The MRL Mitochondrial Genome Decreases Murine Muscular Dystrophy Severity
Abstract
1. Introduction
2. Results
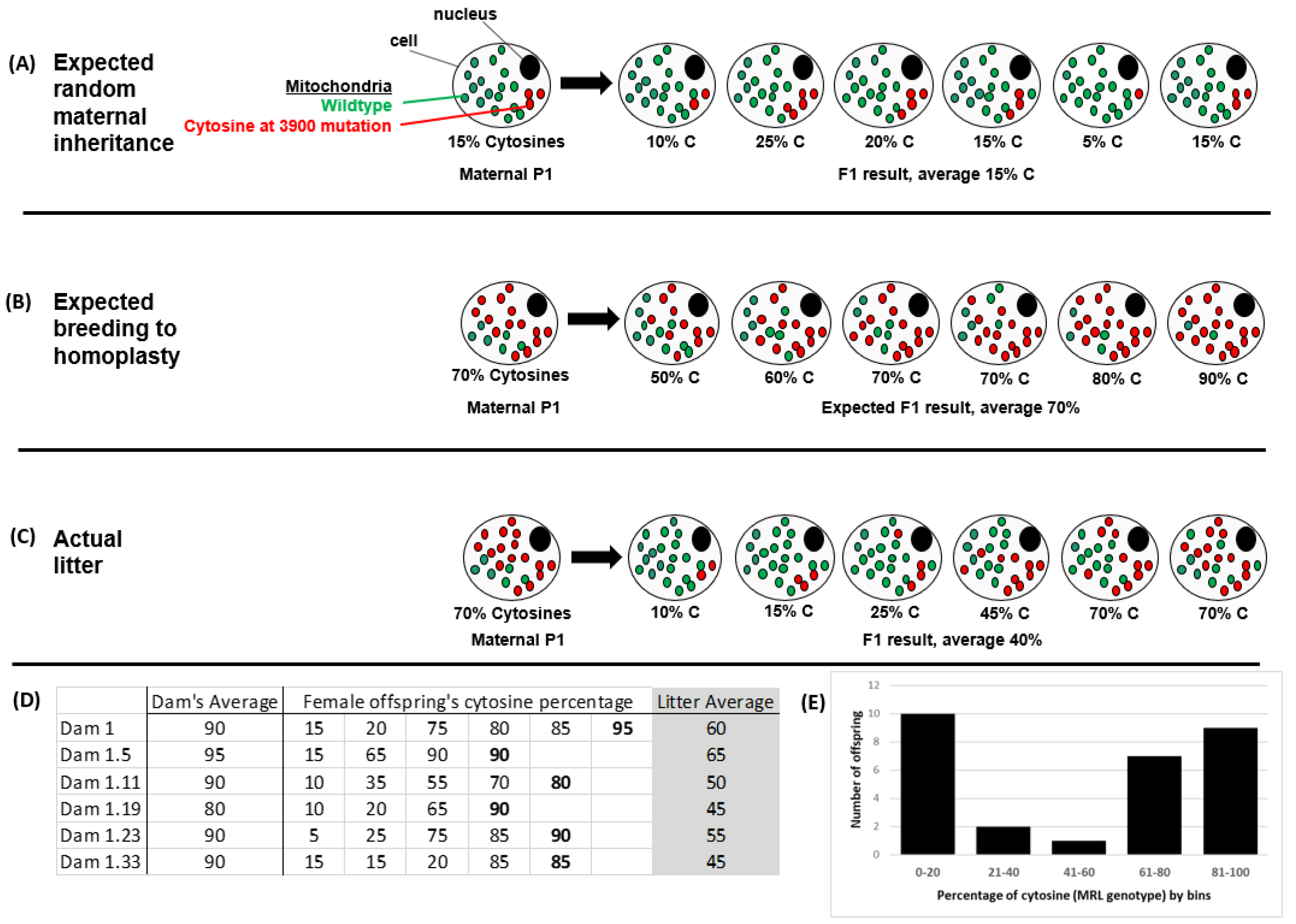
2.1. Non-Random Mitochondrial Inheritance
2.2. The MRL Mitochondrial Genome Contributes to the Muscular Dystrophy Super-Healing Phenotype
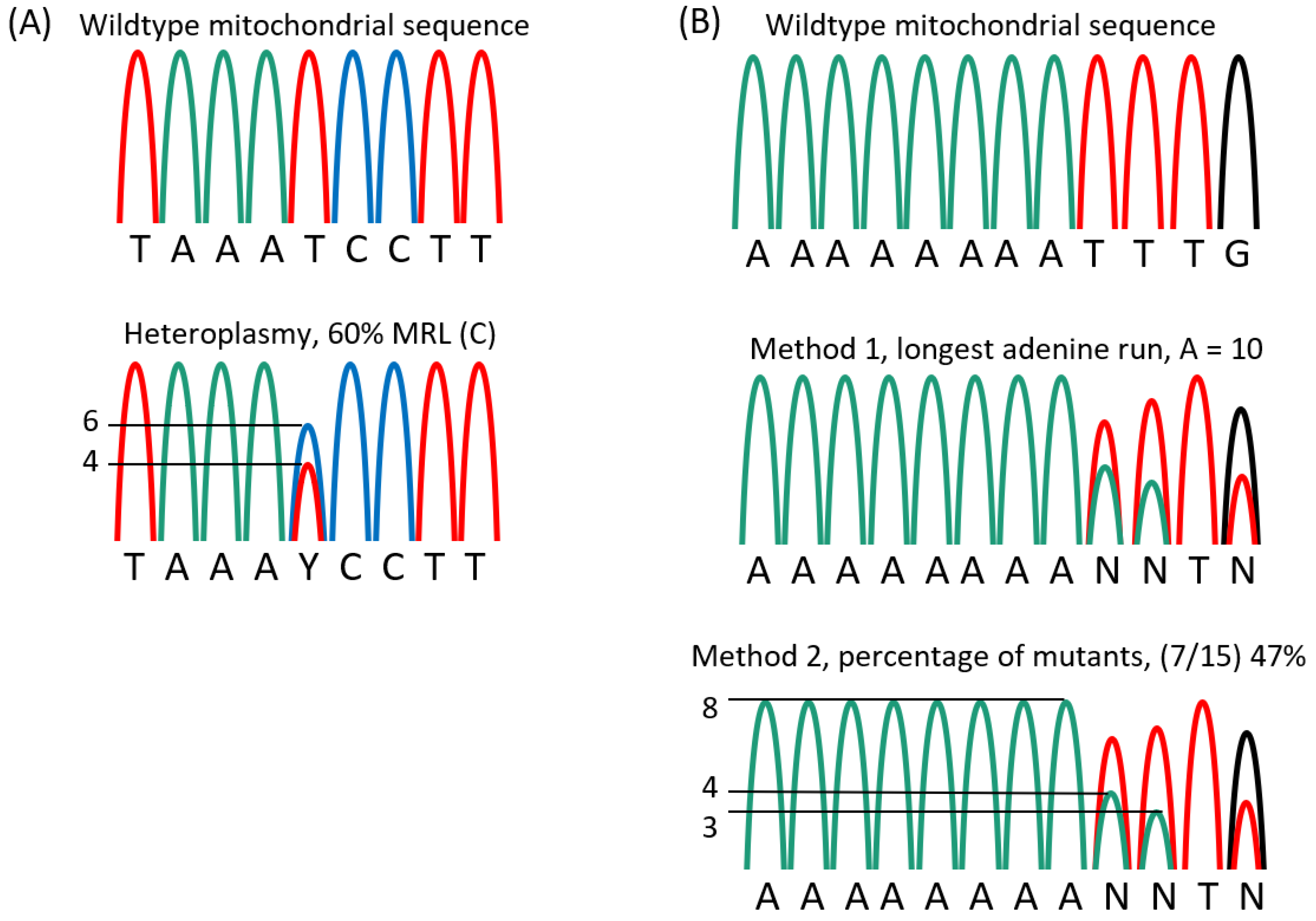
2.3. Results of Mitotyping Analysis
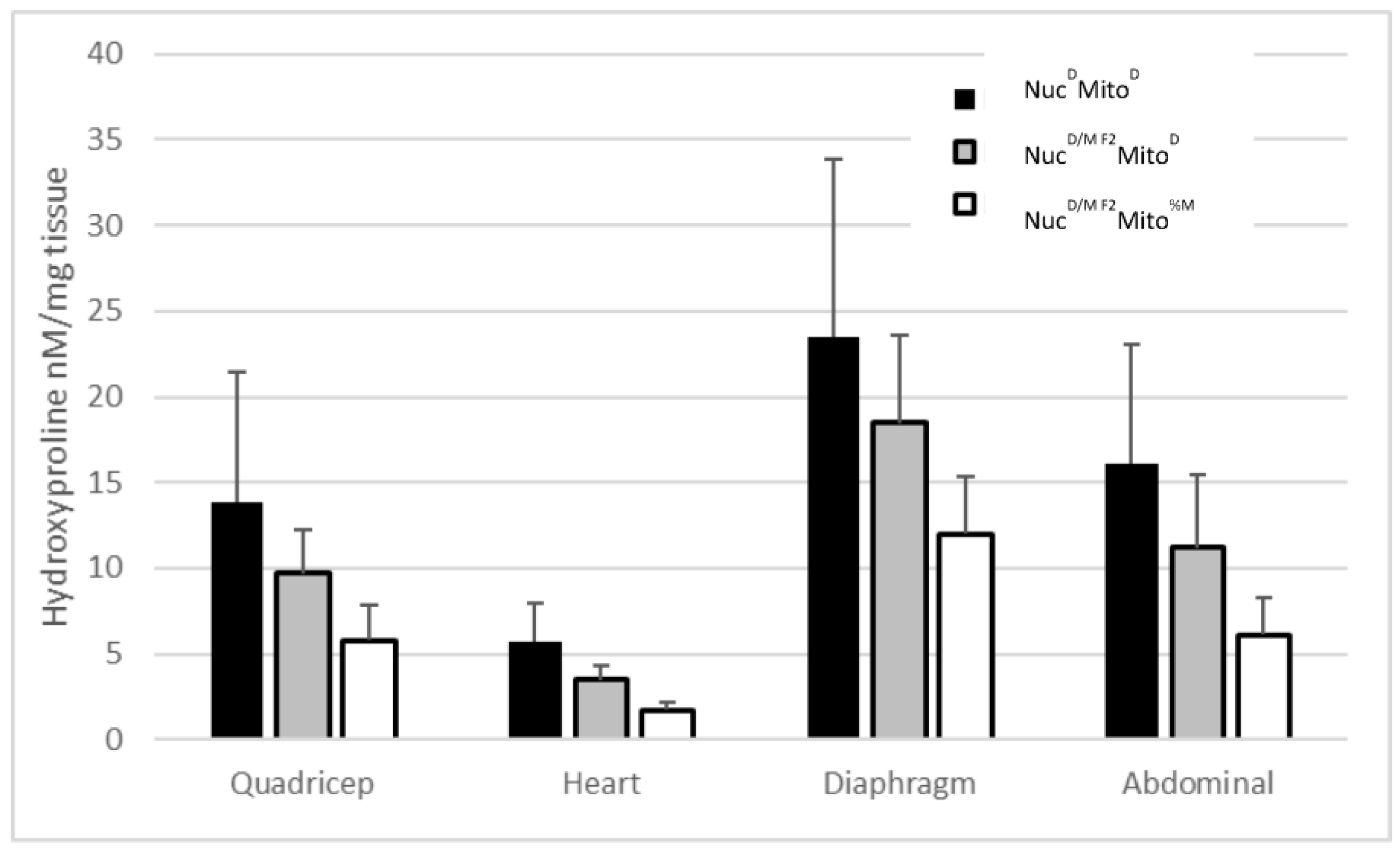
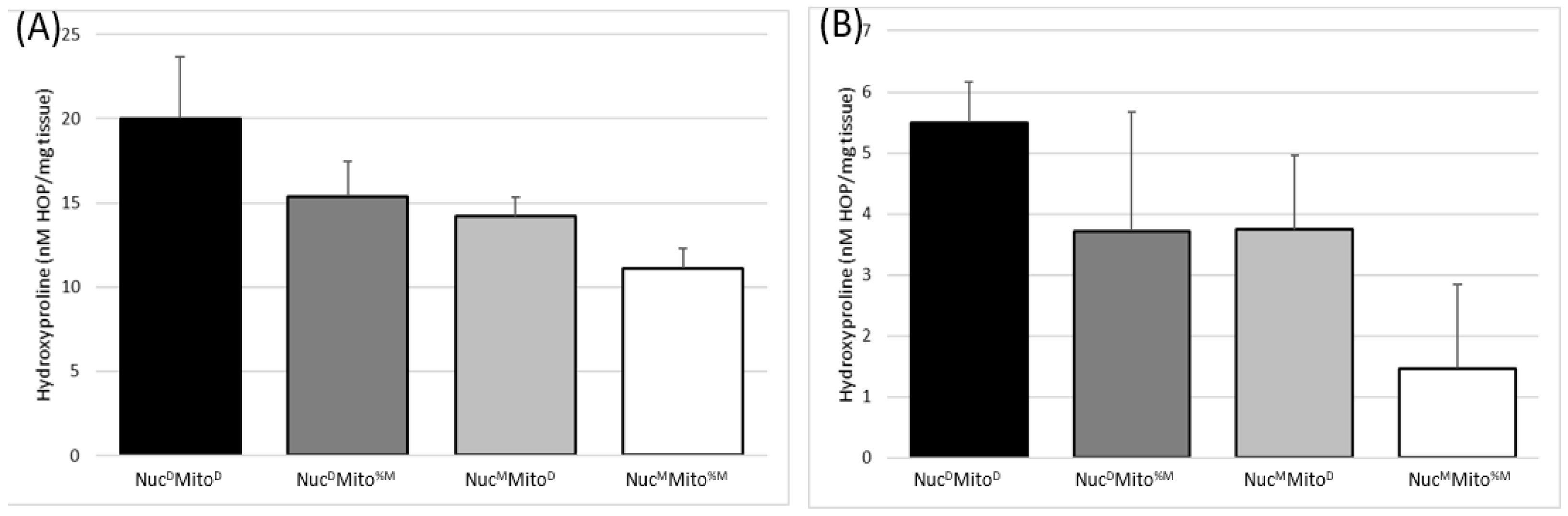
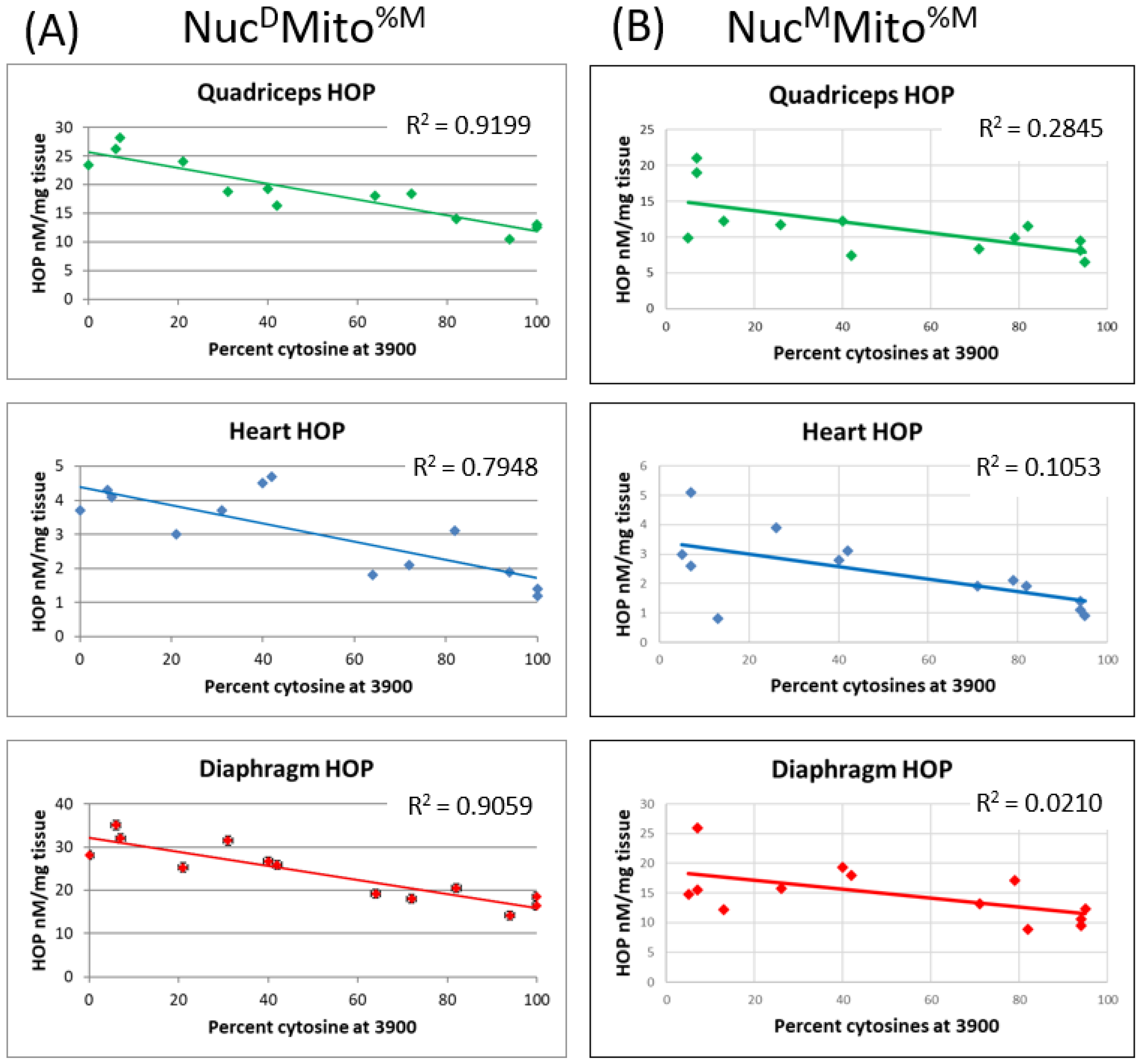
2.4. The Two Mitochondrial Heteroplasmies Differentially Affect MD Fibrosis Pathology
2.5. The MRL Mitochondrial C3900T Heteroplasmy also Correlates with pAMPK Levels and Inversely Correlates with Fibronectin Levels
3. Discussion
4. Conclusion
5. Materials and Methods
5.1. DNA Purification and Mitotyping
5.2. Immunoblot
5.3. Evan’s Blue Dye
5.4. Hydroxyproline Assay
5.5. Statistics
5.6. Ethical Approval
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Hack, A.A.; Ly, C.T.; Jiang, F.; Clendenin, C.J.; Sigrist, K.S.; Wollmann, R.L.; McNally, E.M. Gamma-sarcoglycan deficiency leads to muscle membrane defects and apoptosis independent of dystrophin. J. Cell Biol. 1998, 142, 1279–1287. [Google Scholar] [CrossRef]
