Abstract
Single Nucleotide Polymorphisms (SNPs) are variations that occur at single nucleotides in the genome and are present at an appreciable level in a population. SNPs can be linked to phenotypes of interest, for example diseases, recent adaptations, or species hybridization. They can also be used to study phylogeny and evolutionary history. Technologies that rapidly identify and catalog the presence of SNPs in a DNA sample are known as SNP genotyping panels, and they continue to undergo rapid development. Such methods have great utility across the agricultural sciences in diverse areas such as plant and animal breeding, pathogen and pesticide resistance identification, outbreak tracing, and hybridization detection. Here, we provide an overview of 14 different SNP genotyping technologies and weigh some of the pros and cons associated with each platform. This review is not comprehensive or technical, nor does it aim to be. Rather, the objective is to provide an introduction to the landscape of genotyping technologies for researchers who do not have experience with these methods. Three classes of SNP genotyping methods are Polymerase Chain Reaction (PCR)-based (nine different methods), microarray-based (one method), and Next-Generation Sequencing (NGS)-based (four different methods). We discuss how each genotyping class is suited for different niches; PCR-based has a low SNP count and high sample number, microarray-based has a very high SNP count and a moderate sample number, and Next-Generation Sequencing-based has a moderate SNP count and moderate number of samples. Included are basics about how the methods function and example use cases of each method. Additionally, we introduce and discuss the potential for the MinION sequencer in SNP genotyping. For each technology, we provide insights into cost, equipment needs, labor costs, experimental complexity, data output complexity, and accessibility. These considerations address the feasibility of deploying the technologies in an agricultural science environment.
1. Introduction
Single Nucleotide Polymorphisms (SNPs) are variations in DNA that occur at single nucleotides in the genome at a specific site and are present in a population at an appreciable level (typically >1%) [1]. These variations occur naturally, and across the whole genome. Certain SNPs can be diagnostic of conditions or certain phenotypes. For example, some SNPs can be indicative of the propensity to develop cancers or other diseases and are well studied in medical sciences [2]. Technologies that use SNPs to identify targets of interest in DNA samples are known as SNP genotyping panels. In agricultural sciences, SNPs have been harnessed as diagnostic markers for genetic susceptibility to disease, in plant and animal breeding, species hybridization detection, monitoring population dynamics (example: invasive species), as markers for pathogen and pesticide resistance, and in other applications [3,4,5,6,7,8,9,10,11,12,13,14,15]. Below are some specific use cases in agricultural sciences where SNP genotyping has been applied.
In Guo et al. (2021), SNP genotyping was applied for the purpose of the molecular breeding of corn, where genotyping was used to augment seed production quality [5]. In Lorenzini et al. (2020), SNP genotyping was used to develop methods to differentiate domestic pigs, wild pigs, and their hybrids, where hybridization has been difficult to monitor [6]. In Sato et al. (2019), SNP genotyping was applied to Fugu pufferfish, a type of fish commonly farmed in Japan with the objective of improving breeding methods for the species [9]. In Matukumalli et al. (2009), SNP genotyping methods were developed for cattle with the purpose of assisting in breeding, disease outbreak detection and monitoring, parentage determination, animal identification, and milk production traits in dairy cows [12]. In Wu et al. (2018), genotyping methods were applied in bananas in order to differentiate species in the interests of conserving cryptic wild-type relatives of commercially grown banana varieties [13]. Evolutionary lineages can also be traced in cases like this [13]. In Susi et al. (2020), SNP genotyping was applied to analyze the population dynamics and diversity of a flax plant rust pathogen in an epidemiological study of an important plant pathogen [11]. These above examples were chosen to demonstrate the variety of different purposes that SNP genotyping can be used for in the agricultural sciences.
There are three basic classes of SNP genotyping methods, each having variations of their own. These are Polymerase Chain Reaction (PCR)-based SNP genotyping, microarray-based SNP genotyping, and Next-Generation Sequencing (NGS)-based SNP genotyping (this can also be referred to as genotyping by sequencing (GBS)). Each of these methods will be described and compared below. This article seeks to provide an overview of SNP genotyping technologies and how they may be applied in agricultural sciences, and some methods have been developed for medical sciences but could easily be translated to agrigenomic projects. Some of the technologies we discuss have been developed very recently, while others have been commercialized for many years. We provide examples of new genotyping technologies and technologies that are still commonly used in 2023, the goal being to help research groups interested in genotyping select which methodologies are best suited for their needs. This is not comprehensive or technical, nor does it aim to be. Rather, the objective is to provide an introduction to the landscape of genotyping technologies to researchers who do not have experience with these methods. Pros and cons of each method are also discussed, including number of targetable SNPs and equipment needs.
2. PCR-Based SNP Genotyping
There are many different PCR-based applications for SNP genotyping that have been developed. Currently, there are three methods, Kompetitive Allele Specific PCR (KASP), RNase H2 enzyme-based amplification (rhAMP), and TaqMan, that are prominent in today’s market, but other methods do exist [16]. PCR SNP genotyping generally functions similarly across technologies, where PCR primers are designed specific to SNPs of interest, then assessed for quantitative amplification with qPCR. Each variation of PCR genotyping will be described below. Traditionally, PCR-based SNP genotyping methods are ideal for a small number of target SNPs in a large set of samples that need to be genotyped.
2.1. KASP Method
Kompetitive Allele Specific (KASP) PCR was originally developed as a method to detect SNP markers simply, rapidly, and economically [1]. The KASP method has been applied in applied sciences research many times for a variety of purposes [17,18,19,20]. The method functions basically as follows. The KASP method functions by introducing fluorescence resonance energy transfer (FRET) to cause signal generation. Two fluorescent cassettes are used to identify allele-specific amplification for a single bi-allelic SNP [1]. The primers are designed to flank the target region of interest. The first step in KASP is one round of PCR, where 2 allele-specific forward primers and one common reverse primer are used, and the first forward primer is fluorescently labelled with Fluorescein amidites (FAM) oligos while the second is labelled with Hexachlorofluorescein (HEX) oligos (FAM and HEX are fluorophores) [1]. The first forward primer (specific to variant) will have an allele sequence complementary to the SNP(s) of interest, while the second forward primer (specific to wild-type) will not be complementary to the SNP(s). In the DNA sample of interest, depending on which allele type is present (variant or wild-type), one of the forward primers will bind. When the sequences containing the fluorescent tag (HEX or FAM) are bound, they will no longer be quenched and will produce either type of fluorescence. Depending on which forward primer binds, either the FAM or HEX fluorescent signal will be produced, thereby signaling whether the SNP(s) of interest are present or not in a given target region in a DNA sample [1]. Amplification will only occur if the primer is fully complementary. Next, two additional rounds of PCR will be run to amplify the region of interest and generate fluorescence [1]. If the SNP of interest is homozygous, only one of the fluorescent signals will be generated; if the SNP is heterozygous, both signals will be generated [1]. In Alvarez-Fernandez et al. (2021), the KASP method was applied to identify malaria drug resistance in Plasmodium falciparum, the cause of most malaria cases in humans [1]. This particular example is not directly related to agriculture but does help demonstrate the utility of the KASP method. Alvarez-Fernandez et al. (2021) reported high accuracy, repeatability, and consistency between expected and actual results in the identification of malarial drug resistance in Plasmodium [1]. Additional example cases for the KASP method include Pradhan et al. (2023), where the KASP method was applied to detect genetic resistance traits to the Stem Rust fungus in wheat plants [18]. This group was able to identify novel resistance-associated loci by genotyping resistant wheat strains with the KASP method [18]. In Ilie et al. (2023), KASP genotyping was applied in Romanian cattle breeds to study cattle resistance to pathogens [20]. In that study, a specific SNP in the BOLA-DRB3 gene was identified as a suitable genetic marker for mastitis resistance [20]. Figure 1 shows the workflow for the KASP method assay to help visualize this method (Figure 1).
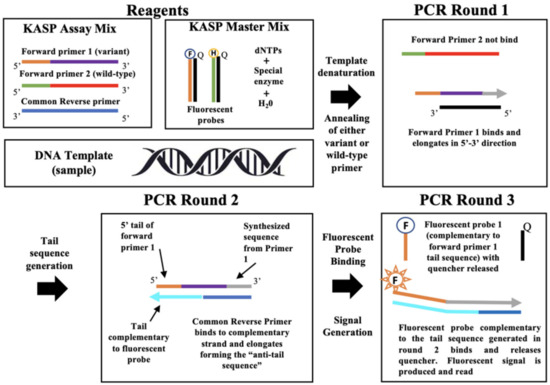
Figure 1.
Pipeline of KASP genotyping mode of action. Figure adapted from Alvarez-Fernandez et al. (2021) [1]. Shown from left to right are the reagents and workflow for the KASP genotyping method. The color purple corresponds to the primer for the variant of interest with the orange 5′ tail sequence complementary to the FAM containing fluorescent probe. The green/red color indicates the primer corresponding to the wild-type sequence of interest, where green is the 5′ tail, which is complementary to the HEX containing fluorescent probe. For each of the forward primers, the only difference is the SNP of interest. The common reverse primer is shown in blue. The gray arrows indicate the direction and location of synthesis during PCR. The black is template DNA containing the SNP of interest. Turquoise indicates the tail sequence generated during PCR step 2, which is complementary to the fluorescent probe F. The specialized enzyme is a Taq polymerize designed specifically for the KASP assay. The letters F, H, and Q are abbreviations for F (FAM fluorophore), H (HEX fluorophore), and Q (Quencher). The fluorophores do not fluoresce until the quencher is released during the 3rd round of PCR. One fluorophore is specific to the variant condition, while the other is specific to the wild-type. In the first round of PCR, the forward primer that binds will be elongated in the 5′ to 3′ direction. In the second round of PCR, the generation of the “anti-tail” sequence will occur, which is complementary to the 5′ tail of the forward primer that was elongated in PCR round 1. In the third round of PCR, the fluorescent probe will bind to its complementary “anti-tail” sequence, releasing the quencher. Fluorescence is then produced and read to determine the genotype.
