Pioneering Arterial Hypertension Phenotyping on Nationally Aggregated Electronic Health Records
Abstract
1. Introduction
2. Results
| Diag-Only (%) | Diag+Med (%) | |||
|---|---|---|---|---|
| Cohort | Random Cohort (n = 1619) | AF Cohort (n = 1194) | Random Cohort (n = 1619) | AF Cohort (n = 1194) |
| Sensitivity | 68.2 | 66.5 | 83.8 | 87.6 |
| Specificity | 95.8 | 85.9 | 92.8 | 67.7 |
| PPV | 91.4 | 94.2 | 88.2 | 90.3 |
| NPV | 82.3 | 42.7 | 89.9 | 61.3 |
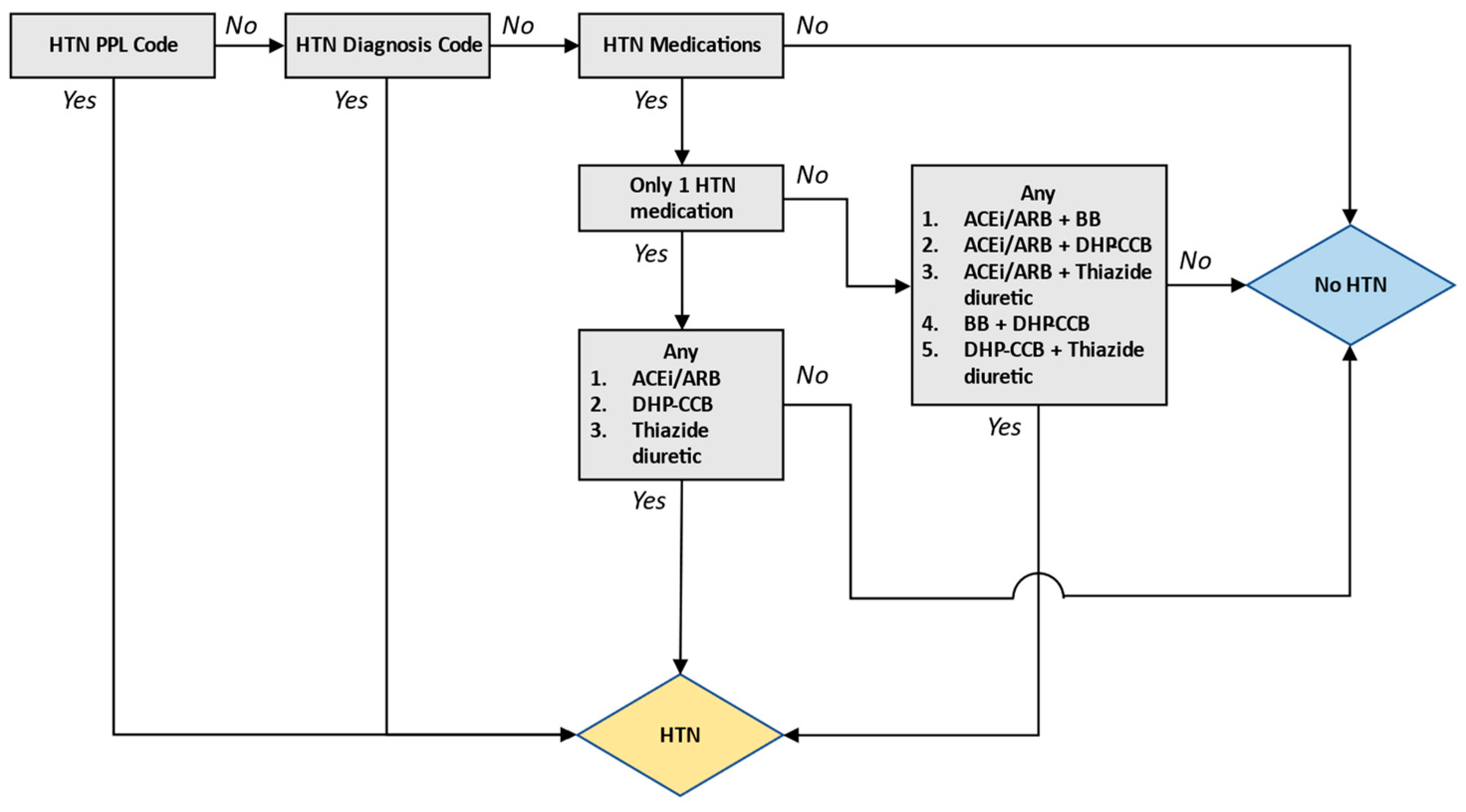
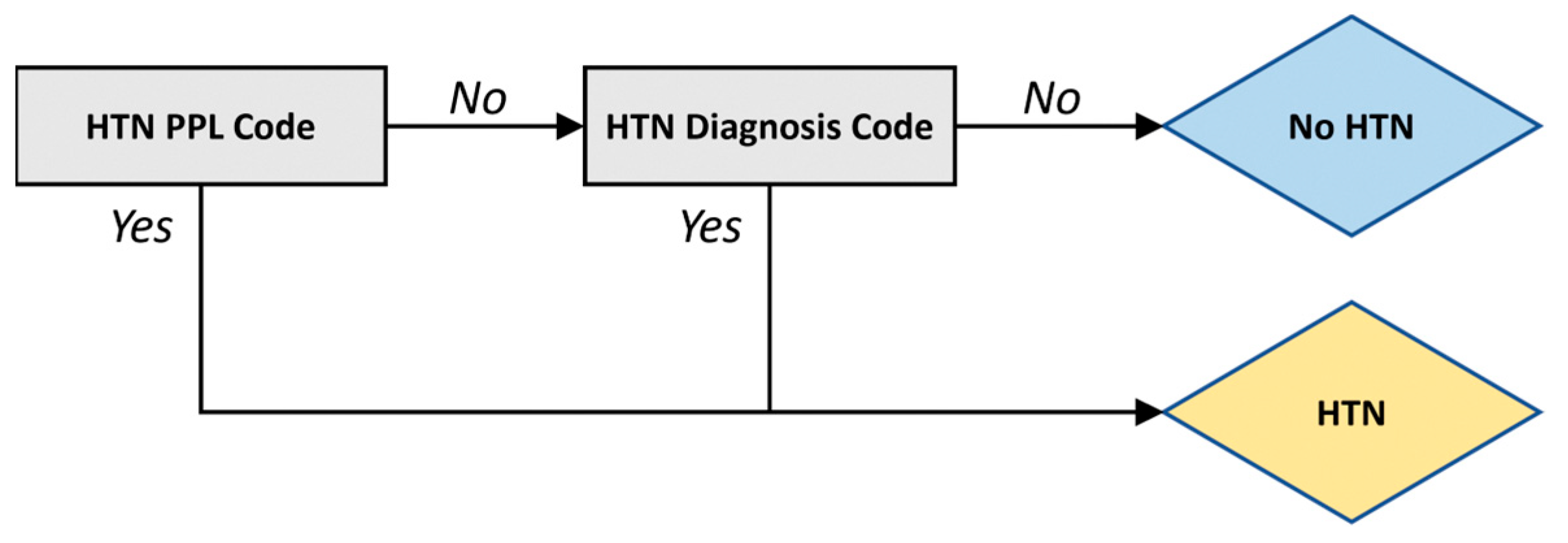
3. Discussion
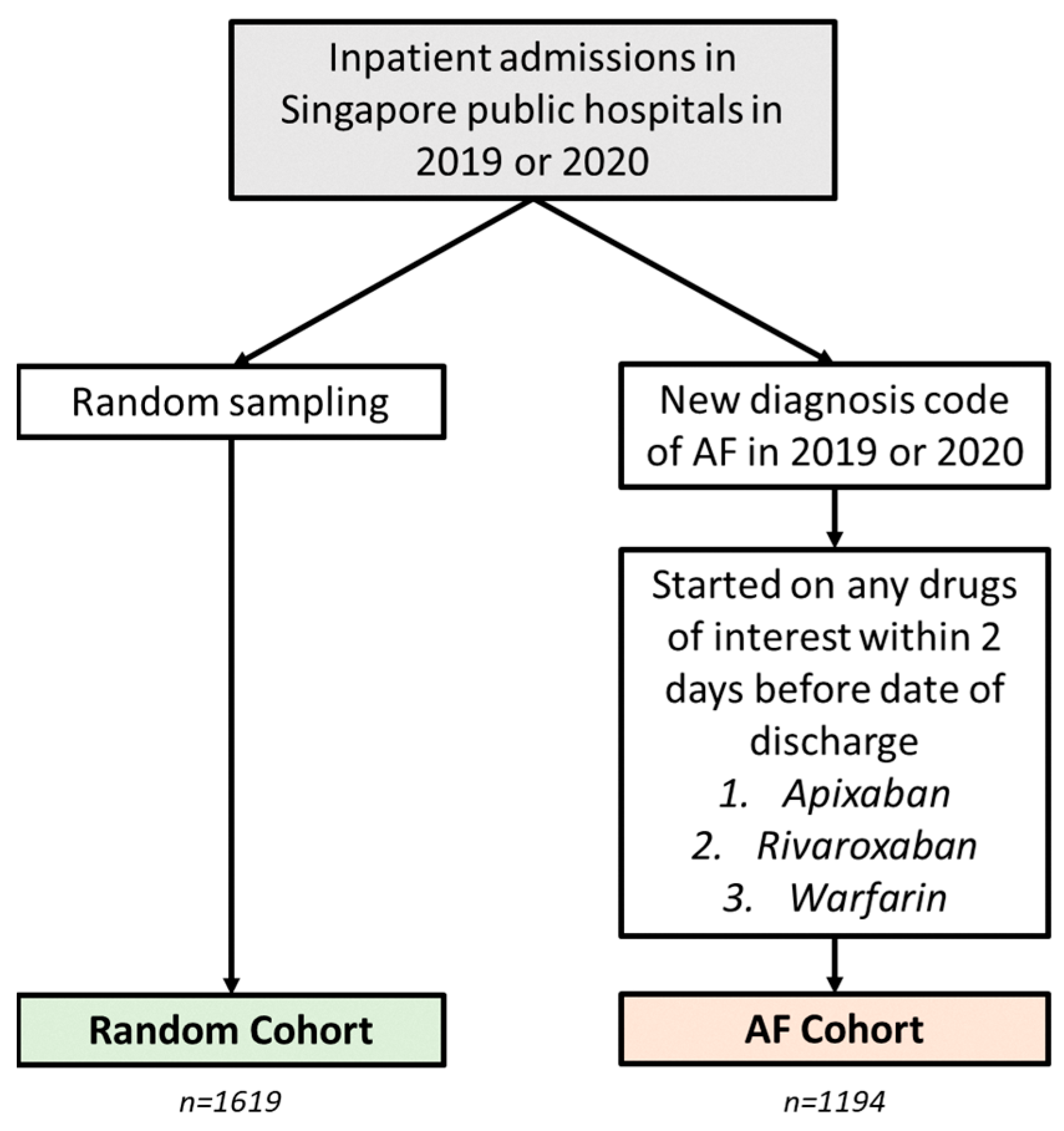
4. Materials and Methods
4.1. Data Sources
4.2. Algorithm Development and Validation
4.3. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Random Cohort (n = 1619) | AF Cohort (n = 1194) | ||||
|---|---|---|---|---|---|
| 2019 (n = 808) | 2020 (n = 811) | 2019 (n = 608) | 2020 (n = 586) | ||
| Hypertension | Yes | 335 (41.5%) | 301 (37.1%) | 461 (75.8%) | 464 (79.2%) |
| No | 473 (58.5%) | 510 (62.9%) | 147 (24.2%) | 122 (20.8%) | |
| Gender | Male | 380 (47.0%) | 401 (49.4%) | 305 (50.2%) | 310 (52.9%) |
| Female | 428 (53.0%) | 410 (50.6%) | 303 (49.8%) | 276 (47.1%) | |
| Race | Chinese | 514 (63.6%) | 489 (60.3%) | 451 (74.2%) | 458 (78.2%) |
| Malay | 139 (17.2%) | 137 (16.8%) | 92 (15.1%) | 81 (13.8%) | |
| Indian | 84 (10.4%) | 99 (12.3%) | 29 (4.8%) | 25 (4.3%) | |
| Others | 71 (8.8%) | 86 (10.6%) | 36 (5.9%) | 22 (3.8%) | |
| Age (Mean, SD) | Overall | 47.5 (28.8) | 45.8 (27.5) | 72.2 (11.8) | 72.4 (12.0) |
| Male | 47.5 (29.9) | 47.0 (28.4) | 69.1 (11.6) | 69.7 (12.0) | |
| Female | 47.5 (27.9) | 44.6 (26.4) | 75.3 (11.2) | 75.3 (11.4) | |
| Chinese | 52.8 (28.1) | 52.9 (27.8) | 73.8 (11.0) | 73.7 (10.9) | |
