Gene Drive: Past, Present and Future Roads to Vertebrate Biocontrol
Abstract
1. Introduction
2. Selfish Genetic Elements
2.1. Homing Endonuclease Genes
2.2. Meiotic Drive
3. Harnessing Evolution
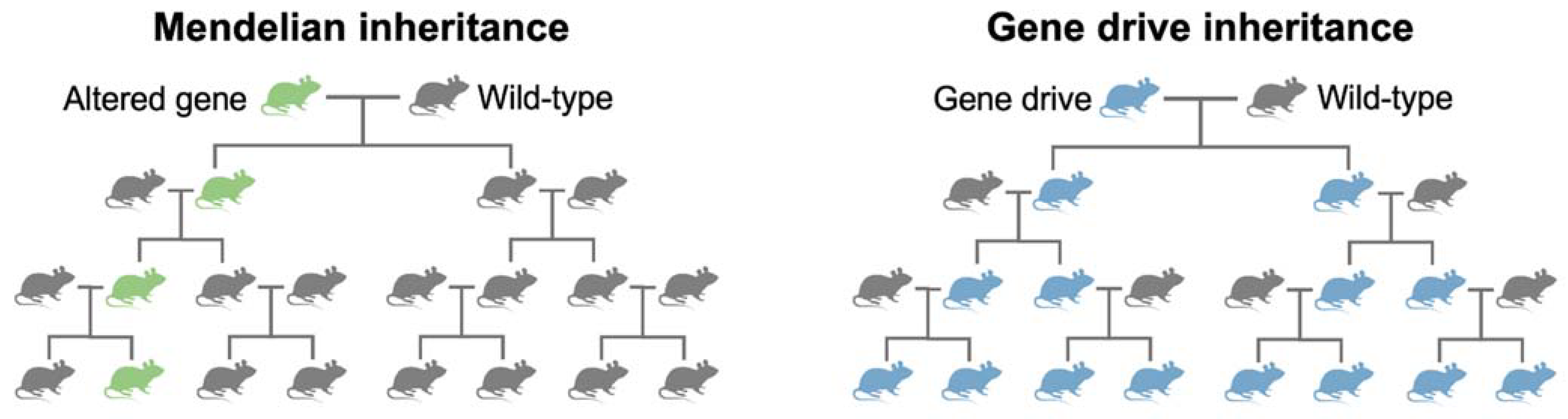
4. What Are Gene Drives?
5. Genetic Engineering
6. Nuclease-Assisted Genetic Engineering
7. ZFNs and TALENs