- Heydemann, A.; Huber, J.M.; Demonbreun, A.; Hadhazy, M.; McNally, E.M. Genetic background influences muscular dystrophy. Neuromuscul. Disord. 2005, 15, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Heydemann, A.; Ceco, E.; Lim, J.E.; Hadhazy, M.; Ryder, P.; Moran, J.L.; Beier, D.R.; Palmer, A.A.; McNally, E.M. Latent TGF-beta-binding protein 4 modifies muscular dystrophy in mice. J. Clin. Investig. 2009, 119, 3703–3712. [Google Scholar] [CrossRef] [PubMed]
- Heydemann, A.; Swaggart, K.A.; Kim, G.H.; Holley-Cuthrell, J.; Hadhazy, M.; McNally, E.M. The superhealing MRL background improves muscular dystrophy. Skelet Muscle 2012, 2, 26. [Google Scholar] [CrossRef]
- Heydemann, A. The super super-healing MRL mouse strain. Front. Biol. 2012, 7, 522–538. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.D.; Clark, R.K.; Heber-Katz, E. A new murine model for mammalian wound repair and regeneration. Clin. Immunol. Immunopathol. 1998, 88, 35–45. [Google Scholar] [CrossRef] [PubMed]
- McBrearty, B.A.; Clark, L.D.; Zhang, X.M.; Blankenhorn, E.P.; Heber-Katz, E. Genetic analysis of a mammalian wound-healing trait. Proc. Natl. Acad. Sci. USA 1998, 95, 11792–11797. [Google Scholar] [CrossRef]
- Gourevitch, D.L.; Clark, L.; Bedelbaeva, K.; Leferovich, J.; Heber-Katz, E. Dynamic changes after murine digit amputation: The MRL mouse digit shows waves of tissue remodeling, growth, and apoptosis. Wound Repair Regen. 2009, 17, 447–455. [Google Scholar] [CrossRef]