The limiting factor with the KASP method is that only a few SNPs can be targeted in a single assay. Amplification will only occur in the regions that the two primer pairs flank. However, the KASP method is one of the most rapid, simple, and economical tests available. For projects that are in the process of characterizing SNPs for a new species, other methods could be used to narrow down overall SNPs in a genome, with a later return to a method like KASP once a small number of “key SNPs” have been identified.
2.2. PACE Method
The PCR Allele Competitive Extension (PACE) genotyping method, described in von Maydell (2023), is a recently developed method for qPCR-based SNP genotyping [21]. Though recently developed, this method has been applied in agricultural sciences before [22,23]. This method functions similarly to the KASP method described above. Two allele-specific forward primers with different fluorescent signals (also HEX and FAM) and one common reverse primer are used to target an SNP of interest. An advantage of the PACE method is that SNPs can be determined to be either homozygous or heterozygous; if one fluorescent signal is produced, the SNP is homozygous, and if both signals are produced then the SNP is heterozygous [21]. The main difference between this method and the KASP method is the type of reagent used for PCR. This method is manufactured by the Integrated DNA Technologies company who design primers and reagents for PACE genotyping reactions. The method is limited by the number of SNPs that it can target per assay (1–2); however, it is a cheap and easy genotyping method. Additional examples of the PACE method include that in Somyong et al. (2022), where the PACE method was applied to help identify genetic variation relative to oil palm height [23]. The authors report that in comparing SNP and indel variation in five different genes linked to oil palm height, only one of the genes had sufficient genotyping consistency to be considered suitable to be a marker for oil palm tree height [23].
2.3. TaqMan Method
One of the most common methods of SNP genotyping with qPCR is the TaqMan method [16,24]. This method remains one of the most commonly used in genotyping today [25,26,27]. This method is ideal for genotyping known polymorphisms that are allele specific in a given genome. The PCR primers used for TaqMan genotyping will flank a target region of interest, and therefore each assay can only target one single region per assay. One advantage of the TaqMan assay is that it can target multiple type variants, insertions/deletions, and presence/absence markers. The TaqMan method is ideal for genotyping projects with a small number of SNPs (1–50) and a large number of samples, since it is fast, cost-effective, and simple to perform [16,24]. The TaqMan method functions by using the 5′ nuclease activity of Taq polymerase. For each assay, there are two probes designed that are only different at the location of the SNP of interest. Like the KASP method, two forward primers (one variant, one wild-type) and one common reverse primer are designed. Depending on which primer hybridizes to a DNA sample of interest, a unique fluorescent signal will be produced [24]. The differences in fluorescence are then used to determine the genotype during the analysis step. This method is similar in design and function to KASP, only differing in reaction reagents and cost. The TaqMan method does have the limitation that it can only target a small number of SNPs in one given panel [16,24]. One option research groups could consider is constructing multiple different TaqMan assays and comparing the efficiency of multiple different panels. Comparing the TaqMan method to the KASP method reveals that these methods function quite similarly and have comparable rates of accuracy and precision [16]. The main difference is the type of reagent used for the actual qPCR reactions [16]. For example, the reaction mastermix solutions for KASP and TaqMan genotyping are different [16]. Additional example cases include Shumate et al. (2023), where the TaqMan method was applied to identify acaricide resistance in Tetranychus urticae [25]. In the study, the authors were able to accurately genotype Bifenazate, Bifenthrin, and Etoxazole resistance in the mites [25]. However, they note that certain populations of T. urticae had varying diversity of the SNPs, relative to these resistances [25]. They also note that one of the selected SNPs (G126S, a potential indicator of bifenazate resistance) was not an accurate identifier of resistance [25]. This outlines the importance of SNP selection in assay design, as not all SNPs will be relative to a research question or ubiquitous in a species. Another example is in Li et al. (2023), where TaqMan genotyping was applied to diagnose tembusu virus in ducks and astrovirus in geese [26]. In that study, the authors report success in designing an accurate diagnostic test for one or both viruses at the same time [26].
2.4. Open Array Method
The OpenArray Method is an SNP genotyping method manufactured by ThermoFischer Scientific (Waltham, MA, USA) [28,29]. The Open Array Method is designed for the high throughput genotyping of a moderate amount of SNPs (12–240 SNPs) using real time PCR, and it allows for combining multiple TaqMan method assays into one single experiment (the OpenArray plate) [28]. Up to 240 TaqMan assays and 480 samples can be screened per run with the OpenArray platform [29]. This method is an advanced real-time method using an array (plate) of 3000 through-holes on the special QuantStudio 12K (a real-time PCR machine) with an OpenArray Block (the genotyping array) [29]. The method functions basically as follows. An OpenArray plate is designed, using target SNPs of interest (up to about 240 SNPs). The Open Array plate is loaded with sample DNA and OpenArray Genotyping Master Mix, and an OpenArray AccuFill System is used to transfer the mix to the OpenArray Plate. Amplification is then performed using the QuantStudio 12K PCR system. Results are analyzed using the TaqMan Genotyper Software (v5.2), where each individual SNP targeted can be analyzed for call rate and concordance rate (concordance is the percentage of SNPs with identical results from OpenArray and sequencing) [28]. In Ragazzo et al. (2021), this platform was used to target 60 different SNPs pertinent to eye color in humans [28]. The authors reported a successful call rate of 96.9% [28]. Research groups would need to purchase the special QuantStudio 12K real-time PCR machine, as the OpenArray plates are specifically designed for this machine. This method is advantageous, as well, in that the analysis pipeline is simple and no bioinformatics are required. Additional example use cases in agricultural sciences include in Noce et al. (2016), where the OpenArray method was applied to analyze milk protein gene regulation in sheep [30]. The authors were able to successfully identify a candidate gene (CSN1S1) as suitable for marker-assisted selection in sheep breeding for desirable milk traits [30]. Another example is in Chagne et al. (2019), where the OpenArray method was applied to validate fruit quality and disease resistance markers in apples [31]. The authors were able to validate 33 SNP markers for use in apple breeding, including those associated with scab resistances, fire blight resistance, powdery mildew resistance, fruit firmness, skin color, flavor intensity, and acidity [31].
2.5. rhAMP SNP Genotyping
The rhAmp SNP genotyping method is a recently developed method based on RNase H2-dependent PCR (rhPCR) [32]. rhAMP genotyping has been applied in the following additional uses for agricultural research [33,34]. This method provides a high level of signal (for the actual qPCR step) and high specificity for the SNP analysis, and it functions basically as follows. Similar to other qPCR-based SNP genotyping methods, this functions with two different forward primers (one containing the target SNP and one not) and one common reverse primer. The rhAmp system works by combining two unique enzymes with special 3′ end blocked DNA-RNA hybrid primers to identify SNP loci [32]. When the blocked primers hybridize, they perfectly match the target template DNA, thereby reducing the chance for primer dimers [32]. The hot start Rnase H2 (rh) cleaves the primer at the 5′ end of the ribose sugar, which releases the blocking group and allows for primer extension. A reporter signal is generated by using two different reporter probes, one for the first forward primer (variant) and one for the second forward primer (wild-type), and a common reverse primer is used [32]. Depending on the genotype of the DNA sample, differing fluorescence is produced and read to determine the genotype. Using this method, Beltz et al. (2018) were able to achieve a 98% call rate with 99% accuracy [32]. This method is also compatible with multiple different qPCR instruments and has a high level of success with small amounts of template DNA [32]. This method is supplied by the Integrated DNA Technologies company. The method is highly accurate and repeatable, simple, and easy to interpret. However, it is limited in the number of SNPs (1–50) that it is able to target in a single given assay. One possibility is designing multiple rhAmp assays to target many different SNPs; however, the feasibility of this would depend on the cost for designing each individual assay. One study compared KASP, TaqMan, and rhAMP to each other, since they are very similar. It was reported that of the three, rhAMP was slightly cheaper and had a higher rate of success during amplification [16]. In Esposito et al. (2022), the rhAMP method and also the High Resolution Melting method were applied to examine the stem thickness in wheat [33]. The authors note that both genotyping methods were able to identify the genetic marker they used, and the variation in success was due to differences in the samples [33]. In Giglioti et al. (2020), the rhAmp method was applied to differentiate cow milk types associated with gastrointestinal effects [34]. The authors report that the rhAmp method was more sensitive for SNP detection in comparison to high resolution melting, though both methods were able to differentiate milk types [34].
2.6. Amplifluor-Based SNP Genotyping
This method, described in Jatayev et al. (2017) and Mohanrao et al. (2023), is a method of allele-specific PCR that is used for SNP genotyping [7,35]. This method has been applied for agricultural research before [36,37]. The method functions basically as follows. Similar in principle to KASP and TaqMan genotyping, this method differs in that it uses special probes and reagents that differ from KASP and TaqMan reagents; it is also reported to be cheaper and easier to design [7]. The Amplifluor method functions by using special universal probes and gene-specific primers which correspond to the SNPs of interest. The universal probes differ from KASP probes in structure, and they also contain fluorophores (such as FAM). The gene-specific primers (which contain the target SNPs) are designed to possess 3′ tails that correspond to the universal probe sequence (visualized in Figure 2). During PCR, the universal probe will bind to the gene specific primers and fluoresce if the amplification of the target SNP occurs [7]. Following primer design, which Jatayev et al. (2017) outsourced to a primer synthesizing company, PCR was conducted using standard reagents and conditions and the analysis was performed with the melt curves produced [7]. An advantage here is that beyond the special universal probes, the PCR mastermix is generic, unlike KASP and TaqMan, which require specific mastermixes [7]. Figure 2 visualizes the universal probes described above. The universal probe complementary to the forward primer that binds during PCR will produce either type of fluorescence indicating genotype in a sample (Figure 2).
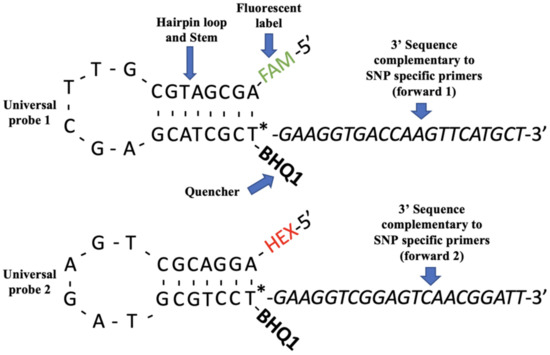
Figure 2.