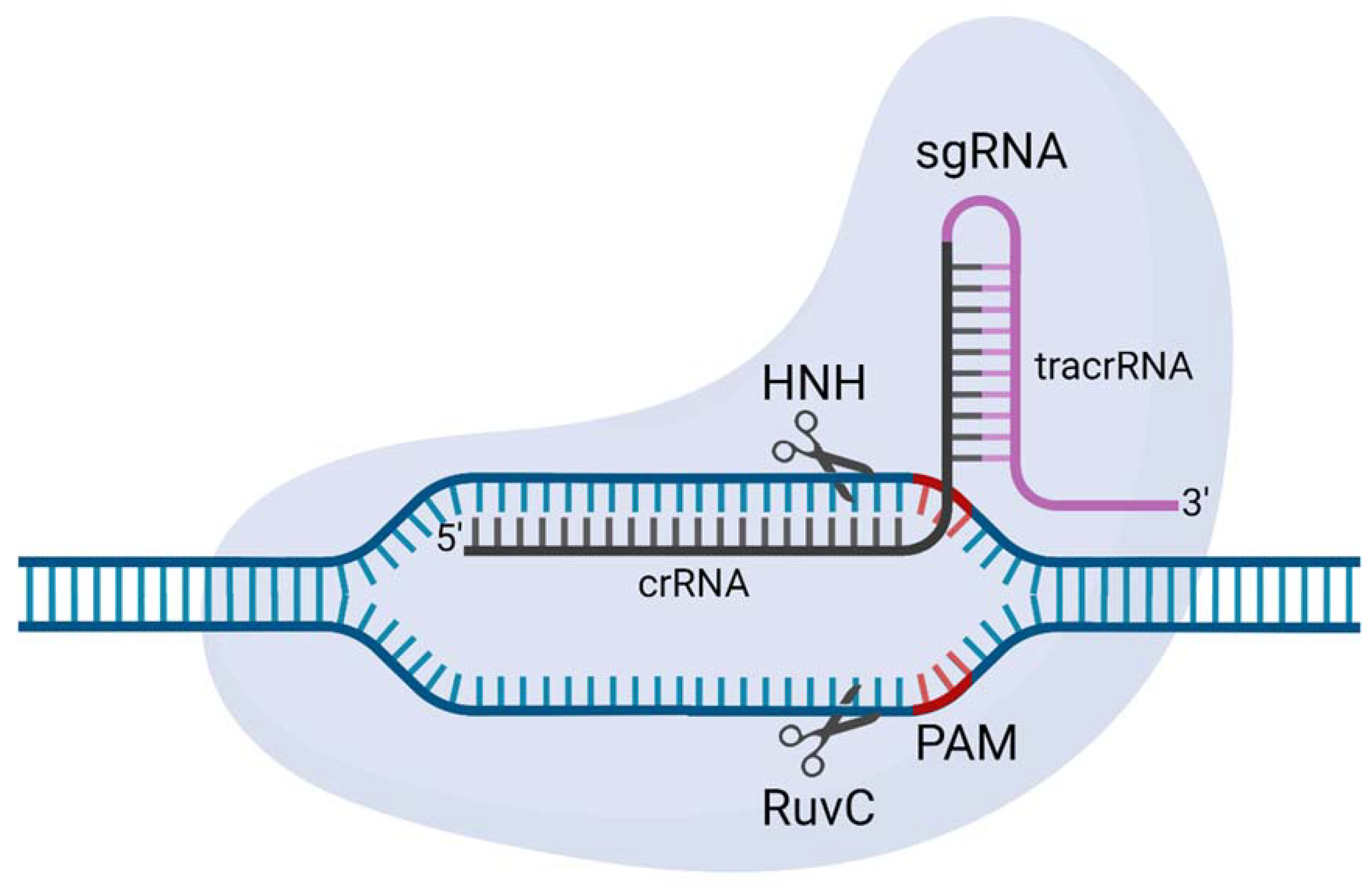
8. CRISPR-Cas
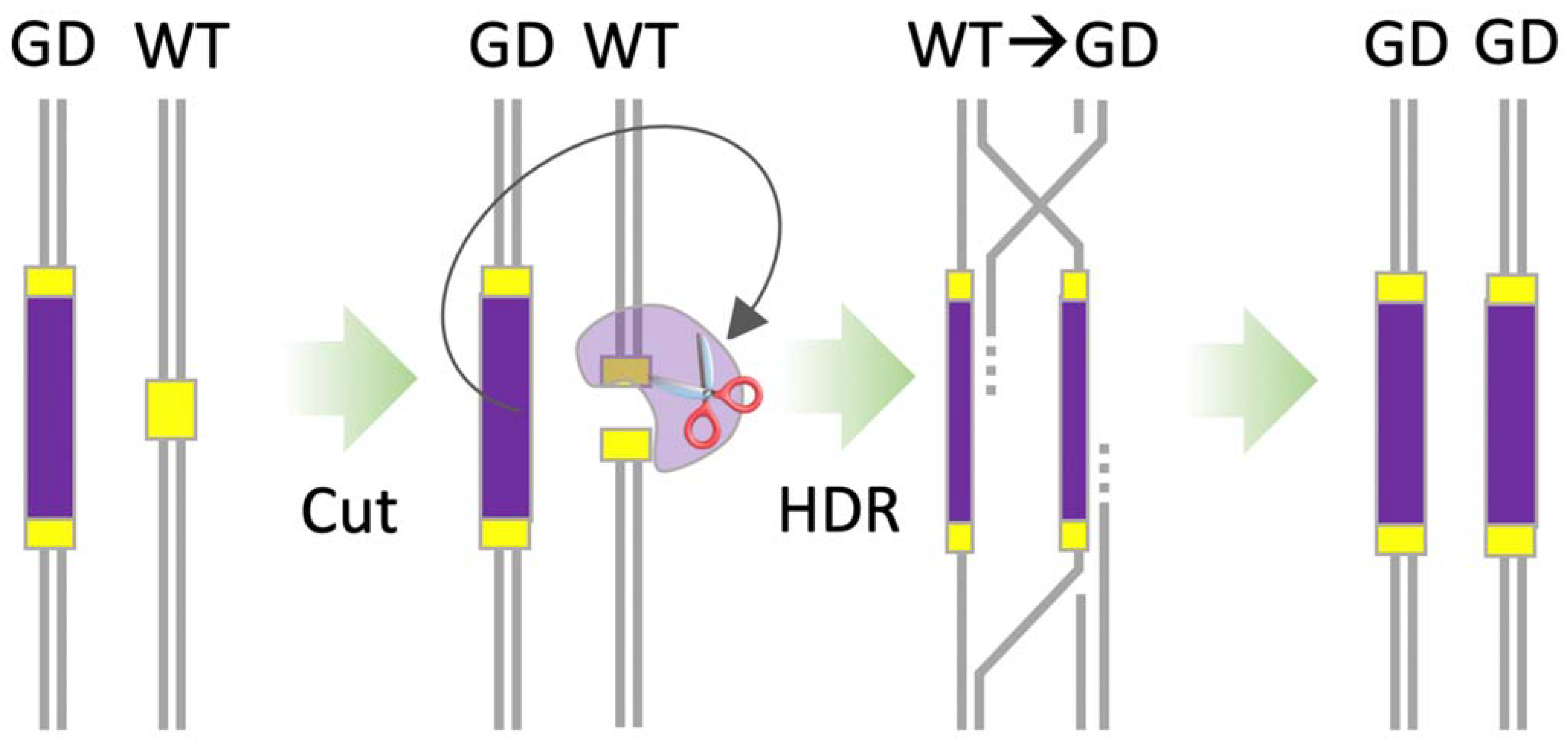
9. CRISPR-Based Gene Drives
- Population suppression: the spread of a genetic element that causes the number of individuals in a population to decrease.
- Population replacement: the spread of a genetic element that causes a population’s genotype to change.
