Impact of Structural Relaxation on Protein–Protein Docking in Large Macromolecular Complexes
Abstract
1. Introduction
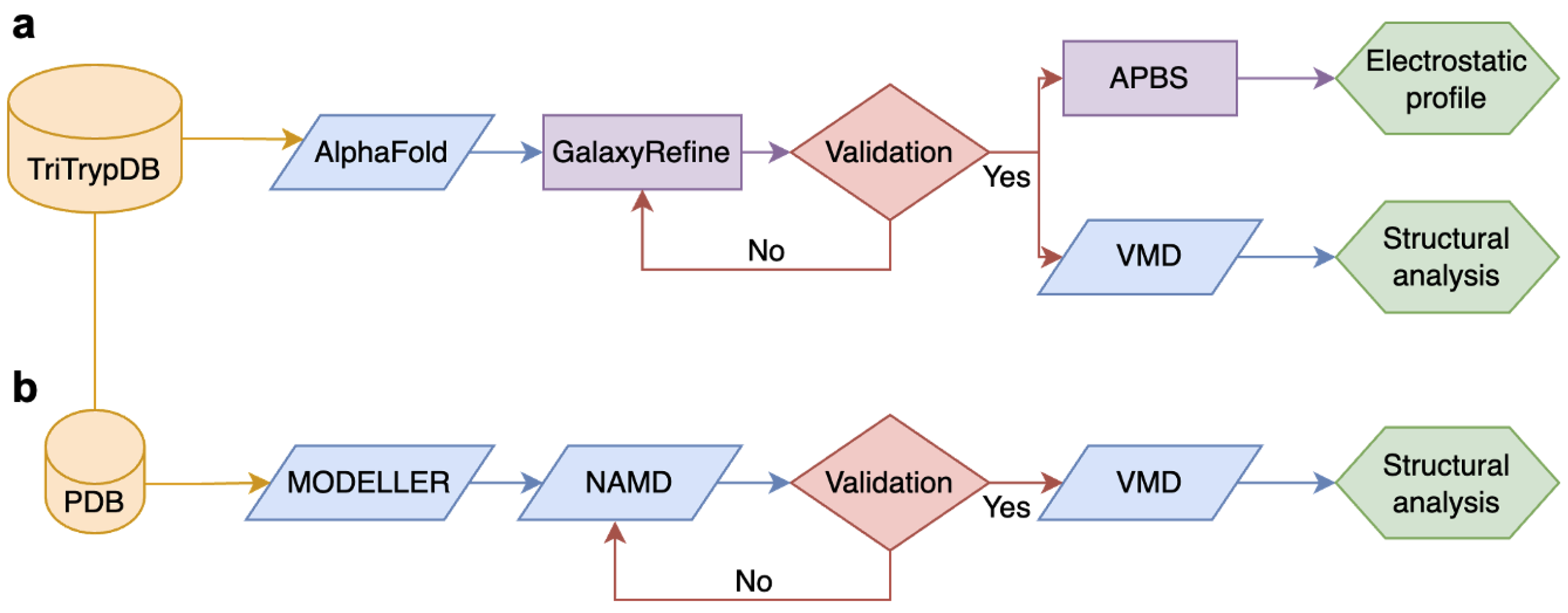
2. Materials and Methods
2.1. Modeling Refinements
2.1.1. Modeling Conformational States
2.1.2. Molecular Dynamics Simulations
2.2. Protein-Protein Docking
2.3. Analysis
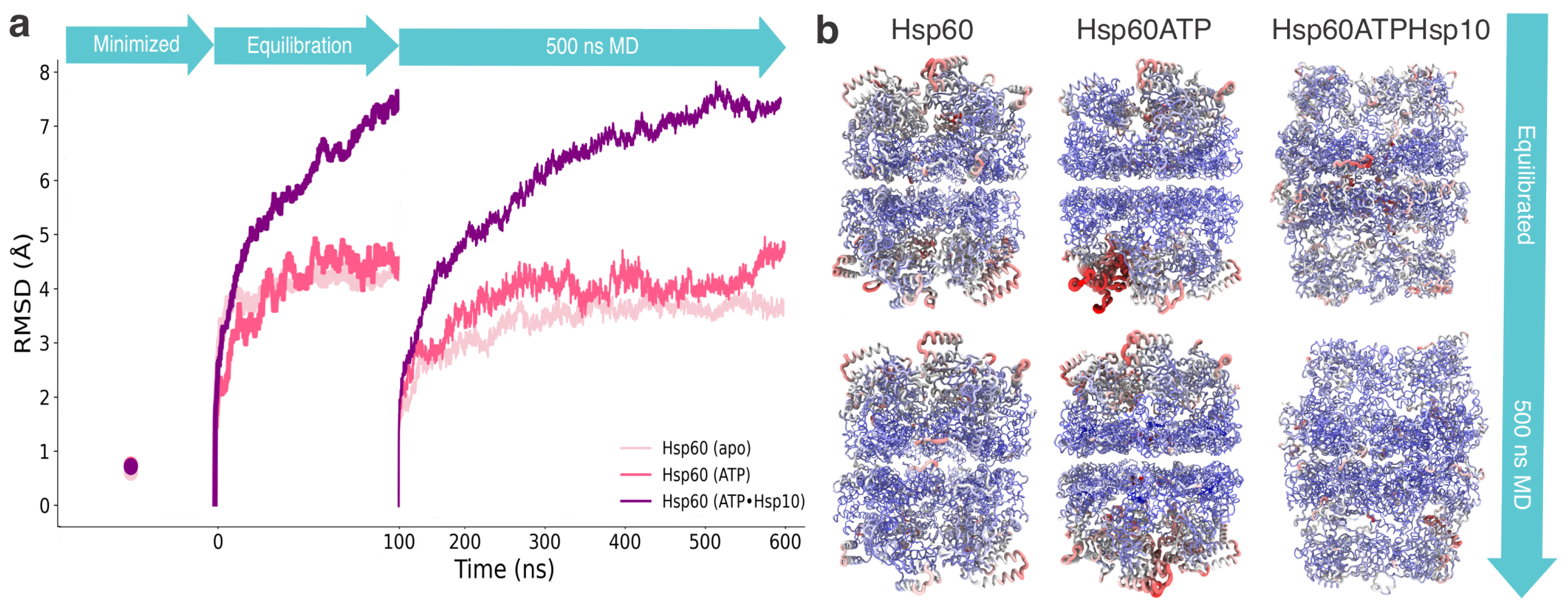
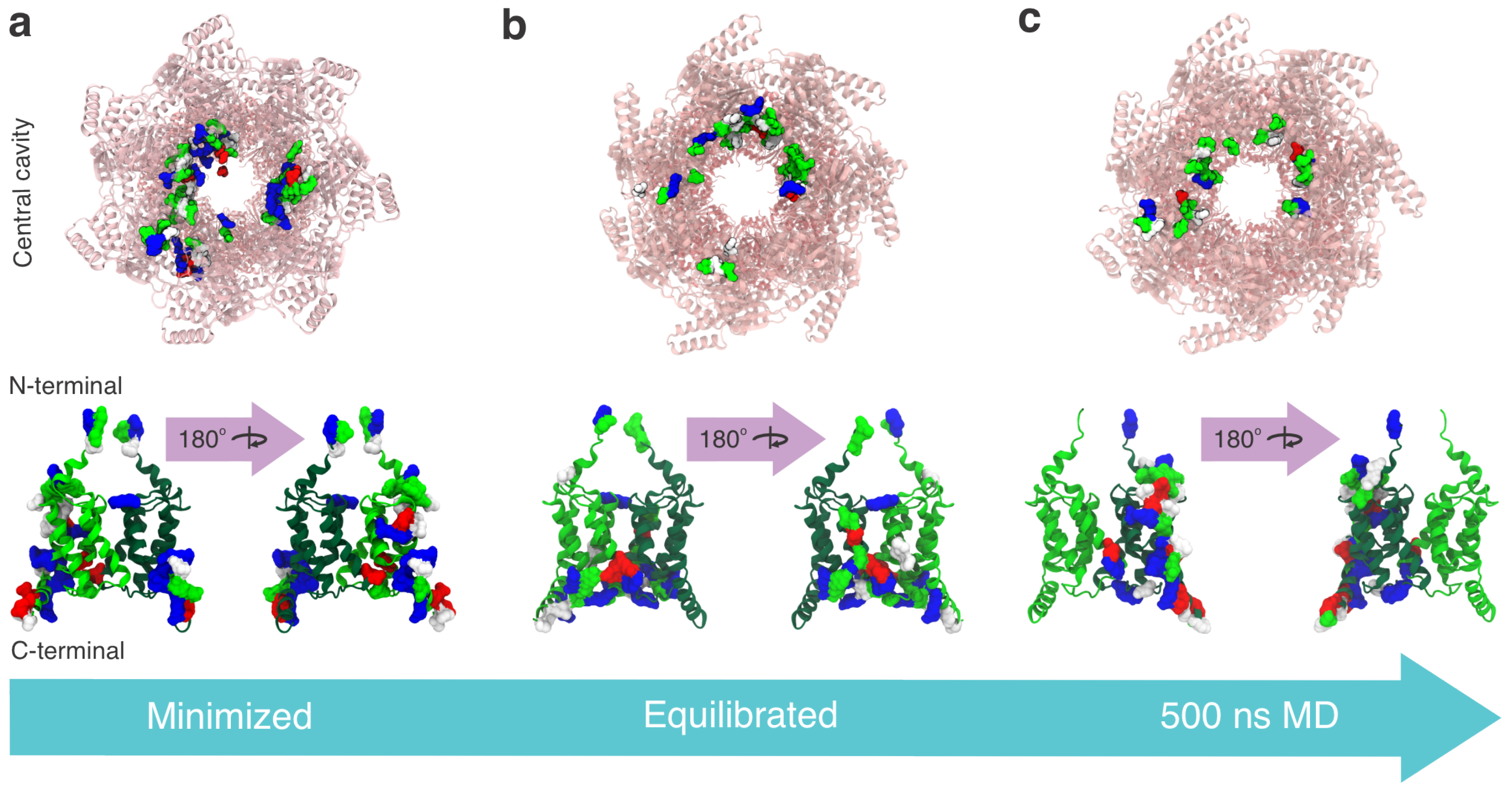
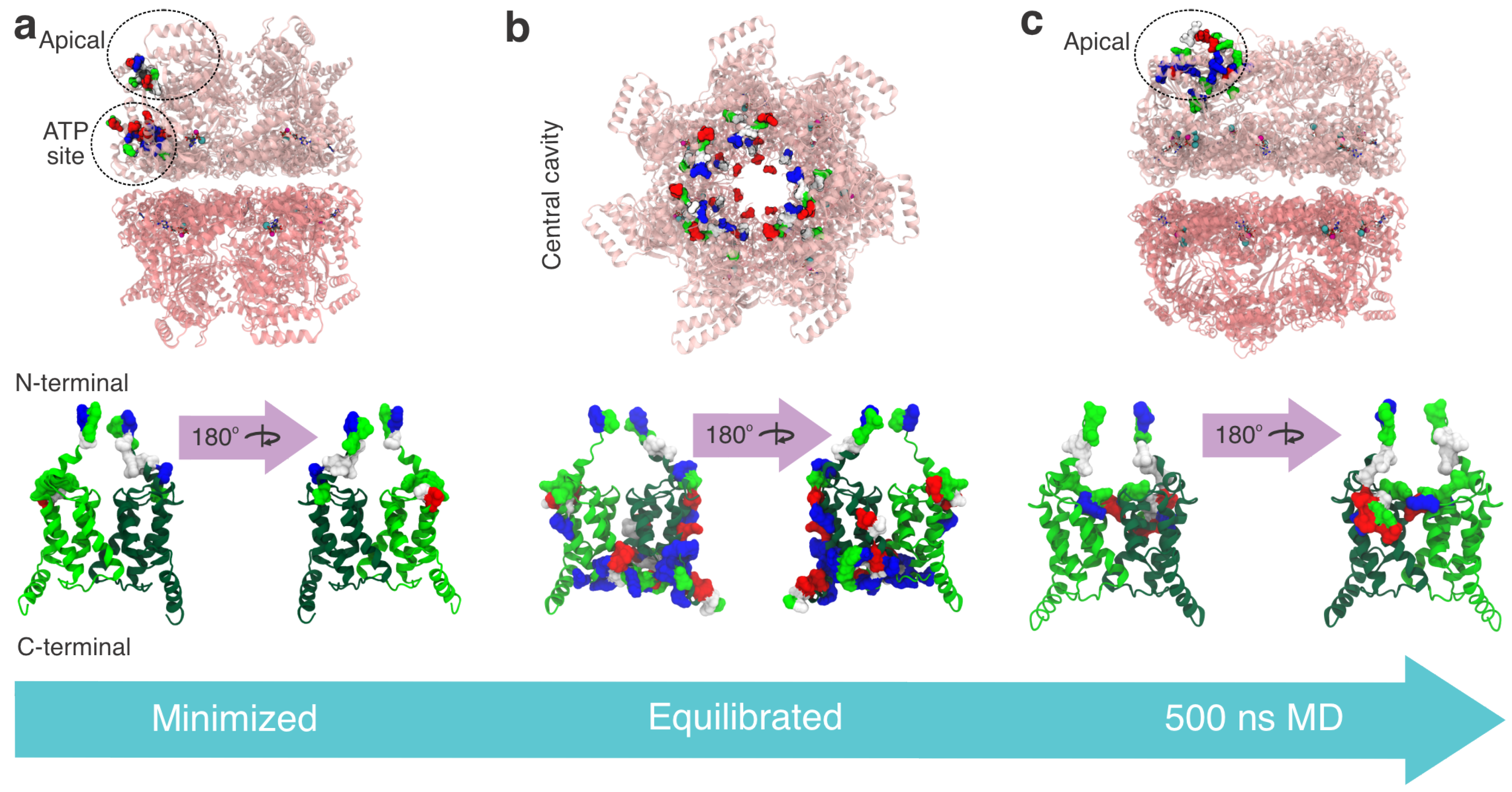
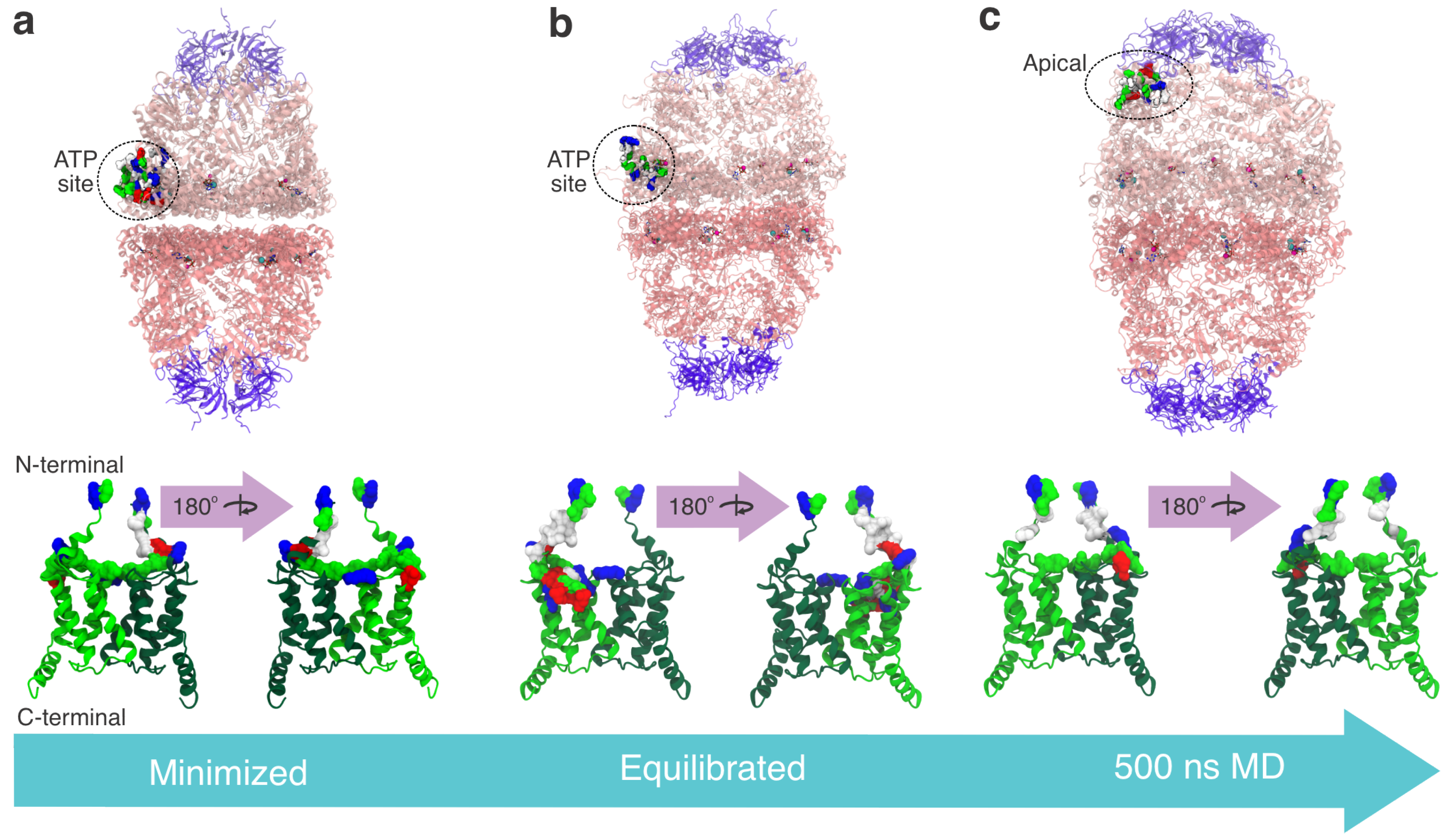
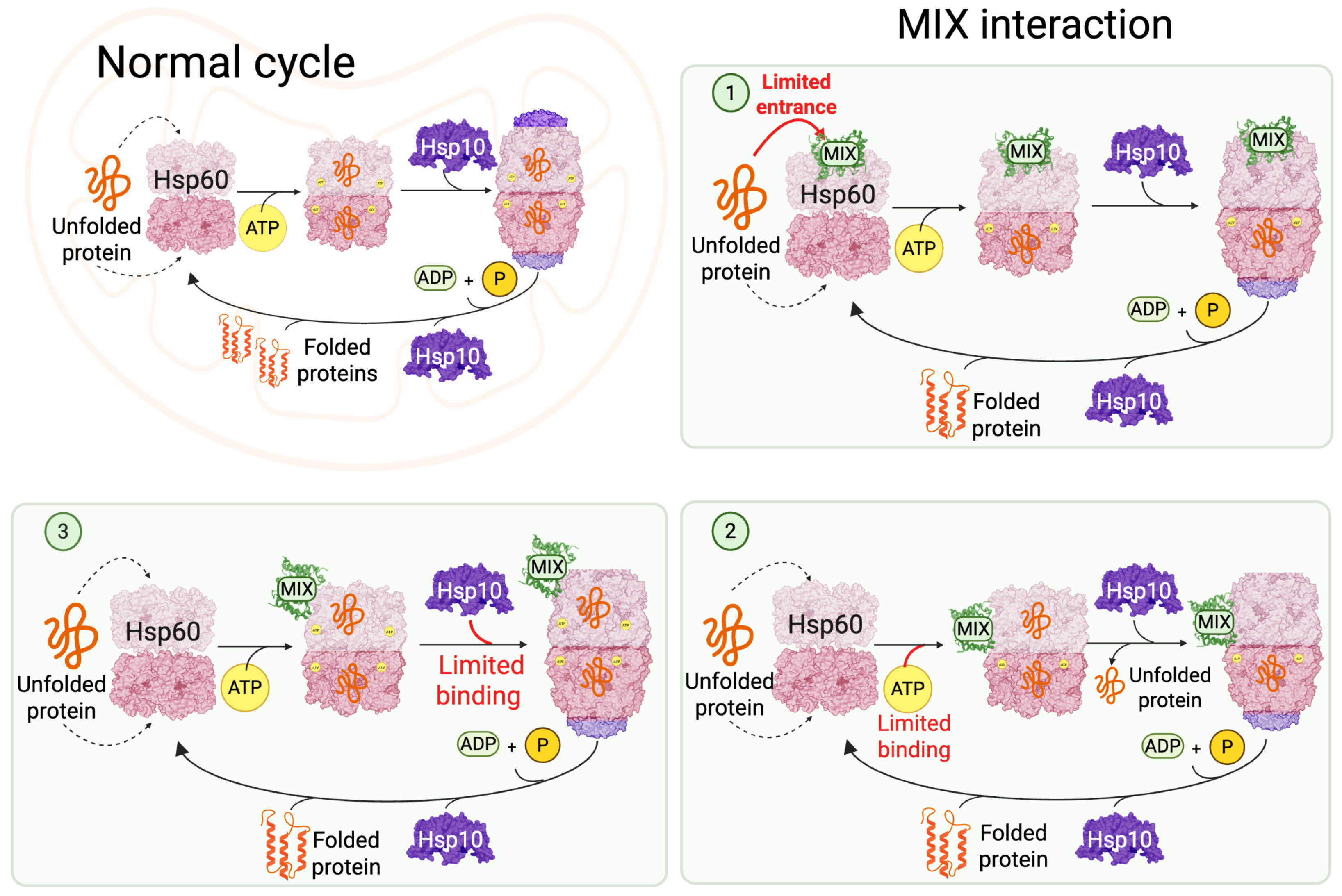
3. Results
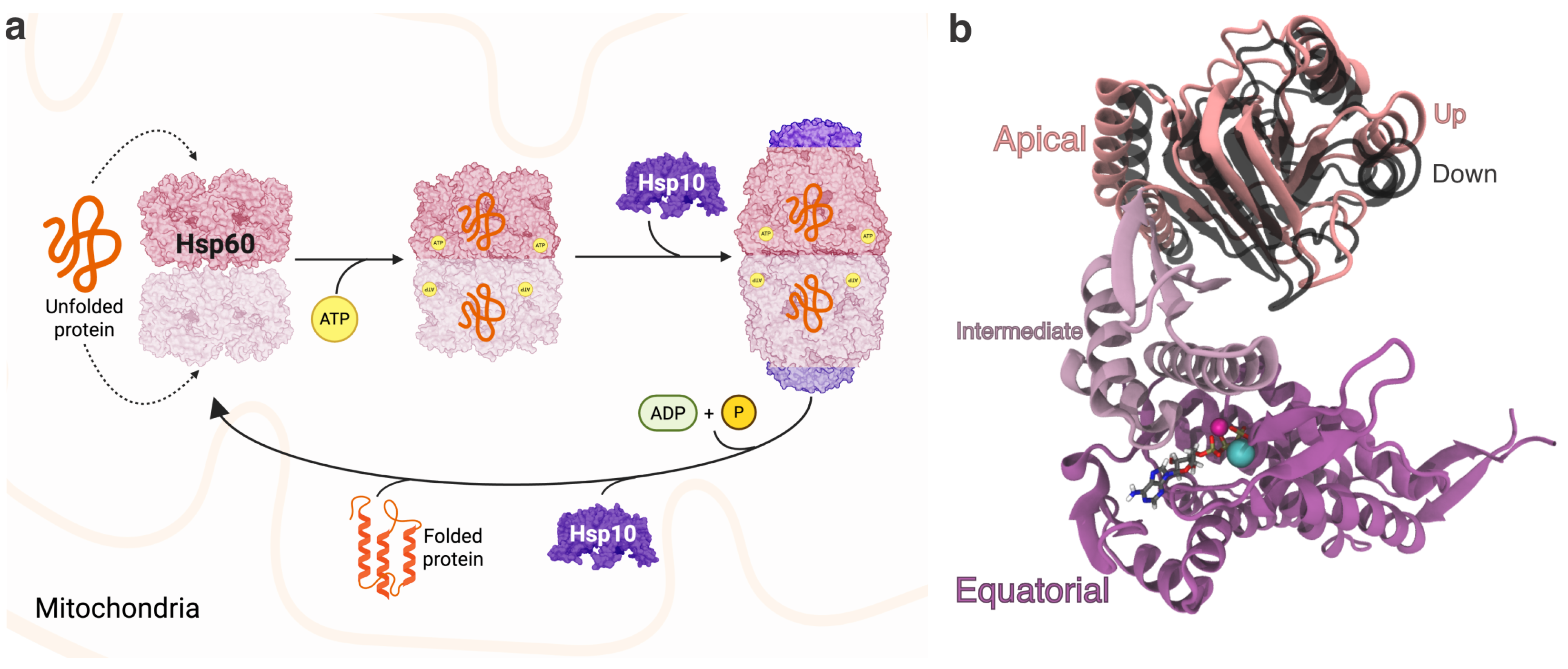
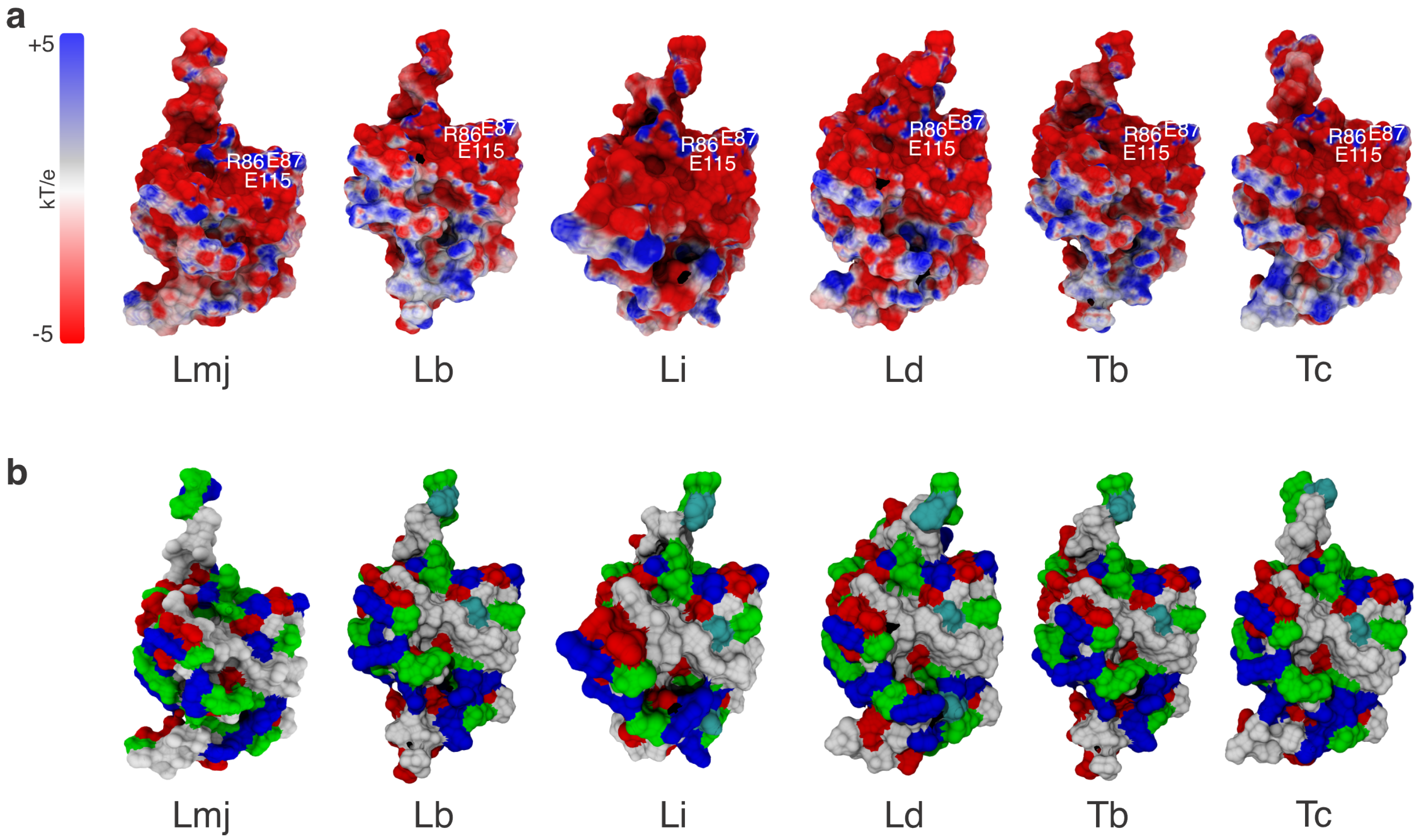
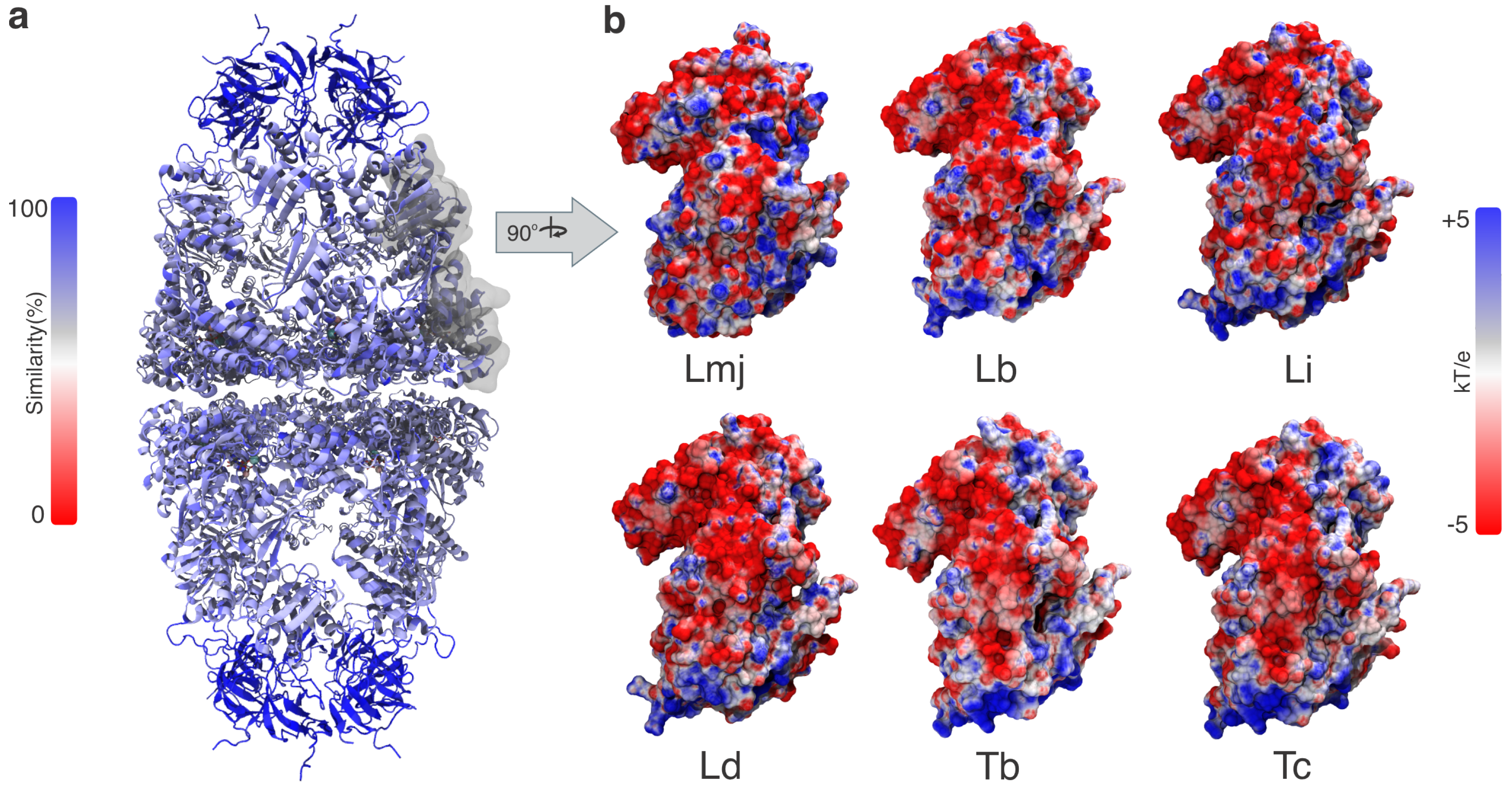
3.1. Structural and Evolutionary Insights Hsp60 and MIX
3.2. Docking Studies of MIX and Hsp60
3.3. Interaction Sites MIX:Hsp60
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ATP | Adenosine triphosphate |
| Hsp10 | Heat shock protein 10 |
| Hsp60 | Heat shock protein 60 |
| Lbr | Leishmania braziliensis |
| Ld | Leishmania donovani |
| Li | Leishmania infantum |
| Lmj | Leishmania major |
| Mix | mt protein X |
| MD | Molecular dynamic |
| RMSD | Root-mean-square deviation |
| Tb | Trypanosoma brucei |
| Tc | Trypanosoma cruzi |
| VMD | Visual Molecular Dynamics |
References
- Maheshwari, S.; Brylinski, M. Across-proteome modeling of dimer structures for the bottom-up assembly of protein-protein interaction networks. BMC Bioinform. 2017, 18, 257. [Google Scholar] [CrossRef]
- Launay, G.; Simonson, T. Homology modelling of protein-protein complexes: A simple method and its possibilities and limitations. BMC Bioinform. 2008, 9, 427. [Google Scholar] [CrossRef] [PubMed]
- Rosa, R.S.; Leal da Silva, M.; Bernardi, R.C. Atomistic Insights into gp82 Binding: A Microsecond, Million-Atom Exploration of Trypanosoma cruzi Host-Cell Invasion. Biochemistry 2025, 64, 2476–2488. [Google Scholar] [CrossRef]
- Porter, K.A.; Desta, I.; Kozakov, D.; Vajda, S. What method to use for protein–protein docking? Curr. Opin. Struct. Biol. 2019, 55, 1–7. [Google Scholar] [CrossRef]
- Harmalkar, A.; Gray, J.J. Advances to tackle backbone flexibility in protein docking. Curr. Opin. Struct. Biol. 2021, 67, 178–186. [Google Scholar] [CrossRef]
- Wu, J.; Jonniya, N.A.; Hirakis, S.P.; Olivieri, C.; Veglia, G.; Kornev, A.P.; Taylor, S.S. Role of the αC-β4 loop in protein kinase structure and dynamics. eLife 2024, 12, RP91980. [Google Scholar] [CrossRef]
- Wang, C.; Schueler-Furman, O.; Baker, D. Improved side-chain modeling for protein–protein docking. Protein Sci. 2005, 14, 1328–1339. [Google Scholar] [CrossRef]
- Goh, B.C.; Hadden, J.A.; Bernardi, R.C.; Singharoy, A.; McGreevy, R.; Rudack, T.; Cassidy, C.K.; Schulten, K. Computational methodologies for real-space structural refinement of large macromolecular complexes. Annu. Rev. Biophys. 2016, 45, 253–278. [Google Scholar] [CrossRef] [PubMed]