| Malay | 33.2 (26.7) | 31.6 (24.8) | 67.6 (15.6) | 69.6 (13.7) | |
| Indian | 45.8 (27.4) | 39.3 (22.9) | 69.0 (11.0) | 66.7 (13.6) | |
| Others | 39.1 (28.7) | 35.2 (19.5) | 63.9 (14.6) | 67.8 (18.3) | |
| Total | 808 (100.0%) | 811 (100.0%) | 608 (100.0%) | 586 (100.0%) | |

| Diag-Only (%) | Diag+Med (%) | ||||
|---|---|---|---|---|---|
| Random Cohort (n = 1619) | AF Cohort (n = 1194) | Random Cohort (n = 1619) | AF Cohort (n = 1194) | ||
| Sensitivity | Overall | 68.2 | 66.5 | 83.8 | 87.6 |
| 2019 | 65.1 | 63.1 | 82.4 | 85.9 | |
| 2020 | 71.8 | 69.8 | 85.4 | 89.2 | |
| Male | 67.1 | 68.3 | 82.4 | 87.7 | |
| Female | 69.7 | 64.6 | 85.5 | 87.4 | |
| Chinese | 68.7 | 68.1 | 84.3 | 88.6 | |
| Malay | 71.2 | 58.4 | 86.4 | 84.7 | |
| Indian | 76.5 | 68.6 | 88.2 | 88.6 | |
| Others | 45.2 | 63.4 | 66.7 | 78.0 | |
| Specificity | Overall | 95.8 | 85.9 | 92.8 | 67.7 |
| 2019 | 95.3 | 85.7 | 92.0 | 66.7 | |
| 2020 | 96.3 | 86.1 | 93.5 | 68.9 | |
| Male | 95.0 | 84.1 | 91.9 | 65.6 | |
| Female | 96.5 | 88.1 | 93.5 | 70.3 | |
| Chinese | 93.9 | 85.3 | 90.2 | 67.0 | |
| Malay | 98.1 | 86.1 | 95.7 | 75.0 | |
| Indian | 98.3 | 84.2 | 95.7 | 57.9 | |
| Others | 98.3 | 94.1 | 96.5 | 70.6 | |
| PPV | Overall | 91.4 | 94.2 | 88.2 | 90.3 |
| 2019 | 90.8 | 93.3 | 87.9 | 89.0 | |
| 2020 | 91.9 | 95.0 | 88.6 | 91.6 | |
| Male | 91.3 | 93.0 | 88.8 | 88.7 | |
| Female | 91.4 | 95.5 | 87.6 | 92.0 | |
| Chinese | 90.5 | 94.4 | 88.0 | 90.7 | |
| Malay | 92.2 | 94.1 | 86.4 | 92.8 | |
| Indian | 96.3 | 88.9 | 92.3 | 79.5 | |
| Others | 90.5 | 96.3 | 87.5 | 86.5 | |
| NPV | Overall | 82.3 | 42.7 | 89.9 | 61.3 |
| 2019 | 79.4 | 42.6 | 88.1 | 60.1 | |
| 2020 | 85.2 | 42.9 | 91.6 | 62.7 | |
| Male | 78.7 | 46.4 | 87.0 | 63.5 | |
| Female | 85.5 | 39.0 | 92.3 | 58.9 | |
| Chinese | 78.0 | 42.5 | 87.2 | 62.0 | |