10. Driving Population Suppression
10.1. tCRISPR
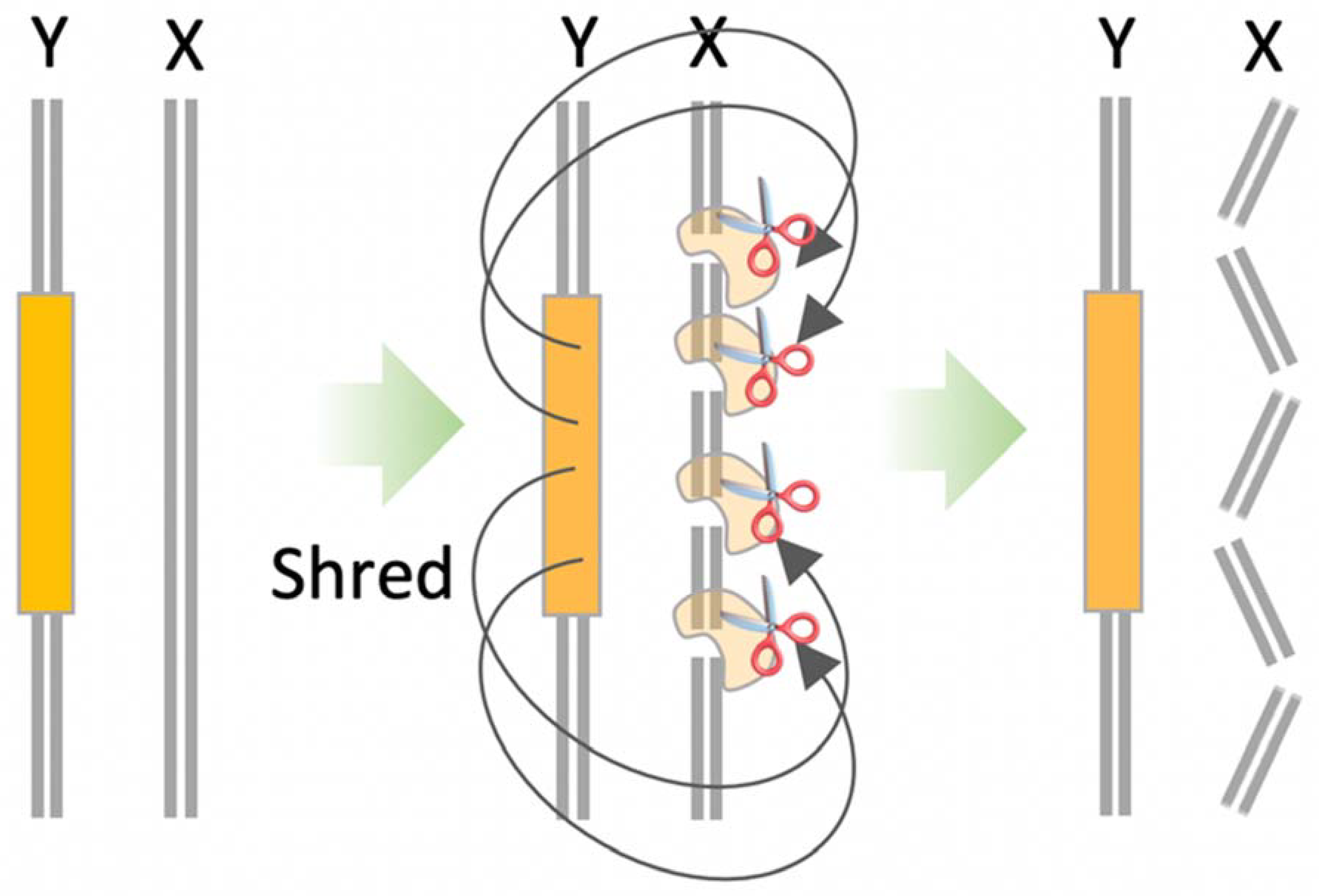
10.2. X-Shredder
10.3. Homing-Based Gene Drives
11. Containment Strategies
11.1. Daisy-Chain System
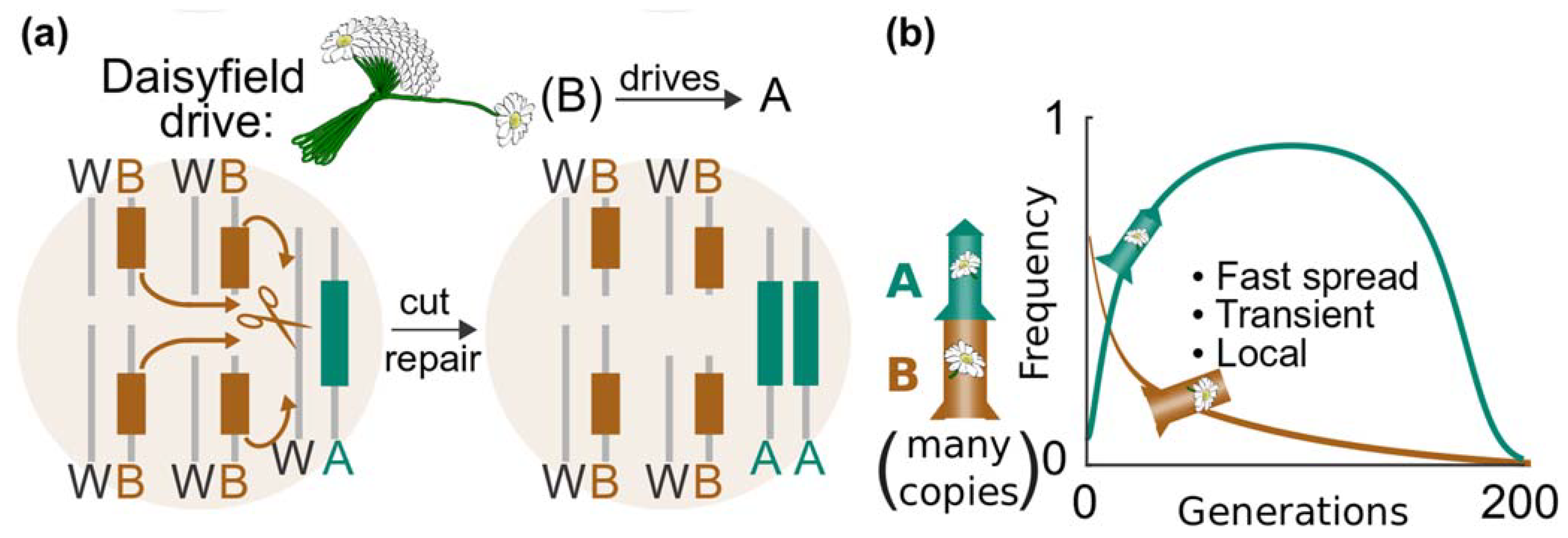
11.2. Daisyfield System
12. Public Perception
13. Governance and Regulations
14. Conclusions
- The road towards gene drive technology has traced centuries of scientific investigation and technological breakthroughs. The study and understanding of naturally occurring selfish genetic elements, such as homing endonuclease genes and meiotic drive, has been essential to conceptualising synthetic gene drives.