- Ueno, M.; Lyons, B.L.; Burzenski, L.M.; Gott, B.; Shaffer, D.J.; Roopenian, D.C.; Shultz, L.D. Accelerated wound healing of alkali-burned corneas in MRL mice is associated with a reduced inflammatory signature. Invest. Ophthalmol. Vis. Sci. 2005, 46, 4097–4106. [Google Scholar] [CrossRef]
- Leferovich, J.M.; Bedelbaeva, K.; Samulewicz, S.; Zhang, X.M.; Zwas, D.; Lankford, E.B.; Heber-Katz, E. Heart regeneration in adult MRL mice. Proc. Natl. Acad. Sci. USA 2001, 98, 9830–9835. [Google Scholar] [CrossRef]
- Naseem, R.H.; Meeson, A.P.; Michael Dimaio, J.; White, M.D.; Kallhoff, J.; Humphries, C.; Goetsch, S.C.; De Windt, L.J.; Williams, M.A.; Garry, M.G.; et al. Reparative myocardial mechanisms in adult C57BL/6 and MRL mice following injury. Physiol. Genomics. 2007, 30, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Heber-Katz, E. Oxygen, Metabolism, and Regeneration: Lessons from Mice. Trends Mol. Med. 2017, 23, 1024–1036. [Google Scholar] [CrossRef] [PubMed]
- Gourevitch, D.; Kossenkov, A.V.; Zhang, Y.; Clark, L.; Chang, C.; Showe, L.C.; Heber-Katz, E. Inflammation and Its Correlates in Regenerative Wound Healing: An Alternate Perspective. Adv. Wound Care 2014, 3, 592–603. [Google Scholar] [CrossRef]