| Malay | 91.6 | 35.2 | 95.7 | 56.2 | |
| Indian | 87.6 | 59.3 | 93.2 | 73.3 | |
| Others | 83.1 | 51.6 | 88.8 | 57.1 | |
| No. | Diagnosis Code | Diagnosis Description | Format |
|---|---|---|---|
| 1 | 38341003 | Hypertensive disorder | SNOMED |
| 2 | 59621000 | Essential hypertension | SNOMED |
| 3 | 10725009 | Benign hypertension | SNOMED |
| 4 | 38481006 | Hypertensive renal disease | SNOMED |
| 5 | 1201005 | Benign essential hypertension | SNOMED |
| 6 | 6962006 | Hypertensive retinopathy | SNOMED |
| 7 | 64715009 | Hypertensive heart disease | SNOMED |
| 8 | 56218007 | Systolic hypertension | SNOMED |
| 9 | 170578008 | Poor hypertension control | SNOMED |
| 10 | I10 | Essential (primary) hypertension | ICD-10 |
| 11 | 86041002 | Pre-existing hypertension in obstetric context | SNOMED |
| 12 | 86234004 | Hypertensive heart AND renal disease | SNOMED |
| 13 | 473392002 | Hypertensive nephrosclerosis | SNOMED |
| 14 | 266287006 | (Hypertensive disease) or (hypertension) | SNOMED |
| 15 | 8762007 | Chronic hypertension in obstetric context | SNOMED |
| 16 | 712832005 | Supine hypertension | SNOMED |
| 17 | 5148006 | Hypertensive heart disease with congestive heart failure | SNOMED |
| 18 | 65402008 | Pre-existing hypertension complicating AND/OR reason for care during pregnancy | SNOMED |
| 19 | 78975002 | Malignant essential hypertension | SNOMED |
| 20 | 194779001 | Hypertensive heart and renal disease with (congestive) heart failure | SNOMED |
| 21 | 46113002 | Hypertensive heart failure | SNOMED |
| 22 | 48146000 | Diastolic hypertension | SNOMED |
| 23 | 194767001 | Benign hypertensive heart disease with congestive cardiac failure | SNOMED |
| 24 | 397748008 | Hypertension with albuminuria | SNOMED |