- To engineer synthetic gene drives, scientists required precision genetic engineering technologies. Due to their complex protein designs, initial attempts using early genome engineering nucleases struggled. The repurposing of CRISPR-SpCas9 into a genome engineering tool overcame many of these challenges and allowed synthetic gene drives to be engineered in a standard molecular laboratory.
- CRISPR-based gene drives have been engineered in yeast, D. melanogaster, three species of mosquitoes and most recently in mice. The gene drives developed in mice have had lower transmission rates than previous invertebrate gene drives. Studies of synthetic mouse gene drives have revealed the challenges of engineering gene drives in mammals.
- Future research efforts aim to optimise vertebrate gene drive designs to improve transmission rates. For homing gene drives, this includes identifying or engineering promoters that provide robust and specific SpCas9 expression during meiosis I. In the immediate future, non-homing gene drive designs, such as tCRISPR or X-shredder may be more feasible in vertebrate species.
- Gene drives for real-world application should include built-in molecular containment strategies to help contain the spread and stop unforeseen impacts. There is a range of molecular-safeguarding options for vertebrates, including targeting private alleles and daisy drives. These molecular containment approaches should be used in parallel to efforts that physically isolate the target population.
- Public acceptance and governance will determine the speed and extent of application. Public acceptance will hinge on the trustworthiness of research, the problem the technology addresses and the outcomes of initial trials. Before gene drives can be applied as a vertebrate biocontrol tool, updating governance and regulations to include the environmental release of gene-drive organisms is required.
- The application of gene drive for vertebrate biocontrol will have profound implications for society, our economy and the environment. With technological development progressing swiftly, public and political engagement must move in parallel to ensure applications align with society’s values.
Author Contributions
Funding
Conflicts of Interest
References
- Krull, C.R.; Galbraith, J.A.; Glen, A.S.; Nathan, H.W. Invasive vertebrates in Australia and New Zealand. In Austral Ark: The State of Wildlife in Australia and New Zealand; Stow, A., Holwell, G.I., Maclean, N., Eds.; Cambridge University Press: Cambridge, UK, 2014; pp. 197–226. [Google Scholar]
- Robertson, P.A.; Adriaens, T.; Lambin, X.; Mill, A.; Roy, S.; Shuttleworth, C.M.; Sutton-Croft, M. The large-scale removal of mammalian invasive alien species in Northern Europe. Pest Manag. Sci. 2017, 73, 273–279. [Google Scholar] [CrossRef]
- Witmer, G.W.; Fuller, P.L. Vertebrate species introductions in the United States and its territories. Curr. Zool. 2011, 57, 559–567. [Google Scholar] [CrossRef]
- Early, R.; Bradley, B.A.; Dukes, J.S.; Lawler, J.J.; Olden, J.D.; Blumenthal, D.M.; Tatem, A.J. Global threats from invasive alien species in the twenty-first century and national response capacities. Nat. Commun. 2016, 7, 12485. [Google Scholar] [CrossRef] [PubMed]
- Singleton, G. Integrated management of rodents: A Southeast Asian and Australian perspective. Belg. J. Zool. 1997, 127, 157–169. [Google Scholar]
- Singleton, G. Impacts of Rodents on Rice Production in Asia; IRRI Discussion Paper Series; IRRI: Los Baños, Philippines, 2003. [Google Scholar]
- Howal, G.; Donald, J.; Galvan, J.P.; Russel, J.; Parkes, J.; Samaniego, A.; Tershy, B. Invasive Rodent Eradication on Islands. Conserv. Biol. 2007, 21, 1258–1268. [Google Scholar] [CrossRef]
- Towns, D.R.; Atkinson, I.A.E.; Daugherty, C.H. Have the Harmful Effects of Introduced Rats on Islands been Exaggerated? Biol. Invasions 2006, 8, 863–891. [Google Scholar] [CrossRef]
- Meerburg, B.G.; Brom, F.W.; Kijlstra, A. The ethics of rodent control. Pest Manag. Sci. 2008, 64, 1205–1211. [Google Scholar] [CrossRef]
- Lorvelec, O.; Pascal, M. French attempts to eradicate non-indigenous mammals and their consequences for native biota. Biol. Invasions 2005, 7, 135–140. [Google Scholar] [CrossRef]
- Burt, A.; Trivers, R. Genes in Conflict: The Biology of Selfish Genetic Elements; Harvard University Press: Cambrideg, MA, USA, 2016. [Google Scholar]
- Esvelt, K.M.; Smidler, A.L.; Catteruccia, F.; Church, G.M. Concerning RNA-guided gene drives for the alteration of wild populations. Elife 2014, 3, e03401. [Google Scholar] [CrossRef]
- Fraser, M.J., Jr. Insect transgenesis: Current applications and future prospects. Annu. Rev. Entomol. 2012, 57, 267–289. [Google Scholar] [CrossRef]