- May, A.; Zacharias, M. Accounting for global protein deformability during protein–protein and protein–ligand docking. Biochim. Biophys. Acta BBA Proteins Proteom. 2005, 1754, 225–231. [Google Scholar] [CrossRef]
- Kapoor, K.; Thangapandian, S.; Tajkhorshid, E. Extended-ensemble docking to probe dynamic variation of ligand binding sites during large-scale structural changes of proteins. Chem. Sci. 2022, 13, 4150–4169. [Google Scholar] [CrossRef] [PubMed]
- Gomes, D.E.; Yang, B.; Vanella, R.; Nash, M.A.; Bernardi, R.C. Integrating dynamic network analysis with AI for enhanced epitope prediction in PD-L1: Affibody interactions. J. Am. Chem. Soc. 2024, 146, 23842–23853. [Google Scholar] [CrossRef]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and its limits in rigid body protein-protein docking. Structure 2020, 28, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Melquiond, A.S.; Karaca, E.; Kastritis, P.L.; Bonvin, A.M. Next challenges in protein–protein docking: From proteome to interactome and beyond. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2012, 2, 642–651. [Google Scholar] [CrossRef]
- Vakser, I.A. Challenges in protein docking. Curr. Opin. Struct. Biol. 2020, 64, 160–165. [Google Scholar] [CrossRef]
- Ocetkiewicz, K.M.; Czaplewski, C.; Krawczyk, H.; Lipska, A.G.; Liwo, A.; Proficz, J.; Sieradzan, A.K.; Czarnul, P. Multi-GPU UNRES for scalable coarse-grained simulations of very large protein systems. Comput. Phys. Commun. 2024, 298, 109112. [Google Scholar] [CrossRef]
- Ocetkiewicz, K.M.; Czaplewski, C.; Krawczyk, H.; Lipska, A.G.; Liwo, A.; Proficz, J.; Sieradzan, A.K.; Czarnul, P. UNRES-GPU for physics-based coarse-grained simulations of protein systems at biological time-and size-scales. Bioinformatics 2023, 39, btad391. [Google Scholar] [CrossRef]
- Yue, Y.; Li, S.; Cheng, Y.; Wang, L.; Hou, T.; Zhu, Z.; He, S. Integration of molecular coarse-grained model into geometric representation learning framework for protein-protein complex property prediction. Nat. Commun. 2024, 15, 9629. [Google Scholar] [CrossRef]
- Baaden, M.; Marrink, S.J. Coarse-grain modelling of protein–protein interactions. Curr. Opin. Struct. Biol. 2013, 23, 878–886. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.G.; Wiehe, K.; Hwang, H.; Kim, B.H.; Vreven, T.; Weng, Z. ZDOCK server: Interactive docking prediction of protein–protein complexes and symmetric multimers. Bioinformatics 2014, 30, 1771–1773. [Google Scholar] [CrossRef] [PubMed]
- Honorato, R.V.; Trellet, M.E.; Jiménez-García, B.; Schaarschmidt, J.J.; Giulini, M.; Reys, V.; Koukos, P.I.; Rodrigues, J.P.; Karaca, E.; van Zundert, G.C.; et al. The HADDOCK2. 4 web server for integrative modeling of biomolecular complexes. Nat. Protoc. 2024, 19, 3219–3241. [Google Scholar] [CrossRef]
- Lyskov, S.; Gray, J.J. The RosettaDock server for local protein–protein docking. Nucleic Acids Res. 2008, 36, W233–W238. [Google Scholar] [CrossRef]