Universal probe design for the amplifluor-like method. Adapted from Jatayev et al. (2017) [7]. The figure visualizes the structure of the universal probes used in the amplifluor-like genotyping method. Universal probe 1 contains a hairpin loop and stem, a fluorescent label (FAM), a quencher, and a 3′ tail specific to the PCR primers specific to the SNPs of interest. Universal probe 2 contains the same, with a different fluorescent label (HEX) and a 3′ tail specific to the second forward primer. The colors green and red for the FAM and HEX fluorescent labels are simply to highlight the labels location in the universal probe. The asterisk indicates the location in the probe where the quencher (black hole quencher (BHQ1)) is attached. This quencher absorbs at maximum wavelengths for all fluorophores and fluorescence until the quencher is released. Sequence variations in the universal probes are part of the probe design to optimize the efficiency of the probe for the different fluorophores and 3′ sequences. The procedure for amplifluor-like genotyping is similar to other PCR-based genotyping methods, and what is unique is the structure of the probes.
This method appears to be a cheap, accurate method of genotyping a small number of SNPs at a large scale. While it is reportedly very cheap and easy to design and use, it is still limited by the number of SNPs it can target in a given assay [36,37]. In Shavrukov et al. (2016), the amplifluor method was applied to study gene expression differences relative to drought resistance in wheat [37]. The authors were able to accurately identify a protein related to drought resistance, and also the SNPs that they examined did not change the protein amino acid sequence [37]. Therefore, they hypothesize that in that case SNPs were more relative to gene regulation than protein changes [37].
2.7. Variable Fragment Length Allele Specific PCR (VFLASP)
Variable Fragment Length Allele Specific PCR (VFLASP) is a method of PCR SNP genotyping developed by Toth et al. (2023) [38]. This method is another variation on allele-specific qPCR genotyping. It functions similarly to KASP and TaqMan genotyping, in that PCR amplification is used for genotyping. However, in the VFLASP method, the variation in amplicon length determines the genotype, rather than the type of fluorescent signal produced [38]. Genotyping is conducted by analyzing differences in DNA fragments on an agarose gel. The forward and reverse primers are designed specifically for a VFLASP genotyping project. The sequence of the forward primers will differ by three bases [38]. With the VFLASP method, instead of using a FRET dye-based system like the KASP method, one dye (FAM) is used, and each of the allele-specific primers will produce a different length amplicon. For example, there is one common reverse primer and two forward primers, one containing the SNP of interest, with 29 bases in length, and the other forward primer not containing the SNP of interest and being 26 bases long [38]. After PCR is complete and a gel has been run, the genotype can be determined by analyzing which primer is amplified based on the fragment length read on the gel [38]. Figure 3 depicts the primers designed for the VFLASP method to help visualize these specialized primers (Figure 3).

Figure 3.
Variable length primers used for the VFLASP genotyping method. Adapted from Toth et al. (2023) [38]. Two forward primers of different lengths and one reverse primer sequence are shown. The two forward primers vary at the last base at the 3′ end, which is where the SNP of interest is located. Turquoise indicates a PIGtail adapter added to the 5’ end of the 2 forward primers to increase PCR specifity. Purple is a short adapter added to increase the primer length for the 2nd forward primer. Green indicates an M13 adapter added to the common reverse primer. PCR results will yield different fragment lengths on a gel depending on genotype.
The advantage with this VFLASP method is that the genotyping results are very easy to interpret; simple gel electrophoresis can be used to interpret results. However, this assay is limited once again in the number of SNPs (1) that are targetable with a single assay. This method only has one use case, by Toth et al. (2023), who validated this method in 2023 in human models [38]. However, the authors report reliable success and a high ease of analysis [38].
2.8. High Resolution Melting SNP Genotyping
High Resolution Melting is a genotyping method that uses DNA melt curves to identify the genotype of a sample [39]. This method has been applied several times before in agricultural sciences and functions basically as follows [33,40,41]. This method functions by multiplexing multiple primers specific to target genes, conducting real-time PCR, and interpreting melt curve results. A single reaction mix can be made to conduct this experiment, containing Master Mix, LC green dye, water, primer mix, and DNA [39]. PCR is then conducted on 384 well plates and the melt curve is read to determine the genotype [39]. In Slomka et al. (2017), 10 different genes were targeted as well as their SNP containing variants [39]. In that particular experiment, direct sequencing was used to validate the real-time PCR results [39]. Slomka et al. (2017) provide a good review of troubleshooting methods for High Resolution Melting Genotyping, and note that optimizing primer (sensitivity to primer length) and melt curve efficacy can be difficult [39]. The authors also note that variations in template DNA (for example, due to the extraction method) can produce variations in results [39]. This method does have the capability to rapidly identify SNPs, and the primer design and experimental design appear to be quite easy. An issue with this methodology is with the interpretation of results. In theory, multiplexing a high number of target SNPs is possible; however, a higher number of targets included on a single melt curve becomes rapidly difficult to interpret. Therefore, this genotyping method is only suitable for a small number of target SNPs (1–5). Additional example cases of this method include in Chou et al. (2020), where the High Resolution Melting Analysis method was applied to study chilling-requirement-associated markers in peaches to assist with breeding selection [40]. The authors report that this method was successful for a low-cost, high-throughput genotyping method that does not require a gel [40].
2.9. MassArray SNP Genotyping System
MassArray SNP genotyping is a method of genotyping that combines multiplex PCR with mass spectrometry to genotype SNPs [2,42]. This method has been applied in agricultural sciences before and functions basically as follows [43,44,45]. The method functions by designing multiple primers specific to target SNPs, performing multiplex PCR to amplify a DNA sample, and then conducting MALDI-TOF mass spectrometry to genotype the samples [2]. In Shah et al. (2020), this method was used to identify 12 different SNPs pertinent to esophageal cancer [2]. A DNA sample is multiplexed with PCR primer mixes and standard reagents, and following amplification, a second round of PCR is performed with shrimp alkaline phosphatase and modified ddNTPs and pooled single extension primers (adding one single base to the first PCR products) [2]. The final PCR product is then analyzed with an Agena MASSarray mass spectrometer. When the second PCR product has been read by the mass spectrometer, the molecular weight of each PCR product is measured, which indicates the genotype [2]. This method of genotyping is different from the other PCR-based genotyping methods, in that fluorescence or gels are not used to genotype. This method is limited by the number of SNPs and special equipment requirements (Agena MASSarray platform). However, it is overall a rapid, accurate genotyping method [43,44,45]. In Zhao et al. (2023), to identify temperature resistance genes in abalone fish [44], the authors report repeatable and accurate genotyping for seven different SNPs that were determined to be relevant to heat resistance [44]. In Ji et al. (2023), the MassArray method was applied to identify candidate marker genes in sheep associated with litter size [45]. The authors were able to identify three suitable candidate genes that are viable for marker-assisted selection in sheep litter size [45].
2.10. PCR-Based Genotyping Conclusions
PCR-based SNP genotyping methods are by far the most simple and cost-effective methods for SNP genotyping. For these reasons, they are the most accessible methods for SNP genotyping available when considering equipment and reagent needs (PCR machines, PCR reagents), the complexity of performing the genotyping assays, and the interpretation of results. The major limiting factor with these types of technologies is the number of SNPs that can be targeted in a single given assay. One possibility for species with a large number of uncharacterized SNPs is to use either computational algorithms (some options are described below), sequencing, or microarrays to identify a core set of SNPs that are key to the species of interest, and then to use those results to design a PCR-based genotyping method for use en masse.
3. Microarray SNP Genotyping
The second class of technology that can be used for SNP genotyping is DNA microarrays. Microarray-based SNP genotyping methods are ideal for analyzing a large number of SNPs in a large number of overall samples. For large scale projects, these methods have proved suitable many times [46,47,48,49,50]. Typically, microarray-based SNP genotyping is used when studies require the analysis of SNPs across an entire genome. This method works by constructing a synthetic chip with thousands of probes (each probe is specific to a target SNP) attached to the chip. If a target SNP is present in a given DNA sample, hybridization at the probe will occur [51,52]. The plate is then read by a plate reader to determine whether hybridization occurred at each individual probe. Many studies use this type of technology for a variety of purposes, including medical research, agricultural research, breeding, and more [53,54,55]. Due to the nature of this technology, microarray chips must be designed by working with one of the major manufactures, such as Illumina (San Diego, CA, USA) or Affymetrix (Santa Clara, CA, USA).
3.1. SNP Arrays
There are two major manufacturers that produce microarray-based SNP genotyping technologies. These are the Illumina Infinium and Affymetrix Axiom platforms, which function similarly [56]. These platforms function basically as follows: first, the chip is designed with either Illumina or Affymetrix using the pre-identified SNPs and reference genome data. This step can be difficult due to trouble with selecting important SNPs. It is possible to take a “shotgun” approach, but some SNPs on the chip may be more informative than others and once designed the chip cannot be changed. The microarray chip is designed with up to millions of different “BeadChips”, which are microscopic beads containing multiple probes, one complementary to the wild-type and one to the variant form in an SNP-containing region of interest. After chip design, the DNA sample of interest is amplified non-specifically by PCR to produce ample copies of sample DNA. The amplified DNA sample is then incubated and enzymatically fragmented to prepare for hybridization. Fragmented DNA is then precipitated and resuspended (to concentrate the sample) and introduced to the flow-through cells on the microarray (the actual chip) and incubated overnight while probe hybridization occurs. Lastly, the hybridized probes are extended enzymatically and fluorescently stained with either red or green stains. Each individual “BeadChip” within a microarray chip has two channels, one for the wild-type probes and one for the variant. The fluorescent stain will only stain the channels where extension has occurred. The DNA array scanner will then indicate the presence or absence of hybridization via a difference in stain color and intensity. Depending on the fluorescence produced, the presence or absence of millions of different SNPs can be analyzed in a sample simultaneously. Data output (stain intensities which correlate to hybridization) is then read with Genome Studio software (v2.05) [56]. This particular software was designed only for the Illumina microarray platforms. The Affymetrix platforms use a different SNP calling software called Axiom Analysis Suite (v4.03). Figure 4 helps to visualize the workflow for microarray-based genotyping, and each major step is depicted (Figure 4). The SNP calling step can be performed by using the GenomeStudio software (v2.05) and the intensity data file and SNP information file produced from the DNA microarray scanner [56]. The calling involves normalization, sample grouping, quality control, and SNP filtering [56]. In Vervalle et al. (2022), a microarray chip was applied to conduct wine grapevine population mapping [46]. Here, the authors were able successfully to map 6697 SNP markers relative to different varieties of grapevine [46]. They note that discrepancies occurred in 88 of the target markers, primarily due to locus proximity [46]. This outlines the importance of SNP loci during microarray design. In Balog et al. (2023), microarray genotyping was applied to study the efficacy of a commercial microarray chip designed for domestic chicken research in pigeon breeding [47]. They report a low call rate due to species difference, but were able to identify 356 conserved SNPs in pigeons using the chicken microarray chip [47]. In Singh et al. (2023), microarray genotyping was applied to assist in heat tolerance breeding in rice [48]. The authors were able to successfully identify four robust candidate markers for heat tolerance in rice [48].