- Jasin, M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996, 12, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Belfort, M.; Bonocora, R.P. Homing endonucleases: From genetic anomalies to programmable genomic clippers. Methods Mol. Biol. 2014, 1123, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Windbichler, N.; Papathanos, P.A.; Catteruccia, F.; Ranson, H.; Burt, A.; Crisanti, A. Homing endonuclease mediated gene targeting in Anopheles gambiae cells and embryos. Nucleic Acids Res. 2007, 35, 5922–5933. [Google Scholar] [CrossRef] [PubMed]
- McDermott, S.R.; Noor, M.A.F. The role of meiotic drive in hybrid male sterility. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 1265–1272. [Google Scholar] [CrossRef]
- Ardlie, K.G. Putting the brake on drive: Meiotic drive of t haplotypes in natural populations of mice. Trends Genet. 1998, 14, 189–193. [Google Scholar] [CrossRef]
- Silver, L.M. The peculiar journey of a selfish chromosome: Mouse t haplotypes and meiotic drive. Trends Genet. 1993, 9, 250–254. [Google Scholar] [CrossRef]
- Bauer, H.; Schindler, S.; Charron, Y.; Willert, J.; Kusecek, B.; Herrmann, B.G. The nucleoside diphosphate kinase gene Nme3 acts as quantitative trait locus promoting non-Mendelian inheritance. PLoS Genet. 2012, 8, e1002567. [Google Scholar] [CrossRef]
- Bauer, H.; Véron, N.; Willert, J.; Herrmann, B.G. The t-complex-encoded guanine nucleotide exchange factor Fgd2 reveals that two opposing signaling pathways promote transmission ratio distortion in the mouse. Genes Dev. 2007, 21, 143–147. [Google Scholar] [CrossRef]
- Herrmann, B.G.; Bauer, H. The mouse t—Haplotype: A selfish chromosome—Genetics, molecular mechanism, and evolution. Evol. House Mouse 2012, 3, 297. [Google Scholar]
- Hickey, W.A.; Craig, G.B. Genetic Distortion of Sex Ratio in a Mosquito Aedes aegypti. Genetics 1966, 53, 1177. [Google Scholar] [CrossRef]
- Craig, G.B.; Hickey, W.A.; Vandehey, R.C. Inherited Male-Producing Factor in Aedes aegypti. Science 1960, 132, 1887–1889. [Google Scholar] [CrossRef] [PubMed]
- Burt, A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. R. Soc. B Biol. Sci. 2003, 270, 921–928. [Google Scholar] [CrossRef] [PubMed]
- National Academies of Sciences, Engineering, and Medicine. Gene Drives on the Horizon: Advancing Science, Navigating Uncertainty, and Aligning Research with Public Values; The National Academies Press: Washington, DC, USA, 2016. [Google Scholar]
- McFarlane, G.R.; Whitelaw, C.B.A.; Lillico, S.G. CRISPR-Based Gene Drives for Pest Control. Trends Biotechnol. 2018, 36, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Adelman, Z.N.; Tu, Z. Control of Mosquito-Borne Infectious Diseases: Sex and Gene Drive. Trends Parasitol. 2016, 32, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Galizi, R.; Doyle, L.; Menichelli, M.; Bernardini, F.; Deredec, A.; Burt, A.; Crisanti, A. A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat. Commun. 2014, 5, 3977. [Google Scholar] [CrossRef]
- Hammond, A.; Galizi, R.; Kyrou, K.; Simoni, A.; Siniscalchi, C.; Katsanos, D.; Nolan, T. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 2016, 34, 78–83. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. World Malaria Report 2019; World Health Organization: Geneva, Switzerland, 2019.
- Whitelaw, B.; McFarlane, G.R. Accelerating evolution. Biologist 2019, 66, 18–21. [Google Scholar]
- Smithies, O.; Gregg, R.G.; Boggs, S.S.; Koralewski, M.A.; Kucherlapati, R.S. Insertion of DNA sequences into the human chromosomal β-globin locus by homologous recombination. Nature 1985, 317, 230–234. [Google Scholar] [CrossRef]
- Capecchi, M.R. Altering the genome by homologous recombination. Science 1989, 244, 1288–1292. [Google Scholar] [CrossRef]
- Rouet, P.; Smih, F.; Jasin, M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 1994, 14, 8096–8106. [Google Scholar] [CrossRef]
- Smih, F.; Rouet, P.; Romanienko, P.J.; Jasin, M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995, 23, 5012–5019. [Google Scholar] [CrossRef]
- Liu, M.; Rehman, S.; Tang, X.; Gu, K.; Fan, Q.; Chen, D.; Ma, W. Methodologies for Improving HDR Efficiency. Front. Genet. 2019, 9, 691. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, T.; Nakade, S.; Sakane, Y.; Suzuki, K.-I.T.; Yamamoto, T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat. Protoc. 2016, 11, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Yanik, M.; Ponnam, S.P.G.; Wimmer, T.; Trimborn, L.; Müller, C.; Gambert, I.; Stieger, K. Development of a Reporter System to Explore MMEJ in the Context of Replacing Large Genomic Fragments. Mol. Ther. Nucleic Acid 2018, 11, 407–415. [Google Scholar] [CrossRef]
- Windbichler, N.; Menichelli, M.; Papathanos, P.A.; Thyme, S.B.; Li, H.; Ulge, U.Y.; Crisanti, A. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature 2011, 473, 212–215. [Google Scholar] [CrossRef]
- Burt, A.; Koufopanou, V. Homing endonuclease genes: The rise and fall and rise again of a selfish element. Curr. Opin. Genet. Dev. 2004, 14, 609–615. [Google Scholar] [CrossRef]
- Rocha-Martins, M.; Cavalheiro, G.R.; Matos-Rodrigues, G.E.; Martins, R.A. From Gene Targeting to Genome Editing: Transgenic animals applications and beyond. Acad. Bras. Cienc. 2015, 87 (Suppl. S2), 1323–1348. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef]