- Sircar, A.; Gray, J.J. SnugDock: Paratope structural optimization during antibody-antigen docking compensates for errors in antibody homology models. PLoS Comput. Biol. 2010, 6, e1000644. [Google Scholar] [CrossRef]
- Alam, N.; Schueler-Furman, O. Modeling peptide-protein structure and binding using Monte Carlo sampling approaches: Rosetta FlexPepDock and FlexPepBind. In Modeling Peptide-Protein Interactions: Methods and Protocols; Springer: New York, NY, USA, 2017; pp. 139–169. [Google Scholar]
- Cavasotto, C.N.; Kovacs, J.A.; Abagyan, R.A. Representing receptor flexibility in ligand docking through relevant normal modes. J. Am. Chem. Soc. 2005, 127, 9632–9640. [Google Scholar] [CrossRef]
- Cavasotto, C.N. Normal mode-based approaches in receptor ensemble docking. In Computational Drug Discovery and Design; Springer: New York, NY, USA, 2011; pp. 157–168. [Google Scholar]
- Jamali, K.; Käll, L.; Zhang, R.; Brown, A.; Kimanius, D.; Scheres, S.H. Automated model building and protein identification in cryo-EM maps. Nature 2024, 628, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cai, Y.; Zhang, B.; Zheng, W.; Freddolino, L.; Zhang, G.; Zhou, X. EM2: Assembling protein complex structures from cryo-EM maps through intertwined chain and domain fitting. Brief. Bioinform. 2024, 25, bbae113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, L.; Zhang, S.; Peng, C.; Zhang, G.; Zhou, X. EMol: Modeling protein-nucleic acid complex structures from cryo-EM maps by coupling chain assembly with map segmentation. Nucleic Acids Res. 2025, 53, W228–W237. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, H.; Terashi, G.; Taluja, M.; Kihara, D. DiffModeler: Large macromolecular structure modeling for cryo-EM maps using a diffusion model. Nat. Methods 2024, 21, 2307–2317. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Zheng, W.; Wuyun, Q.; Li, Y.; Zhang, C.; Freddolino, L.; Zhang, Y. Improving deep learning protein monomer and complex structure prediction using DeepMSA2 with huge metagenomics data. Nat. Methods 2024, 21, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5–6. [Google Scholar] [CrossRef]
- Braxton, J.R.; Shao, H.; Tse, E.; Gestwicki, J.E.; Southworth, D.R. Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly. Nat. Struct. Mol. Biol. 2024, 31, 1848–1858. [Google Scholar] [CrossRef]
- Caruso Bavisotto, C.; Alberti, G.; Vitale, A.M.; Paladino, L.; Campanella, C.; Rappa, F.; Gorska, M.; Conway de Macario, E.; Cappello, F.; Macario, A.J.; et al. Hsp60 post-translational modifications: Functional and pathological consequences. Front. Mol. Biosci. 2020, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Zíková, A.; Panigrahi, A.K.; Uboldi, A.D.; Dalley, R.A.; Handman, E.; Stuart, K. Structural and functional association of Trypanosoma brucei MIX protein with cytochrome c oxidase complex. Eukaryot. Cell 2008, 7, 1994–2003. [Google Scholar] [CrossRef]