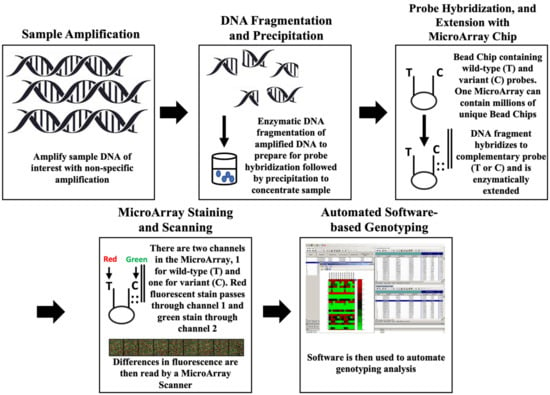
Figure 4.
Workflow for microarray genotyping assay. Adapted from the workflow for Illumina Infinium Assay (Illumina Inc., San Diego, CA, USA, 2023). From left to right, each major step of microarray genotyping is outlined for visualization. The non-specific amplification of sample DNA in step 1 is conducted via PCR. DNA fragmentation is achieved using enzymatic digestion, followed by propanol precipitation to concentrate the fragmented DNA. Probe hybridization occurs when a DNA fragment from step 2 becomes hybridized to either the wild-type or variant probe attached to the BeadChip, and this will depend on the genotype (and therefore which probe it is complementary to) of the original DNA sample. Enzymatic extension (with TEM reagents) involves adding single bases to the probes on the BeadChip that have been successfully hybridized, and this will allow for staining in step 4. The channel color is arbitrary; it is only important that all wild-type probes are in the same channel while all variant probes in the microarray panel are also in the same color channel. Staining involves introducing a series of different fluorescent stains that will produce different color intensities which indicate to the plate reader which BeadChips were hybridized. Analyzing which color intensities occurred at each BeadChip (and therefore which probe was hybridized) will determine genotype.
3.2. Microarray-Based Genotyping Conclusions
This method is compatible with large-scale genotyping projects; up to millions of genome wide SNPs can be targeted simultaneously, and the technology is also readily available commercially. The workflow and analysis are also relatively simple in comparison to other methods due to the analysis being facilitated by array genotyping software. However, these methods would require microarray scanners, which can be quite expensive. An issue with this method is that Illumina (iScan) and Affymetrix market specific microarray scanners to be compatible with the chips they design, which can be quite expensive. However, after the high initial cost, genotyping at a huge scale (up to 2.5 million SNPs can be targeted in one DNA sample) can be performed rather simply. No other genotyping methods, apart from whole genome sequencing, are able to target such a high number of SNPs simultaneously. Another limitation is that once the array panels are designed, they cannot be changed, limiting experimental flexibility.
4. Next-Generation Sequencing of SNP Genotyping
SNP genotyping by NGS is the most recently developed class of SNP genotyping methods. Traditionally, whole genome sequencing was used to identify SNPs present in a given genome [57]. However, over the years, sequencing technologies have become cheaper and more precise, in that they can also be used in some cases for the genotyping process itself (targeted sequencing). However, the monetary and time costs of this method for SNP genotyping must be considered in each individual project. Traditionally, genotyping by sequencing methods is ideal for genotyping a medium to large number of SNPs in a moderately sized sample with high accuracy. The below methods function similarly, in that multiplexed PCR is used to amplify target SNPs in a DNA sample, and the amplicons are indexed and then sequenced. For this reason, the following sections focus more specifically on the described example cases and how each group selected SNPs, designed primer pools, ran multiplex PCR, and performed sequencing. Variations of these methodologies have been applied in agricultural sciences many times [58,59,60,61,62,63,64]. Below are four different pipelines developed for targeted sequencing genotyping. The methods described below differ in reagents (and manufacturer), sequencing platform, the number of targetable SNPs, the approach to target SNP selection, and cost. Figure 5 shows the general workflow for genotyping using NGS (Figure 5).
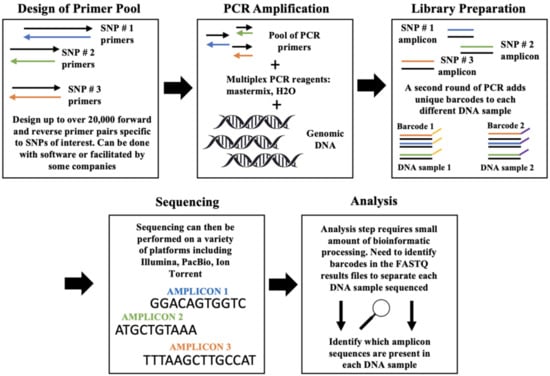
Figure 5.
Simplified workflow for genotyping by next generation sequencing. Left to right, the workflow is outlined for the basics of each step in targeted sequencing-based genotyping. The pool of primers consists of up to over 20,000 different forward and reverse primer pairs, and each pair will flank a SNP of interest. The blue-, green-, and orange-colored arrows, lines, and words each correspond to a different target SNP included in the design of an assay. The yellow and purple lines in window #3 indicate barcode indices. Multiplex PCR is then performed with genomic DNA samples and the primer pool. Each different DNA sample must then be barcoded in a second round of PCR, in order to differentiate each DNA sample that will be sequenced. This allows for the pooling of multiple different DNA samples to be screened for SNPs at one time. Sequencing will produce raw reads for all primers in the primer pool that were successfully amplified in each DNA sample that has been tested. Sorting the raw reads for barcode type and amplicon sequence will identify which DNA samples and which target SNPs were amplified in a given sample.
4.1. Target SNP-Seq
A recently developed type of NGS-based SNP-genotyping is called Target SNP-Seq. In Zhang et al. (2020), it was developed for cucumber genotyping (where over 4 million SNPs had been identified), and this method aimed to combine the advantages of multiplex PCR and high-throughput sequencing by yielding a high accuracy for a large number of samples and genome-wide SNPs, while still maintaining cost efficiency [10]. The method was developed in response to other high-throughput SNP genotyping methods, such as gene chip microarrays or the KASP method, because they can be quite expensive to perform in the laboratory and can sometimes produce false positives or negatives [10]. Target SNP-seq can target between 100 and 2000 SNPs with high accuracy and low cost [10]. For the project of Zhang et al. (2020) a core set of SNPs (out of >4 million SNPs total) for cucumber were selected using the MinimalMarker method described in [10,65]. This is a Perl-based computer program which can be used to identify a core set of SNPs from a larger number of genomes for a particular species or variant [65]. In this study, 298 “perfect” SNPs found across the genome were selected for the identification of cucumber variants. This method functions by conducting two rounds of PCR for target SNP-seq library construction. For the first round, the target SNP locus is captured by PCR-amplifying 200–280 base pair sequences containing the SNPs of interest in the DNA samples [10]. The second round of PCR distinguishes each DNA sample by adding a unique barcode adaptor, which is the library preparation step prior to sequencing [10]. This step allows for multiple separate DNA samples to be sequenced at one time, increasing the overall through-put. The DNA library is then sequenced using the Illumina High-Seq platform. The reagents used in this pipeline are suitable for Illumina sequencing; however, there are other similar methods described below that are compatible with other sequencing platforms. The raw reads produced from sequencing are then analyzed for the specific barcodes added in the second PCR step to identify the SNP genotype of interest [10]. For the purposes of the original study, the SNP genotyping accuracy was reported as 98.7% with a low cost of only 3 days and 7 USD per DNA sample [10].
For research groups with a large set of SNPs, one option with this method is to use the MinimalMarker method to identify a core set of SNPs. Then, between 100 and 2000 of these core SNPs are selected to use for designing the SNP PCR primers. This requires specialized PCR reagents and SNP primer design; Zhang et al. (2020) used Molbreeding Biotechnology Company (Shijiazhuang, China) to purchase reagents and design the multiplex PCR SNP primer mix [10]. However, there are many companies around the globe that facilitate similar purchases.
4.2. Genotyping by Target Sequencing (GBTS)
This genotyping pipeline is a modified method for target sequencing to genotype samples of interest with a large number of target SNPs. In this method, target sequencing is combined with a “capture in solution” or a liquid chip to capture multiple SNPs with one amplicon [4]. This method is a modification of the method previously described in Xu et al. (2018) [66], where 55,000 SNPs were selected from the total SNPs in corn by first randomly selecting 30,000 genome-wide SNPs within at least 100 kb of each other, and then 25,000 other SNPs based on other genotyping panels already developed for corn were added to the random 30 K to make a 55 K SNP panel microarray [66]. Guo et al. (2019) used and improved this panel from the 55,000 SNPs designed in Xu et al. (2017). Guo et al. (2019) selected 24,000 SNP loci from the previous 55,000 SNP panel, designing probes of 120 nt in length to cover each SNP loci [4].