- Heydemann, A. Skeletal Muscle Metabolism in Duchenne and Becker Muscular Dystrophy-Implications for Therapies. Nutrients 2018, 10, 796. [Google Scholar] [CrossRef]
- Reid, A.L.; Alexander, M.S. The Interplay of Mitophagy and Inflammation in Duchenne Muscular Dystrophy. Life 2021, 11, 648. [Google Scholar] [CrossRef] [PubMed]
- Ljubicic, V.; Khogali, S.; Renaud, J.M.; Jasmin, B.J. Chronic AMPK stimulation attenuates adaptive signaling in dystrophic skeletal muscle. Am. J. Physiol. Cell Physiol. 2012, 302, C110–C121. [Google Scholar] [CrossRef]
- Pauly, M.; Daussin, F.; Burelle, Y.; Li, T.; Godin, R.; Fauconnier, J.; Koechlin-Ramonatxo, C.; Hugon, G.; Lacampagne, A.; Coisy-Quivy, M.; et al. AMPK activation stimulates autophagy and ameliorates muscular dystrophy in the mdx mouse diaphragm. Am. J. Pathol. 2012, 181, 583–592. [Google Scholar] [CrossRef]
- Winder, W.W.; Holmes, B.F. Insulin stimulation of glucose uptake fails to decrease palmitate oxidation in muscle if AMPK is activated. J. Appl. Physiol. 2000, 89, 2430–2437. [Google Scholar] [CrossRef]
- Niu, W.; Wang, H.; Wang, B.; Mao, X.; Du, M. Resveratrol improves muscle regeneration in obese mice through enhancing mitochondrial biogenesis. J. Nutr. Biochem. 2021, 98, 108804. [Google Scholar] [CrossRef]