| 25 | 49220004 | Hypertensive renal failure | SNOMED |
| 26 | 443482000 | Hypertensive urgency | SNOMED |
| 27 | 62275004 | Hypertensive episode | SNOMED |
| 28 | 50490005 | Hypertensive encephalopathy | SNOMED |
| 29 | 706882009 | Hypertensive crisis | SNOMED |
| 30 | 70272006 | Malignant hypertension | SNOMED |
| 31 | 31992008 | Secondary hypertension | SNOMED |
| 32 | 161501007 | H/O: hypertension * | SNOMED |
| 33 | 52698002 | Transient hypertension | SNOMED |
| 34 | 123799005 | Renovascular hypertension | SNOMED |
| 35 | 28119000 | Renal hypertension | SNOMED |
| 36 | 193003 | Benign hypertensive renal disease (disorder) | SNOMED |
| 37 | 194785008 | Benign secondary hypertension | SNOMED |
| 38 | 449759005 | Hypertensive complication | SNOMED |
| 39 | 428163005 | Hypertensive left ventricular hypertrophy | SNOMED |
| 40 | 89242004 | Malignant secondary hypertension | SNOMED |
| 41 | 37618003 | Chronic hypertension complicating AND/OR reason for care during pregnancy | SNOMED |
| No. | Class of Medicine Included | ATC L4 Code | Included Drugs (Not Exclusive) | Excluded Drugs |
|---|---|---|---|---|
| 1 | Dihydropyridine derivatives | C08CA C08GA | Amlodipine Nifedipine Felodipine Lacidipine Cilnidipine Nimodipine | |
| 2 | Angiotensin II antagonists, plain | C09CA | Losartan Valsartan Telmisartan Irbesartan Candesartan Olmesartan medoxomil | |
| 3 | ACE inhibitors, plain | C09AA | Enalapril Lisinopril Perindopril Captopril Ramipril Imidapril | |
| 4 | Beta blocking agents, selective | C07AB | Atenolol Bisoprolol Metoprolol Nebivolol | Sotalol Timolol Betaxolol Esmolol |
| 5 | Angiotensin II antagonists and calcium channel blockers | C09DB C09DX | Valsartan and amlodipine Telmisartan and amlodipine Olmesartan, medoxomil, and amlodipine | |