- Aslett, M.; Aurrecoechea, C.; Berriman, M.; Brestelli, J.; Brunk, B.P.; Carrington, M.; Depledge, D.P.; Fischer, S.; Gajria, B.; Gao, X.; et al. TriTrypDB: A functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010, 38, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- McGuffin, L.J.; Bryson, K.; Jones, D.T. The PSIPRED protein structure prediction server. Bioinformatics 2000, 16, 404–405. [Google Scholar] [CrossRef]
- Lee, G.R.; Won, J.; Heo, L.; Seok, C. GalaxyRefine2: Simultaneous refinement of inaccurate local regions and overall protein structure. Nucleic Acids Res. 2019, 47, W451–W455. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Ramachandran, G.N.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef]
- Lüthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Uboldi, A.D.; Lueder, F.B.; Walsh, P.; Spurck, T.; McFadden, G.I.; Curtis, J.; Likic, V.A.; Perugini, M.A.; Barson, M.; Lithgow, T.; et al. A mitochondrial protein affects cell morphology, mitochondrial segregation and virulence in Leishmania. Int. J. Parasitol. 2006, 36, 1499–1514. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Sali, A.; Blundell, T. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Ribeiro, J.V.; Bernardi, R.C.; Rudack, T.; Stone, J.E.; Phillips, J.C.; Freddolino, P.L.; Schulten, K. QwikMD—Integrative molecular dynamics toolkit for novices and experts. Sci. Rep. 2016, 6, 26536. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Spivak, M.; Stone, J.E.; Ribeiro, J.; Saam, J.; Freddolino, P.L.; Bernardi, R.C.; Tajkhorshid, E. VMD as a platform for interactive small molecule preparation and visualization in quantum and classical simulations. J. Chem. Inf. Model. 2023, 63, 4664–4678. [Google Scholar] [CrossRef]
- Scheurer, M.; Rodenkirch, P.; Siggel, M.; Bernardi, R.C.; Schulten, K.; Tajkhorshid, E.; Rudack, T. PyContact: Rapid, customizable, and visual analysis of noncovalent interactions in MD simulations. Biophys. J. 2018, 114, 577–583. [Google Scholar] [CrossRef]
- Barreto, C.A.; Baptista, S.J.; Preto, A.J.; Matos-Filipe, P.; Mourao, J.; Melo, R.; Moreira, I. Prediction and targeting of GPCR oligomer interfaces. Prog. Mol. Biol. Transl. Sci. 2020, 169, 105–149. [Google Scholar] [PubMed]
- Parate, S.; Rampogu, S.; Lee, G.; Hong, J.C.; Lee, K.W. Exploring the binding interaction of Raf kinase inhibitory protein with the N-terminal of C-Raf through molecular docking and molecular dynamics simulation. Front. Mol. Biosci. 2021, 8, 655035. [Google Scholar] [CrossRef]
- Li, Y.; Mohanty, S.; Nilsson, D.; Hansson, B.; Mao, K.; Irbäck, A. When a foreign gene meets its native counterpart: Computational biophysics analysis of two PgiC loci in the grass Festuca ovina. Sci. Rep. 2020, 10, 18752. [Google Scholar] [CrossRef] [PubMed]
- Gomes, P.S.; Gomes, D.E.; Bernardi, R.C. Protein structure prediction in the era of AI: Challenges and limitations when applying to in silico force spectroscopy. Front. Bioinform. 2022, 2, 983306. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Dunbrack, R.L., Jr. ProtCID: A data resource for structural information on protein interactions. Nat. Commun. 2020, 11, 711. [Google Scholar] [CrossRef]