- Park, C.; Ji, S.Y.; Lee, H.; Choi, S.H.; Kwon, C.Y.; Kim, S.Y.; Lee, E.T.; Choo, S.T.; Kim, G.Y.; Choi, Y.H.; et al. Mori Ramulus Suppresses Hydrogen Peroxide-Induced Oxidative Damage in Murine Myoblast C2C12 Cells through Activation of AMPK. Int. J. Mol. Sci. 2021, 22, 11729. [Google Scholar] [CrossRef]
- Tian, L.; Cao, W.; Yue, R.; Yuan, Y.; Guo, X.; Qin, D.; Xing, J.; Wang, X. Pretreatment with Tilianin improves mitochondrial energy metabolism and oxidative stress in rats with myocardial ischemia/reperfusion injury via AMPK/SIRT1/PGC-1 alpha signaling pathway. J. Pharmacol. Sci. 2019, 139, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Haycock, J.W.; MacNeil, S.; Jones, P.; Harris, J.B.; Mantle, D. Oxidative damage to muscle protein in Duchenne muscular dystrophy. Neuroreport 1996, 8, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Ljubicic, V.; Jasmin, B.J. AMP-activated protein kinase at the nexus of therapeutic skeletal muscle plasticity in Duchenne muscular dystrophy. Trends Mol. Med. 2013, 19, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Berhanu, T.K.; Holley-Cuthrell, J.; Roberts, N.W.; Mull, A.J.; Heydemann, A. Increased AMP-activated protein kinase in skeletal muscles of Murphy Roth Large mice and its potential role in altered metabolism. Physiol. Rep. 2014, 2, e00252. [Google Scholar] [CrossRef]
