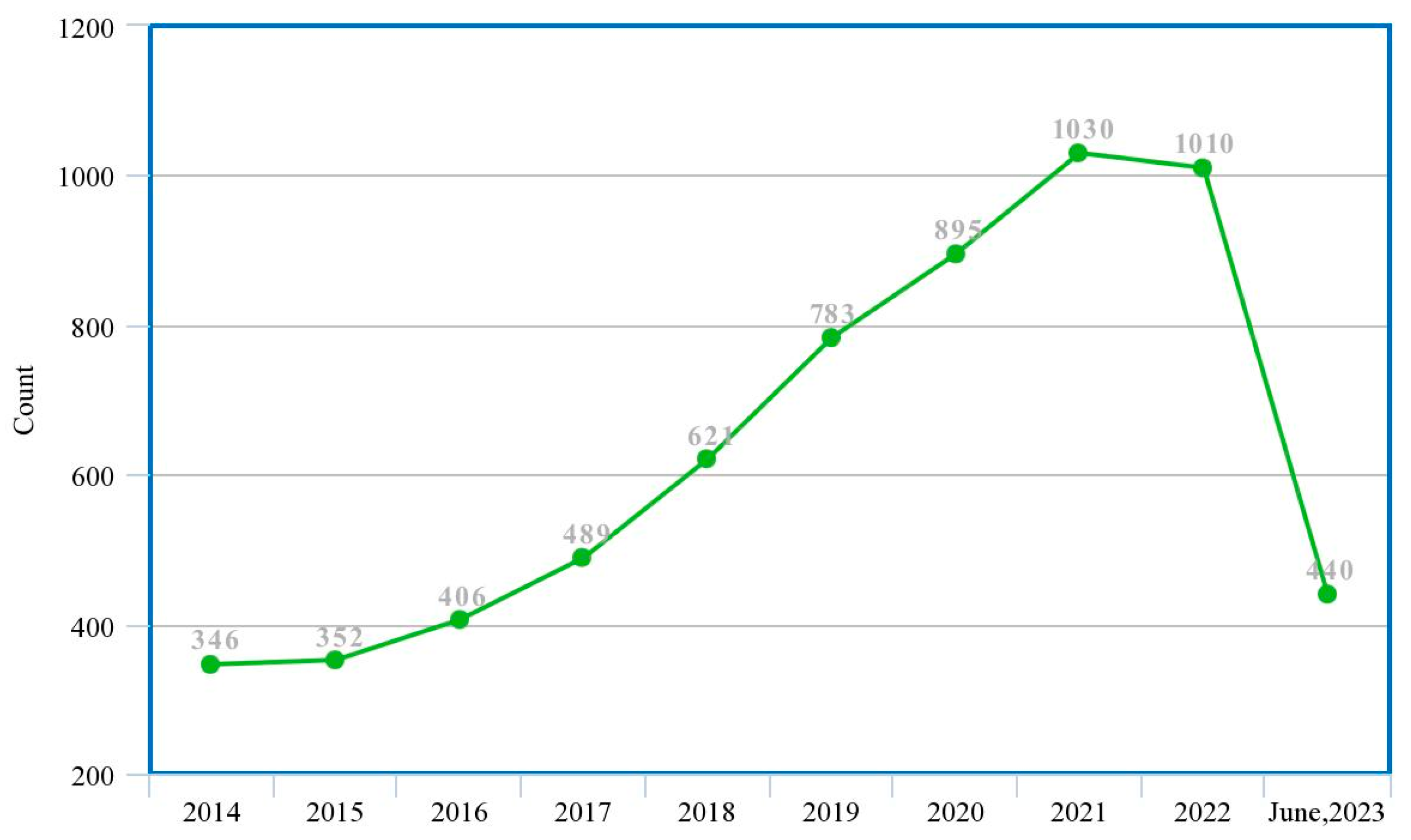
This study presents a guided systematic review of the application of these ML and DL techniques in the diagnosis of PCa, especially their applications in the process of segmentation, cancer detection, the assessment of lesion aggressiveness, local staging and pre-treatment assessment, among others. We present, evaluate and summarize various studies from our selected databases, give insights into the use of different datasets and different imaging modalities, explore the trends in this area, analyze the state-of-the-art deep learning architectures and provide derivations, taxonomies and summaries based on these observations and some limitations, open challenges and possible future directions. Machine learning specialists, medics and decision makers can benefit from this study as it will help them determine which machine learning model is appropriate for which characteristics of the dataset as well as gain insights into future directions for research and development. Figure 2 shows the trend of publications on the subject matter from the previous ten years to date, which was obtained from a tailored search on Google Scholar (https://scholar.google.com accessed on 4 July 2023) with the query ‘machine learning deep learning “prostate cancer”-review’, and filtered by year.
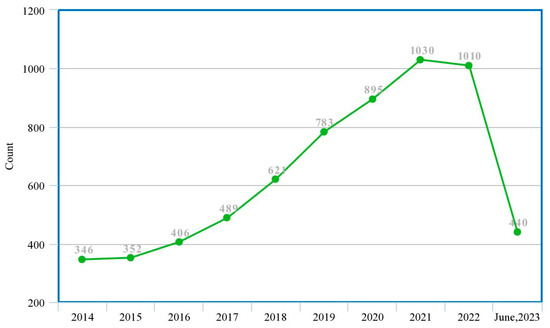
Figure 2.
Trend of research in ML/DL models for PCa diagnosis (actual experimental study).
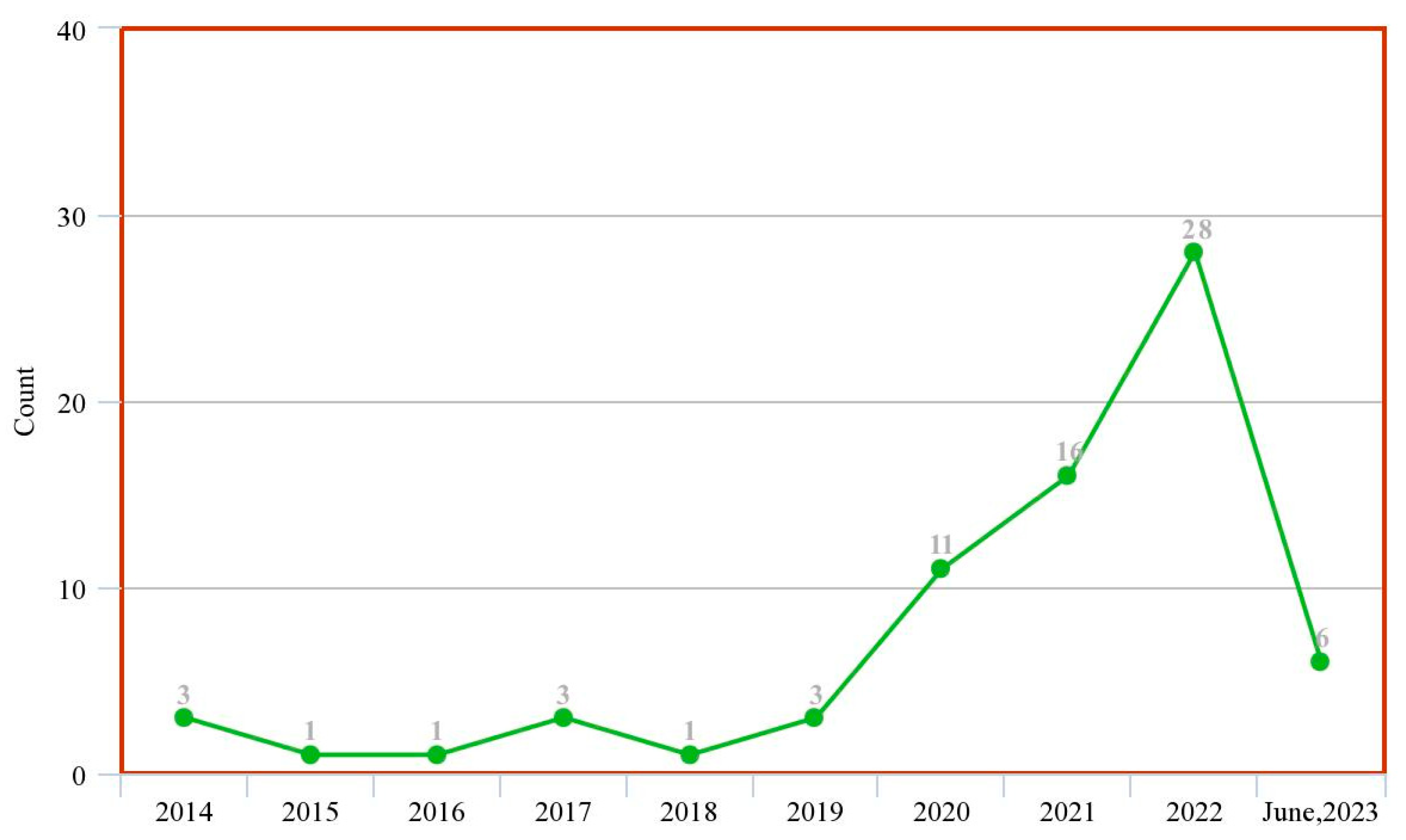
Figure 3 shows the trend of published review papers on the subject matter over the previous ten years to date. This was obtained from tailored search on Google Scholar (https://scholar.google.com accessed on 4 July 2023) with the query ‘machine learning deep learning intitle: “prostate cancer” intitle: “review”’ and filtered by year.
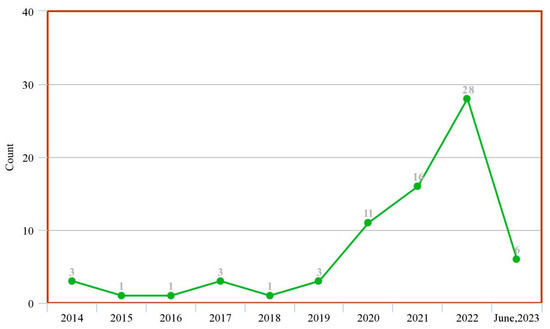
Figure 3.
Trend of research in ML/DL models for PCa diagnosis (systematic review study).
These two figures show that although there is an increasing wave of research in this subject matter, there are not enough systematic review studies to match up with this ever-rising trend. This justifies that this study is highly relevant given the experiment/review study ratio in the past decade.
1.1. Related Works
Many review and survey papers have investigated the application of machine learning and deep learning models to support the diagnosis and decision-making process of PCa. These papers have addressed the use of several deep learning models on various datasets and image modalities and presented the findings of the authors in those respective papers. Table 1 summarizes the review papers identified as relevant to our aim in this study as well as their findings so far.