- Shoemaker, B.A.; Panchenko, A.R.; Bryant, S.H. Finding biologically relevant protein domain interactions: Conserved binding mode analysis. Protein Sci. 2006, 15, 352–361. [Google Scholar] [CrossRef]
- Alsop, J.D.; Mitchell, J.C. Interolog interfaces in protein–protein docking. Proteins Struct. Funct. Bioinform. 2015, 83, 1940–1946. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, X.; Jiang, Z.; You, Q. Modulation of protein fate decision by small molecules: Targeting molecular chaperone machinery. Acta Pharm. Sin. B 2020, 10, 1904–1925. [Google Scholar] [CrossRef]
- Cheng, M.Y.; Hartl, F.U.; Martin, J.; Pollock, R.A.; Kalousek, F.; Neuper, W.; Hallberg, E.M.; Hallberg, R.L.; Horwich, A.L. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature 1989, 337, 620–625. [Google Scholar] [CrossRef]
- Ansari, M.Y.; Mande, S.C. A glimpse into the structure and function of atypical type I chaperonins. Front. Mol. Biosci. 2018, 5, 31. [Google Scholar] [CrossRef]
- Hayer-Hartl, M.K.; Martin, J.; Hartl, F.U. Asymmetrical interaction of GroEL and GroES in the ATPase cycle of assisted protein folding. Science 1995, 269, 836–841. [Google Scholar] [CrossRef]
- Syed, A.; Zhai, J.; Guo, B.; Zhao, Y.; Wang, J.C.Y.; Chen, L. Cryo-EM structure and molecular dynamic simulations explain the enhanced stability and ATP activity of the pathological chaperonin mutant. Structure 2024, 32, 575–584. [Google Scholar] [CrossRef]
- Gorman, M.A.; Uboldi, A.D.; Walsh, P.J.; Tan, K.S.; Hansen, G.; Huyton, T.; Ji, H.; Curtis, J.; Kedzierski, L.; Papenfuss, A.T.; et al. Crystal structure of the Leishmania major MIX protein: A scaffold protein that mediates protein–protein interactions. Protein Sci. 2011, 20, 1060–1068. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, M.J.; Gabb, H.A.; Jackson, R.M.; Moont, G. Protein-protein docking: Generation and filtering of complexes. In Protein Structure Prediction: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2000; pp. 399–415. [Google Scholar]
- Shen, M.y.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef] [PubMed]
- Gomes, P.S.; Forrester, M.; Pace, M.; Gomes, D.E.; Bernardi, R.C. May the force be with you: The role of hyper-mechanostability of the bone sialoprotein binding protein during early stages of Staphylococci infections. Front. Chem. 2023, 11, 1107427. [Google Scholar] [CrossRef]
- Chantraine, C.; Gomes, P.S.F.C.; Mathelié-Guinlet, M.; Gomes, D.E.B.; Zheng, Z.; Clowry, J.; Turley, M.B.; Irvine, A.D.; Geoghegan, J.A.; Bernardi, R.C.; et al. Ultrastrong Staphylococcus aureus adhesion to Human Skin: Calcium as a key regulator of noncovalent interactions. Sci. Adv. 2025, 11, eadu7457. [Google Scholar] [CrossRef]