| 6 | Thiazides, plain | C03AA | Hydrochlorothiazide | |
| 7 | Sulfonamides, plain | C03BA C03CA | Furosemide Indapamide Metolazone Bumetanide | Verapamil |
| 8 | Alpha and beta blocking agents | C07AG | Carvedilol Labetalol | |
| 9 | Organic nitrates | C01DA C01DB C01DX | Isosorbide dinitrate Isosorbide mononitrate | Glyceryl trinitrate |
| 10 | Beta blocking agents, non-selective | C07AA | Propranolol Nadolol | |
| 11 | Angiotensin II antagonists and diuretics | C09DA | Valsartan and diuretics Losartan and diuretics Irbesartan and diuretics | |
| 12 | Benzothiazepine derivatives | C08DB | Diltiazem | |
| 13 | Aldosterone antagonists | C03DA | Spironolactone Eplerenone | |
| 14 | Beta blocking agents, selective, and other antihypertensives | C07FB C07FX | Atenolol and other antihypertensives | |
| 15 | ACE inhibitors and calcium channel blockers | C09BB | Perindopril and amlodipine | |
| 16 | Low-ceiling diuretics and potassium-sparing agents | C03EA | Hydrochlorothiazide and potassium-sparing agents | |
| 17 | ACE inhibitors, other combinations | C09BX | Perindopril, amlodipine and indapamide Cosyrel | |
| 18 | Angiotensin II antagonists, other combinations | C09DX | Sacubitril-valsartan | |
| NA | Other excluded medicines | C03XA C01DX | Tolvaptan Nicorandil |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.99 | 0.99 | 0.96 | 0.97 | 0.95 | 0.97 | 0.97 | 0.92 | 0.96 | 0.98 | 0.93 | 0.96 | 0.99 | 0.96 | |
| 2 | 0.99 | 0.98 | 0.95 | 0.96 | 0.96 | 0.98 | 0.96 | 0.93 | 0.95 | 0.99 | 0.94 | 0.95 | 0.98 | 0.97 | |
| 3 | 0.99 | 0.98 | 0.97 | 0.98 | 0.94 | 0.96 | 0.98 | 0.93 | 0.95 | 0.97 | 0.94 | 0.97 | 0.98 | 0.95 | |
| 4 | 0.96 | 0.95 | 0.97 | 0.95 | 0.91 | 0.93 | 0.95 | 0.9 | 0.92 | 0.96 | 0.95 | 0.96 | 0.95 | 0.92 | |