Table 1.
Some selected related systematic review and survey articles for deep learning diagnosis of PCa in clinical patients.
Review articles have conducted tremendous work in the investigation of the roles of ML and DL models in clinically significant prostate cancer (csPCa). However, some limitations are identified. First, the review articles that met most authors’ final inclusion criteria are very small compared to the hundreds of articles released on a weekly basis. Second, most studies focused on a single image modality, whereas there are other imaging modalities that should be included. Some studies also used a single database as a reference search, which we know cannot provide a representative study of the subject matter. Also, some studies did not discuss major considerations such as the choice of dataset, the choice of image modalities, the choice of ML/DL models, hyperparameter tuning and optimization, among others. These are some of the lapses that our work seeks to address.
1.2. Scope of Review
This study aims to address the following research questions in the context of diagnosing PCa with ML and DL techniques. This can be utilized by researchers and medics to obtain a comprehensive view of the evolution of these techniques, datasets and imaging modalities and the effectiveness of these techniques in PCa diagnosis. The following research questions (RQs) are considered in this study:
| RQ1: | What are the trends and evolutions of this study? |
| RQ2: | Which ML and DL models are used for this study? |
| RQ3: | Which datasets are publicly available? |
| RQ4: | What are the necessary considerations for the application of these artificial intelligence (AI) techniques in PCa diagnosis? |
| RQ5: | What are the limitations that were identified so far by the authors? |
| RQ6: | What are the future directions for this research? |
We also investigated the verifiability of these studies by checking whether a medic or radiologist was one of the contributors or if it was stated that the results of the model were verified by one. We also included a citation metric and impact index in our work to measure the impact of the reviewed articles.
1.3. High-Level Structure of This Study
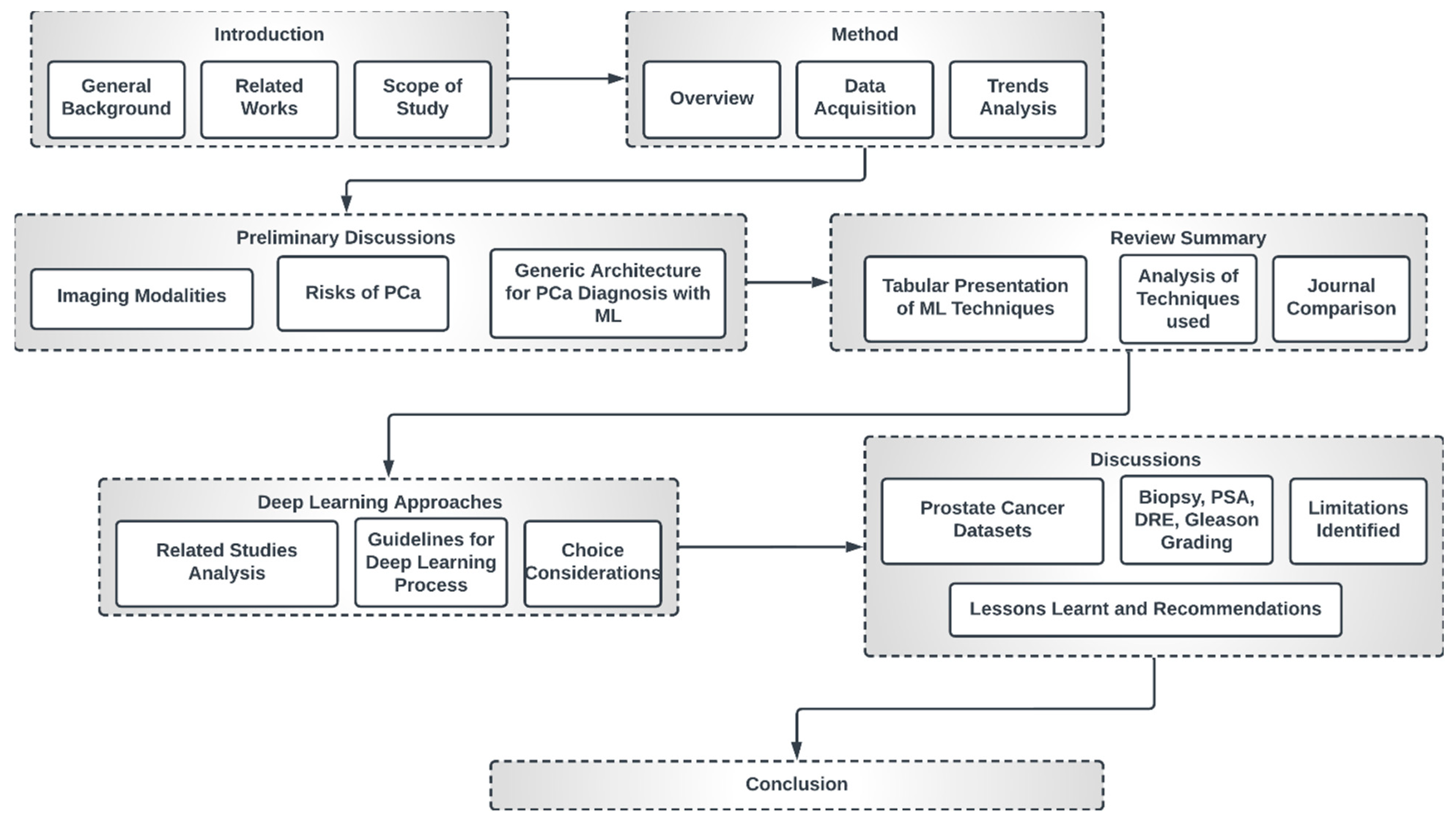
This study is organized as indicated in Figure 4. The first section presents a general overview of this study, the related review works and the scope of this study. Section 2 discusses the method of review employed in this paper. Section 3 engages in preliminary discussions concerning imaging modalities, the risks of PCa and general deep learning architecture for PCa diagnosis. Section 4 presents a summary table of papers that meet the inclusion criteria of this study with a comparative analysis of the trends, datasets, methods, techniques and journals.
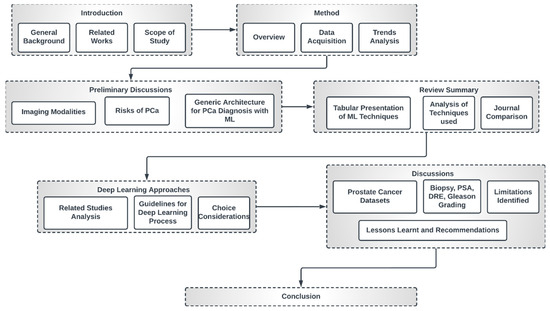
Figure 4.
High-level structure of this research.
Section 5 discusses some popular deep learning approaches and gives guidelines for the choice of individual techniques and optimization considerations as well as the choice of loss function. Section 6 presents a discussion of the findings. We also discuss the identified limitations, lessons learned and recommendations. The final section concludes this study.
2. Methods
This review paper explores, investigates, evaluates and summarizes findings in the literature that discuss PCa diagnosis with ML and DL techniques and image datasets, thereby equipping readers with a wholistic view of the subject matter, summaries of different techniques, datasets and models, as well as various optimization techniques available for model training. The authors will conduct various possible comparisons, discuss challenges and limitations and suggest future work directions and areas of improvement. The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) [23] guideline was used for conducting this review.
2.1. Database Search and Eligibility Criteria
In this systematic review, we constructed a search strategy and used it to scout four major databases (ScienceDirect, PubMed, Springer and IEEE) in search of up-to-date, relevant research publications on the research study of using ML and DL models to clinically diagnose csPCa. Google Scholar was used as the secondary resource in the preliminary and expository discussions. The timeframe for the investigation is the 2015–2023 period. These sources were selected because of their extensive publications of research in this area of study.
2.2. Review Strategy
The review process involved study selection, research design, search strategy, information sources and data collection techniques. It also involved an evaluation of papers that complied with the initial inclusion and exclusion criteria. Editorials, comments, letters, preprints and databases were not included in the four categories, and other types of manuscripts were not accepted. The search strategy was composed as follows: (a) construct search terms by identifying major keywords, required action and expected results; (b) determine the synonyms or alternative words for the major keywords; (c) establish exclusion criteria to make exclusions during search and (d) apply Boolean operators to construct the required search term.
| Results for (a): | Deep Learning Machine Learning Significant Prostate Cancer Artificial Intelligence Prediction Diagnosis; |
| Results for (b): | Prediction/Diagnosis/Classification Machine/Deep Prostate Cancer/PCa/csPCa; |
| Results for (c): | Review, systematic review, preprint, risk factor, treatment, biopsy, Gleason grading, DRE; |
| Results for (d): | a, b and c combined using AND OR. |
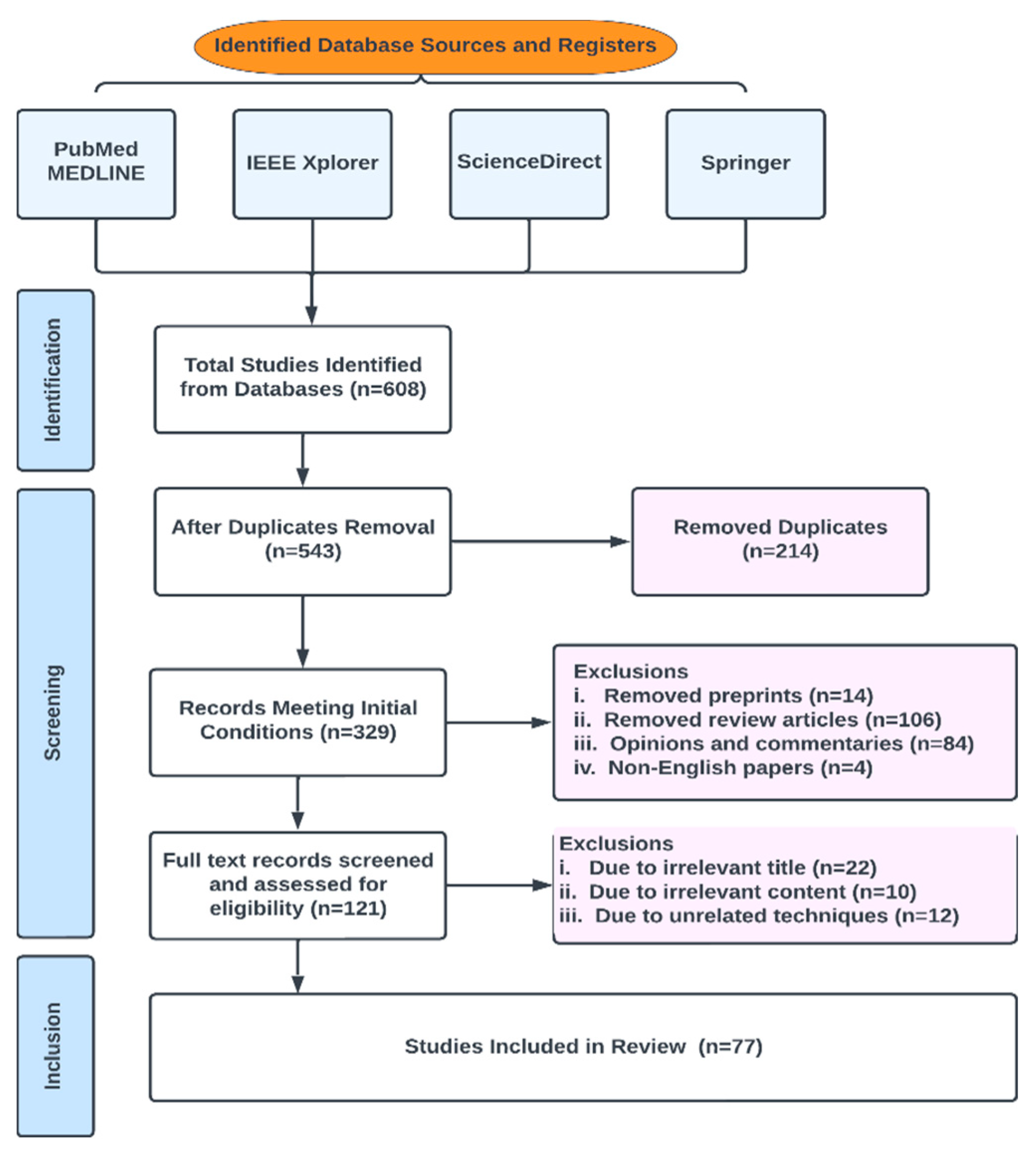
In this review, publications were chosen from peer-reviewed works in the literature by conducting a search using a generated search phrase (the final search terms used in this study to query our database sources were (“ Multiparametric-MRI “ OR Machine Learning ” OR “Deep Learning” OR “ANN” OR “AI” OR “Prostate Cancer”) intitle:”Prostate Cancer” source: “<Springer/IEEE/PubMed/ScienceDirect>“) in Science Direct, Springer, IEEE and PubMed. Conference proceedings, journals, book chapters and whole books are all examples of vetted resources. The initial number of results returned was 608; of those, 543 fulfilled the initial selection criteria and 77 fulfilled the final requirements. The studies were appropriately grouped. Figure 5 shows the Preferred Reporting Items for Systematic Reviews and Meta-Analyses for scoping reviews (PRISMA-ScR) flowchart for study selection.
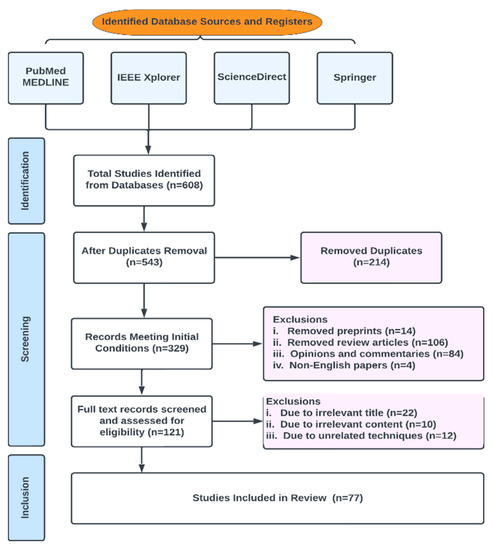
Figure 5.
PRISMA-Scr numerical flow guideline for systematic review employed in this study.
Our exclusion criteria included duplicates, preprints, review articles, opinions and commentaries, editorials, non-English papers, irrelevant titles, irrelevant contents, irrelevant techniques and date of publication.
2.3. Characteristics of Studies
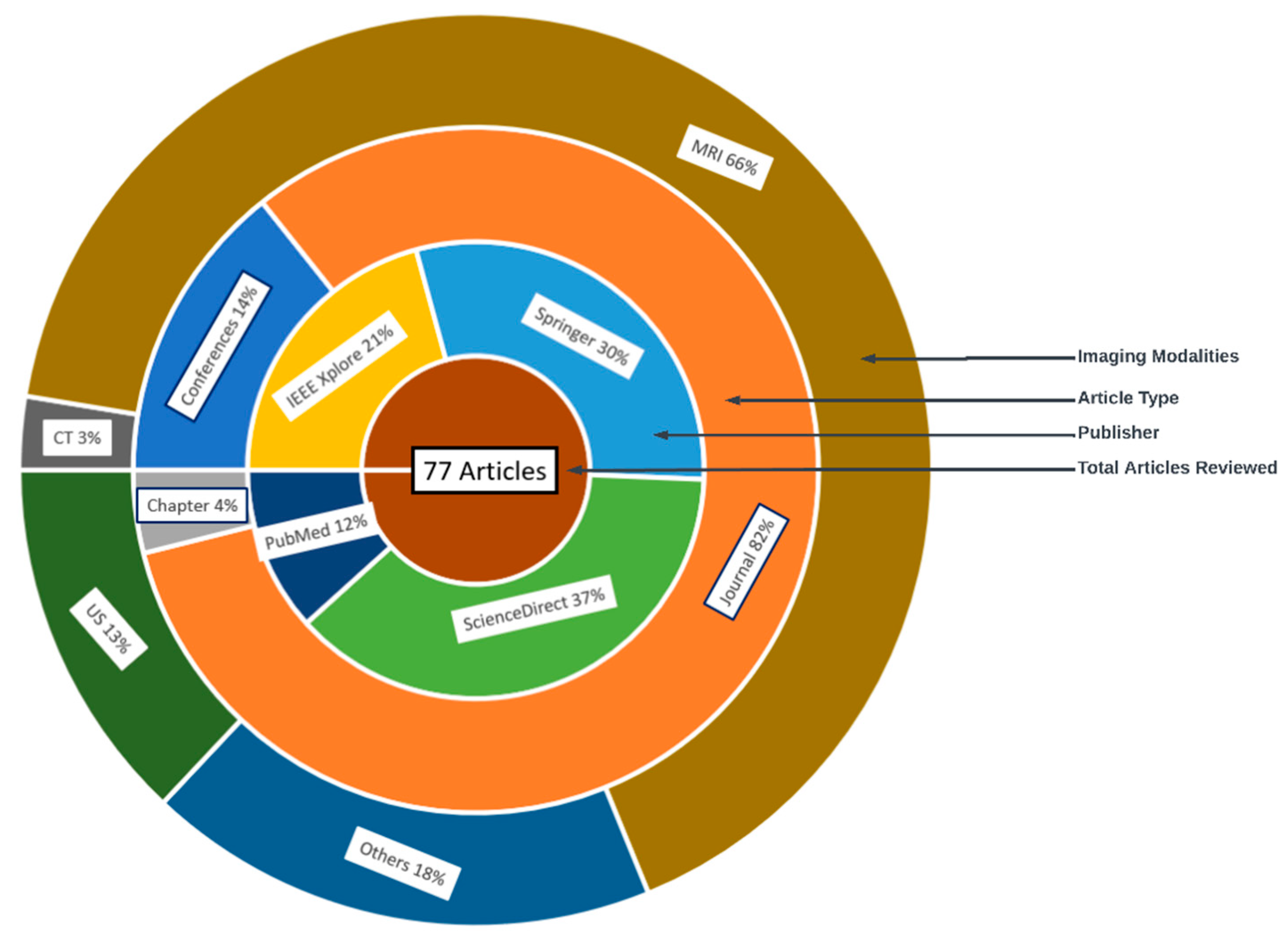
The characteristics of the 77 reviewed articles are given in Figure 6. The outer later is the distribution of the image modalities, followed by the article type, database and total number of articles reviewed.
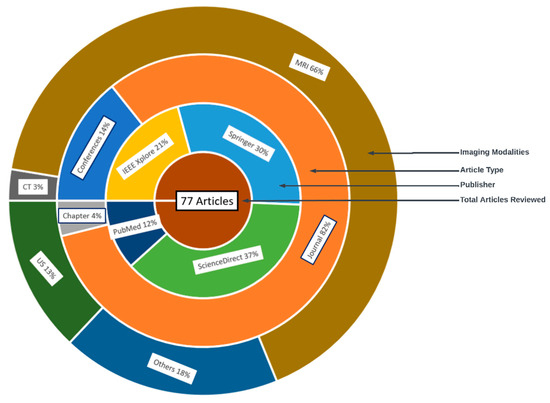
Figure 6.
Characteristics of reviewed articles in the literature.
2.4. Quality Assessment
Most studies failed to satisfy standards in at least one of the six quality criteria examined. Limited sample size, an inadequate scientific strategy and failure to disclose results for computational techniques were the most frequently observed issues regarding lack of quality throughout the investigations.
2.5. Data Sources and Search Strategy
We searched the four selected databases for studies published before July 2023 but not earlier than 2015. Keywords from subject headings or titles or abstracts of the studies were searched for with the help of Boolean operators (and/or) with language restricted to English. In addition, we reviewed the reference lists of primary studies and review articles.
2.6. Inclusion and Exclusion Criteria
Research papers in which ML and DL approaches were applied to predict and characterize PCa were reported. The included publications document the AI technique(s) used and PCa image analysis problem addressed in this article. Articles dealing with PCa key datasets and associated analysis techniques were also included in the study selection. Articles in preprints, not published in our selected databases, opinions, commentaries and non-English papers were all excluded. Editorials, narrative review articles, case studies, conference abstracts and duplicate publications were all discarded from the analysis. Articles that discuss similar techniques and results were ignored.
2.7. Data Extraction
The full texts of the qualified papers chosen for review were acquired, and the reviewers independently collected all study data, resolving disagreements via consensus. The references, year of publication, study setting, ML approach, the imaging modality used or recommended, performance measures used and accuracy attained were all extracted for every included paper, and comparative analyses were conducted on the extracted dataset.
2.8. Data Synthesis
The included studies were analyzed with respect to the types of models employed, datasets used, preprocessing techniques, features extracted and performance metrics reported. Some ML/DL models such as Convolutional Neural Networks (CNNs) perform better, are scalable and are more adaptive than others, especially in terms of the different modalities in medical image analysis. Performance evaluation was conducted through a spectrum of metrics including sensitivity, specificity, accuracy and area under the receiver operating characteristic curve (AUC-ROC).
2.9. Risk of Bias Assessment
Our study assessment aims to evaluate the methodological quality and potential sources of bias that could influence the reported findings. For instance, studies that rely solely on single-center datasets or imbalanced class distributions may introduce biases that affect the model generalizability. Additionally, the lack of clear documentation of preprocessing steps, considerations for model-fitting problems and hyperparameter tuning could hinder reproducibility.
3. Preliminary Discussions
3.1. Imaging Modalities
Prostate imaging refers to various techniques and procedures used to visualize the prostate gland for diagnostic and treatment purposes. These imaging methods help in evaluating the size, shape and structure of the prostate, as well as detecting any abnormalities or diseases, such as prostate cancer [24,25], and they include Transrectal Ultrasound (TRUS) [26], Magnetic Resonance Imaging (MRI) [27], Computed Tomography (CT) [28], Prostate-Specific Antigen (PSA) [29], Prostate-Specific Membrane Antigen (PET/CT) [30] and bone scans [31]. A TRUS involves inserting a small probe into the rectum, which emits high-frequency sound waves to create real-time images of the prostate gland. A TRUS is commonly used to guide prostate biopsies and assess the size of the prostate [26,32]. MRI, one of the most common prostate imaging methods, uses a powerful magnetic field and radio waves to generate detailed images of the prostate gland. It can provide information about the size, location and extent of tumors or other abnormalities. A multiparametric MRI (mpMRI) combines different imaging sequences to improve the accuracy of prostate cancer detection [33,34]. A CT scan uses X-ray technology to produce cross-sectional images of the prostate gland. It may be utilized to evaluate the spread of prostate cancer to nearby lymph nodes or other structures. PSMA PET/CT imaging is a relatively new technique that uses a radioactive tracer targeting PSMA, a protein that is highly expressed in prostate cancer cells [35]. It provides detailed information about the location and extent of prostate cancer, including metastases. Bone scans are often performed in cases where prostate cancer has spread to the bones. A small amount of radioactive material is injected into the bloodstream, which is then detected by a scanner [31]. The scan can help to identify areas of bone affected by cancer. PSA (density mapping) combines the results of PSA blood tests with transrectal ultrasound measurements to estimate the risk of prostate cancer. It helps to assess the likelihood of cancer based on the size of the prostate and the PSA level [36]. The choice of imaging technique depends on various factors, including the specific clinical scenario, the availability of resources and the goals of the evaluation [37,38].
3.2. Risks of PCa
The risk of PCa varies in men depending on several factors, and identifying these factors can aid in the prevention and early detection of PCa, personalized healthcare, research and public health policies, genetic counseling and testing and lifestyle modifications. The most common clinically and scientifically verified risk factors include age, obesity and family history [39,40]. In low-risk vulnerable populations, the risk factors include benign prostatic hyperplasia (BPH), smoking, diet and alcohol consumption [41]. Although PCa is found to be rare in people below 40 years of age, an autopsy study on China, Israel, Germany, Jamaica, Sweden and Uganda showed that 30% of men in their fifties and 80% of men in their seventies had PCa [42]. Studies also found that genetic factors, a lack of exercise and sedentary lifestyles are cogent risk factors of PCa, including obesity and an elevated blood testosterone level [43,44,45,46]. The consumption of fruits and vegetables, the frequency of high-fat meat consumption, the level of Vitamin D in blood streams, cholesterol level, infections and other environmental factors are deemed to contribute to PCa occurrence in men [47,48].
3.3. Generic Overview of Deep Learning Architecture for PCa Diagnosis
Deep learning (DL) architectures have shown promising effectiveness and relative efficiency in PCa diagnosis due to their ability to analyze complex patterns and extract features from medical imaging data [13]. One commonly used deep learning architecture for cancer diagnosis is Convolutional Neural Networks (CNNs). CNNs are particularly effective in image analysis tasks, including medical image classification, segmentation, prognosis and detection [49]. Deep learning, given its ever-advancing variations, has recorded significant advancements in the analysis of cancer images including histopathology slides, mammograms, CT scans and other medical imaging modalities. DL models can automatically learn hierarchical representations of images, enabling them to detect patterns and features that are indicative of cancer. They are also trained to classify PCa images into different categories or subtypes. By learning from labeled training data, these models can accurately classify new images, aiding in cancer diagnosis and subtyping [50].
Transfer learning is often employed in PCa image analysis. Pre-trained models, such as CNNs trained on large-scale datasets like ImageNet, are fine-tuned or used as feature extractors for PCa-related tasks. This approach leverages the learned features from pre-training, improving performance even with limited annotated medical image data. One image dataset augmentation framework is a Generative Adversarial Network (GAN). GANs can generate realistic synthetic images, which can be used to supplement training data, enhance model generalization and improve the performance of cancer image analysis models. The performance and effectiveness of deep learning models for PCa image analysis, however, depend on various factors, including the quantity and quality of labeled data, the choice of architecture, the training methodology and careful validation of diverse datasets.
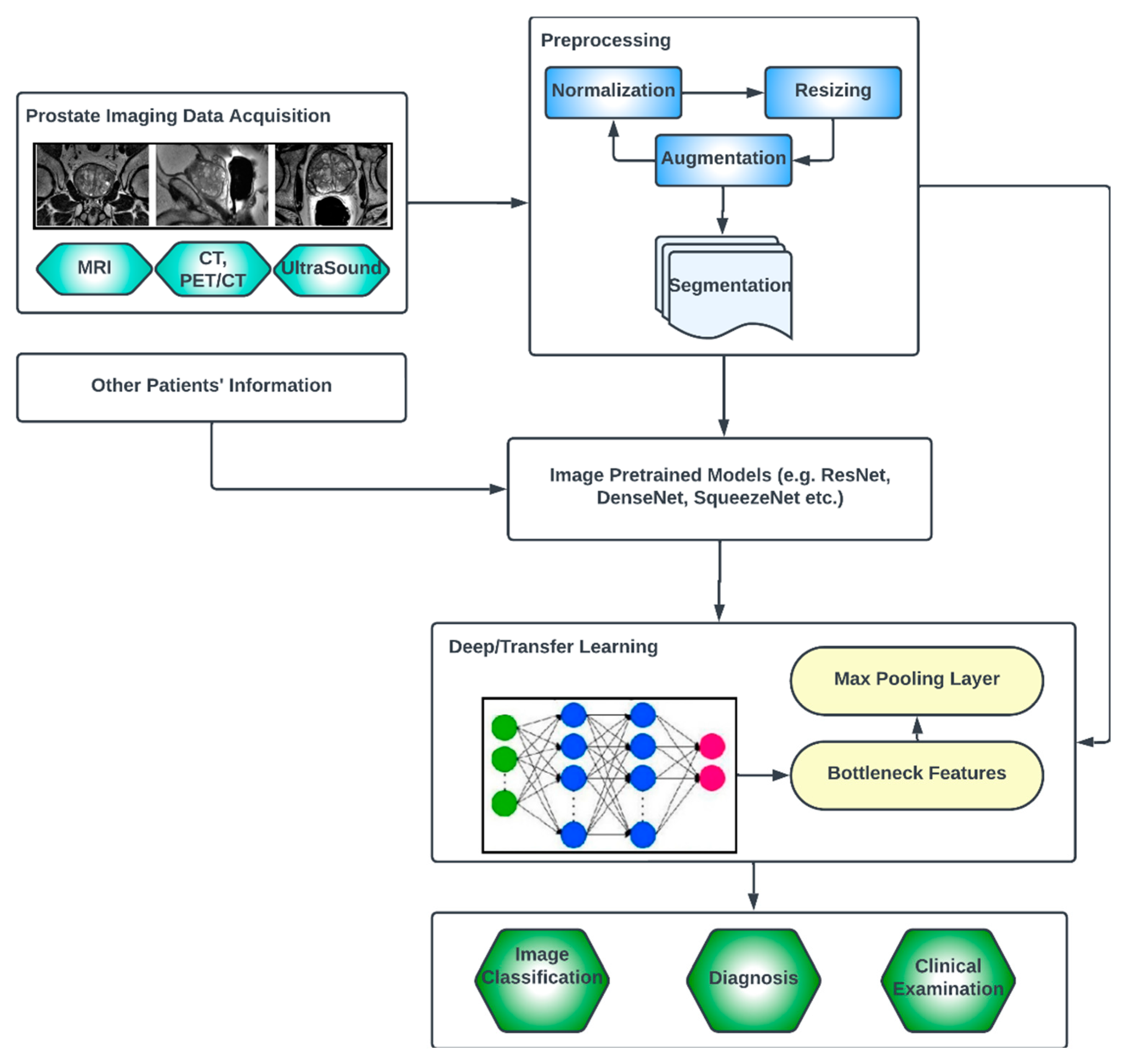
The key compartments in a typical deep CNN model for PCa diagnosis, as shown in Figure 7, include the convolutional layers, the pooling layers, the fully connected layers, the activation functions, the data augmentation and the attention mechanisms [51,52]. The convolutional layers are the fundamental building blocks of CNNs. They apply filters or kernels to input images to extract relevant features. These filters detect patterns at different scales and orientations, allowing for the network to learn meaningful representations from the input data. The pooling layers downsample feature maps, reducing the spatial dimensions while retaining important features. Max pooling is a commonly used pooling technique, where the maximum value in each pooling window is selected as the representative value [53]. The fully connected layers are used at the end of CNN architectures to make predictions based on the extracted features. These layers connect all the neurons from the previous layer to the subsequent layer, allowing for the network to learn complex relationships and make accurate classifications. Activation functions introduce non-linearity into the CNN architecture, enabling the network to model more complex relationships. Common activation functions include ReLU (Rectified Linear Unit), sigmoid and tanh [54,55]. Transfer learning involves leveraging pre-trained CNN models on large datasets (such as ImageNet, ResNet, VGG-16, VGG-19, Inception-v3, ShuffleNet, EfficientNet, GoogleNet, ResNet-50, SqueezeNet, etc.) and adapting them to specific medical imaging tasks. By using pre-trained models, which have learned general features from extensive data, the model construction time can be saved, as well as computational resources, and can achieve good performance even on smaller medical datasets. Data augmentation techniques, such as rotation, scaling and flipping, can be employed to artificially increase the diversity of the training data. Data augmentation helps to improve the generalization of a CNN model by exposing it to variations and reducing overfitting. Attention mechanisms allow for the network to focus on relevant regions or features within the image. These mechanisms assign weights or importance to different parts of the input, enabling the network to selectively attend to salient information [56,57].
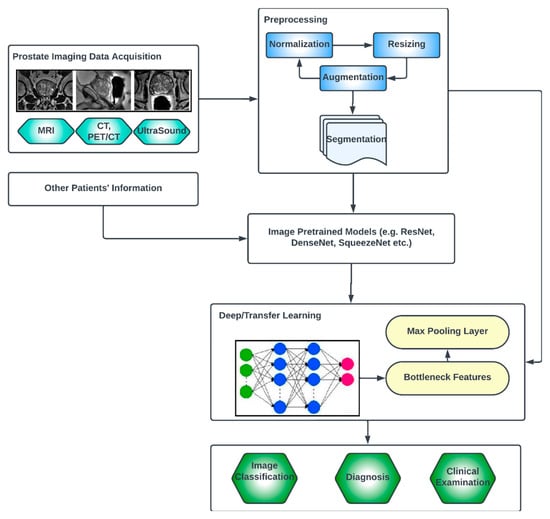
Figure 7.
Generic deep learning architecture for PCa image analysis.
Vision Transformers (ViTs) [58,59,60] are a special type of deep learning architecture, which, although originally designed for natural language processing (NLP) tasks, has shown promising performances for medical image processing. They consist of an encoder, which typically comprises multiple transformer layers. The authors of [61] studied the use of ViTs to perform prostate cancer prediction using Whole Slide Images (WSIs). A patch extraction from the detected region of interest (ROI) was first performed, and the performance showed promising results. A novel artificial intelligent transformer U-NET was proposed in a recent study [62]. The authors found that inserting a Visual Transformer block between the encoder and decoder of the U-Net architecture was ideal to achieve the lowest loss value, which is an indicator of better performance. In another study [63], a 3D ViT stacking ensemble model was presented for assessing PCa aggressiveness from T2w images with state-of-the-art results in terms of AUC and precision. Similar work was presented by other authors [64,65].
4. Results
Review Summary of Relevant Papers
In this section, we present a table summarizing the core contents of the papers that met our final inclusion criteria. The overall search captured 77 papers. The distribution of these publications among the four databases consulted is indicated in Table 2. PubMed serves as a mop-up database for the other three because some papers that were published elsewhere are also indexed in PubMed, which form a part of the removed duplicates, which was explained in our PRISMA-ScR in Figure 5. The figure indicates that ScienceDirect has the most papers on the subject matter.