- Klemm, P.; Schembri, M.A. Bacterial adhesins: Function and structure. Int. J. Med. Microbiol. 2000, 290, 27–35. [Google Scholar] [CrossRef]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Höök, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef]
- dos Santos Natividade, R.; Koehler, M.; Gomes, P.S.; Simpson, J.D.; Smith, S.C.; Gomes, D.E.; de Lhoneux, J.; Yang, J.; Ray, A.; Dermody, T.S.; et al. Deciphering molecular mechanisms stabilizing the reovirus-binding complex. Proc. Natl. Acad. Sci. USA 2023, 120, e2220741120. [Google Scholar] [CrossRef]
- Zhang, Q.; Rosa, R.S.; Ray, A.; Durlet, K.; Dorrazehi, G.M.; Bernardi, R.C.; Alsteens, D. Probing SARS-CoV-2 membrane binding peptide via single-molecule AFM-based force spectroscopy. Nat. Commun. 2025, 16, 6. [Google Scholar] [CrossRef] [PubMed]
- Iannacone, M.; Guidotti, L.G. Immunobiology and pathogenesis of hepatitis B virus infection. Nat. Rev. Immunol. 2022, 22, 19–32. [Google Scholar] [CrossRef]
- Seppälä, J.; Bernardi, R.C.; Haataja, T.J.; Hellman, M.; Pentikäinen, O.T.; Schulten, K.; Permi, P.; Ylänne, J.; Pentikäinen, U. Skeletal dysplasia mutations effect on human filamins’ structure and mechanosensing. Sci. Rep. 2017, 7, 4218. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Weber, A.; Maly, K.; Manjaly, G.; Deek, J.; Tsvyetkova, O.; Stulić, M.; Toca-Herrera, J.L.; Jantsch, M.F. A-to-I RNA editing of Filamin A regulates cellular adhesion, migration and mechanical properties. FEBS J. 2022, 289, 4580–4601. [Google Scholar] [CrossRef]
- Johnson, J.; González-Morales, N. Computational Screening of Filamin Mechanical Binding Proteins using AlphaFold2. Biochem. Cell Biol. 2025, 103, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cann, I.; Pereira, G.V.; Abdel-Hamid, A.M.; Kim, H.; Wefers, D.; Kayang, B.B.; Kanai, T.; Sato, T.; Bernardi, R.C.; Atomi, H.; et al. Thermophilic degradation of hemicellulose, a critical feedstock in the production of bioenergy and other value-added products. Appl. Environ. Microbiol. 2020, 86, e02296-19. [Google Scholar] [CrossRef]
- Melo, M.C.; Bernardi, R.C. AI Uncovers the Rapid Activation of Catch-Bonds under Force. J. Chem. Theory Comput. 2025; online ahead of print. [Google Scholar] [CrossRef]
- Wang, T.; Ahmad, S.; Cruz-Lebrón, A.; Ernst, S.E.; Olivos Caicedo, K.Y.; Jeong, Y.; Binion, B.; Mbuvi, P.; Dutta, D.; Fernandez-Materan, F.V.; et al. An expanded metabolic pathway for androgen production by commensal bacteria. Nat. Microbiol. 2025, 10, 1084–1098. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Gomes, D.E.; Liu, Z.; Santos, M.S.; Li, J.; Bernardi, R.C.; Nash, M.A. Engineering the Mechanical Stability of a Therapeutic Complex between Affibody and Programmed Death-Ligand 1 by Anchor Point Selection. ACS Nano 2024, 18, 31912–31922. [Google Scholar] [CrossRef]
- Lim, H.; Cankara, F.; Tsai, C.J.; Keskin, O.; Nussinov, R.; Gursoy, A. Artificial intelligence approaches to human-microbiome protein–protein interactions. Curr. Opin. Struct. Biol. 2022, 73, 102328. [Google Scholar] [CrossRef]