- Pratt, J.; Venkatraman, N.; Brinker, A.; Xiao, Y.; Blasberg, J.; Thompson, D.C.; Bourner, M. Use of zinc finger nuclease technology to knock out efflux transporters in C2BBe1 cells. Curr. Protoc. Toxicol. 2012, 52, 23. [Google Scholar] [CrossRef]
- Boch, J.; Bonas, U. Xanthomonas AvrBs3 Family-Type III Effectors: Discovery and Function. Annu. Rev. Phytopathol. 2010, 48, 419–436. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Zhang, H.; Xu, N.; Zhang, B.; Gou, F.; Zhu, J.-K. Application of the CRISPR–Cas system for efficient genome engineering in plants. Mol. Plant 2013, 6, 2008–2011. [Google Scholar] [CrossRef]
- Simoni, A.; Siniscalchi, C.; Chan, Y.-S.; Huen, D.S.; Russell, S.; Windbichler, N.; Crisanti, A. Development of synthetic selfish elements based on modular nucleases in Drosophila melanogaster. Nucleic Acids Res. 2014, 42, 7461–7472. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Zhang, F. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Lander, E.S. The heroes of CRISPR. Cell 2016, 164, 18–28. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef]
- Mojica, F.J.M.; Montoliu, L. On the Origin of CRISPR-Cas Technology: From Prokaryotes to Mammals. Trends Microbiol. 2016, 24, 811–820. [Google Scholar] [CrossRef]
- Yao, R.; Liu, D.; Jia, X.; Zheng, Y.; Liu, W.; Xiao, Y. CRISPR-Cas9/Cas12a biotechnology and application in bacteria. Synth. Syst. Biotechnol. 2018, 3, 135–149. [Google Scholar] [CrossRef]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef]
- Garneau, J.E.; Dupuis, M.-È.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.M.; Musunuru, K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J. Clin. Investig. 2014, 124, 4154–4161. [Google Scholar] [CrossRef] [PubMed]
- DiCarlo, J.E.; Chavez, A.; Dietz, S.L.; Esvelt, K.M.; Church, G.M. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol. 2015, 33, 1250–1255. [Google Scholar] [CrossRef]
- Gantz, V.M.; Bier, E. The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science 2015, 348, 442. [Google Scholar] [CrossRef]
- Gantz, V.M.; Jasinskiene, N.; Tatarenkova, O.; Fazekas, A.; Macias, V.M.; Bier, E.; James, A.A. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. USA 2015, 112, E6736–E6743. [Google Scholar] [CrossRef]
- Kyrou, K.; Hammond, A.M.; Galizi, R.; Kranjc, N.; Burt, A.; Beaghton, A.K.; Crisanti, A. A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 2018, 36, 1062–1066. [Google Scholar] [CrossRef]
- Gierus, L.; Birand, A.; Bunting, M.D.; Godahewa, G.I.; Piltz, S.G.; Oh, K.P.; Thomas, P.Q. Leveraging a natural murine meiotic drive to suppress invasive populations. Proc. Natl. Acad. Sci. USA 2022, 119, e2213308119. [Google Scholar] [CrossRef]
- Grunwald, H.A.; Gantz, V.M.; Poplawski, G.; Xu, X.-R.S.; Bier, E.; Cooper, K.L. Super-Mendelian inheritance mediated by CRISPR–Cas9 in the female mouse germline. Nature 2019, 566, 105–109. [Google Scholar] [CrossRef]
- Weitzel, A.J.; Grunwald, H.A.; Weber, C.; Levina, R.; Gantz, V.M.; Hedrick, S.M.; Cooper, K.L. Meiotic Cas9 expression mediates gene conversion in the male and female mouse germline. PLoS Biol. 2021, 19, e3001478. [Google Scholar] [CrossRef]
- Galizi, R.; Hammond, A.; Kyrou, K.; Taxiarchi, C.; Bernardini, F.; O’Loughlin, S.M.; Crisanti, A. A CRISPR-Cas9 sex-ratio distortion system for genetic control. Sci. Rep. 2016, 6, 31139. [Google Scholar] [CrossRef]
- Oberhofer, G.; Ivy, T.; Hay, B.A. Behavior of homing endonuclease gene drives targeting genes required for viability or female fertility with multiplexed guide RNAs. Proc. Natl. Acad. Sci. USA 2018, 115, E9343–E9352. [Google Scholar] [CrossRef]
- Yang, E.; Metzloff, M.; Langmüller, A.M.; Xu, X.; Clark, A.G.; Messer, P.W.; Champer, J. A homing suppression gene drive with multiplexed gRNAs maintains high drive conversion efficiency and avoids functional resistance alleles. G3 Genes Genomes Genet. 2022, 12, jkac081. [Google Scholar] [CrossRef]
- Berry, R.; Scriven, P. The house mouse: A model and motor for evolutionary understanding. Biol. J. Linn. Soc. 2005, 84, 335–347. [Google Scholar] [CrossRef]
- Pocock, M.J.; Hauffe, H.C.; Searle, J.B. Dispersal in house mice. Biol. J. Linn. Soc. 2005, 84, 565–583. [Google Scholar] [CrossRef]
- Champer, J.; Buchman, A.; Akbari, O.S. Cheating evolution: Engineering gene drives to manipulate the fate of wild populations. Nat. Rev. Genet. 2016, 17, 146–159. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, W.D. Extraordinary sex ratios. A sex-ratio theory for sex linkage and inbreeding has new implications in cytogenetics and entomology. Science 1967, 156, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Backus, G.A.; Gross, K. Genetic engineering to eradicate invasive mice on islands: Modeling the efficiency and ecological impacts. Ecosphere 2016, 7, e01589. [Google Scholar] [CrossRef]