The probes were synthesized using semiconductor-based in situ synthesis [4]. From the 24,000 SNP panel, a 20 K SNP, a 10 K SNP, a 5 K SNP, and a 1 K SNP panel were next constructed one after another by narrowing down the number of SNPs to make a “core set”. The core set was determined by performing the genotyping by sequencing with the 24 K panel, then ranking SNP loci by average missing rate per locus and average sequencing depth, then removing 4000 of the lowest rank, then selecting those SNPs left that were uniformly distributed. This was repeated with the 20 K panel, the 10 K, and the 5 K until a core set of 1000 SNPs were left [4]. After SNP selection and probe design, the DNA samples of interest must be processed into a DNA library with the following sequence of steps: DNA fragmentation by ultrasound, end repair and poly-A tailing, adapter ligation, and library amplification with barcoded primers [4]. Probes are hybridized by mixing the DNA libraries with special blockers, then using PCR amplification with the probes and hybridization buffer [4]. Target capture is performed by adding Streptavidin C1 and binding buffer to select only the DNA fragments that have been hybridized with SNP probes, and all other fragments in the library are digested with a special mutant enzyme. The library is then amplified using PCR, purified with AMPure beads and sequenced with Illumina HiSeq [4]. The analysis is performed by mapping reads to the reference genome for corn, then converting the alignments to BAM files and using the FreeBayes program to identify SNP variants [4]. This procedure was used for each of the 55 K, 24 K, 20 K, 10 K, 5 K, and 1 K panels. An average SNP call rate of between 93 and 98% accuracy was reported for the panels [4]. An issue with this method is that selecting the core set of SNPs requires several steps of panel design and sequencing in order to narrow down the overall number of SNPs wanted as targets. Since they created their panels in-house using semiconductor in situ synthesis, this is more time- and cost-effective. Using a company to outsource these steps would likely be expensive and time consuming. However, the actual genotyping step appears to be suitable for genotyping a large number of SNPs with high accuracy. This pipeline also appears to be quite labor intensive and complicated. While suitable for accurately targeting a large number of genome-wide SNPs, the accessibility of this method seems low.
4.3. AmpliSeq
A genotyping method using NGS is described in Sato et al. (2019), consisting of a modified method for the amplicon-sequencing (AmpliSeq) platform [9]. This method was developed in Sato et al. (2019) for aquaculture for the purpose of economically targeting 3000 genome wide SNPs [9]. For this particular protocol, to construct an Ampliseq primer panel, first selected 3000 SNPs from a pool of 3,232,903 SNPs identified in the genome of pufferfish (Fugu rubripes) [9]. In order to select SNPs, the authors filtered the total number of SNPs for their species of interest by excluding SNPs near known repeat elements in the reference sequence, and excluding SNPs with minor allele frequency less than 0.2, low average read depth, and those SNPs near homo-nucleotides or microsatellite regions [9]. A total of 3000 SNPs with 20,000 bp of space between neighboring SNPs were then selected. The custom AmpliSeq panel targeting each of the 3000 different SNP loci was designed using the Ion AmpliSeq Designer [9]. PCR was then performed using genomic DNA, multiplex PCR reagents, and the custom primer pool (the price of the primer pool will vary depending on how many primer pairs are in the pool) [9]. Library preparation was then conducted with the ion ampliseq library kit on the PCR products, where each sample was barcoded and subsequently sequenced using the Illumina MiSeq platform [9]. The resulting FASTQ files were then quality trimmed and mapped with SAMtools [9]. The genotyping itself was carried out using the GATK program to identify barcoding [9]. Ampliseq genotyping kits are also compatible with either Ion Torrent or Illumina NGS platforms.
This method is suitable for large-scale genotyping projects. Thousands of genome-wide SNPs are targetable with high accuracy (99% of the SNPs targeted in the Sato et al. (2019) custom panel were detected) [9]. A drawback of this method is that it does require a few steps of bioinformatic analysis in the last phase of genotyping, which can be a barrier for some research groups. Reagents for this method are also readily available, and manufacturers quote up to 24,000 primer pairs able to be multiplexed with the Ampliseq method.
4.4. MTA-Seq
Multiplex PCR Targeted Amplicon Sequencing (MTA-Seq) is a method described in Onda et al. (2018) [67]. In this method, multiplex PCR targeted amplicon sequencing, or MTA-Seq, was developed as a means for high throughput genotyping by sequencing. The method allows for targeted genotyping with a high number of primer pairs (in a multiplex) that are specific to target SNPs. Following multiplexed PCR, sequencing is required to identify which primer pairs are amplified in the template DNA [67]. In Onda et al. (2018), this method was performed by first designing primers using Primer3: each primer targeted a 150–200 bp amplicon specific to one SNP locus and they were 18–32 bases in length [67]. A total of 443 primer pairs were designed to target 443 individual SNPs in the sample DNA [67]. However, the genotyping of more than 400 SNP markers is possible with this method [67]. Next, multiplex PCR was performed using a multiplex PCR kit, and PCR products were then prepared for sequencing with library preparation kits for Ion Torrent [67]. The DNA library then had to be amplified and purified using PrimeStar GXL and AMPure kits [67]. Sequencing was then performed on the Ion Proton platform, and the resulting reads were analyzed via SNP calling with VarScan Software (v2.4.5) [67]. Using this pipeline, 95% of the target 443 SNPs were adequately amplified and called from the nine different DNA samples that were tested using the MTA-Seq pipeline [67]. This method allows for high throughput genotyping at relatively low cost; however, the end step does require bioinformatic assembly and analysis after sequencing is complete, which is a barrier to the genotype interpretation. However, a large number of SNPs can be targeted using this method. This method can also be sequenced on the Ion Torrent or Illumina sequencing platforms, allowing flexibility depending on availability for individual research groups.
4.5. Next-Generation Sequencing-Based Genotyping Conclusions
NGS-based methods for SNP genotyping are a good fit for projects where there are a moderate amount of target SNPs (500–24,000). These methods have the advantage of being highly accurate and also being able to target a high number of SNPs. They are also flexible, in that primer pairs can be added or removed after an initial primer set is designed. Equipment and reagent needs include DNA extraction equipment and PCR equipment and reagents. Most laboratories will not be able to conduct the sequencing themselves and will need to outsource to a sequencing company, which adds to the overall time and monetary cost. Library preparation is also a required part of these protocols, which could be carried out at a sequencing lab or “in-house” depending on the capabilities and needs of each lab. In some situations, it may be more advantageous for some groups to outsource the steps of library preparation and sequencing. A small amount of analysis and bioinformatics would be required in-lab to interpret results [58,59,60,61,62,63,64]. Additional example use-cases of sequencing-based genotyping include the following. In Sekine et al. (2022), amplicon sequencing was applied to compile a genome-wide dataset of markers that would be useful for onion agriculture and marker-assisted breeding [58]. The authors selected 480 different markers which could later be used to select for desirable traits in onions, and [58] report that amplicon sequencing provided cost-effective, reliable, and flexible genotyping [58]. In Lee et al. (2023), sequencing-based genotyping was applied in elk to identify SNPs suitable to diagnose a susceptibility to chronic wasting disease [64]. The authors report success in identifying a new SNP marker suitable for this disease [64]. In Gashururu et al. (2023), sequencing-based genotyping was applied in Tsetse flies to screen for endosymbionts associated with disease [63]. The authors were able to link an endosymbiont presence in the flies with specific host species (buffalo) [63]. This result outlines the potential for genotyping in epidemiological studies, where disease origins or reservoirs are difficult to identify.
4.6. Potential for MinION in SNP Genotyping to Improve Sequencing-Based Methods
One of the drawbacks of NGS-based genotyping methods is that most laboratories are not equipped to perform the sequencing step of genotyping with these methods. Therefore, most groups must outsource to a sequencing company to perform the sequencing. This does have the benefit that most labs are able to outsource the DNA library preparation or sequencing steps of these genotyping methods if, for example, they do not have the time or resources to perform this in their own labs. However, it does involve a significant time lag, which can be problematic for some projects that require rapid results. An optional improvement to a sequencing-based genotyping pipeline is a MinION sequencing platform. A way to rapidly sequence “in-house” would be with MinION (Oxford NanoPore Technologies, Oxford, UK). This platform allows for affordable sequencing without the need for outsourcing to a sequencing company. The platform is a relatively small, portable and affordable sequencer which can easily be installed or moved from lab to lab. The platform uses nanopore sequencing, which functions differently from Illumina sequencing, where nucleotides or DNA strands are read sequentially through a pore. The end result is the same, where genotyping data can be analyzed via an analysis of resulting fastq files after sequencing. This has huge potential in that a large number of SNPs can be targeted quickly and economically in one laboratory. Oftentimes, the results of SNP genotyping panels are needed as quickly as possible in order to make important decisions. For example, in plant and animal breeding or hybridization and cryptic species detection, oftentimes there is a short window of time to receive genotyping results.
The MinION has been applied for SNP genotyping applications several times before [68,69,70,71,72,73,74]. However, it must be noted that, in comparison to other NGS platforms, the MinION can produce both higher rate of errors and higher length reads [68,69]. In comparing MinION to Ion Torrent sequencing, Singh and Bhatia (2020) report that the MinION produced significantly longer reads (~100–7000 bp reads vs. ~25–800 bp reads), though the MinION error rate was much higher (20% vs. 1.5%) [68]. However, the authors conclude that the research objective of microbial species identification was still accomplished using MinION [68]. In comparing MinION to Illumina sequencing, Nygaard et al. (2020) also reported a longer read and higher error rate using MinION in comparison to Illumina MiSeq [69]. However, the authors also state that they were able identify microbial species with the MinION sequencing results [69]. In Cornelis et al. (2017), MinION was used to sequence a multiplex of 52 different target SNPs in forensic genotyping [74]. The authors reported that several different SNP loci were problematic to identify using MinION, but the majority were successfully genotyped, indicating that target SNP selection must be carried out carefully when using MinION to augment genotyping via a sequencing pipeline [74]. In Tabata et al. (2022), MinION was used to compare the SNP loci detection frequency between MinION and conventional dideoxy sequencing for six different SNP loci, finding consistent results between the two types of sequencing [73]. In Ren at al. (2021), 94 different target SNP loci were successfully genotyped to 99% overall accuracy [71]. In applying MinION for SNP genotyping, it is important to take into account the problems that may occur in targeting some SNP loci [70,71,72,73,74]. Therefore, one possible use for MinION in SNP genotyping would be to improve on a set of characterized SNPs that had been successfully genotyped before using traditional sequencing-based genotyping methods; MinION can be used to improve and streamline these existing pipelines. Each individual research group must weigh the monetary cost, time requirements, level of SNP characterization, and error rates when choosing a genotyping by sequencing method.