- Naviaux, R.K.; Le, T.P.; Bedelbaeva, K.; Leferovich, J.; Gourevitch, D.; Sachadyn, P.; Zhang, X.M.; Clark, L.; Heber-Katz, E. Retained features of embryonic metabolism in the adult MRL mouse. Mol. Genet. Metab. 2009, 96, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Mull, A.J.; Berhanu, T.K.; Roberts, N.W.; Heydemann, A. The Murphy Roths Large (MRL) mouse strain is naturally resistant to high fat diet-induced hyperglycemia. Metabolism 2014, 63, 1577–1586. [Google Scholar] [CrossRef][Green Version]
- Heydemann, A.; Gonzalez-Vega, M.; Berhanu, T.K.; Mull, A.J.; Sharma, R.; Holley-Cuthrell, J. Hepatic Adaptations to a High Fat Diet in the MRL Mouse Strain are Associated with an Inefficient Oxidative Phosphorylation System. Jacobs J. Diabetes Endocrinol. 2016, 2, 13. [Google Scholar]
- Roberts, N.W.; Gonzalez-Vega, M.; Berhanu, T.K.; Mull, A.; Garcia, J.; Heydemann, A. Successful metabolic adaptations leading to the prevention of high fat diet-induced murine cardiac remodeling. Cardiovasc. Diabetol. 2015, 14, 127. [Google Scholar] [CrossRef]
- Sachadyn, P.; Zhang, X.M.; Clark, L.D.; Naviaux, R.K.; Heber-Katz, E. Naturally occurring mitochondrial DNA heteroplasmy in the MRL mouse. Mitochondrion 2008, 8, 358–366. [Google Scholar] [CrossRef]