Table 2.
Distribution of publications included in the study according to databases consulted after screening.
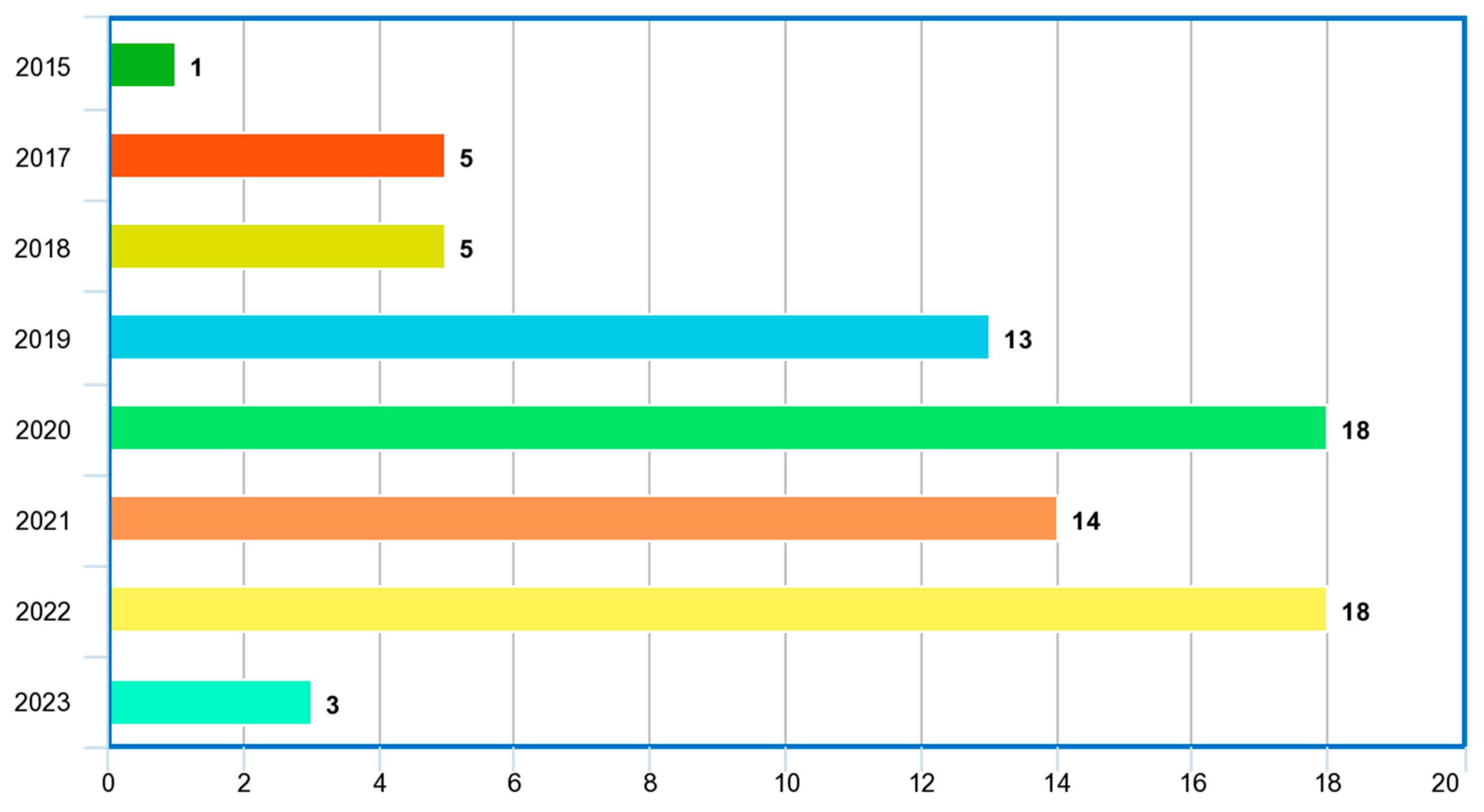
Table 3, Table 4, Table 5 and Table 6 highlight, for each database considered, the year in which the study was conducted, the imaging modalities, the ML/DL models employed in the study, the problem addressed, the reported performance metrics and scores, the reported hyperparameter tuning, the country in which the study was conducted, the citations received for each paper at the time of the study, whether the study was verified by a medical personnel or a radiologist, the number of observations or images considered for the study and the machine learning type, whether supervised or unsupervised. The distribution of the included publications by the years in which the studies were conducted, as given in Figure 8, shows that this study paid more attention to recent publications in the application of ML/DL for PCa diagnosis.

Table 3.
Springer papers on prostate cancer detection using machine learning, deep learning or artificial intelligence methods.

Table 4.
ScienceDirect papers on prostate cancer detection using machine learning, deep learning or artificial intelligence methods.

Table 5.
IEEE papers on prostate cancer detection using machine learning, deep learning or artificial intelligence methods.

Table 6.
PubMed papers on prostate cancer detection using machine learning, deep learning or artificial intelligence methods.
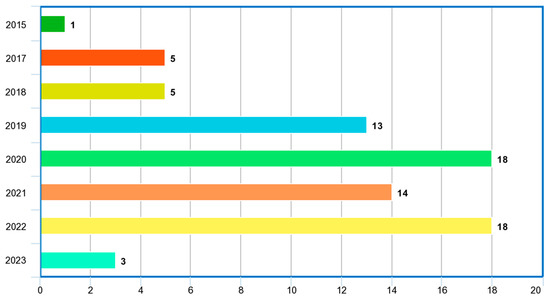
Figure 8.
Distribution of included papers by year.
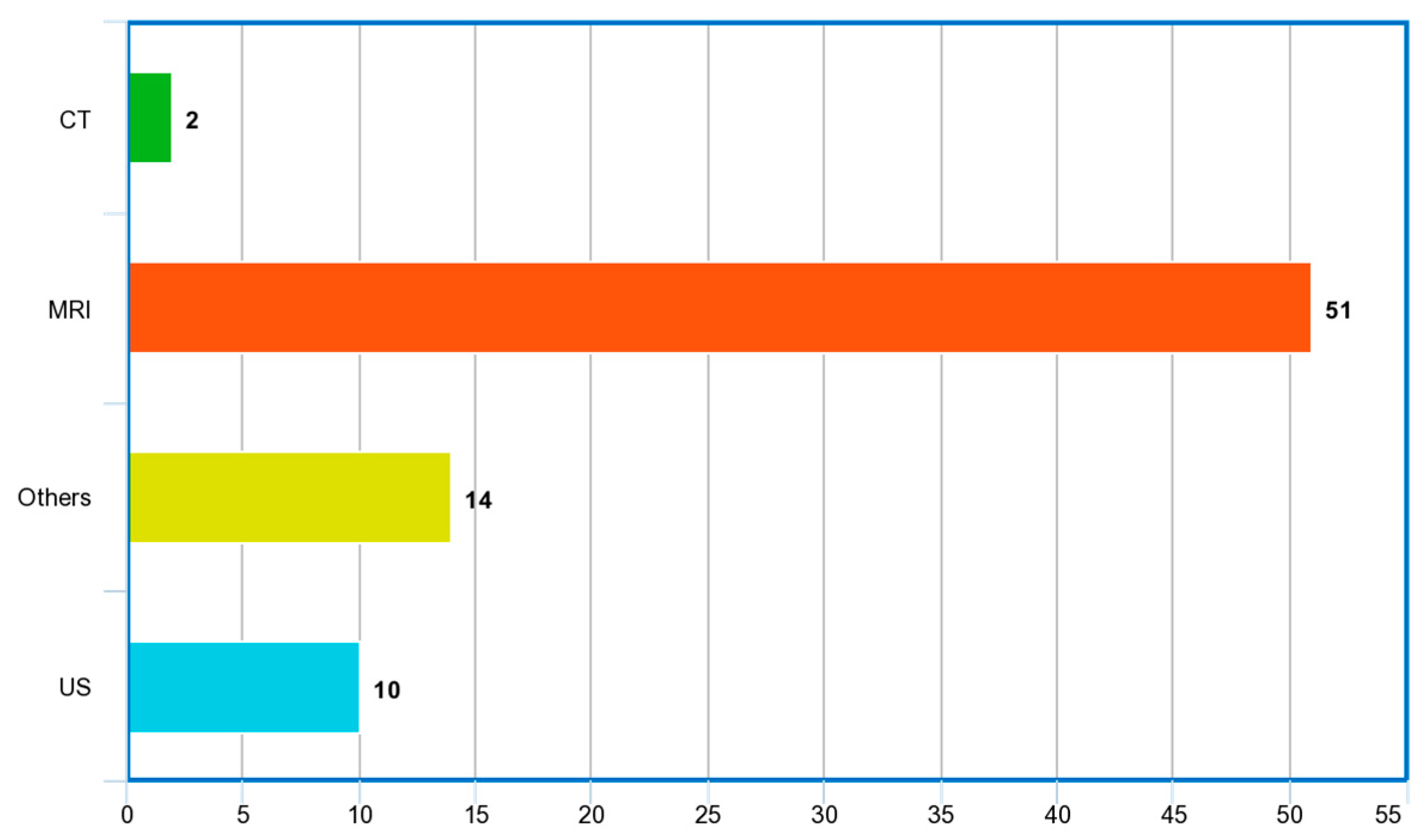
Figure 9 and Figure 10 show the word cloud of topics and word cloud of keywords of reviewed papers as generated by their word frequencies. They show that this study focuses on the image-based detection of prostate cancer using deep learning techniques. Figure 11 shows the image modalities used in the diagnosis of PCa. A total of 2 papers used Computed Tomography (CT), 51 papers used Magnetic Resonance Imaging (MRI) and 10 papers used ultrasound (US), while 14 papers used other imaging methods such as Whole Slide Images (WSIs), histopathological images and biopsy images.
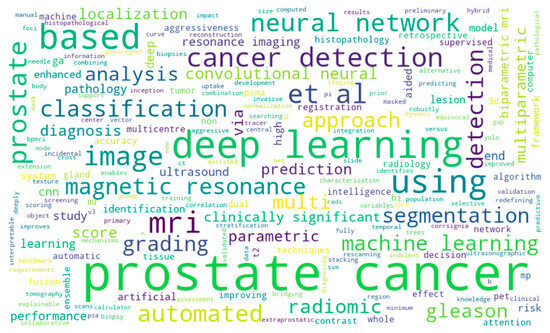
Figure 9.
Word cloud of topics of reviewed papers.