| 5 | 0.97 | 0.96 | 0.98 | 0.95 | 0.92 | 0.94 | 0.96 | 0.91 | 0.95 | 0.95 | 0.92 | 0.95 | 0.96 | 0.93 | |
| 6 | 0.95 | 0.96 | 0.94 | 0.91 | 0.92 | 0.94 | 0.92 | 0.91 | 0.91 | 0.95 | 0.92 | 0.95 | 0.96 | 0.93 | |
| 7 | 0.97 | 0.98 | 0.96 | 0.93 | 0.94 | 0.94 | 0.94 | 0.91 | 0.93 | 0.97 | 0.92 | 0.93 | 0.96 | 0.95 | |
| 8 | 0.97 | 0.96 | 0.98 | 0.95 | 0.96 | 0.92 | 0.94 | 0.91 | 0.93 | 0.95 | 0.92 | 0.95 | 0.96 | 0.93 | |
| 9 | 0.92 | 0.93 | 0.93 | 0.90 | 0.91 | 0.91 | 0.91 | 0.91 | 0.90 | 0.92 | 0.89 | 0.92 | 0.91 | 0.90 | |
| 10 | 0.96 | 0.95 | 0.95 | 0.92 | 0.95 | 0.91 | 0.93 | 0.93 | 0.90 | 0.94 | 0.89 | 0.92 | 0.95 | 0.92 | |
| 11 | 0.98 | 0.99 | 0.97 | 0.96 | 0.95 | 0.95 | 0.97 | 0.95 | 0.92 | 0.94 | 0.95 | 0.94 | 0.97 | 0.96 | |
| 12 | 0.93 | 0.94 | 0.94 | 0.95 | 0.92 | 0.92 | 0.92 | 0.92 | 0.89 | 0.89 | 0.95 | 0.91 | 0.94 | 0.91 | |
| 13 | 0.96 | 0.95 | 0.97 | 0.96 | 0.95 | 0.95 | 0.93 | 0.95 | 0.92 | 0.92 | 0.94 | 0.91 | 0.95 | 0.92 | |
| 14 | 0.99 | 0.98 | 0.98 | 0.95 | 0.96 | 0.96 | 0.96 | 0.96 | 0.91 | 0.95 | 0.97 | 0.94 | 0.95 | 0.95 | |
| 15 | 0.96 | 0.97 | 0.95 | 0.92 | 0.93 | 0.93 | 0.95 | 0.93 | 0.90 | 0.92 | 0.96 | 0.91 | 0.92 | 0.95 |
References
- Brouwers, S.; Sudano, I.; Kokubo, Y.; Sulaica, E.M. Arterial hypertension. Lancet 2021, 10296, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Ta, C.N.; Weng, C. Detecting Systemic Data Quality Issues in Electronic Health Records. Stud. Health Technol. Inform. 2019, 264, 383–387. [Google Scholar] [CrossRef] [PubMed]
- D’Amore, J. Electronic Health Record Data Governance and Data Quality in the Real World. Healthcare Information and Management Systems Society. 2023. Available online: https://www.himss.org/resources/electronic-health-record-data-governance-and-data-quality-real-world (accessed on 16 February 2023).