- Johnson, K.R.; Zheng, Q.Y.; Bykhovskaya, Y.; Spirina, O.; Fischel-Ghodsian, N. A nuclear-mitochondrial DNA interaction affecting hearing impairment in mice. Nat. Genet. 2001, 27, 191–194. [Google Scholar] [CrossRef]
- Tuppen, H.A.; Hogan, V.E.; He, L.; Blakely, E.L.; Worgan, L.; Al-Dosary, M.; Saretzki, G.; Alston, C.L.; Morris, A.A.; Clarke, M.; et al. The p.M292T NDUFS2 mutation causes complex I-deficient Leigh syndrome in multiple families. Brain 2010, 133, 2952–2963. [Google Scholar] [CrossRef] [PubMed]
- Seed, L.M.; Dean, A.; Krishnakumar, D.; Phyu, P.; Horvath, R.; Harijan, P.D. Molecular and neurological features of MELAS syndrome in paediatric patients: A case series and review of the literature. Mol. Genet. Genomic Med. 2022, 10, e1955. [Google Scholar] [CrossRef] [PubMed]
- Abbott, J.A.; Francklyn, C.S.; Robey-Bond, S.M. Transfer RNA and human disease. Front. Genet. 2014, 5, 158. [Google Scholar] [CrossRef] [PubMed]
- Sharpley, M.S.; Marciniak, C.; Eckel-Mahan, K.; McManus, M.; Crimi, M.; Waymire, K.; Lin, C.S.; Masubuchi, S.; Friend, N.; Koike, M.; et al. Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell. 2012, 151, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Waymire, K.G.; Narula, N.; Li, P.; Rocher, C.; Coskun, P.E.; Vannan, M.A.; Narula, J.; Macgregor, G.R.; Wallace, D.C. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science 2008, 319, 958–962. [Google Scholar] [CrossRef] [PubMed]