Figure 10.
Word cloud of keywords of included papers.
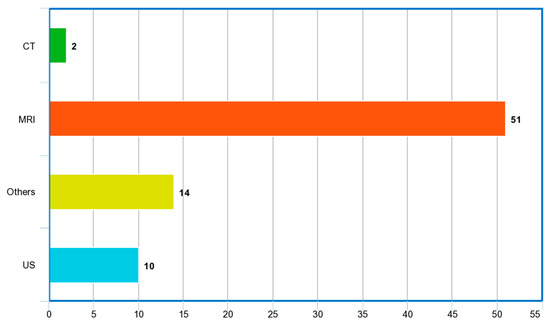
Figure 11.
Image modalities used in reviewed papers.
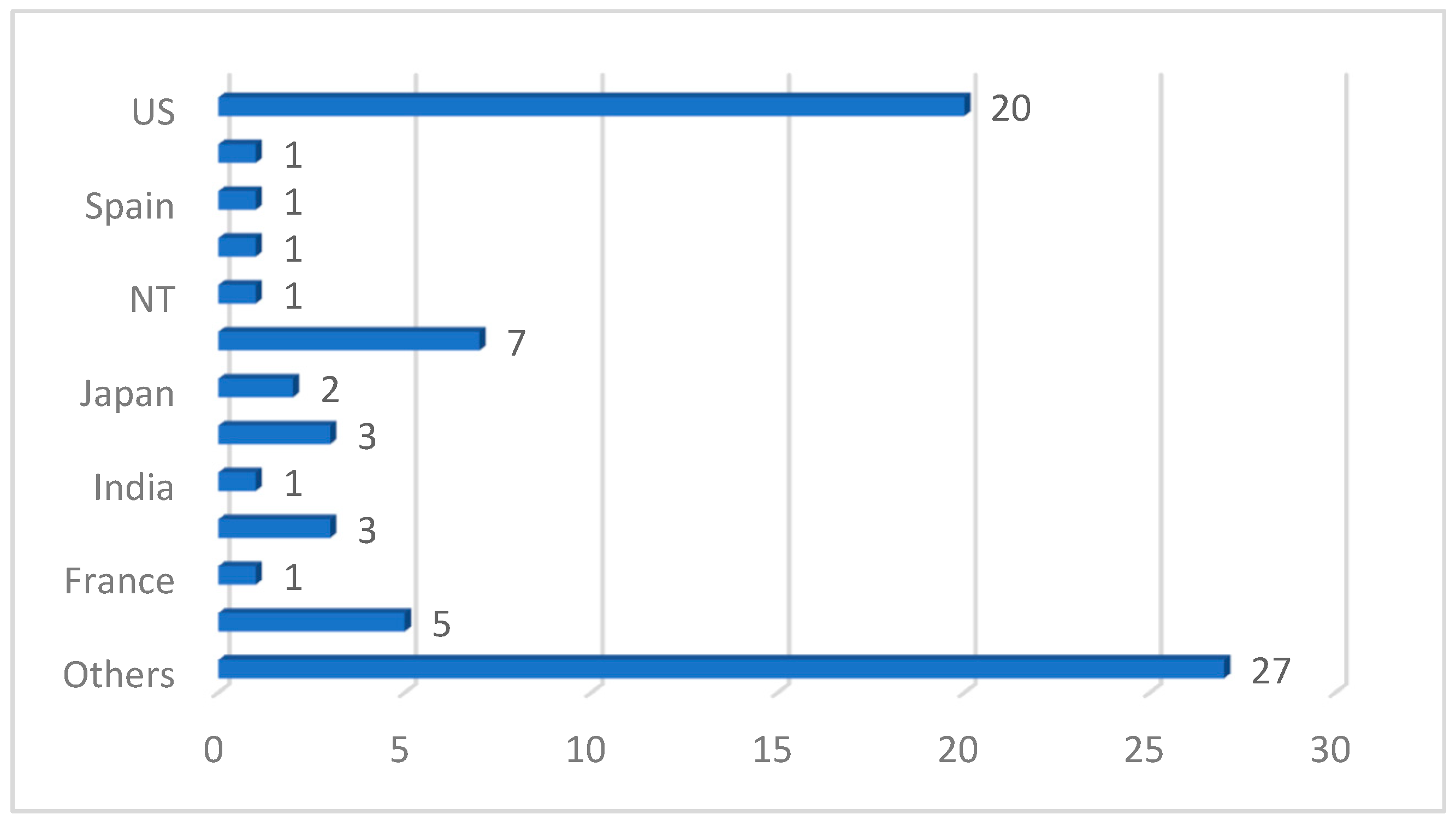
We also discovered from the review that a deep CNN is the most used ML/DL model, spanning about 49 studies out of the 77 reviewed papers. It is also noteworthy that transfer-learning-based DL architecture dominated the studies reviewed in this work, with UNet, ResNet, GoogleNet and DenseNet being the topmost frameworks used in the analysis of PCa images. This is understandable because transfer learning offers a range of advantages, including a reduced training time, improved generalization, effective feature extraction, addressing data imbalance and facilitating domain adaptation. In the aspect of the performance of the models reported in the reviewed papers, the area under curve (AUC) is the most used metric, followed by accuracy and sensitivity. Most studies also used supervised learning (classification) methods. The images were manually annotated by medical professionals and radiologists for an adequate performance evaluation of the models. However, the absence of sufficient data for model training forced most authors into using secondary data and pretrained (transfer learning) models. In terms of the countries where the reviewed studies were conducted, Figure 12 shows that the USA has the highest number of studies. Table 7 shows the topmost impactful papers included in our study. The impact index is calculated using Equation (1) below:
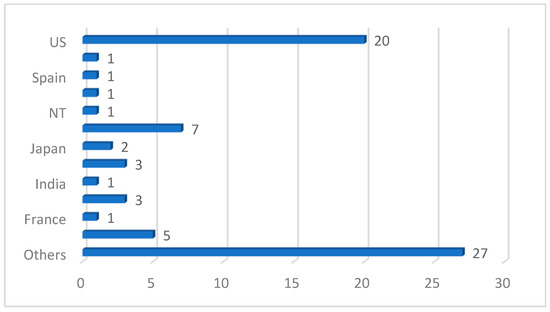
Figure 12.
Country distribution of reviewed studies.

Table 7.
Top 10 most impactful papers.
This gives readers an overview of where to publish related research. It is evident from this table that Nature, Elsevier and Springer are the top publishers to consider.
5. Discussion
The included papers in Table 3, Table 4, Table 5 and Table 6 gave a synthesized overview of the state-of-the-art machine learning and deep learning applications in the detection and analysis of PCa using medical images vis-à-vis the various problems addressed, the techniques applied, the datasets used, feature extraction (if any), hyperparameter tuning and their respective reported performance metrics. The expanded summary of the included papers is given in Appendix A, Appendix B, Appendix C and Appendix D. The use of deep neural nets and transfer learning architecture with outperforming accuracies is imminent when compared to non-deep models [8,15,66,67,68]. Also, MRIs (parametric and non-parametric) were the most used imaging modality in the detection of prostate cancer. The authors have also shown that histological data can be combined with MRI, CT or US images to better improve the accuracy of automatic detection systems of PCa [72,123,136]. CNNs have also proven to have a strong ability to learn and extract features from medical images and have also demonstrated remarkable accuracy in telling malignant and benign prostate regions apart. The reviewed papers have also demonstrated that a multi-modality integration of medical images into DL architectures can allow for a more comprehensive assessment and potentially improve diagnostic accuracy.
5.1. Considerations for Choice of Deep Learning for PCa Image Data Analysis
The choice of which deep learning models to use for PCa detection in clinical images must be guided by a thorough exploration of their context of usages and associated strengths and weaknesses. Table 8 gives a summary of some specialties of each of the deep learning models to guide researchers’ choices of experimenting with PCa image datasets.

Table 8.
Summary of considerations for choice of deep learning models for PCa diagnosis using medical images.
5.2. Considerations for Choice of Loss Functions for PCa Image Data Analysis
One specific and very important concept in the training of deep learning models for PCa diagnosis is the choice of loss functions, which plays a significant role in training and optimizing the performance of the models [148,149]. Loss functions guide the optimization process by quantifying the discrepancy between the predicted output of the model and the ground truth labels or targets. The choice of loss function affects how the model learns and updates its parameters during training. A carefully selected loss function helps the model converge to an optimal solution efficiently [150]. Loss functions are also helpful in handling imbalance datasets—this is a common challenge where certain classes or abnormalities are rare compared to others. In such cases, loss functions need to address the imbalance to prevent the model from being biased towards the majority class. It also helps to handle noise and outliers in model interpretability as well as in gradient stability [151]. Although the choice of loss function depends largely on the specific task, the nature of the problem and the characteristics of the dataset, Table 9 summarizes some of the most used loss functions in deep learning and their best-suited context of usage. This does not replace the need for necessary experimentation and evaluation while choosing the applicable and appropriate loss function.

Table 9.
Considerations for choice of loss functions in deep learning.
5.3. Prostate Cancer Datasets
Prostate cancer datasets consist of clinical and pathological information collected from patients diagnosed with prostate cancer and may include various types of data, such as patient demographics, clinical features, laboratory test results, imaging data (e.g., MRI, US or CT scans), histopathology slides (WSI) and treatment outcomes. They are useful for developing and evaluating machine learning and deep learning models for prostate cancer detection, diagnosis, prognosis and treatment prediction. Table 10 presents some publicly available databases of PCa datasets.

Table 10.
Some publicly available databases for PCa datasets.
5.4. Some Important Limitations Discussed in the Literature
In this section, we harvest some crucial limitations identified by the authors of the reviewed works in the literature (see Appendix A, Appendix B, Appendix C and Appendix D). This will aid readers in understanding the challenges encountered by researchers in conducting experiments in the application of deep learning to PCa diagnosis. The authors of [94,101] identified limitations, which included small and highly unbalanced datasets [98] with unavoidable undersampling. They also noted that in an ultrasound-guided biopsy’s registration, similar to other manual pathological–radiological strategies, a personal bias in the selection of regions of interest (ROIs) cannot be avoided. A study has also shown that when explainability and interpretability are taken into account in PCa prediction model construction, the runtime becomes a critical issue, and a conscious trade-off decision must be made [98]. CNN engines have also been reported to have poor interpretability. This is because the last convolutional layer of a classical CNN model contains the richest spatial and semantic information through multiple convolutions and pooling, and the next layer is fully connected with SoftMax layers, which contain information that is difficult for humans to understand and difficult to visualize [109]. Some authors noted that models that behave like feed-forward Long Short-Term Memory (LSTM), for instance, have a bit of a parity issue if not augmented with deep and transfer learning methods to classify PCa and non-PCa subjects [118]. In the summary tables, the studies identified that multi-modal and multi-center studies can deflect the performance of a model that is adjudged to be good enough in a unimodal and single-center study [129].
5.5. Lessons Learned and Recommendations
The application of deep learning for prostate cancer detection has made significant advancements in recent years, and this study will expose readers to the trends in the techniques, models, datasets and some other critical considerations when venturing into similar studies. The data quality and availability have been major limitations of the existing studies. The data for PCa are scanty and often small and imbalanced, leading to issues with the model’s generalizability and performance. Interpretability is of great concern in deep learning models, especially because the models reviewed in this study are meant to be utilized by medics and radiologists as a decision support system (DSS).
Deep learning models are often referred to as “black boxes” because they lack explainability. While they can accurately make predictions, understanding the underlying factors or features that contribute to those predictions can be difficult. This lack of interpretability is a significant limitation when it comes to clinical decision making and explaining the rationale behind a model’s predictions. Given the need for an accurate and efficient radiologic interpretation, PCa detection systems must serve as a decision-making aid to clinicians through its explainability. Explainable artificial intelligence (XAI) models can enable more accurate and informed decision making for csPCa, thereby fulfilling the need for improved workflow efficiency [88,176,177].
Clinical validation, as seen in the summary tables, should be given attention in CAD-related studies. Many deep learning studies for prostate cancer focus on retrospective analyses using archival data. While these studies can provide valuable insights, there is a need for robust clinical validation to assess the real-world performance and impact of these models. Clinical validation requires multi-modal and multi-center applicability.
Also, PCa datasets are often limited, and the complex nature of the disease makes it challenging to build models that can be generalized effectively. Regularization techniques and careful validation are required to mitigate the risk of overfitting and improve generalization. Finally, for deep learning models to have a real impact on prostate cancer diagnosis, prognosis or treatment, they need to be seamlessly integrated into the clinical workflow. This requires addressing practical challenges such as compatibility with existing electronic health record systems, establishing trust among healthcare professionals and addressing regulatory and ethical considerations.
6. Conclusions
This study wholistically investigated the application of machine learning and deep learning models in prostate cancer detection and diagnosis. We also conducted a publisher-based comparison to give readers a view of some possible tendencies such as the potential impact. Considerations regarding ML/DL models, PCa datasets and loss functions were also discussed. We found that although the trend curves of systematic reviews (Figure 3) and actual experimental studies (Figure 2) look similar, there is a need for a thorough systematic study to investigate the trend, challenges and future directions in the application of ML/DL models to the ravaging disease. Although one of the advantages of deep learning models for segmentation is that they are fully automatic, requiring no intervention, the studies showed that their performance can be improved by having some method to improve the initial organ localization, which would allow for a relatively smaller, higher-resolution sub-volume to be extracted instead of using the entire image, which contains noise. We conclude that transfer learning models are recommended for PCa diagnosis. This is because transfer learning offers significant advantages for prostate cancer diagnosis by leveraging pre-trained models, reducing data requirements, improving model performance, enabling faster training, capturing complex features, enhancing generalization and expediting deployment in clinical practice. Clinical verification is also required in these studies to ensure the usability and responsibility of these studies. This will ensure that CAD-related studies do not just end up as papers, but are integrated into existing clinical systems.
Author Contributions
Conceptualization, O.O. (Olusola Olabanjo) and A.W.; methodology, O.O. (Olusola Olabanjo) and O.F.; validation, O.O. (Olusola Olabanjo), M.A. and M.M.; investigation, M.M.; resources, M.A. and O.O. (Olufemi Olabanjo); data curation, B.O.; writing—original draft preparation, O.O. (Olufemi Olabanjo), M.A. and A.W.; writing—review and editing, O.O. (Olufemi Olabanjo), M.M., O.A. and A.W.; visualization, O.O. (Olusola Olabanjo) and A.W.; supervision, O.A., O.F. and M.M.; project administration, A.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A. Springer Papers on Prostate Cancer Detection Using Machine Learning, Deep Learning or Artificial Intelligence Methods