- Koopman, P.; Gubbay, J.; Vivian, N.; Goodfellow, P.; Lovell-Badge, R. Male development of chromosomally female mice transgenic for Sry. Nature 1991, 351, 117–121. [Google Scholar] [CrossRef]
- Manser, A.; Cornell, S.J.; Sutter, A.; Blondel, D.V.; Serr, M.; Godwin, J.; Price, T.A.R. Controlling invasive rodents via synthetic gene drive and the role of polyandry. Proc. R. Soc. B Biol. Sci. 2019, 286, 20190852. [Google Scholar] [CrossRef] [PubMed]
- Manser, A.; Lindholm, A.K.; Simmons, L.W.; Firman, R.C. Sperm competition suppresses gene drive among experimentally evolving populations of house mice. Mol. Ecol. 2017, 26, 5784–5792. [Google Scholar] [CrossRef]
- Sutter, A.; Lindholm, A.K. Detrimental effects of an autosomal selfish genetic element on sperm competitiveness in house mice. Proc. R. Soc. B Biol. Sci. USA 2015, 282, 20150974. [Google Scholar] [CrossRef] [PubMed]
- Manser, A.; Lindholm, A.K.; König, B.; Bagheri, H.C. Polyandry and the decrease of a selfish genetic element in a wild house mouse population. Evol. Int. J. Org. Evol. 2011, 65, 2435–2447. [Google Scholar] [CrossRef] [PubMed]
- Hay, B.A.; Guo, M. Gene drive-mediated population elimination for biodiversity conservation. When you come to a fork in the road, take it. Proc. Natl. Acad. Sci. USA 2022, 119, e2218020119. [Google Scholar] [CrossRef] [PubMed]
- Prowse, T.A.A.; Cassey, P.; Ross, J.V.; Pfitzner, C.; Wittmann, T.A.; Thomas, P. Dodging silver bullets: Good CRISPR gene-drive design is critical for eradicating exotic vertebrates. Proc. R. Soc. B Biol. Sci. 2017, 284, 20170799. [Google Scholar] [CrossRef] [PubMed]
- Pfitzner, C.; White, M.A.; Piltz, S.G.; Scherer, M.; Adikusuma, F.; Hughes, J.N.; Thomas, P.Q. Progress Toward Zygotic and Germline Gene Drives in Mice. CRISPR J. 2020, 3, 388–397. [Google Scholar] [CrossRef]
- Prowse, T.A.A.; Adikusuma, F.; Cassey, P.; Thomas, P.; Ross, J.V. A Y-chromosome shredding gene drive for controlling pest vertebrate populations. Elife 2019, 8, e41873. [Google Scholar] [CrossRef]
- Bull, J. Evolutionary decay and the prospects for long-term disease intervention using engineered insect vectors. Evol. Med. Public Health 2015, 2015, 152–166. [Google Scholar] [CrossRef]
- Champer, J.; Kim, I.K.; Champer, S.E.; Clark, A.G.; Messer, P.W. Performance analysis of novel toxin-antidote CRISPR gene drive systems. BMC Biol. 2020, 18, 27. [Google Scholar] [CrossRef] [PubMed]
- Hammond, A.; Kyrou, K.; Bruttini, M.; North, A.; Galizi, R.; Karlsson, X.; Crisanti, A. The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLoS Genet. 2017, 13, e1007039. [Google Scholar] [CrossRef] [PubMed]
- Sudweeks, J.; Hollingsworth, B.; Blondel, D.V.; Campbell, K.J.; Dhole, S.; Eisemann, J.D.; Lloyd, A.L. Locally Fixed Alleles: A method to localise gene drive to island populations. Sci. Rep. 2019, 9, 15821. [Google Scholar] [CrossRef] [PubMed]
- Willis, K.; Burt, A. Double drives and private alleles for localised population genetic control. PLoS Genet. 2021, 17, e1009333. [Google Scholar] [CrossRef]
- Champer, J.; Liu, J.; Oh, S.Y.; Reeves, R.; Luthra, A.; Oakes, N.; Messer, P. Reducing resistance allele formation in CRISPR gene drive. Proc. Natl. Acad. Sci. USA 2018, 115, 5522–5527. [Google Scholar] [CrossRef]
- Champer, S.; Oh, S.Y.; Liu, C.; Wen, Z.; Clark, A.; Messer, P.; Champer, J. Computational and experimental performance of CRISPR homing gene drive strategies with multiplexed gRNAs. Sci. Adv. 2020, 6, eaaz0525. [Google Scholar] [CrossRef]
- Champer, J.; Reeves, R.; Oh, S.Y.; Liu, C.; Liu, J.; Clark, A.G.; Messer, P.W. Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLoS Genet. 2017, 13, e1006796. [Google Scholar] [CrossRef]
- Metzloff, M.; Yang, E.; Dhole, S.; Clark, A.G.; Messer, P.W.; Champer, J. Experimental demonstration of tethered gene drive systems for confined population modification or suppression. BMC Biol. 2022, 20, 119. [Google Scholar] [CrossRef]
- Min, J.; Noble, C.; Najjar, D.; Esvelt, K.M. Daisy quorum drives for the genetic restoration of wild populations. BioRxiv 2017, 115618. [Google Scholar] [CrossRef]
- Min, J.; Noble, C.; Najjar, D.; Esvelt, K.M. Daisyfield gene drive systems harness repeated genomic elements as a generational clock to limit spread. BioRxiv 2017, 104877. [Google Scholar] [CrossRef]
- Noble, C.; Min, J.; Olejarz, J.; Buchthal, J.; Chavez, A.; Smidler, A.L.; Esvelt, K.M. Daisy-chain gene drives for the alteration of local populations. Proc. Natl. Acad. Sci. USA 2019, 116, 8275–8282. [Google Scholar] [CrossRef]
- Oberhofer, G.; Ivy, T.; Hay, B.A. Cleave and Rescue, a novel selfish genetic element and general strategy for gene drive. Proc. Natl. Acad. Sci. USA 2019, 116, 6250–6259. [Google Scholar] [CrossRef]
- Esvelt, K.M.; Gemmell, N. Conservation demands safe gene drive. PLoS Biol. 2017, 15, e2003850. [Google Scholar] [CrossRef] [PubMed]
- Esvelt, K.M. Daisy Drive: Safe Genes Research Plan. Responsive Science. Available online: https://www.responsivescience.org/pub/daisy-drive (accessed on 20 November 2020).