- Kim, D.N.; McNaughton, A.D.; Kumar, N. Leveraging artificial intelligence to expedite antibody design and enhance antibody–antigen interactions. Bioengineering 2024, 11, 185. [Google Scholar] [CrossRef]
- Cockell, S.J.; Oliva, B.; Jackson, R.M. Structure-based evaluation of in silico predictions of protein–protein interactions using Comparative Docking. Bioinformatics 2007, 23, 573–581. [Google Scholar] [CrossRef]
- Bayarsaikhan, B.; Zsidó, B.Z.; Börzsei, R.; Hetényi, C. Efficient Refinement of Complex Structures of Flexible Histone Peptides Using Post-Docking Molecular Dynamics Protocols. Int. J. Mol. Sci. 2024, 25, 5945. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Nakayoshi, T.; Kurimoto, E.; Oda, A. Molecular dynamics simulations for the protein–ligand complex structures obtained by computational docking studies using implicit or explicit solvents. Chem. Phys. Lett. 2021, 781, 139022. [Google Scholar] [CrossRef]
- Vreven, T.; Moal, I.H.; Vangone, A.; Pierce, B.G.; Kastritis, P.L.; Torchala, M.; Chaleil, R.; Jiménez-García, B.; Bates, P.A.; Fernandez-Recio, J.; et al. Updates to the integrated protein–protein interaction benchmarks: Docking benchmark version 5 and affinity benchmark version 2. J. Mol. Biol. 2015, 427, 3031–3041. [Google Scholar] [CrossRef]
- Santos, L.H.; Ferreira, R.S.; Caffarena, E.R. Integrating molecular docking and molecular dynamics simulations. In Docking Screens for Drug Discovery; Springer: New York, NY, USA, 2019; pp. 13–34. [Google Scholar]
- Sahu, S.N.; Pattanayak, S.K. Molecular docking and molecular dynamics simulation studies on PLCE1 encoded protein. J. Mol. Struct. 2019, 1198, 126936. [Google Scholar] [CrossRef]
- Vidal-Limon, A.; Aguilar-Toalá, J.E.; Liceaga, A.M. Integration of molecular docking analysis and molecular dynamics simulations for studying food proteins and bioactive peptides. J. Agric. Food Chem. 2022, 70, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, R.C.; Melo, M.C.; Schulten, K. Enhanced sampling techniques in molecular dynamics simulations of biological systems. Biochim. Biophys. Acta BBA Gen. Subj. 2015, 1850, 872–877. [Google Scholar] [CrossRef]
- Perilla, J.R.; Goh, B.C.; Cassidy, C.K.; Liu, B.; Bernardi, R.C.; Rudack, T.; Yu, H.; Wu, Z.; Schulten, K. Molecular dynamics simulations of large macromolecular complexes. Curr. Opin. Struct. Biol. 2015, 31, 64–74. [Google Scholar] [CrossRef]
- Bryant, P.; Pozzati, G.; Zhu, W.; Shenoy, A.; Kundrotas, P.; Elofsson, A. Predicting the structure of large protein complexes using AlphaFold and Monte Carlo tree search. Nat. Commun. 2022, 13, 6028. [Google Scholar] [CrossRef]
- Hegedűs, T.; Geisler, M.; Lukács, G.L.; Farkas, B. Ins and outs of AlphaFold2 transmembrane protein structure predictions. Cell. Mol. Life Sci. 2022, 79, 73. [Google Scholar] [CrossRef]