| Ref. | Problem Addressed | Imaging Modality | Machine Learning Type | Data Collection | Medic-Verified | Discussion | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MRI | US | Others | Transfer | SL | UL | Primary | Secondary | Yes | No | Strengths | Weaknesses | ||
| [15] | Comparison between deep learning and non-deep learning classifier for performance evaluation in classification of PCa | ✓ | ✓ | ✓ | ✓ | Convolution features learned from morphologic images (axial 2D T2-weighted imaging) of the prostate were used to classify PCa | One image from each patient was used, assuming independence among them | ||||||
| [69] | Classifying PCa tissue with weakly semi-supervised technique | ✓ | ✓ | ✓ | ✓ | Pseudo-labeled regions in the task of prostate cancer classification | Increase in time to label the training data | ||||||
| [75] | Predicting csPCa with a deep learning approach | ✓ | ✓ | ✓ | ✓ | Significantly reduce unnecessary biopsies and aid in the precise diagnosis of csPCa | It was difficult to achieve a complete balance between the training and external validation cohorts | ||||||
| [66] | Classification of patient’s overall risk with ML on high or low lesion in PCa | ✓ | ✓ | ✓ | ✓ | Lesion characterization and risk prediction in PCa | Model built on a single-center cohort and included only patients with confirmed PCa | ||||||
| [81] | Localization of PCa lesion using multiparametric ML on transrectal US | ✓ | ✓ | ✓ | ✓ | Visibility of a multiparametric classifier to improve single US modalities for the localization of PCa | Data collected in a single center and 2D imaging were used | ||||||
| [67] | Clinically significant PCa detection using CNN | ✓ | ✓ | ✓ | ✓ | Automated deep learning pipeline for slice-level and patient-level PCa diagnosis with DWI | Data are inherently biased | ||||||
| [76] | ML model capable of predicting PI-RADS score 3 lesions, differentiating between non-csPCa and csPCa | ✓ | ✓ | ✓ | ✓ | Solid feature extraction techniques were used | Relatively small dataset for training developed model | ||||||
| [68] | PCa risk classification using ML techniques | ✓ | ✓ | ✓ | ✓ | PCa risk based on PSA, free PSA and age in patients | Dataset was collected retrospectively, and thus, patient management was not consistent and oncological outcome was absent | ||||||
| [8] | Prostate detection, segmentation and localization in MRI | ✓ | ✓ | ✓ | ✓ | Ability to segment and diagnose prostate images | Lack of availability of manually annotated data | ||||||
| [70] | Impact of scanning systems and cycle-GAN-based normalization on performance of DL algorithms in detecting PCa | ✓ | ✓ | ✓ | ✓ | Model was developed on multi-center cohort | Significant class imbalance occurred with the data | ||||||
| [83] | Transfer learning approach using breast histopathological images for detection of PCa | ✓ | ✓ | ✓ | ✓ | Transfer learning approach for cross cancer domains was demonstrated | No extensive pre-training of the models | ||||||
| [82] | Developed a feature extraction framework from US prostate tissues | ✓ | ✓ | ✓ | ✓ | High-dimensional temporal ultrasound features were used to detect PCa | All originally labeled data are seen as suspicious PCa | ||||||
| [77] | Multimodality to improve detection of PCa in cancer foci during biopsy | ✓ | ✓ | ✓ | ✓ | Improved targeting of PCa biopsies through generation of cancer likelihood maps | Transfer learning network was not used | ||||||
| [85] | Image-based PCa staging support system | ✓ | ✓ | ✓ | ✓ | Expert assessment for identification and anatomical location classification of suspicious uptake sites in whole-body for PCa | A limited number of subjects with advanced prostate cancer were included | ||||||
| [78] | Risk assessment of csPCa using mpMRI | ✓ | ✓ | ✓ | ✓ | Established that using risk estimates from built 3D CNN is a better strategy | Single-center study on a heterogeneous cohort and the size was still limited | ||||||
| [79] | Proposed a better segmentation technique for csPCa | ✓ | ✓ | ✓ | ✓ | Automatic segmentation of csPCa combined with radiomics modeling | Low number of patients used | ||||||
| [80] | Lesion detection and novel segmentation method for both local and global image features | ✓ | ✓ | ✓ | ✓ | Novel panoptic model for PCa lesion detection | Method was used for a single lesion only | ||||||
| [86] | Incident detection of csPCa on CT scan | ✓ | ✓ | ✓ | ✓ | CT scans for detection of prostate cancer through deep learning pipeline | Only CT data were used | ||||||
| [71] | Gleason grading of whole-slide images of prostatectomies | ✓ | ✓ | ✓ | ✓ | Gleason scoring of whole-slide images with millions of images | Grade group informs postoperative treatment decision only | ||||||
| [72] | Detection of PCa tissue in whole-slide images | ✓ | ✓ | ✓ | ✓ | Solid analysis of histological images in patients with PCa | Needs more datasets to train the model for better accuracy | ||||||
| [73] | Segmentation and grading of epithelial tissue for PCa region detection | ✓ | ✓ | ✓ | ✓ | High performance characteristics of a multi-task algorithm for PCa interpretation | Misclassifications were occasionally discovered in the output | ||||||
| [74] | Image analysis AI support for PCa and tissue region detection | ✓ | ✓ | ✓ | ✓ | High accuracy in image examination | Increase in time to label the dataset | ||||||
| [84] | Gleason grading for PCa in biopsy tissues | ✓ | ✓ | ✓ | ✓ | Strength in determining the stage of PCa | Availability of relatively small data | ||||||
Appendix B. ScienceDirect Papers on Prostate Cancer Detection Using Machine Learning, Deep Learning or Artificial Intelligence Methods

| Ref. | Problem Addressed | Imaging Modality | Machine Learning Type | Data | Medic Verified | Discussion | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MRI | US | Others | Transfer | SL | UL | Primary | Secondary | Yes | No | Strengths | Weaknesses | ||
| [87] | Effect of labeling strategies on performance of PCa detection | ✓ | ✓ | ✓ | ✓ | Identification of aggressive and indolent prostate cancer on MRI | Number of samples used is relatively small and they were obtained from a single institution | ||||||
| [88] | Detection of PCa with an explainable early detection classification model | ✓ | ✓ | ✓ | ✓ | Improved the classification accuracy of prostate cancer from MRI and US images with fusion algorithm models | Faced difficulty in selecting which MRI to be fed as input for the fusion model | ||||||
| [89] | Radiomics and machine learning techniques to detect PCa aggressiveness biopsy | ✓ | ✓ | ✓ | ✓ | Image-derived radiomics features integrated with automatic machine learning approaches for PCa detection gave high accuracy | Relatively small-sized samples were used | ||||||
| [92] | Segmentation of prostate glands with an ensemble deep and classical learning method | ✓ | ✓ | ✓ | ✓ | Detect prostate glands accurately and assist the pathologists in making accurate diagnosis | Study was based on stroma segmentation only | ||||||
| [93] | An automated grading PCa detection model with YOLO | ✓ | ✓ | ✓ | ✓ | Grading of prostate biopsies with high performance | Relatively small amount of data used | ||||||
| [90] | Textual analysis and machine learning models to detect extra prostatic cancer | ✓ | ✓ | ✓ | ✓ | Combined TA and ML approaches for predicting presence of EPE in PCa patients | Low number of patients was used | ||||||
| [94] | Diagnosis of PCa with integration of multiple deep learning approaches | ✓ | ✓ | ✓ | ✓ | Improve the detection of PCa without significantly increasing the complexity model | Limited dataset and use of only bilinear interpolation algorithm | ||||||
| [91] | Detection of PCa with an improved feature extraction method with ensemble machine learning | ✓ | ✓ | ✓ | ✓ | Combined machine learning techniques to improve GrowCut algorithm and Zernik feature selection algorithm | Limited dataset used | ||||||
| [95] | Prostate biopsy calculator using an automated machine learning technique | ✓ | ✓ | ✓ | ✓ | First report of ML approach to formulae PBCG RC | No external validation for the experimentation | ||||||
| [96] | Upgrading a patient from MRI-targeted biopsy to active surveillance with machine learning model | ✓ | ✓ | ✓ | ✓ | Machine learning with the ability to give diagnostic assessments for PCa patients was developed | A lot of missing values in the dataset and small dataset | ||||||
| [97] | A pathological grading of PCa on single US image | ✓ | ✓ | ✓ | ✓ | High accuracy in grading of PCa from single ultrasound images without puncture biopsy | Low detection of PCa lesion region and imbalance of data | ||||||
| [99] | A radiomics deeply supervised segmentation method for prostate gland and lesion | ✓ | ✓ | ✓ | ✓ | Prostate lesion detection and prostate gland delineation with the inclusion of local and global features | Small sample size | ||||||
| [100] | Performance comparison of promising machine learning models on typical PCa radiomics | ✓ | ✓ | ✓ | ✓ | GBDT model implemented with CatBoost that gave consistent high performance | Only radiomic features with whole prostate in the T2-w MRI were used | ||||||
| [101] | SVM on Gleason grading of PCa-based image features (mpMRI) | ✓ | ✓ | ✓ | ✓ | Accurate and automatic discrimination of low-grade and high-grade prostate cancer in the central gland | The number of study patients was relatively small and highly unbalanced | ||||||
| [102] | Deep learning model to simplify PCa image registration in order to map regions of interest | ✓ | ✓ | ✓ | ✓ | Image alignment in developing radiomic and deep learning approaches for early detection of PCa | Segmentation on MRI, histopathology images and gross rotation were not captured | ||||||
| [98] | An interpretable PCa ensemble deep learning model to enhance decision making for clinicians | ✓ | ✓ | ✓ | ✓ | Stacking-based tree ensemble method used | Relatively small sample size was used | ||||||
| [103] | Ensemble feature extraction methods for PCa aggressiveness and indolent detection | ✓ | ✓ | ✓ | ✓ | Radiology–pathology fusion-based algorithm for PCa detection from adolescence and aggressiveness | Training cohort was relatively small and it was taken from a single institution | ||||||
| [104] | Detection of PCa using 3D CAD in bpMR images | ✓ | ✓ | ✓ | ✓ | Demonstration of a deep learning-based 3D detection and diagnosis system for csPCa | Prostate scans were acquired using MRI scanners developed by the same vendor | ||||||
| [106] | PCa localization and classification with ML | ✓ | ✓ | ✓ | ✓ | Automatic classification of 3D PCa | There is a need to increase the dataset | ||||||
| [105] | Segmentation of MR images tested on DL methods | ✓ | ✓ | ✓ | ✓ | Automatic classification of PCa in MRI | 3D images are relatively small | ||||||
| [108] | Segmenting MRI of PCa using deep learning techniques | ✓ | ✓ | ✓ | ✓ | Established that ensemble DCNNs initialized with pre-trained weights substantially improve segmentation accuracy | Approach is time-consuming | ||||||
| [109] | Detection of PCa leveraging on the strength of multi-modality of MR images | ✓ | ✓ | ✓ | ✓ | Novel model that detects PCa with different modalities of MRI and still maintains its robustness | Dual-attention model in depth was not considered | ||||||
| [110] | GANs were investigated for detection of PCa with MRI | ✓ | ✓ | ✓ | ✓ | GAN models in an end-to-end pipeline for automated PCa detection on T2W MRI | T2-weighted scans were used in this study | ||||||
| [111] | Gleason grading for PCa detection with deep learning techniques | ✓ | ✓ | ✓ | ✓ | Classify PCa belonging to different grade groups | More datasets needed for higher accuracy and diagnostic accuracy also needs further improvement | ||||||
| [112] | HC for early diagnosis of PCa | ✓ | ✓ | ✓ | ✓ | Detection of PCa with unsupervised HC in mpMRI | Relatively small patients used and they do not include other quantitative parameters and clinical information | ||||||
| [107] | Ensemble method of mpMRI and PHI for diagnosis of early PCa | ✓ | ✓ | ✓ | ✓ | The presence of PCa is automatically identified | Only present the design of co-trained CNNs for fusing ADC and T2w images, and their performance is based on two image modalities | ||||||
| [113] | Ensemble method of mpMRI and PHI for diagnosis of early PCa | ✓ | ✓ | ✓ | ✓ | Combined PHI and mpMRI to obtain higher csPCa detection | Relatively small amount of data for training | ||||||
| [114] | An improved CAD MRI for significant PCa detection | ✓ | ✓ | ✓ | ✓ | An improved inter-reader agreement and diagnostic performance for PCa detection | Lack of reproducibility of prostate MRI interpretations | ||||||
| [115] | Compared deep learning models for classification of PCa with GG | ✓ | ✓ | ✓ | ✓ | combining strongly and weakly supervised models | Labeling of data consumes time | ||||||
Appendix C. IEEE Xplore Papers on Prostate Cancer Detection Using Machine Learning, Deep Learning or Artificial Intelligence Methods

| Ref. | Problem Addressed | Imaging Modality | Machine Learning Type | Data | Medic Verified | Discussion | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MRI | US | Others | Transfer | SL | UL | Primary | Secondary | Yes | No | Strengths | Weaknesses | ||
| [14] | Classification of MRI for diagnosis of PCa. | ✓ | ✓ | ✓ | ✓ | Model was trained steadily which results in high accuracy. | Only diffusion-weighted images were used. | ||||||
| [116] | Prediction of PCa using machine learning classifiers. | ✓ | ✓ | ✓ | ✓ | Improved LR for better prediction. | mpMRI was not considered. | ||||||
| [120] | PCa detection in CEUS images through deep learning framework. | ✓ | ✓ | ✓ | ✓ | Captured dynamic information through 3D convolution operations. | Availability of limited dataset. | ||||||
| [117] | Deep learning regression analysis for PCa detection and Gleason scoring. | ✓ | ✓ | ✓ | ✓ | Improvement of PCa grading and detection with soft-label ordinal regression. | Fixed sized box in the middle of the image was used for segmentation. | ||||||
| [118] | PCa detection with classical and deep learning models. | ✓ | ✓ | ✓ | ✓ | Feature extraction through hand-crafted and non-hand-crafted methods and comparison in performance. | Only LSTM with possible bit parity was used. | ||||||
| [122] | PCa detection with WSI using CNN. | ✓ | ✓ | ✓ | ✓ | Developed an excellent patch-scoring model. | Model was limited with heatmap. | ||||||
| [124] | An improved Gleason score and PCa detection with a better feature extraction technique. | ✓ | ✓ | ✓ | ✓ | Enhancing radiomics with deep entropy feature generation through pre-trained CNN. | Only one feature extraction technique was utilized. | ||||||
| [125] | csPCa detection using deep neural network. | ✓ | ✓ | ✓ | ✓ | The neural network was optimized with different loss functions, which resulted in high accuracy in detecting PCa. | 2D network was used in their work. | ||||||
| [123] | Epithelial cell detection and Gleason grading in histological images. | ✓ | ✓ | ✓ | ✓ | Developed a model with the ability to perform multi-task prediction. | Experiment was not based on patient-wise validation. | ||||||
| [119] | Detection of PCa lesions with transfer learning. | ✓ | ✓ | ✓ | ✓ | Compared three (3) CNN models and suggested the best model. | Limited dataset used for testing the model developed. | ||||||
| [127] | Early diagnosis of Pca using CNN-CAD. | ✓ | ✓ | ✓ | ✓ | PCa segmentation, feature extraction and classification were performed with an improved CNN-CAD. | Classification was found only on one b-value. | ||||||
| [126] | Prediction of PCa lesions and their aggressiveness through Gleason grading. | ✓ | ✓ | ✓ | ✓ | A multi-class CNN and Focal-Net was developed in order to predict PCa. | No inclusion of non-visible MRI lesions. | ||||||
| [128] | Detection of PCa with CNN. | ✓ | ✓ | ✓ | ✓ | Transferred learning with reduction in MRI size to reduce complexity gave high accuracy in PCa detection. | Minimal dataset to work with. | ||||||
| [129] | Classification of Pca lesions into high-grade and low-grade through evaluation of radiomics. | ✓ | ✓ | ✓ | ✓ | Established that radiomics has high tendency to distinguish between high-grade and low-grade Pca tumor. | Tendency to have some wrong cases in the ground truth data. | ||||||
| [130] | Pca MRI segmentation improvement. | ✓ | ✓ | ✓ | ✓ | Developed an improved 2D PCa segmentation network. | They only focused on MRI segmentation of PCa. | ||||||
| [121] | Improved TRUS for csPCa detection. | ✓ | ✓ | ✓ | ✓ | Combined acoustic radiation force impulse (ARFI) imaging and shear wave elasticity imaging (SWEI) to give an improved csPCa detection. | Limited number of patients were used during the experiment. | ||||||
Appendix D. PubMed Papers on Prostate Cancer Detection Using Machine Learning, Deep Learning or Artificial Intelligence Methods