5. Discussion
The above information provides an overview of 14 different genotyping technologies. All of the technologies have their pros and cons, and are more suitable for certain niches in comparison to others. PCR-based methods are simple, cheap, and easy to design and perform and can be performed en masse. However, they are limited in the number of SNP markers that they can target (about 1–200, depending on the method). Additionally, PCR-based methods must have pre-identified SNPs for genotyping. For projects where SNPs have not been well characterized for conditions of interest, the use of PCR-based methods may be difficult. However, PCR-based genotyping methods are the most effective for projects where the species of interest has well-characterized SNPs. The degree of SNP characterization is important in assay design, and the success or failure (and accuracy) of a genotyping test is reliant on which SNPs are included in a given assay. For example, SNPs with low allele frequencies or from unknown genomic regions are not appropriate for genotyping assays [9]. Microarray technologies are a suitable method for targeting up to 2.5 million SNP markers from one DNA sample, and they are also relatively simple to perform and interpret. Their design is also facilitated by well-established companies like Illumina or Affymetrix. However, these technologies have a high initial cost in reagent design and equipment requirements. The high initial cost is a barrier; however, the ability to target millions of genome-wide SNPs in a genotyping project is most accessible with microarrays. The NGS-based approaches are able to accurately target a large number of SNPs (500–24,000); however, the pipelines for this type of genotyping are more complex and difficult to perform/analyze in comparison to the PCR and microarray-based methods. There is also the added complexity of sequencing and bioinformatics for these types of genotyping methods, which adds to the time requirement for genotyping. However, NGS-based genotyping is highly accurate, flexible, and affordable. Research groups interested in performing SNP genotyping must consider many variables during method selection: the level of characterization of SNPs in the species of interest, budget, time cost, labor cost, experimental accuracy, equipment needs, and the skill sets of lab personnel. Table 1 provides a summary of the advantages and disadvantages for each technology in this review and also includes referenced use cases where each method has been applied in practice (Table 1). Included in Table 1 is an estimate of experimental accuracy for each assay, as reported from the included use case references. However, it is important to note that accuracy will be heavily reliant on which SNPs are targeted in a given species, rather than the assay itself. For example, a certain SNP that is highly conserved in a species will yield more accurate results than a target SNP with low conservation due to the characteristics of the SNP rather than the functionality of the assay. Also, the accuracy of an identity marker in determining species is different from its accuracy in determining whether a particular SNP is present. This highlights the importance of careful SNP selection during the assay design process.

Table 1.
Conclusions for 14 genotyping technologies and use cases.
6. Conclusions
This article covers 14 different methodologies of SNP genotyping; however, other methods have been developed and may be more suitable for specific niche use-cases. Many of the methods described above have been developed recently. The category of NGS-based genotyping is the most “modern” class of genotyping. However, PCR-based methods are still being developed in 2023, showing that all types of genotyping can be further improved. In the future, SNP genotyping will continue to be an effective method in applied biosciences.
Author Contributions
R.D.L. conducted the review and constructed figures to help visualize workflows. R.D.L. and S.E.M. prepared and edited the review manuscript for publication. Conceptualization, R.D.L. and S.E.M.; investigation, R.D.L.; resources, S.E.M.; writing—original draft preparation, R.D.L.; writing—review and editing, R.D.L. and S.E.M.; visualization, R.D.L.; supervision, S.E.M.; project administration, S.E.M.; funding acquisition, S.E.M. All authors have read and agreed to the published version of the manuscript.
Funding
The authors are funded by a USDA-APHIS grant #AP21PPQ&T00C069. This grant also provided funds for publication costs.
Acknowledgments
The authors would like to thank Consuelo Estevez de Jensen, who provides administrative support and guidance for all projects in the Center for Excellence in Quarantine and Invasive Species.
Conflicts of Interest
The authors declare no conflict of interest. Neither of the authors, the University of Puerto Rico, or USDA-APHIS have any financial interest or endorsement for any of the methods, technologies, companies, or platforms discussed in this review.
References
- Alvarez-Fernandez, A.; Bernal, M.J.; Fradejas, I.; Martin Ramírez, A.; Md Yusuf, N.A.; Lanza, M.; Hisam, S.; Pérez de Ayala, A.; Rubio, J.M. KASP: A Genotyping Method to Rapid Identification of Resistance in Plasmodium falciparum. Malar. J. 2021, 20, 16. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Sharma, V.; Bhat, A.; Singh, H.; Sharma, I.; Verma, S.; Bhat, G.R.; Sharma, B.; Bakshi, D.; Kumar, R.; et al. MassARRAY Analysis of Twelve Cancer Related SNPs in Esophageal Squamous Cell Carcinoma in J&K, India. BMC Cancer 2020, 20, 497. [Google Scholar] [CrossRef]
- Darrier, B.; Russell, J.; Milner, S.G.; Hedley, P.E.; Shaw, P.D.; Macaulay, M.; Ramsay, L.D.; Halpin, C.; Mascher, M.; Fleury, D.L.; et al. A Comparison of Mainstream Genotyping Platforms for the Evaluation and Use of Barley Genetic Resources. Front. Plant Sci. 2019, 10, 544. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, H.; Tao, J.; Ren, Y.; Xu, C.; Wu, K.; Zou, C.; Zhang, J.; Xu, Y. Development of Multiple SNP Marker Panels Affordable to Breeders through Genotyping by Target Sequencing (GBTS) in Maize. Mol. Breed. 2019, 39, 37. [Google Scholar] [CrossRef]
- Guo, Z.; Yang, Q.; Huang, F.; Zheng, H.; Sang, Z.; Xu, Y.; Zhang, C.; Wu, K.; Tao, J.; Prasanna, B.M.; et al. Development of High-Resolution Multiple-SNP Arrays for Genetic Analyses and Molecular Breeding through Genotyping by Target Sequencing and Liquid Chip. Plant Commun. 2021, 2, 100230. [Google Scholar] [CrossRef] [PubMed]
- Lorenzini, R.; Fanelli, R.; Tancredi, F.; Siclari, A.; Garofalo, L. Matching STR and SNP Genotyping to Discriminate between Wild Boar, Domestic Pigs and Their Recent Hybrids for Forensic Purposes. Sci. Rep. 2020, 10, 3188. [Google Scholar] [CrossRef]
- Jatayev, S.; Kurishbayev, A.; Zotova, L.; Khasanova, G.; Serikbay, D.; Zhubatkanov, A.; Botayeva, M.; Zhumalin, A.; Turbekova, A.; Soole, K.; et al. Advantages of Amplifluor-like SNP Markers over KASP in Plant Genotyping. BMC Plant Biol. 2017, 17, 254. [Google Scholar] [CrossRef]
- Kim, N.; Kwon, J.-S.; Kang, W.-H.; Yeom, S.-I. High-Resolution Melting (HRM) Genotyping. Methods Mol Biol. 2023, 2638, 337–349. [Google Scholar] [CrossRef]
- Sato, M.; Hosoya, S.; Yoshikawa, S.; Ohki, S.; Kobayashi, Y.; Itou, T.; Kikuchi, K. A Highly Flexible and Repeatable Genotyping Method for Aquaculture Studies Based on Target Amplicon Sequencing Using Next-Generation Sequencing Technology. Sci. Rep. 2019, 9, 6904. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, J.; Zhang, L.; Luo, J.; Zhao, H.; Zhang, J.; Wen, C. A New SNP Genotyping Technology Target SNP-Seq and Its Application in Genetic Analysis of Cucumber Varieties. Sci. Rep. 2020, 10, 5623. [Google Scholar] [CrossRef]
- Susi, H.; Burdon, J.J.; Thrall, P.H.; Nemri, A.; Barrett, L.G. Genetic Analysis Reveals Long-Standing Population Differentiation and High Diversity in the Rust Pathogen Melampsora lini. PLoS Pathog. 2020, 16, e1008731. [Google Scholar] [CrossRef] [PubMed]
- Matukumalli, L.K.; Lawley, C.T.; Schnabel, R.D.; Taylor, J.F.; Allan, M.F.; Heaton, M.P.; O’Connell, J.; Moore, S.S.; Smith, T.P.L.; Sonstegard, T.S.; et al. Development and Characterization of a High Density SNP Genotyping Assay for Cattle. PLoS ONE 2009, 4, e5350. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Ng, W.-L.; Yang, J.-X.; Li, W.-M.; Ge, X.-J. High Cryptic Species Diversity Is Revealed by Genome-Wide Polymorphisms in a Wild Relative of Banana, Musa Itinerans, and Implications for Its Conservation in Subtropical China. BMC Plant Biol. 2018, 18, 194. [Google Scholar] [CrossRef] [PubMed]