- Esvelt, K.M. Daisy Drive Systems. MIT Media Lab. Available online: http://www.sculptingevolution.org/daisydrives (accessed on 20 November 2020).
- Emerson, C.; James, S.; Littler, K.; Randazzo, F. Principles for gene drive research. Science 2017, 358, 1135–1136. [Google Scholar] [CrossRef]
- Long Kanya, C.; Alphey, L.; Annas George, J.; Bloss Cinnamon, S.; Campbell Karl, J.; Champer, J.; Akbari Omar, S. Core commitments for field trials of gene drive organisms. Science 2020, 370, 1417–1419. [Google Scholar] [CrossRef]
- Stelmach, A.; Nerlich, B.; Hartley, S. Gene Drives in the UK, US, and Australian Press (2015–2019): How a New Focus on Responsibility Is Shaping Science Communication. Sci. Commun. 2022, 44, 143–168. [Google Scholar] [CrossRef] [PubMed]
- Buchthal, J.; Evans, S.W.; Lunshof, J.; Telford, S.R.; Esvelt, K.M. Mice Against Ticks: An experimental community-guided effort to prevent tick-borne disease by altering the shared environment. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180105. [Google Scholar] [CrossRef] [PubMed]
- Funk, C.; Hefferon, M. Most Americans Accept Genetic Engineering of Animals That Benefits Human Health, but Many Oppose Other Uses. Available online: https://www.pewinternet.org/wp-content/uploads/sites/9/2018/08/PS_2018.08.16_biotech-animals_FINAL.pdf (accessed on 18 January 2022).
- Teem, J.L.; Alphey, L.; Descamps, S.; Edgington, M.P.; Edwards, O.; Gemmell, N.; Roberts, A. Genetic Biocontrol for Invasive Species. Front. Bioeng. Biotechnol. 2020, 8, 452. [Google Scholar] [CrossRef]
- Brossard, D.; Belluck, P.; Gould, F.; Wirz, C.D. Promises and perils of gene drives: Navigating the communication of complex, post-normal science. Proc. Natl. Acad. Sci. USA 2019, 116, 7692. [Google Scholar] [CrossRef]
- Kelsey, A.; Stillinger, D.; Pham, T.B.; Murphy, J.; Firth, S.; Carballar-Lejarazú, R. Global Governing Bodies: A Pathway for Gene Drive Governance for Vector Mosquito Control. Am. J. Trop. Med. Hyg. 2020, 103, 976–985. [Google Scholar] [CrossRef]
- UNCBD Ad Hoc Technical Expert Group (AHTEG). Report of the Ad Hoc Technical Expert Group on Risk Assessment. Available online: https://www.cbd.int/doc/c/a763/e248/4fa326e03e3c126b9615e95d/cp-ra-ahteg-2020-01-05-en.pdf (accessed on 12 May 2021).



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McFarlane, G.R.; Whitelaw, C.B.A.; Lillico, S.G. Gene Drive: Past, Present and Future Roads to Vertebrate Biocontrol. Appl. Biosci. 2023, 2, 52-70. https://doi.org/10.3390/applbiosci2010006
McFarlane GR, Whitelaw CBA, Lillico SG. Gene Drive: Past, Present and Future Roads to Vertebrate Biocontrol. Applied Biosciences. 2023; 2(1):52-70. https://doi.org/10.3390/applbiosci2010006
Chicago/Turabian StyleMcFarlane, Gus R., C. Bruce A. Whitelaw, and Simon G. Lillico. 2023. "Gene Drive: Past, Present and Future Roads to Vertebrate Biocontrol" Applied Biosciences 2, no. 1: 52-70. https://doi.org/10.3390/applbiosci2010006
APA StyleMcFarlane, G. R., Whitelaw, C. B. A., & Lillico, S. G. (2023). Gene Drive: Past, Present and Future Roads to Vertebrate Biocontrol. Applied Biosciences, 2(1), 52-70. https://doi.org/10.3390/applbiosci2010006