- Angelow, A.; Reber, K.C.; Schmidt, C.O.; Baumeister, S.E.; Chenot, J.-F. Prevalence of Cardiovascular Risk Factors at The Population Level: A Comparison of Ambulatory Physician-Coded Claims Data with Clinical Data from A Population-Based Study. Gesundheitswesen 2019, 81, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Chen, G.; Kaplan, G.G.; Lix, L.M.; Drummond, N.; Lucyk, K.; Garies, S.; Lowerison, M.; Weibe, S.; Quan, H. Methods of defining hypertension in electronic medical records: Validation against national survey data. J. Public Health 2016, 38, e392–e399. [Google Scholar] [CrossRef] [PubMed]
- Nadkarni, G.N.; Gottesman, O.; Linneman, J.G.; Chase, H.; Berg, R.L.; Farouk, S.; Nadukuru, R.; Lotay, V.; Ellis, S.; Hripcsak, G.; et al. Development and validation of an electronic phenotyping algorithm for chronic kidney disease. AMIA Annu. Symp. Proc. 2014, 2014, 907–916. [Google Scholar] [PubMed]
- Teixeira, P.L.; Wei, W.-Q.; Cronin, R.M.; Mo, H.; VanHouten, J.P.; Carroll, R.J.; LaRose, E.; Bastarache, L.A.; Rosenbloom, S.T.; Edwards, T.L.; et al. Evaluating electronic health record data sources and algorithmic approaches to identify hypertensive individuals. J. Am. Med. Inform. Assoc. 2016, 24, 162–171. [Google Scholar] [CrossRef] [PubMed]
- McDonough, C.W.; Babcock, K.; Chucri, K.; Crawford, D.C.; Bian, J.; Modave, F.; Cooper-DeHoff, R.M.; Hogan, W.R. Optimizing identification of resistant hypertension: Computable phenotype development and validation. Pharmacoepidemiol. Drug Saf. 2020, 29, 1393–1401. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.C.; Lam, C.S.P.; Matchar, D.B.; Zee, Y.K.; Wong, J.E.L. Singapore’s health-care system: Key features, challenges, and shifts. Lancet 2021, 398, 1091–1104. [Google Scholar] [CrossRef] [PubMed]
- MOH Clinical Practice Guidelines on Hypertension. Ministry of Health, Singapore. 2023. Available online: https://www.moh.gov.sg/hpp/doctors/guidelines/GuidelineDetails/cpgmed_hypertension (accessed on 16 February 2023).