| Ref. | Problem Addressed | Imaging Modality | Machine Learning Type | Data | Medic Verified | Discussion | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MRI | US | Others | Transfer | SL | UL | Primary | Secondary | Yes | No | Strengths | Weaknesses | ||
| [131] | Aggressiveness of PCa was predicted using ML and DL frameworks | ✓ | ✓ | ✓ | ✓ | Characterization of PCa according to their aggressiveness level | Sample size was relatively small and study was monocentric | ||||||
| [178] | Survival analysis of localized PCa | ✓ | ✓ | ✓ | ✓ | Large cohort of localized prostate cancer patients were used | Lack of independent external validation | ||||||
| [132] | Transfer learning approach with CNN framework for detecting PCa | ✓ | ✓ | ✓ | ✓ | Compared the performances of machine learning and deep learning in detecting PCa with multimodal feature extraction | Better results could be achieved with more datasets | ||||||
| [135] | Detection of csPCa with deep learning-based imaging prediction using PI-RADS scoring and clinical variables | ✓ | ✓ | ✓ | ✓ | Models built were validated on different external sites | Manual delineations of the prostate gland were used with possibility of inter-reader variability | ||||||
| [134] | PCa detection using UNet | ✓ | ✓ | ✓ | ✓ | DL-based AI approach can predict prostate cancer lesions | Only one highly experienced genitourinary radiologist was involved in annotation, and histopathology verification was based on targeted biopsies but not surgical specimens | ||||||
| [133] | UNet architecture for PCa detection with minimal dataset | ✓ | ✓ | ✓ | ✓ | Detection of csPCa with prior knowledge on DL-based zonal segmentation | All data came from one MRI vendor (Siemens) | ||||||
| [136] | Bi-modal deep learning model fusion of pathology–radiology data for PCa diagnostic classification | ✓ | ✓ | ✓ | ✓ | Complementary information from biopsy report and MRI used to improve prediction of PCa | Axial T2w MRI only was used in this study and MRI was labeled using pathology labels, which may include inaccurate histological findings | ||||||
| [137] | ANN was used to accurately predict PCa | ✓ | ✓ | ✓ | ✓ | Accurately predicted PCa on prostate biopsy | The sample size was limited | ||||||
References
- Litwin, M.S.; Tan, H.-J. The diagnosis and treatment of prostate cancer: A review. JAMA 2017, 317, 2532–2542. [Google Scholar] [CrossRef] [PubMed]
- Akinnuwesi, B.A.; Olayanju, K.A.; Aribisala, B.S.; Fashoto, S.G.; Mbunge, E.; Okpeku, M.; Owate, P. Application of support vector machine algorithm for early differential diagnosis of prostate cancer. Data Sci. Manag. 2023, 6, 1–12. [Google Scholar] [CrossRef]
- Ayenigbara, I.O. Risk-Reducing Measures for Cancer Prevention. Korean J. Fam. Med. 2023, 44, 76. [Google Scholar] [CrossRef] [PubMed]
- Musekiwa, A.; Moyo, M.; Mohammed, M.; Matsena-Zingoni, Z.; Twabi, H.S.; Batidzirai, J.M.; Singini, G.C.; Kgarosi, K.; Mchunu, N.; Nevhungoni, P. Mapping evidence on the burden of breast, cervical, and prostate cancers in Sub-Saharan Africa: A scoping review. Front. Public Health 2022, 10, 908302. [Google Scholar] [CrossRef]
- Walsh, P.C.; Worthington, J.F. Dr. Patrick Walsh’s Guide to Surviving Prostate Cancer; Grand Central Life & Style: New York, NY, USA, 2010. [Google Scholar]
- Hayes, R.; Pottern, L.; Strickler, H.; Rabkin, C.; Pope, V.; Swanson, G.; Greenberg, R.; Schoenberg, J.; Liff, J.; Schwartz, A. Sexual behaviour, STDs and risks for prostate cancer. Br. J. Cancer 2000, 82, 718–725. [Google Scholar] [CrossRef]
- Plym, A.; Zhang, Y.; Stopsack, K.H.; Delcoigne, B.; Wiklund, F.; Haiman, C.; Kenfield, S.A.; Kibel, A.S.; Giovannucci, E.; Penney, K.L. A healthy lifestyle in men at increased genetic risk for prostate cancer. Eur. Urol. 2023, 83, 343–351. [Google Scholar] [CrossRef]
- Alkadi, R.; Taher, F.; El-Baz, A.; Werghi, N. A deep learning-based approach for the detection and localization of prostate cancer in T2 magnetic resonance images. J. Digit. Imaging 2019, 32, 793–807. [Google Scholar] [CrossRef]
- Ishioka, J.; Matsuoka, Y.; Uehara, S.; Yasuda, Y.; Kijima, T.; Yoshida, S.; Yokoyama, M.; Saito, K.; Kihara, K.; Numao, N. Computer-aided diagnosis of prostate cancer on magnetic resonance imaging using a convolutional neural network algorithm. BJU Int. 2018, 122, 411–417. [Google Scholar] [CrossRef]
- Reda, I.; Shalaby, A.; Abou El-Ghar, M.; Khalifa, F.; Elmogy, M.; Aboulfotouh, A.; Hosseini-Asl, E.; El-Baz, A.; Keynton, R. A new NMF-autoencoder based CAD system for early diagnosis of prostate cancer. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016. [Google Scholar]
- Wildeboer, R.R.; van Sloun, R.J.; Wijkstra, H.; Mischi, M. Artificial intelligence in multiparametric prostate cancer imaging with focus on deep-learning methods. Comput. Methods Programs Biomed. 2020, 189, 105316. [Google Scholar] [CrossRef]
- Aribisala, B.; Olabanjo, O. Medical image processor and repository–mipar. Inform. Med. Unlocked 2018, 12, 75–80. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.-I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Liu, Y.; An, X. A classification model for the prostate cancer based on deep learning. In Proceedings of the 2017 10th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Shanghai, China, 13–16 April 2016. [Google Scholar]
- Wang, X.; Yang, W.; Weinreb, J.; Han, J.; Li, Q.; Kong, X.; Yan, Y.; Ke, Z.; Luo, B.; Liu, T. Searching for prostate cancer by fully automated magnetic resonance imaging classification: Deep learning versus non-deep learning. Sci. Rep. 2017, 7, 15415. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Ibarrola, R.; Hein, S.; Reis, G.; Gratzke, C.; Miernik, A. Current and future applications of machine and deep learning in urology: A review of the literature on urolithiasis, renal cell carcinoma, and bladder and prostate cancer. World J. Urol. 2020, 38, 2329–2347. [Google Scholar] [CrossRef]
- Almeida, G.; Tavares, J.M.R. Deep learning in radiation oncology treatment planning for prostate cancer: A systematic review. J. Med. Syst. 2020, 44, 179. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Yahya, N.; Alsaih, K.; Al-Hiyali, M.I.; Meriaudeau, F. Recent automatic segmentation algorithms of MRI prostate regions: A review. IEEE Access 2021, 9, 97878–97905. [Google Scholar] [CrossRef]
- Roest, C.; Fransen, S.J.; Kwee, T.C.; Yakar, D. Comparative Performance of Deep Learning and Radiologists for the Diagnosis and Localization of Clinically Significant Prostate Cancer at MRI: A Systematic Review. Life 2022, 12, 1490. [Google Scholar] [CrossRef]
- Castillo, T.J.M.; Arif, M.; Niessen, W.J.; Schoots, I.G.; Veenland, J.F. Automated classification of significant prostate cancer on MRI: A systematic review on the performance of machine learning applications. Cancers 2020, 12, 1606. [Google Scholar] [CrossRef]
- Michaely, H.J.; Aringhieri, G.; Cioni, D.; Neri, E. Current value of biparametric prostate MRI with machine-learning or deep-learning in the detection, grading, and characterization of prostate cancer: A systematic review. Diagnostics 2022, 12, 799. [Google Scholar] [CrossRef]
- Naik, N.; Tokas, T.; Shetty, D.K.; Hameed, B.Z.; Shastri, S.; Shah, M.J.; Ibrahim, S.; Rai, B.P.; Chłosta, P.; Somani, B.K. Role of Deep Learning in Prostate Cancer Management: Past, Present and Future Based on a Comprehensive Literature Review. J. Clin. Med. 2022, 11, 3575. [Google Scholar] [CrossRef] [PubMed]
- Sarkis-Onofre, R.; Catalá-López, F.; Aromataris, E.; Lockwood, C. How to properly use the PRISMA Statement. Syst. Rev. 2021, 10, 117. [Google Scholar] [CrossRef]
- Hricak, H.; Choyke, P.L.; Eberhardt, S.C.; Leibel, S.A.; Scardino, P.T. Imaging prostate cancer: A multidisciplinary perspective. Radiology 2007, 243, 28–53. [Google Scholar] [CrossRef] [PubMed]
- Kyle, K.Y.; Hricak, H. Imaging prostate cancer. Radiol. Clin. North Am. 2000, 38, 59–85. [Google Scholar]
- Cornud, F.; Brolis, L.; Delongchamps, N.B.; Portalez, D.; Malavaud, B.; Renard-Penna, R.; Mozer, P. TRUS–MRI image registration: A paradigm shift in the diagnosis of significant prostate cancer. Abdom. Imaging 2013, 38, 1447–1463. [Google Scholar] [CrossRef]
- Reynier, C.; Troccaz, J.; Fourneret, P.; Dusserre, A.; Gay-Jeune, C.; Descotes, J.L.; Bolla, M.; Giraud, J.Y. MRI/TRUS data fusion for prostate brachytherapy. Preliminary results. Med. Phys. 2004, 31, 1568–1575. [Google Scholar] [CrossRef] [PubMed]
- Rasch, C.; Barillot, I.; Remeijer, P.; Touw, A.; van Herk, M.; Lebesque, J.V. Definition of the prostate in CT and MRI: A multi-observer study. Int. J. Radiat. Oncol. 1999, 43, 57–66. [Google Scholar] [CrossRef]
- Pezaro, C.; Woo, H.H.; Davis, I.D. Prostate cancer: Measuring PSA. Intern. Med. J. 2014, 44, 433–440. [Google Scholar] [CrossRef]
- Takahashi, N.; Inoue, T.; Lee, J.; Yamaguchi, T.; Shizukuishi, K. The roles of PET and PET/CT in the diagnosis and management of prostate cancer. Oncology 2008, 72, 226–233. [Google Scholar] [CrossRef]
- Sturge, J.; Caley, M.P.; Waxman, J. Bone metastasis in prostate cancer: Emerging therapeutic strategies. Nat. Rev. Clin. Oncol. 2011, 8, 357. [Google Scholar] [CrossRef]
- Raja, J.; Ramachandran, N.; Munneke, G.; Patel, U. Current status of transrectal ultrasound-guided prostate biopsy in the diagnosis of prostate cancer. Clin. Radiol. 2006, 61, 142–153. [Google Scholar] [CrossRef]
- Bai, H.; Xia, W.; Ji, X.; He, D.; Zhao, X.; Bao, J.; Zhou, J.; Wei, X.; Huang, Y.; Li, Q. Multiparametric magnetic resonance imaging-based peritumoral radiomics for preoperative prediction of the presence of extracapsular extension with prostate cancer. J. Magn. Reson. Imaging 2021, 54, 1222–1230. [Google Scholar] [CrossRef]
- Jansen, B.H.; Nieuwenhuijzen, J.A.; Oprea-Lager, D.E.; Yska, M.J.; Lont, A.P.; van Moorselaar, R.J.; Vis, A.N. Adding multiparametric MRI to the MSKCC and Partin nomograms for primary prostate cancer: Improving local tumor staging? In Urologic Oncology: Seminars and Original Investigations; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Maurer, T.; Eiber, M.; Schwaiger, M.; Gschwend, J.E. Current use of PSMA–PET in prostate cancer management. Nat. Rev. Urol. 2016, 13, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Stavrinides, V.; Papageorgiou, G.; Danks, D.; Giganti, F.; Pashayan, N.; Trock, B.; Freeman, A.; Hu, Y.; Whitaker, H.; Allen, C. Mapping PSA density to outcome of MRI-based active surveillance for prostate cancer through joint longitudinal-survival models. Prostate Cancer Prostatic Dis. 2021, 24, 1028–1031. [Google Scholar] [CrossRef] [PubMed]
- Fuchsjäger, M.; Shukla-Dave, A.; Akin, O.; Barentsz, J.; Hricak, H. Prostate cancer imaging. Acta Radiol. 2008, 49, 107–120. [Google Scholar] [CrossRef]
- Ghafoor, S.; Burger, I.A.; Vargas, A.H. Multimodality imaging of prostate cancer. J. Nucl. Med. 2019, 60, 1350–1358. [Google Scholar] [CrossRef] [PubMed]
- Rohrmann, S.; Roberts, W.W.; Walsh, P.C.; Platz, E.A. Family history of prostate cancer and obesity in relation to high-grade disease and extraprostatic extension in young men with prostate cancer. Prostate 2003, 55, 140–146. [Google Scholar] [CrossRef]
- Porter, M.P.; Stanford, J.L. Obesity and the risk of prostate cancer. Prostate 2005, 62, 316–321. [Google Scholar] [CrossRef]
- Gann, P.H. Risk factors for prostate cancer. Rev. Urol. 2002, 4 (Suppl. 5), S3. [Google Scholar]
- Tian, W.; Osawa, M. Prevalent latent adenocarcinoma of the prostate in forensic autopsies. J. Clin. Pathol. Forensic Med. 2015, 6, 11–13. [Google Scholar]
- Marley, A.R.; Nan, H. Epidemiology of colorectal cancer. Int. J. Mol. Epidemiol. Genet. 2016, 7, 105. [Google Scholar]
- Kumagai, H.; Zempo-Miyaki, A.; Yoshikawa, T.; Tsujimoto, T.; Tanaka, K.; Maeda, S. Lifestyle modification increases serum testosterone level and decrease central blood pressure in overweight and obese men. Endocr. J. 2015, 62, 423–430. [Google Scholar] [CrossRef]
- Moyad, M.A. Is obesity a risk factor for prostate cancer, and does it even matter? A hypothesis and different perspective. Urology 2002, 59, 41–50. [Google Scholar] [CrossRef]
- Parikesit, D.; Mochtar, C.A.; Umbas, R.; Hamid, A.R.A.H. The impact of obesity towards prostate diseases. Prostate Int. 2016, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Tse, L.A.; Lee, P.M.Y.; Ho, W.M.; Lam, A.T.; Lee, M.K.; Ng, S.S.M.; He, Y.; Leung, K.-S.; Hartle, J.C.; Hu, H. Bisphenol A and other environmental risk factors for prostate cancer in Hong Kong. Environ. Int. 2017, 107, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, V.; Naidu, V.; Kao, C.H.-J.; Karunasinghe, N.; Bishop, K.S.; Wang, A.; Pallati, R.; Shepherd, P.; Masters, J.; Zhu, S. Environmental factors and risk of aggressive prostate cancer among a population of New Zealand men–a genotypic approach. Mol. BioSystems 2017, 13, 681–698. [Google Scholar] [CrossRef]
- Minaee, S.; Boykov, Y.; Porikli, F.; Plaza, A.; Kehtarnavaz, N.; Terzopoulos, D. Image segmentation using deep learning: A survey. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 3523–3542. [Google Scholar] [CrossRef]
- Zhang, X.; Li, H.; Wang, C.; Cheng, W.; Zhu, Y.; Li, D.; Jing, H.; Li, S.; Hou, J.; Li, J. Evaluating the accuracy of breast cancer and molecular subtype diagnosis by ultrasound image deep learning model. Front. Oncol. 2021, 11, 623506. [Google Scholar] [CrossRef]
- Tammina, S. Transfer learning using vgg-16 with deep convolutional neural network for classifying images. Int. J. Sci. Res. Publ. (IJSRP) 2019, 9, 143–150. [Google Scholar] [CrossRef]
- Abbas, A.; Abdelsamea, M.M.; Gaber, M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell. 2021, 51, 854–864. [Google Scholar] [CrossRef]
- Christlein, V.; Spranger, L.; Seuret, M.; Nicolaou, A.; Král, P.; Maier, A. Deep generalized max pooling. In Proceedings of the 2019 International Conference on Document Analysis and Recognition (ICDAR), Sydney, NSW, Australia, 20–25 September 2019. [Google Scholar]
- Sharma, S.; Sharma, S.; Athaiya, A. Activation functions in neural networks. Towards Data Sci. 2017, 6, 310–316. [Google Scholar] [CrossRef]
- Sibi, P.; Jones, S.A.; Siddarth, P. Analysis of different activation functions using back propagation neural networks. J. Theor. Appl. Inf. Technol. 2013, 47, 1264–1268. [Google Scholar]
- Fu, J.; Zheng, H.; Mei, T. Look closer to see better: Recurrent attention convolutional neural network for fine-grained image recognition. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Yin, W.; Schütze, H.; Xiang, B.; Zhou, B. Abcnn: Attention-based convolutional neural network for modeling sentence pairs. Trans. Assoc. Comput. Linguist. 2016, 4, 259–272. [Google Scholar] [CrossRef]
- Ranftl, R.; Bochkovskiy, A.; Koltun, V. Vision transformers for dense prediction. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021. [Google Scholar]
- Fan, H.; Xiong, B.; Mangalam, K.; Li, Y.; Yan, Z.; Malik, J.; Feichtenhofer, C. Multiscale vision transformers. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021. [Google Scholar]
- Khan, S.; Naseer, M.; Hayat, M.; Zamir, S.W.; Khan, F.S.; Shah, M. Transformers in vision: A survey. ACM Comput. Surv. (CSUR) 2022, 54, 1–41. [Google Scholar] [CrossRef]
- Ikromjanov, K.; Bhattacharjee, S.; Hwang, Y.-B.; Sumon, R.I.; Kim, H.-C.; Choi, H.-K. Whole slide image analysis and detection of prostate cancer using vision transformers. In Proceedings of the 2022 International Conference on Artificial Intelligence in Information and Communication (ICAIIC), Jeju Island, Republic of Korea, 21–24 February 2022. [Google Scholar]