- Lucas, E.R.; Rockett, K.A.; Lynd, A.; Essandoh, J.; Grisales, N.; Kemei, B.; Njoroge, H.; Hubbart, C.; Rippon, E.J.; Morgan, J.; et al. A High Throughput Multi-Locus Insecticide Resistance Marker Panel for Tracking Resistance Emergence and Spread in Anopheles gambiae. Sci. Rep. 2019, 9, 13335. [Google Scholar] [CrossRef]
- Pujolar, J.M.; Limborg, M.T.; Ehrlich, M.; Jaspers, C. High Throughput SNP Chip as Cost Effective New Monitoring Tool for Assessing Invasion Dynamics in the Comb Jelly Mnemiopsis leidyi. Front. Mar. Sci. 2022, 9, 1019001. [Google Scholar] [CrossRef]
- Ayalew, H.; Tsang, P.W.; Chu, C.; Wang, J.; Liu, S.; Chen, C.; Ma, X.-F. Comparison of TaqMan, KASP and RhAmp SNP Genotyping Platforms in Hexaploid Wheat. PLoS ONE 2019, 14, e0217222. [Google Scholar] [CrossRef]
- Zhang, Z.; Cao, Y.; Wang, Y.; Yu, H.; Wu, H.; Liu, J.; An, D.; Zhu, Y.; Feng, X.; Zhang, B.; et al. Development and Validation of KASP Markers for Resistance to Phytophthora capsici in Capsicum annuum L. Mol. Breed. 2023, 43, 20. [Google Scholar] [CrossRef]
- Pradhan, A.K.; Budhlakoti, N.; Chandra Mishra, D.; Prasad, P.; Bhardwaj, S.C.; Sareen, S.; Sivasamy, M.; Jayaprakash, P.; Geetha, M.; Nisha, R.; et al. Identification of Novel QTLs/Defense Genes in Spring Wheat Germplasm Panel for Seedling and Adult Plant Resistance to Stem Rust and Their Validation Through KASP Marker Assays. Plant Dis. 2023, 107, 1847–1860. [Google Scholar] [CrossRef]
- Verma, N.; Garcha, K.S.; Sharma, A.; Sharma, M.; Bhatia, D.; Khosa, J.S.; Kaur, B.; Chuuneja, P.; Dhatt, A.S. Identification of a Major-Effect Quantitative Trait Loci Associated with Begomovirus Resistance in Cucurbita moschata. Phytopathology 2023, 113, 824–835. [Google Scholar] [CrossRef]
- Ilie, D.E.; Gavojdian, D.; Kusza, S.; Neamț, R.I.; Mizeranschi, A.E.; Mihali, C.V.; Cziszter, L.T. Kompetitive Allele Specific PCR Genotyping of 89 SNPs in Romanian Spotted and Romanian Brown Cattle Breeds and Their Association with Clinical Mastitis. Animals 2023, 13, 1484. [Google Scholar] [CrossRef]
- Von Maydell, D. PCR Allele Competitive Extension (PACE). Methods Mol. Biol. 2023, 2638, 263–271. [Google Scholar] [CrossRef] [PubMed]
- von Maydell, D.; Brandes, J.; Lehnert, H.; Junghanns, W.; Marthe, F. Breeding Synthetic Varieties in Annual Caraway: Observations on the Outcrossing Rate in a Polycross Using a High-Throughput Genotyping System. Euphytica 2021, 217, 1. [Google Scholar] [CrossRef]
- Somyong, S.; Phetchawang, P.; Bihi, A.K.; Sonthirod, C.; Kongkachana, W.; Sangsrakru, D.; Jomchai, N.; Pootakham, W.; Tangphatsornruang, S. A SNP Variation in an Expansin (EgExp4) Gene Affects Height in Oil Palm. PeerJ 2022, 10, e13046. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.-Q.; Abdullah, K.G.; Wang, Q.K. The TaqMan Method for SNP Genotyping. Methods Mol. Biol. 2009, 578, 293–306. [Google Scholar] [CrossRef]
- Shumate, S.; Haylett, M.; Nelson, B.; Young, N.; Lamour, K.; Walsh, D.; Bradford, B.; Clements, J. Using Targeted Sequencing and TaqMan Approaches to Detect Acaricide (Bifenthrin, Bifenazate, and Etoxazole) Resistance Associated SNPs in Tetranychus urticae Collected from Peppermint Fields and Hop Yards. PLoS ONE 2023, 18, e0283211. [Google Scholar] [CrossRef]
- Li, H.; Wan, C.; Wang, Z.; Tan, J.; Tan, M.; Zeng, Y.; Huang, J.; Huang, Y.; Su, Q.; Kang, Z.; et al. Rapid Diagnosis of Duck Tembusu Virus and Goose Astrovirus with TaqMan-Based Duplex Real-Time PCR. Front. Microbiol. 2023, 14, 1146241. [Google Scholar] [CrossRef]
- Zhang, L.; Jiang, Z.; Zhou, Z.; Sun, J.; Yan, S.; Gao, W.; Shao, Y.; Bai, Y.; Wu, Y.; Yan, Z.; et al. A TaqMan Probe-Based Multiplex Real-Time PCR for Simultaneous Detection of Porcine Epidemic Diarrhea Virus Subtypes G1 and G2, and Porcine Rotavirus Groups A and C. Viruses 2022, 14, 1819. [Google Scholar] [CrossRef]
- Ragazzo, M.; Puleri, G.; Errichiello, V.; Manzo, L.; Luzzi, L.; Potenza, S.; Strafella, C.; Peconi, C.; Nicastro, F.; Caputo, V.; et al. Evaluation of OpenArrayTM as a Genotyping Method for Forensic DNA Phenotyping and Human Identification. Genes 2021, 12, 221. [Google Scholar] [CrossRef]
- Broccanello, C.; Gerace, L.; Stevanato, P. QuantStudioTM 12K Flex OpenArray® System as a Tool for High-Throughput Genotyping and Gene Expression Analysis. In Quantitative Real-Time PCR; Humana: New York, NY, USA, 2020; pp. 199–208. [Google Scholar]
- Noce, A.; Pazzola, M.; Dettori, M.L.; Amills, M.; Castelló, A.; Cecchinato, A.; Bittante, G.; Vacca, G.M. Variations at Regulatory Regions of the Milk Protein Genes Are Associated with Milk Traits and Coagulation Properties in the Sarda Sheep. Anim. Genet. 2016, 47, 717–726. [Google Scholar] [CrossRef]
- Chagné, D.; Vanderzande, S.; Kirk, C.; Profitt, N.; Weskett, R.; Gardiner, S.E.; Peace, C.P.; Volz, R.K.; Bassil, N.V. Validation of SNP Markers for Fruit Quality and Disease Resistance Loci in Apple (Malus × domestica Borkh.) Using the OpenArray® Platform. Hortic. Res. 2019, 6, 30. [Google Scholar] [CrossRef]
- Beltz, K.; Tsang, D.; Wang, J.; Rose, S.; Bao, Y.; Wang, Y.; Larkin, K.; Rupp, S.; Schrepfer, D.; Datta, K.; et al. A High-Performing and Cost-Effective SNP Genotyping Method Using RhPCR and Universal Reporters. Adv. Biosci. Biotechnol. 2018, 9, 497–512. [Google Scholar] [CrossRef]
- Esposito, S.; Taranto, F.; Vitale, P.; Ficco, D.B.M.; Colecchia, S.A.; Stevanato, P.; De Vita, P. Unlocking the Molecular Basis of Wheat Straw Composition and Morphological Traits through Multi-Locus GWAS. BMC Plant Biol. 2022, 22, 519. [Google Scholar] [CrossRef] [PubMed]
- Giglioti, R.; Gutmanis, G.; Katiki, L.M.; Okino, C.H.; de Sena Oliveira, M.C.; Vercesi Filho, A.E. New High-Sensitive RhAmp Method for A1 Allele Detection in A2 Milk Samples. Food Chem. 2020, 313, 126167. [Google Scholar] [CrossRef] [PubMed]
- Mohanrao, M.D.; Senthilvel, S.; Reddy, Y.R.; Kumar, C.A.; Kadirvel, P. Amplifluor-Based SNP Genotyping. Methods Mol. Biol. 2023, 2638, 191–200. [Google Scholar] [CrossRef]
- Khassanova, G.; Khalbayeva, S.; Serikbay, D.; Mazkirat, S.; Bulatova, K.; Utebayev, M.; Shavrukov, Y. SNP Genotyping with Amplifluor-Like Method. Methods Mol. Biol. 2023, 2638, 201–219. [Google Scholar] [CrossRef] [PubMed]
- Shavrukov, Y.; Zhumalin, A.; Serikbay, D.; Botayeva, M.; Otemisova, A.; Absattarova, A.; Sereda, G.; Sereda, S.; Shvidchenko, V.; Turbekova, A.; et al. Expression Level of the DREB2-Type Gene, Identified with Amplifluor SNP Markers, Correlates with Performance, and Tolerance to Dehydration in Bread Wheat Cultivars from Northern Kazakhstan. Front. Plant Sci. 2016, 7, 1736. [Google Scholar] [CrossRef] [PubMed]
- Tóth, T.; Csaba, Á.; Bokor, A.; Ács, N. Variable Fragment Length Allele-Specific Polymerase Chain Reaction (VFLASP), a Method for Simple and Reliable Genotyping. Mol. Cell. Probes 2023, 69, 101910. [Google Scholar] [CrossRef]
- Słomka, M.; Sobalska-Kwapis, M.; Wachulec, M.; Bartosz, G.; Strapagiel, D. High Resolution Melting (HRM) for High-Throughput Genotyping—Limitations and Caveats in Practical Case Studies. Int. J. Mol. Sci. 2017, 18, 2316. [Google Scholar] [CrossRef]
- Chou, L.; Huang, S.-J.; Hsieh, C.; Lu, M.-T.; Song, C.-W.; Hsu, F.-C. A High Resolution Melting Analysis-Based Genotyping Toolkit for the Peach (Prunus persica) Chilling Requirement. Int. J. Mol. Sci. 2020, 21, 1543. [Google Scholar] [CrossRef]
- Chatzidimopoulos, M.; Ganopoulos, I.; Moraitou-Daponta, E.; Lioliopoulou, F.; Ntantali, O.; Panagiotaki, P.; Vellios, E.K. High-Resolution Melting (HRM) Analysis Reveals Genotypic Differentiation of Venturia inaequalis Populations in Greece. Front. Ecol. Evol. 2019, 7, 489. [Google Scholar] [CrossRef]
- Ellis, J.A.; Ong, B. The MassARRAY® System for Targeted SNP Genotyping. Methods Mol. Biol. 2017, 1492, 77–94. [Google Scholar] [CrossRef] [PubMed]
- da Costa Lima Moraes, A.; Sforça, D.A.; Mancini, M.C.; Vigna, B.B.Z.; de Souza, A.P. Polyploid SNP Genotyping Using the MassARRAY System. Methods Mol. Biol. 2023, 2638, 93–113. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Gan, H.; Lin, X.; Wang, L.; Yao, Y.; Li, L.; Wang, Y.; Zhang, Z. Genome-Wide Association Screening and MassARRAY for Detection of High-Temperature Resistance-Related SNPs and Genes in a Hybrid Abalone (Haliotis discus hannai ♀ × H. fulgens ♂) Based on Super Genotyping-by-Sequencing. Aquaculture 2023, 573, 739576. [Google Scholar] [CrossRef]