- Parikh, R.; Mathai, A.; Parikh, S.; Chandra Sekhar, G.; Thomas, R. Understanding and using sensitivity, specificity and predictive values. Indian J. Ophthalmol. 2008, 56, 45–50. [Google Scholar] [CrossRef]
- Leeflang, M.M.; Rutjes, A.W.; Reitsma, J.B.; Hooft, L.; Bossuyt, P.M. Variation of a test’s sensitivity and specificity with disease prevalence. CMAJ 2013, 185, E537–E544. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Lee, S.G.S.; How, C.H. Management of the heart failure patient in the primary care setting. Singapore Med. J. 2020, 61, 225–229. [Google Scholar] [CrossRef]
- Whelton, P.K.; Carey, R.M.; Aronow, W.S.; Casey, D.E., Jr.; Collins, K.J.; Himmelfarb, C.D.; DePalma, S.M.; Gidding, S.; Jamerson, K.A.; Jones, D.W.; et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension 2018, 71, e13–e115. [Google Scholar] [CrossRef]
| Random Cohort (n = 1619) | AF Cohort (n = 1194) | ||||
|---|---|---|---|---|---|
| 2019 (n = 808) | 2020 (n = 811) | 2019 (n = 608) | 2020 (n = 586) | ||
| Hypertension | Yes | 335 (41.5%) | 301 (37.1%) | 461 (75.8%) | 464 (79.2%) |
| No | 473 (58.5%) | 510 (62.9%) | 147 (24.2%) | 122 (20.8%) | |
| Gender | Male | 380 (47.0%) | 401 (49.4%) | 305 (50.2%) | 310 (52.9%) |
| Female | 428 (53.0%) | 410 (50.6%) | 303 (49.8%) | 276 (47.1%) | |
| Race | Chinese | 514 (63.6%) | 489 (60.3%) | 451 (74.2%) | 458 (78.2%) |
| Malay | 139 (17.2%) | 137 (16.8%) | 92 (15.1%) | 81 (13.8%) | |
| Indian | 84 (10.4%) | 99 (12.3%) | 29 (4.8%) | 25 (4.3%) | |
| Others | 71 (8.8%) | 86 (10.6%) | 36 (5.9%) | 22 (3.8%) | |
| Age | Mean | 47.5 | 45.8 | 72.2 | 72.4 |
| Standard deviation | 28.8 | 27.5 | 11.8 | 12.0 | |
| Total | 808 (100.0%) | 811 (100.0%) | 608 (100.0%) | 586 (100.0%) | |
| Random Cohort (n = 1619) | AF Cohort (n = 1194) | |||||
|---|---|---|---|---|---|---|
| Statistics | 2019 (%) | 2020 (%) | Overall (%) | 2019 (%) | 2020 (%) | Overall (%) |
| Sensitivity | 82.4 | 85.4 | 83.8 | 85.9 | 89.2 | 87.6 |
| Specificity | 92.0 | 93.5 | 92.8 | 66.7 | 68.9 | 67.7 |
| PPV | 87.9 | 88.6 | 88.2 | 89.0 | 91.6 | 90.3 |
| NPV | 88.1 | 91.6 | 89.9 | 60.1 | 62.7 | 61.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neo, J.W.; Xie, Q.; Ang, P.S.; Tan, H.X.; Foo, B.; Koon, Y.L.; Ng, A.; Tan, S.H.; Teo, D.; Tham, M.Y.; et al. Pioneering Arterial Hypertension Phenotyping on Nationally Aggregated Electronic Health Records. Pharmacoepidemiology 2024, 3, 169-182. https://doi.org/10.3390/pharma3010010
Neo JW, Xie Q, Ang PS, Tan HX, Foo B, Koon YL, Ng A, Tan SH, Teo D, Tham MY, et al. Pioneering Arterial Hypertension Phenotyping on Nationally Aggregated Electronic Health Records. Pharmacoepidemiology. 2024; 3(1):169-182. https://doi.org/10.3390/pharma3010010
Chicago/Turabian StyleNeo, Jing Wei, Qihuang Xie, Pei San Ang, Hui Xing Tan, Belinda Foo, Yen Ling Koon, Amelia Ng, Siew Har Tan, Desmond Teo, Mun Yee Tham, and et al. 2024. "Pioneering Arterial Hypertension Phenotyping on Nationally Aggregated Electronic Health Records" Pharmacoepidemiology 3, no. 1: 169-182. https://doi.org/10.3390/pharma3010010
APA StyleNeo, J. W., Xie, Q., Ang, P. S., Tan, H. X., Foo, B., Koon, Y. L., Ng, A., Tan, S. H., Teo, D., Tham, M. Y., Yap, A., Ng, N., Loke, C. W. P., Peck, L. F., Huang, H., & Dorajoo, S. R. (2024). Pioneering Arterial Hypertension Phenotyping on Nationally Aggregated Electronic Health Records. Pharmacoepidemiology, 3(1), 169-182. https://doi.org/10.3390/pharma3010010