- Singla, D.; Cimen, F.; Narasimhulu, C.A. Novel artificial intelligent transformer U-NET for better identification and management of prostate cancer. Mol. Cell. Biochem. 2023, 478, 1439–1445. [Google Scholar] [CrossRef]
- Pachetti, E.; Colantonio, S. 3D-Vision-Transformer Stacking Ensemble for Assessing Prostate Cancer Aggressiveness from T2w Images. Bioengineering 2023, 10, 1015. [Google Scholar] [CrossRef]
- Pachetti, E.; Colantonio, S.; Pascali, M.A. On the effectiveness of 3D vision transformers for the prediction of prostate cancer aggressiveness. In Image Analysis and Processing; Springer: Cham, Switzerland, 2022. [Google Scholar]
- Li, C.; Deng, M.; Zhong, X.; Ren, J.; Chen, X.; Chen, J.; Xiao, F.; Xu, H. Multi-view radiomics and deep learning modeling for prostate cancer detection based on multi-parametric MRI. Front. Oncol. 2023, 13, 1198899. [Google Scholar] [CrossRef] [PubMed]
- Papp, L.; Spielvogel, C.; Grubmüller, B.; Grahovac, M.; Krajnc, D.; Ecsedi, B.; Sareshgi, R.A.; Mohamad, D.; Hamboeck, M.; Rausch, I. Supervised machine learning enables non-invasive lesion characterization in primary prostate cancer with [68 Ga] Ga-PSMA-11 PET/MRI. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 1795–1805. [Google Scholar] [CrossRef]
- Yoo, S.; Gujrathi, I.; Haider, M.A.; Khalvati, F. Prostate cancer detection using deep convolutional neural networks. Sci. Rep. 2019, 9, 19518. [Google Scholar] [CrossRef]
- Perera, M.; Mirchandani, R.; Papa, N.; Breemer, G.; Effeindzourou, A.; Smith, L.; Swindle, P.; Smith, E. PSA-based machine learning model improves prostate cancer risk stratification in a screening population. World J. Urol. 2021, 39, 1897–1902. [Google Scholar] [CrossRef] [PubMed]
- Otálora, S.; Marini, N.; Müller, H.; Atzori, M. Semi-weakly supervised learning for prostate cancer image classification with teacher-student deep convolutional networks. In Interpretable and Annotation-Efficient Learning for Medical Image Computing, Proceedings of the Third International Workshop, iMIMIC 2020, Second International Workshop, MIL3ID 2020, and 5th International Workshop, LABELS 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4–8 October 2020; Proceedings 3; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Swiderska-Chadaj, Z.; de Bel, T.; Blanchet, L.; Baidoshvili, A.; Vossen, D.; van der Laak, J.; Litjens, G. Impact of rescanning and normalization on convolutional neural network performance in multi-center, whole-slide classification of prostate cancer. Sci. Rep. 2020, 10, 14398. [Google Scholar] [CrossRef]
- Nagpal, K.; Foote, D.; Liu, Y.; Chen, P.-H.C.; Wulczyn, E.; Tan, F.; Olson, N.; Smith, J.L.; Mohtashamian, A.; Wren, J.H. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit. Med. 2019, 2, 48. [Google Scholar] [CrossRef]
- Tolkach, Y.; Dohmgörgen, T.; Toma, M.; Kristiansen, G. High-accuracy prostate cancer pathology using deep learning. Nat. Mach. Intell. 2020, 2, 411–418. [Google Scholar] [CrossRef]
- Singhal, N.; Soni, S.; Bonthu, S.; Chattopadhyay, N.; Samanta, P.; Joshi, U.; Jojera, A.; Chharchhodawala, T.; Agarwal, A.; Desai, M. A deep learning system for prostate cancer diagnosis and grading in whole slide images of core needle biopsies. Sci. Rep. 2022, 12, 3383. [Google Scholar] [CrossRef] [PubMed]
- Del Rio, M.; Lianas, L.; Aspegren, O.; Busonera, G.; Versaci, F.; Zelic, R.; Vincent, P.H.; Leo, S.; Pettersson, A.; Akre, O. AI support for accelerating histopathological slide examinations of prostate cancer in clinical studies. In Image Analysis and Processing. ICIAP 2022 Workshops, Proceedings of the ICIAP International Workshops, Lecce, Italy, 23–27 May 2022; Revised Selected Papers, Part I; Springer: Berlin/Heidelberg, Germany, 2022. [Google Scholar]
- Zhao, L.; Bao, J.; Qiao, X.; Jin, P.; Ji, Y.; Li, Z.; Zhang, J.; Su, Y.; Ji, L.; Shen, J. Predicting clinically significant prostate cancer with a deep learning approach: A multicentre retrospective study. Eur. J. Nucl. Med. Mol. Imaging 2023, 50, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Bao, M.-L.; Wu, C.-J.; Zhang, J.; Zhang, Y.-D.; Shi, H.-B. A radiomics machine learning-based redefining score robustly identifies clinically significant prostate cancer in equivocal PI-RADS score 3 lesions. Abdom. Radiol. 2020, 45, 4223–4234. [Google Scholar] [CrossRef] [PubMed]
- Sedghi, A.; Mehrtash, A.; Jamzad, A.; Amalou, A.; Wells, W.M.; Kapur, T.; Kwak, J.T.; Turkbey, B.; Choyke, P.; Pinto, P. Improving detection of prostate cancer foci via information fusion of MRI and temporal enhanced ultrasound. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1215–1223. [Google Scholar] [CrossRef]
- Deniffel, D.; Abraham, N.; Namdar, K.; Dong, X.; Salinas, E.; Milot, L.; Khalvati, F.; Haider, M.A. Using decision curve analysis to benchmark performance of a magnetic resonance imaging–based deep learning model for prostate cancer risk assessment. Eur. Radiol. 2020, 30, 6867–6876. [Google Scholar] [CrossRef]
- Bleker, J.; Kwee, T.C.; Rouw, D.; Roest, C.; Borstlap, J.; de Jong, I.J.; Dierckx, R.A.; Huisman, H.; Yakar, D. A deep learning masked segmentation alternative to manual segmentation in biparametric MRI prostate cancer radiomics. Eur. Radiol. 2022, 32, 6526–6535. [Google Scholar] [CrossRef]
- Yu, X.; Lou, B.; Zhang, D.; Winkel, D.; Arrahmane, N.; Diallo, M.; Meng, T.; von Busch, H.; Grimm, R.; Kiefer, B. Deep attentive panoptic model for prostate cancer detection using biparametric MRI scans. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2020, Proceedings of the 23rd International Conference, Lima, Peru, 4–8 October 2020; Proceedings, Part IV 23; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Wildeboer, R.R.; Mannaerts, C.K.; van Sloun, R.J.; Budäus, L.; Tilki, D.; Wijkstra, H.; Salomon, G.; Mischi, M. Automated multiparametric localization of prostate cancer based on B-mode, shear-wave elastography, and contrast-enhanced ultrasound radiomics. Eur. Radiol. 2020, 30, 806–815. [Google Scholar] [CrossRef]
- Azizi, S.; Imani, F.; Zhuang, B.; Tahmasebi, A.; Kwak, J.T.; Xu, S.; Uniyal, N.; Turkbey, B.; Choyke, P.; Pinto, P. Ultrasound-based detection of prostate cancer using automatic feature selection with deep belief networks. In Medical Image Computing and Computer-Assisted Intervention--MICCAI 2015, Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part II 18; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Khan, U.A.H.; Stürenberg, C.; Gencoglu, O.; Sandeman, K.; Heikkinen, T.; Rannikko, A.; Mirtti, T. Improving prostate cancer detection with breast histopathology images. In Digital Pathology, Proceedings of the 15th European Congress, Proceedings of the ECDP 2019, Warwick, UK, 10–13 April 2019; Proceedings 15; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Gour, M.; Jain, S.; Shankar, U. Application of Deep Learning Techniques for Prostate Cancer Grading Using Histopathological Images. In Computer Vision and Image Processing, Proceedings of the 6th International Conference, CVIP 2021, Rupnagar, India, 3–5 December 2021; Revised Selected Papers, Part I; Springer: Berlin/Heidelberg, Germany, 2022. [Google Scholar]
- Capobianco, N.; Sibille, L.; Chantadisai, M.; Gafita, A.; Langbein, T.; Platsch, G.; Solari, E.L.; Shah, V.; Spottiswoode, B.; Eiber, M. Whole-body uptake classification and prostate cancer staging in 68 Ga-PSMA-11 PET/CT using dual-tracer learning. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 517–526. [Google Scholar] [CrossRef]
- Korevaar, S.; Tennakoon, R.; Page, M.; Brotchie, P.; Thangarajah, J.; Florescu, C.; Sutherland, T.; Kam, N.M.; Bab-Hadiashar, A. Incidental detection of prostate cancer with computed tomography scans. Sci. Rep. 2021, 11, 7956. [Google Scholar] [CrossRef]
- Bhattacharya, I.; Lim, D.S.; Aung, H.L.; Liu, X.; Seetharaman, A.; Kunder, C.A.; Shao, W.; Soerensen, S.J.; Fan, R.E.; Ghanouni, P. Bridging the gap between prostate radiology and pathology through machine learning. Med. Phys. 2022, 49, 5160–5181. [Google Scholar] [CrossRef]
- Hassan, M.R.; Islam, M.F.; Uddin, M.Z.; Ghoshal, G.; Hassan, M.M.; Huda, S.; Fortino, G. Prostate cancer classification from ultrasound and MRI images using deep learning based Explainable Artificial Intelligence. Future Gener. Comput. Syst. 2022, 127, 462–472. [Google Scholar] [CrossRef]
- Liu, B.; Cheng, J.; Guo, D.; He, X.; Luo, Y.; Zeng, Y.; Li, C. Prediction of prostate cancer aggressiveness with a combination of radiomics and machine learning-based analysis of dynamic contrast-enhanced MRI. Clin. Radiol. 2019, 74, 896.e1–896.e8. [Google Scholar] [CrossRef]
- Stanzione, A.; Cuocolo, R.; Cocozza, S.; Romeo, V.; Persico, F.; Fusco, F.; Longo, N.; Brunetti, A.; Imbriaco, M. Detection of extraprostatic extension of cancer on biparametric MRI combining texture analysis and machine learning: Preliminary results. Acad. Radiol. 2019, 26, 1338–1344. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, L.; Tang, M.; Huan, Y.; Zhang, X.; Zhe, X. A new approach to diagnosing prostate cancer through magnetic resonance imaging. Alex. Eng. J. 2021, 60, 897–904. [Google Scholar] [CrossRef]
- Salvi, M.; Bosco, M.; Molinaro, L.; Gambella, A.; Papotti, M.; Acharya, U.R.; Molinari, F. A hybrid deep learning approach for gland segmentation in prostate histopathological images. Artif. Intell. Med. 2021, 115, 102076. [Google Scholar] [CrossRef]
- Salman, M.E.; Çakar, G.Ç.; Azimjonov, J.; Kösem, M.; Cedimoğlu, İ.H. Automated prostate cancer grading and diagnosis system using deep learning-based Yolo object detection algorithm. Expert Syst. Appl. 2022, 201, 117148. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, C.; Huang, J.; Liu, S.; Zhuo, Y.; Lu, X. Deep learning framework based on integration of S-Mask R-CNN and Inception-v3 for ultrasound image-aided diagnosis of prostate cancer. Future Gener. Comput. Syst. 2021, 114, 358–367. [Google Scholar] [CrossRef]
- Stojadinovic, M.; Milicevic, B.; Jankovic, S. Improved predictive performance of prostate biopsy collaborative group risk calculator when based on automated machine learning. Comput. Biol. Med. 2021, 138, 104903. [Google Scholar] [CrossRef]
- ElKarami, B.; Deebajah, M.; Polk, S.; Peabody, J.; Shahrrava, B.; Menon, M.; Alkhateeb, A.; Alanee, S. Machine learning-based prediction of upgrading on magnetic resonance imaging targeted biopsy in patients eligible for active surveillance. In Urologic Oncology: Seminars and Original Investigations; Elsevier: Amsterdam, The Netherlands, 2022. [Google Scholar]
- Lu, X.; Zhang, S.; Liu, Z.; Liu, S.; Huang, J.; Kong, G.; Li, M.; Liang, Y.; Cui, Y.; Yang, C. Ultrasonographic pathological grading of prostate cancer using automatic region-based Gleason grading network. Comput. Med. Imaging Graph. 2022, 102, 102125. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, D.; Geng, N.; Wang, Y.; Yin, Y.; Jin, Y. Stacking-based ensemble learning of decision trees for interpretable prostate cancer detection. Appl. Soft Comput. 2019, 77, 188–204. [Google Scholar] [CrossRef]
- Hambarde, P.; Talbar, S.; Mahajan, A.; Chavan, S.; Thakur, M.; Sable, N. Prostate lesion segmentation in MR images using radiomics based deeply supervised U-Net. Biocybern. Biomed. Eng. 2020, 40, 1421–1435. [Google Scholar] [CrossRef]
- Isaksson, L.J.; Repetto, M.; Summers, P.E.; Pepa, M.; Zaffaroni, M.; Vincini, M.G.; Corrao, G.; Mazzola, G.C.; Rotondi, M.; Bellerba, F. High-performance prediction models for prostate cancer radiomics. Inform. Med. Unlocked 2023, 37, 101161. [Google Scholar] [CrossRef]
- Li, J.; Weng, Z.; Xu, H.; Zhang, Z.; Miao, H.; Chen, W.; Liu, Z.; Zhang, X.; Wang, M.; Xu, X. Support Vector Machines (SVM) classification of prostate cancer Gleason score in central gland using multiparametric magnetic resonance images: A cross-validated study. Eur. J. Radiol. 2018, 98, 61–67. [Google Scholar] [CrossRef]
- Shao, W.; Banh, L.; Kunder, C.A.; Fan, R.E.; Soerensen, S.J.; Wang, J.B.; Teslovich, N.C.; Madhuripan, N.; Jawahar, A.; Ghanouni, P. ProsRegNet: A deep learning framework for registration of MRI and histopathology images of the prostate. Med. Image Anal. 2021, 68, 101919. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, I.; Seetharaman, A.; Kunder, C.; Shao, W.; Chen, L.C.; Soerensen, S.J.; Wang, J.B.; Teslovich, N.C.; Fan, R.E.; Ghanouni, P. Selective identification and localization of indolent and aggressive prostate cancers via CorrSigNIA: An MRI-pathology correlation and deep learning framework. Med. Image Anal. 2022, 75, 102288. [Google Scholar] [CrossRef]
- Saha, A.; Hosseinzadeh, M.; Huisman, H. End-to-end prostate cancer detection in bpMRI via 3D CNNs: Effects of attention mechanisms, clinical priori and decoupled false positive reduction. Med. Image Anal. 2021, 73, 102155. [Google Scholar] [CrossRef]
- Chen, J.; Wan, Z.; Zhang, J.; Li, W.; Chen, Y.; Li, Y.; Duan, Y. Medical image segmentation and reconstruction of prostate tumor based on 3D AlexNet. Comput. Methods Programs Biomed. 2021, 200, 105878. [Google Scholar] [CrossRef] [PubMed]
- Trigui, R.; Mitéran, J.; Walker, P.M.; Sellami, L.; Hamida, A.B. Automatic classification and localization of prostate cancer using multi-parametric MRI/MRS. Biomed. Signal Process. Control 2017, 31, 189–198. [Google Scholar] [CrossRef]
- Yang, X.; Liu, C.; Wang, Z.; Yang, J.; Le Min, H.; Wang, L.; Cheng, K.-T.T. Co-trained convolutional neural networks for automated detection of prostate cancer in multi-parametric MRI. Med. Image Anal. 2017, 42, 212–227. [Google Scholar] [CrossRef]
- Jia, H.; Xia, Y.; Song, Y.; Cai, W.; Fulham, M.; Feng, D.D. Atlas registration and ensemble deep convolutional neural network-based prostate segmentation using magnetic resonance imaging. Neurocomputing 2018, 275, 1358–1369. [Google Scholar] [CrossRef]
- Li, B.; Oka, R.; Xuan, P.; Yoshimura, Y.; Nakaguchi, T. Robust multi-modal prostate cancer classification via feature autoencoder and dual attention. Inform. Med. Unlocked 2022, 30, 100923. [Google Scholar] [CrossRef]
- Patsanis, A.; Sunoqrot, M.R.; Langørgen, S.; Wang, H.; Selnæs, K.M.; Bertilsson, H.; Bathen, T.F.; Elschot, M. A comparison of Generative Adversarial Networks for automated prostate cancer detection on T2-weighted MRI. Inform. Med. Unlocked 2023, 39, 101234. [Google Scholar] [CrossRef]
- Abraham, B.; Nair, M.S. Automated grading of prostate cancer using convolutional neural network and ordinal class classifier. Inform. Med. Unlocked 2019, 17, 100256. [Google Scholar] [CrossRef]
- Akamine, Y.; Ueda, Y.; Ueno, Y.; Sofue, K.; Murakami, T.; Yoneyama, M.; Obara, M.; Van Cauteren, M. Application of hierarchical clustering to multi-parametric MR in prostate: Differentiation of tumor and normal tissue with high accuracy. Magn. Reson. Imaging 2020, 74, 90–95. [Google Scholar] [CrossRef]
- Gentile, F.; La Civita, E.; Della Ventura, B.; Ferro, M.; Cennamo, M.; Bruzzese, D.; Crocetto, F.; Velotta, R.; Terracciano, D. A combinatorial neural network analysis reveals a synergistic behaviour of multiparametric magnetic resonance and prostate health index in the identification of clinically significant prostate cancer. Clin. Genitourin. Cancer 2022, 20, e406–e410. [Google Scholar] [CrossRef]
- Anderson, M.A.; Mercaldo, S.; Chung, R.; Ulrich, E.; Jones, R.W.; Harisinghani, M. Improving Prostate Cancer Detection With MRI: A Multi-Reader, Multi-Case Study Using Computer-Aided Detection (CAD). Acad. Radiol. 2022, 30, 1340–1349. [Google Scholar] [CrossRef]