- Ji, X.; Cao, Z.; Hao, Q.; He, M.; Cang, M.; Yu, H.; Ma, Q.; Li, X.; Bao, S.; Wang, J.; et al. Effects of New Mutations in BMPRIB, GDF9, BMP15, LEPR, and B4GALNT2 Genes on Litter Size in Sheep. Vet. Sci. 2023, 10, 258. [Google Scholar] [CrossRef] [PubMed]
- Vervalle, J.A.; Costantini, L.; Lorenzi, S.; Pindo, M.; Mora, R.; Bolognesi, G.; Marini, M.; Lashbrooke, J.G.; Tobutt, K.R.; Vivier, M.A.; et al. A High-Density Integrated Map for Grapevine Based on Three Mapping Populations Genotyped by the Vitis18K SNP Chip. Theor. Appl. Genet. 2022, 135, 4371–4390. [Google Scholar] [CrossRef] [PubMed]
- Balog, K.; Mizeranschi, A.E.; Wanjala, G.; Sipos, B.; Kusza, S.; Bagi, Z. Application Potential of Chicken DNA Chip in Domestic Pigeon Species—Preliminary Results. Saudi J. Biol. Sci. 2023, 30, 103594. [Google Scholar] [CrossRef]
- Singh, B.K.; Venkadesan, S.; Ramkumar, M.K.; Shanmugavadivel, P.S.; Dutta, B.; Prakash, C.; Pal, M.; Solanke, A.U.; Rai, A.; Singh, N.K.; et al. Meta-Analysis of Microarray Data and Their Utility in Dissecting the Mapped QTLs for Heat Acclimation in Rice. Plants 2023, 12, 1697. [Google Scholar] [CrossRef]
- Muhu-Din Ahmed, H.G.; Sajjad, M.; Zeng, Y.; Iqbal, M.; Habibullah Khan, S.; Ullah, A.; Nadeem Akhtar, M. Genome-Wide Association Mapping through 90K SNP Array for Quality and Yield Attributes in Bread Wheat against Water-Deficit Conditions. Agriculture 2020, 10, 392. [Google Scholar] [CrossRef]
- Xiao, Q.; Wang, H.; Song, N.; Yu, Z.; Imran, K.; Xie, W.; Qiu, S.; Zhou, F.; Wen, J.; Dai, C.; et al. The Bnapus50K Array: A Quick and Versatile Genotyping Tool for Brassica Napus Genomic Breeding and Research. G3 Genes|Genomes|Genet. 2021, 11, jkab241. [Google Scholar] [CrossRef]
- Gunderson, K.L.; Steemers, F.J.; Lee, G.; Mendoza, L.G.; Chee, M.S. A Genome-Wide Scalable SNP Genotyping Assay Using Microarray Technology. Nat. Genet. 2005, 37, 549–554. [Google Scholar] [CrossRef]
- Perkel, J. SNP Genotyping: Six Technologies That Keyed a Revolution. Nat. Methods 2008, 5, 447–453. [Google Scholar] [CrossRef]
- Gardner, S.N.; Thissen, J.B.; McLoughlin, K.S.; Slezak, T.; Jaing, C.J. Optimizing SNP Microarray Probe Design for High Accuracy Microbial Genotyping. J. Microbiol. Methods 2013, 94, 303–310. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rungroj, N.; Nettuwakul, C.; Sudtachat, N.; Praditsap, O.; Sawasdee, N.; Sritippayawan, S.; Chuawattana, D.; Yenchitsomanus, P. A Whole Genome SNP Genotyping by DNA Microarray and Candidate Gene Association Study for Kidney Stone Disease. BMC Med. Genet. 2014, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Balagué-Dobón, L.; Cáceres, A.; González, J.R. Fully Exploiting SNP Arrays: A Systematic Review on the Tools to Extract Underlying Genomic Structure. Brief. Bioinform. 2022, 23, bbac043. [Google Scholar] [CrossRef] [PubMed]
- You, Q.; Yang, X.; Peng, Z.; Xu, L.; Wang, J. Development and Applications of a High Throughput Genotyping Tool for Polyploid Crops: Single Nucleotide Polymorphism (SNP) Array. Front. Plant Sci. 2018, 9, 104. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Banks, T.W.; Cloutier, S. SNP Discovery through Next-Generation Sequencing and Its Applications. Int. J. Plant Genom. 2012, 2012, 831460. [Google Scholar] [CrossRef]
- Sekine, D.; Oku, S.; Nunome, T.; Hirakawa, H.; Tsujimura, M.; Terachi, T.; Toyoda, A.; Shigyo, M.; Sato, S.; Tsukazaki, H. Development of a Genome-Wide Marker Design Workflow for Onions and Its Application in Target Amplicon Sequencing-Based Genotyping. DNA Res. 2022, 29, dsac020. [Google Scholar] [CrossRef]
- Takeshima, R.; Ogiso-Tanaka, E.; Yasui, Y.; Matsui, K. Targeted Amplicon Sequencing + Next-Generation Sequencing–Based Bulked Segregant Analysis Identified Genetic Loci Associated with Preharvest Sprouting Tolerance in Common Buckwheat (Fagopyrum esculentum). BMC Plant Biol. 2021, 21, 18. [Google Scholar] [CrossRef]
- Nagano, S.; Hirao, T.; Takashima, Y.; Matsushita, M.; Mishima, K.; Takahashi, M.; Iki, T.; Ishiguri, F.; Hiraoka, Y. SNP Genotyping with Target Amplicon Sequencing Using a Multiplexed Primer Panel and Its Application to Genomic Prediction in Japanese Cedar, Cryptomeria japonica (L.f.) D.Don. Forests 2020, 11, 898. [Google Scholar] [CrossRef]
- Shirasawa, K.; Kuwata, C.; Watanabe, M.; Fukami, M.; Hirakawa, H.; Isobe, S. Target Amplicon Sequencing for Genotyping Genome-Wide Single Nucleotide Polymorphisms Identified by Whole-Genome Resequencing in Peanut. Plant Genome 2016, 9, 1–8. [Google Scholar] [CrossRef]
- Ishikawa, G.; Sakai, H.; Mizuno, N.; Solovieva, E.; Tanaka, T.; Matsubara, K. Developing Core Marker Sets for Effective Genomic-Assisted Selection in Wheat and Barley Breeding Programs. Breed. Sci. 2022, 72, 22004. [Google Scholar] [CrossRef] [PubMed]
- Gashururu, R.S.; Maingi, N.; Githigia, S.M.; Getange, D.O.; Ntivuguruzwa, J.B.; Habimana, R.; Cecchi, G.; Gashumba, J.; Bargul, J.L.; Masiga, D.K. Trypanosomes Infection, Endosymbionts, and Host Preferences in Tsetse Flies (Glossina Spp.) Collected from Akagera Park Region, Rwanda: A Correlational Xenomonitoring Study. One Health 2023, 16, 100550. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-R.; Kim, Y.-C.; Won, S.-Y.; Jeong, M.-J.; Park, K.-J.; Park, H.-C.; Roh, I.-S.; Kang, H.-E.; Sohn, H.-J.; Jeong, B.-H. Identification of a Novel Risk Factor for Chronic Wasting Disease (CWD) in Elk: S100G Single Nucleotide Polymorphism (SNP) of the Prion Protein Gene (PRNP). Vet. Res. 2023, 54, 48. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Ogata, T.; Shimada, T.; Endo, T.; Iketani, H.; Shimizu, T.; Yamamoto, T.; Omura, M. Minimal Marker: An algorithm and computer program for the identification of minimal sets of discriminating DNA markers for efficient variety identification. J. Bioinform. Comput. Biol. 2013, 11, 1250022. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Ren, Y.; Jian, Y.; Guo, Z.; Zhang, Y.; Xie, C.; Fu, J.; Wang, H.; Wang, G.; Xu, Y.; et al. Development of a Maize 55 K SNP Array with Improved Genome Coverage for Molecular Breeding. Mol. Breed. 2017, 37, 20. [Google Scholar] [CrossRef]
- Onda, Y.; Takahagi, K.; Shimizu, M.; Inoue, K.; Mochida, K. Multiplex PCR Targeted Amplicon Sequencing (MTA-Seq): Simple, Flexible, and Versatile SNP Genotyping by Highly Multiplexed PCR Amplicon Sequencing. Front. Plant Sci. 2018, 9, 201. [Google Scholar] [CrossRef]
- Singh, A.; Bhatia, P. Comparative Sequencing Data Analysis of Ion Torrent and MinION Sequencing Platforms Using a Clinical Diagnostic Haematology Panel. Int. J. Lab. Hematol. 2020, 42, 833–841. [Google Scholar] [CrossRef]
- Nygaard, A.B.; Tunsjø, H.S.; Meisal, R.; Charnock, C. A Preliminary Study on the Potential of Nanopore MinION and Illumina MiSeq 16S RRNA Gene Sequencing to Characterize Building-Dust Microbiomes. Sci. Rep. 2020, 10, 3209. [Google Scholar] [CrossRef]
- Whitford, W.; Hawkins, V.; Moodley, K.S.; Grant, M.J.; Lehnert, K.; Snell, R.G.; Jacobsen, J.C. Proof of Concept for Multiplex Amplicon Sequencing for Mutation Identification Using the MinION Nanopore Sequencer. Sci. Rep. 2022, 12, 8572. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.-L.; Zhang, J.-R.; Zhang, X.-M.; Liu, X.; Lin, Y.-F.; Bai, H.; Wang, M.-C.; Cheng, F.; Liu, J.-D.; Li, P.; et al. Forensic Nanopore Sequencing of STRs and SNPs Using Verogen’s ForenSeq DNA Signature Prep Kit and MinION. Int. J. Legal Med. 2021, 135, 1685–1693. [Google Scholar] [CrossRef]
- Cornelis, S.; Gansemans, Y.; Vander Plaetsen, A.-S.; Weymaere, J.; Willems, S.; Deforce, D.; Van Nieuwerburgh, F. Forensic Tri-Allelic SNP Genotyping Using Nanopore Sequencing. Forensic Sci. Int. Genet. 2019, 38, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Tabata, Y.; Matsuo, Y.; Fujii, Y.; Ohta, A.; Hirota, K. Rapid Detection of Single Nucleotide Polymorphisms Using the MinION Nanopore Sequencer: A Feasibility Study for Perioperative Precision Medicine. JA Clin. Rep. 2022, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, S.; Gansemans, Y.; Deleye, L.; Deforce, D.; Van Nieuwerburgh, F. Forensic SNP Genotyping Using Nanopore MinION Sequencing. Sci. Rep. 2017, 7, 41759. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).