- Han, X.; Terashi, G.; Christoffer, C.; Chen, S.; Kihara, D. VESPER: Global and local cryo-EM map alignment using local density vectors. Nat. Commun. 2021, 12, 2090. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zhang, Z.; Xu, X.; Xu, L.; Chen, Y.; Zhang, G.; Zhou, X. Fitting atomic structures into Cryo-EM maps by coupling deep learning-enhanced map processing with global-local optimization. J. Chem. Inf. Model. 2025, 65, 3800–3811. [Google Scholar] [CrossRef] [PubMed]
- Wriggers, W.; Milligan, R.A.; McCammon, J.A. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 1999, 125, 185–195. [Google Scholar] [CrossRef]


| Lbr | Li | Ld | Tb | Tc | 3HA4 | |
|---|---|---|---|---|---|---|
| Lbr | 100 | 94 | 93 | 68 | 73 | 75 |
| Li | 94 | 100 | 99 | 67 | 73 | 77 |
| Ld | 93 | 99 | 100 | 66 | 72 | 76 |
| Tb | 68 | 67 | 66 | 100 | 83 | 56 |
| Tc | 73 | 73 | 72 | 83 | 100 | 60 |
| 3HA4 | 75 | 77 | 76 | 56 | 60 | 100 |
| Lb | Li | Ld | Tb | Tc | Lmj | Hs | |
|---|---|---|---|---|---|---|---|
| Lb | 100 | 97.47 | 97.47 | 84.85 | 53.03 | 52.37 | 47.89 |
| Li | 97.47 | 100 | 100 | 84.85 | 55.86 | 55.44 | 48.07 |
| Ld | 97.47 | 100 | 100 | 84.85 | 55.86 | 55.44 | 48.07 |
| Tb | 84.85 | 84.85 | 84.85 | 100 | 55.50 | 54.55 | 46.97 |
| Tc | 56.04 | 55.86 | 55.86 | 55.50 | 100 | 74.24 | 53.03 |
| Lmj | 55.61 | 55.44 | 55.44 | 54.55 | 74.24 | 100 | 52.37 |
| Hs | 47.89 | 48.07 | 48.07 | 46.97 | 53.03 | 52.37 | 100 |
| Metric | Hsp60apo | Hsp60ATP | Hsp60ATP-Hsp10 |
|---|---|---|---|
| Template | 8G7J | 8G7L | 8G7N |
| Template Resolution (Å) | 3.40 | 2.50 | 2.70 |
| RMSD (Template → Pre-Mini) (Å) | 0.228 | 0.332 | 0.485 |
| RMSD (Template → Post-Mini) (Å) | 0.619 | 0.755 | 0.727 |
| DOPE Score | −398,109.16 | −791,101.88 | −621,065.00 |
| MolProbity Score | 1.64 | 0.74 | 2.35 |
| ClashScore | 1.5 | 0.77 | 3.95 |
| Poor Rotamers (%) | 4.8 | 0.26 | 7.93 |
| Ramachandran Analysis | |||
| Favored (%) | 96.5 | 98.67 | 92.1 |
| Allowed (%) | 99.4 | 100 | 98.1 |
| Disallowed (%) | 0.56 | 0 | 1.88 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosa, R.S.d.L.; Bulla, A.C.S.; Bernardi, R.C.; Leal da Silva, M. Impact of Structural Relaxation on Protein–Protein Docking in Large Macromolecular Complexes. Appl. Biosci. 2025, 4, 48. https://doi.org/10.3390/applbiosci4040048
Rosa RSdL, Bulla ACS, Bernardi RC, Leal da Silva M. Impact of Structural Relaxation on Protein–Protein Docking in Large Macromolecular Complexes. Applied Biosciences. 2025; 4(4):48. https://doi.org/10.3390/applbiosci4040048
Chicago/Turabian StyleRosa, Raissa Santos de Lima, Ana Carolina Silva Bulla, Rafael C. Bernardi, and Manuela Leal da Silva. 2025. "Impact of Structural Relaxation on Protein–Protein Docking in Large Macromolecular Complexes" Applied Biosciences 4, no. 4: 48. https://doi.org/10.3390/applbiosci4040048
APA StyleRosa, R. S. d. L., Bulla, A. C. S., Bernardi, R. C., & Leal da Silva, M. (2025). Impact of Structural Relaxation on Protein–Protein Docking in Large Macromolecular Complexes. Applied Biosciences, 4(4), 48. https://doi.org/10.3390/applbiosci4040048