- Otálora, S.; Atzori, M.; Khan, A.; Jimenez-del-Toro, O.; Andrearczyk, V.; Müller, H. Systematic comparison of deep learning strategies for weakly supervised Gleason grading. In Medical Imaging 2020: Digital Pathology; SPIE: Bellingham, WA, USA, 2020. [Google Scholar]
- Alam, M.; Tahernezhadi, M.; Vege, H.K.; Rajesh, P. A machine learning classification technique for predicting prostate cancer. In Proceedings of the 2020 IEEE International Conference on Electro Information Technology (EIT), Chicago, IL, USA, 31 July–1 August 2020. [Google Scholar]
- De Vente, C.; Vos, P.; Hosseinzadeh, M.; Pluim, J.; Veta, M. Deep learning regression for prostate cancer detection and grading in bi-parametric MRI. IEEE Trans. Biomed. Eng. 2020, 68, 374–383. [Google Scholar] [CrossRef]
- Iqbal, S.; Siddiqui, G.F.; Rehman, A.; Hussain, L.; Saba, T.; Tariq, U.; Abbasi, A.A. Prostate cancer detection using deep learning and traditional techniques. IEEE Access 2021, 9, 27085–27100. [Google Scholar] [CrossRef]
- Wiratchawa, K.; Wanna, Y.; Cha-in, S.; Aphinives, C.; Aphinives, P.; Intharah, T. Training Deep CNN’s to Detect Prostate Cancer Lesion with Small Training Data. In Proceedings of the 2022 37th International Technical Conference on Circuits/Systems, Computers and Communications (ITC-CSCC), Phuket, Thailand, 5–8 July 2022. [Google Scholar]
- Feng, Y.; Yang, F.; Zhou, X.; Guo, Y.; Tang, F.; Ren, F.; Guo, J.; Ji, S. A deep learning approach for targeted contrast-enhanced ultrasound based prostate cancer detection. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 16, 1794–1801. [Google Scholar] [CrossRef]
- Morris, D.C.; Chan, D.Y.; Chen, H.; Palmeri, M.L.; Polascik, T.J.; Foo, W.-C.; Huang, J.; Mamou, J.; Nightingale, K.R. Multiparametric Ultrasound for the Targeting of Prostate Cancer using ARFI, SWEI, B-mode, and QUS. In Proceedings of the 2019 IEEE International Ultrasonics Symposium (IUS), Glasgow, UK, 6–9 October 2019. [Google Scholar]
- Duran-Lopez, L.; Dominguez-Morales, J.P.; Conde-Martin, A.F.; Vicente-Diaz, S.; Linares-Barranco, A. PROMETEO: A CNN-based computer-aided diagnosis system for WSI prostate cancer detection. IEEE Access 2020, 8, 128613–128628. [Google Scholar] [CrossRef]
- Li, W.; Li, J.; Sarma, K.V.; Ho, K.C.; Shen, S.; Knudsen, B.S.; Gertych, A.; Arnold, C.W. Path R-CNN for prostate cancer diagnosis and gleason grading of histological images. IEEE Trans. Med. Imaging 2018, 38, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Kucharczyk, M.J.; Desrosiers, C.; Okuwobi, I.P.; Katib, Y.; Zhang, M.; Rathore, S.; Sargos, P.; Niazi, T. Deep radiomic analysis to predict gleason score in prostate cancer. IEEE Access 2020, 8, 167767–167778. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, C.; Cheng, D.; Wang, L.; Yang, X.; Cheng, K.-T. Automated detection of clinically significant prostate cancer in mp-MRI images based on an end-to-end deep neural network. IEEE Trans. Med. Imaging 2018, 37, 1127–1139. [Google Scholar] [CrossRef]
- Cao, R.; Bajgiran, A.M.; Mirak, S.A.; Shakeri, S.; Zhong, X.; Enzmann, D.; Raman, S.; Sung, K. Joint prostate cancer detection and Gleason score prediction in mp-MRI via FocalNet. IEEE Trans. Med. Imaging 2019, 38, 2496–2506. [Google Scholar] [CrossRef]
- Reda, I.; Ayinde, B.O.; Elmogy, M.; Shalaby, A.; El-Melegy, M.; Abou El-Ghar, M.; Abou El-fetouh, A.; Ghazal, M.; El-Baz, A. A new CNN-based system for early diagnosis of prostate cancer. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018. [Google Scholar]
- Mosleh, M.A.; Hamoud, M.H.; Alsabri, A.A. Detection of Prostate Cancer Using MRI Images Classification with Deep Learning Techniques. In Proceedings of the 2022 2nd International Conference on Emerging Smart Technologies and Applications (eSmarTA), Ibb, Yemen, 25–26 October 2022. [Google Scholar]
- Starmans, M.P.; Niessen, W.J.; Schoots, I.; Klein, S.; Veenland, J.F. Classification of prostate cancer: High grade versus low grade using a radiomics approach. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019. [Google Scholar]
- Hassanzadeh, T.; Hamey, L.G.; Ho-Shon, K. Convolutional neural networks for prostate magnetic resonance image segmentation. IEEE Access 2019, 7, 36748–36760. [Google Scholar] [CrossRef]
- Bertelli, E.; Mercatelli, L.; Marzi, C.; Pachetti, E.; Baccini, M.; Barucci, A.; Colantonio, S.; Gherardini, L.; Lattavo, L.; Pascali, M.A. Machine and deep learning prediction of prostate cancer aggressiveness using multiparametric mri. Front. Oncol. 2022, 11, 802964. [Google Scholar] [CrossRef]
- Abbasi, A.A.; Hussain, L.; Awan, I.A.; Abbasi, I.; Majid, A.; Nadeem, M.S.A.; Chaudhary, Q.-A. Detecting prostate cancer using deep learning convolution neural network with transfer learning approach. Cogn. Neurodynamics 2020, 14, 523–533. [Google Scholar] [CrossRef]
- Hosseinzadeh, M.; Saha, A.; Brand, P.; Slootweg, I.; de Rooij, M.; Huisman, H. Deep learning–assisted prostate cancer detection on bi-parametric MRI: Minimum training data size requirements and effect of prior knowledge. Eur. Radiol. 2021, 32, 2224–2234. [Google Scholar] [CrossRef]
- Mehralivand, S.; Yang, D.; Harmon, S.A.; Xu, D.; Xu, Z.; Roth, H.; Masoudi, S.; Kesani, D.; Lay, N.; Merino, M.J. Deep learning-based artificial intelligence for prostate cancer detection at biparametric MRI. Abdom. Radiol. 2022, 47, 1425–1434. [Google Scholar] [CrossRef]
- Hiremath, A.; Shiradkar, R.; Fu, P.; Mahran, A.; Rastinehad, A.R.; Tewari, A.; Tirumani, S.H.; Purysko, A.; Ponsky, L.; Madabhushi, A. An integrated nomogram combining deep learning, Prostate Imaging–Reporting and Data System (PI-RADS) scoring, and clinical variables for identification of clinically significant prostate cancer on biparametric MRI: A retrospective multicentre study. Lancet Digit. Health 2021, 3, e445–e454. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, P.; Lysandrou, M.; Eljalby, M.; Li, Q.; Kazemi, E.; Zisimopoulos, P.; Sigaras, A.; Brendel, M.; Barnes, J.; Ricketts, C. A deep learning approach to diagnostic classification of prostate cancer using pathology–radiology fusion. J. Magn. Reson. Imaging 2021, 54, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Hattori-Kato, M.; Okuno, Y.; Iwai, S.; Mikami, K. Prediction of prostate cancer by deep learning with multilayer artificial neural network. Can. Urol. Assoc. J. 2019, 13, E145. [Google Scholar] [CrossRef] [PubMed]
- Soni, M.; Khan, I.R.; Babu, K.S.; Nasrullah, S.; Madduri, A.; Rahin, S.A. Light weighted healthcare CNN model to detect prostate cancer on multiparametric MRI. Comput. Intell. Neurosci. 2022, 2022, 5497120. [Google Scholar] [CrossRef]
- Azizi, S.; Bayat, S.; Yan, P.; Tahmasebi, A.; Kwak, J.T.; Xu, S.; Turkbey, B.; Choyke, P.; Pinto, P.; Wood, B. Deep recurrent neural networks for prostate cancer detection: Analysis of temporal enhanced ultrasound. IEEE Trans. Med. Imaging 2018, 37, 2695–2703. [Google Scholar] [CrossRef]
- Laabidi, A.; Aissaoui, M. Performance analysis of Machine learning classifiers for predicting diabetes and prostate cancer. In Proceedings of the 2020 1st International Conference on Innovative Research in Applied Science, Engineering and Technology (IRASET), Meknes, Morocco, 16–19 April 2020. [Google Scholar]
- Murakami, Y.; Magome, T.; Matsumoto, K.; Sato, T.; Yoshioka, Y.; Oguchi, M. Fully automated dose prediction using generative adversarial networks in prostate cancer patients. PLoS ONE 2020, 15, e0232697. [Google Scholar] [CrossRef]
- Kohl, S.; Bonekamp, D.; Schlemmer, H.-P.; Yaqubi, K.; Hohenfellner, M.; Hadaschik, B.; Radtke, J.-P.; Maier-Hein, K. Adversarial networks for the detection of aggressive prostate cancer. arXiv 2017, arXiv:1702.08014. [Google Scholar]
- Yu, H.; Zhang, X. Synthesis of prostate MR images for classification using capsule network-based GAN Model. Sensors 2020, 20, 5736. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Hu, M.; Patel, P.; Mao, H.; Liu, T.; Yang, X. Prostate Gleason score prediction via MRI using capsule network. In Medical Imaging 2023: Computer-Aided Diagnosis; SPIE: Bellingham, WA, USA, 2023. [Google Scholar]
- Ding, M.; Lin, Z.; Lee, C.H.; Tan, C.H.; Huang, W. A multi-scale channel attention network for prostate segmentation. IEEE Trans. Circuits Syst. II Express Briefs 2023, 70, 1754–1758. [Google Scholar] [CrossRef]
- Xu, X.; Lian, C.; Wang, S.; Zhu, T.; Chen, R.C.; Wang, A.Z.; Royce, T.J.; Yap, P.-T.; Shen, D.; Lian, J. Asymmetric multi-task attention network for prostate bed segmentation in computed tomography images. Med. Image Anal. 2021, 72, 102116. [Google Scholar] [CrossRef]
- Yuan, Y.; Qin, W.; Buyyounouski, M.; Ibragimov, B.; Hancock, S.; Han, B.; Xing, L. Prostate cancer classification with multiparametric MRI transfer learning model. Med. Phys. 2019, 46, 756–765. [Google Scholar] [CrossRef]
- Janocha, K.; Czarnecki, W.M. On loss functions for deep neural networks in classification. arXiv 2017, arXiv:1702.05659. [Google Scholar] [CrossRef]
- Zhao, H.; Gallo, O.; Frosio, I.; Kautz, J. Loss functions for neural networks for image processing. arXiv 2015, arXiv:1511.08861. [Google Scholar]
- Ghosh, A.; Kumar, H.; Sastry, P.S. Robust loss functions under label noise for deep neural networks. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Xu, C.; Lu, C.; Liang, X.; Gao, J.; Zheng, W.; Wang, T.; Yan, S. Multi-loss regularized deep neural network. IEEE Trans. Circuits Syst. Video Technol. 2015, 26, 2273–2283. [Google Scholar] [CrossRef]
- Kim, T.; Oh, J.; Kim, N.; Cho, S.; Yun, S.-Y. Comparing kullback-leibler divergence and mean squared error loss in knowledge distillation. arXiv 2021, arXiv:2105.08919. [Google Scholar]
- Qi, J.; Du, J.; Siniscalchi, S.M.; Ma, X.; Lee, C.-H. On mean absolute error for deep neural network based vector-to-vector regression. IEEE Signal Process. Lett. 2020, 27, 1485–1489. [Google Scholar] [CrossRef]
- Ruby, U.; Yendapalli, V. Binary cross entropy with deep learning technique for image classification. Int. J. Adv. Trends Comput. Sci. Eng 2020, 9, 5393–5397. [Google Scholar]
- Ho, Y.; Wookey, S. The real-world-weight cross-entropy loss function: Modeling the costs of mislabeling. IEEE Access 2019, 8, 4806–4813. [Google Scholar] [CrossRef]
- Gordon-Rodriguez, E.; Loaiza-Ganem, G.; Pleiss, G.; Cunningham, J.P. Uses and abuses of the cross-entropy loss: Case studies in modern deep learning. arXiv 2020, arXiv:2011.05231. [Google Scholar]
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Jorge Cardoso, M. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Proceedings of the Third International Workshop, DLMIA 2017, and 7th International Workshop, ML-CDS 2017, Held in Conjunction with MICCAI 2017, Québec City, QC, Canada, 14 September 2017; Proceedings 3; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Zhang, Y.; Liu, S.; Li, C.; Wang, J. Rethinking the dice loss for deep learning lesion segmentation in medical images. J. Shanghai Jiaotong Univ. 2021, 26, 93–102. [Google Scholar] [CrossRef]
- Mukhoti, J.; Kulharia, V.; Sanyal, A.; Golodetz, S.; Torr, P.; Dokania, P. Calibrating deep neural networks using focal loss. Adv. Neural Inf. Process. Syst. 2020, 33, 15288–15299. [Google Scholar]
- Lin, T.-Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 42, 318–327. [Google Scholar] [CrossRef]
- Asperti, A.; Trentin, M. Balancing reconstruction error and kullback-leibler divergence in variational autoencoders. IEEE Access 2020, 8, 199440–199448. [Google Scholar] [CrossRef]
- Phan, H.; Mikkelsen, K.; Chén, O.Y.; Koch, P.; Mertins, A.; Kidmose, P.; De Vos, M. Personalized automatic sleep staging with single-night data: A pilot study with Kullback–Leibler divergence regularization. Physiol. Meas. 2020, 41, 064004. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. Review The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol./Współczesna Onkol. 2015, 2015, 68–77. [Google Scholar] [CrossRef]
- Wang, Z.; Jensen, M.A.; Zenklusen, J.C. A practical guide to the cancer genome atlas (TCGA). In Statistical Genomics; Humana Press: New York, NY, USA, 2016; Volume 1418, pp. 111–141. [Google Scholar]
- Hutter, C.; Zenklusen, J.C. The cancer genome atlas: Creating lasting value beyond its data. Cell 2018, 173, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Way, G.P.; Sanchez-Vega, F.; La, K.; Armenia, J.; Chatila, W.K.; Luna, A.; Sander, C.; Cherniack, A.D.; Mina, M.; Ciriello, G. Machine learning detects pan-cancer ras pathway activation in the cancer genome atlas. Cell Rep. 2018, 23, 172–180.e3. [Google Scholar] [CrossRef] [PubMed]
- Ganini, C.; Amelio, I.; Bertolo, R.; Bove, P.; Buonomo, O.C.; Candi, E.; Cipriani, C.; Di Daniele, N.; Juhl, H.; Mauriello, A. Global mapping of cancers: The Cancer Genome Atlas and beyond. Mol. Oncol. 2021, 15, 2823–2840. [Google Scholar] [CrossRef]
- Rosenkrantz, A.B.; Oto, A.; Turkbey, B.; Westphalen, A.C. Prostate Imaging Reporting and Data System (PI-RADS), version 2: A critical look. Am. J. Roentgenol. 2016, 206, 1179–1183. [Google Scholar] [CrossRef] [PubMed]
- Westphalen, A.C.; Rosenkrantz, A.B. Prostate imaging reporting and data system (PI-RADS): Reflections on early experience with a standardized interpretation scheme for multiparametric prostate MRI. Am. J. Roentgenol. 2014, 202, 121–123. [Google Scholar] [CrossRef]
- Deng, K.; Li, H.; Guan, Y. Treatment stratification of patients with metastatic castration-resistant prostate cancer by machine learning. Iscience 2020, 23, 100804. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, K.; Hugh-Jones, C.; Norman, T.; Friend, S.; Stolovitzky, G. The Prostate Cancer DREAM Challenge: A Community-Wide Effort to Use Open Clinical Trial Data for the Quantitative Prediction of Outcomes in Metastatic Prostate Cancer; Oxford University Press: Oxford, UK, 2015; Volume 20, pp. 459–460. [Google Scholar]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Prior, F.; Smith, K.; Sharma, A.; Kirby, J.; Tarbox, L.; Clark, K.; Bennett, W.; Nolan, T.; Freymann, J. The public cancer radiology imaging collections of The Cancer Imaging Archive. Sci. Data 2017, 4, 170124. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, G.; Mirak, S.A.; Hosseiny, M.; Azadikhah, A.; Zhong, X.; Reiter, R.E.; Lee, Y.; Raman, S.S.; Sung, K. Automatic prostate zonal segmentation using fully convolutional network with feature pyramid attention. IEEE Access 2019, 7, 163626–163632. [Google Scholar] [CrossRef]
- Mehrtash, A.; Sedghi, A.; Ghafoorian, M.; Taghipour, M.; Tempany, C.M.; Wells, W.M., III; Kapur, T.; Mousavi, P.; Abolmaesumi, P.; Fedorov, A. Classification of clinical significance of MRI prostate findings using 3D convolutional neural networks. In Medical Imaging 2017: Computer-Aided Diagnosis; SPIE: Bellingham, WA, USA, 2017. [Google Scholar]
- Hamm, C.A.; Baumgärtner, G.L.; Biessmann, F.; Beetz, N.L.; Hartenstein, A.; Savic, L.J.; Froböse, K.; Dräger, F.; Schallenberg, S.; Rudolph, M. Interactive Explainable Deep Learning Model Informs Prostate Cancer Diagnosis at MRI. Radiology 2023, 307, e222276. [Google Scholar] [CrossRef]
- Kraaijveld, R.C.; Philippens, M.E.; Eppinga, W.S.; Jürgenliemk-Schulz, I.M.; Gilhuijs, K.G.; Kroon, P.S.; van der Velden, B.H. Multi-modal volumetric concept activation to explain detection and classification of metastatic prostate cancer on PSMA-PET/CT. In International Workshop on Interpretability of Machine Intelligence in Medical Image Computing; Springer: Berlin/Heidelberg, Germany, 2022. [Google Scholar]
- Dai, X.; Park, J.H.; Yoo, S.; D’Imperio, N.; McMahon, B.H.; Rentsch, C.T.; Tate, J.P.; Justice, A.C. Survival analysis of localized prostate cancer with deep learning. Sci. Rep. 2022, 12, 17821. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).