- Jenuth, J.P.; Peterson, A.C.; Shoubridge, E.A. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat. Genet. 1997, 16, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Sobenin, I.A.; Revin, V.V.; Orekhov, A.N.; Bobryshev, Y.V. Mitochondrial aging and age-related dysfunction of mitochondria. Biomed. Res. Int. 2014, 2014, 238463. [Google Scholar] [CrossRef]
- Silaidos, C.; Pilatus, U.; Grewal, R.; Matura, S.; Lienerth, B.; Pantel, J.; Eckert, G.P. Sex-associated differences in mitochondrial function in human peripheral blood mononuclear cells (PBMCs) and brain. Biol. Sex Differ. 2018, 9, 34. [Google Scholar] [CrossRef]
- Hafner, P.; Bonati, U.; Erne, B.; Schmid, M.; Rubino, D.; Pohlman, U.; Peters, T.; Rutz, E.; Frank, S.; Neuhaus, C.; et al. Improved Muscle Function in Duchenne Muscular Dystrophy through L-Arginine and Metformin: An Investigator-Initiated, Open-Label, Single-Center, Proof-Of-Concept-Study. PLoS ONE 2016, 11, e0147634. [Google Scholar] [CrossRef]

| N | df | Pearson’s Correlation | p< | |
|---|---|---|---|---|
| All Sgcg–/– mice | 28 | 26 | 0.5883 | 0.01 |
| Sgcg–/–NucMMito%M mice | 17 | 15 | 0.6006 | 0.05 |
| Sgcg–/–NucDMito%M mice | 11 | 9 | 0.6173 | 0.05 |
| N | df | Pearson’s Correlation | p< | |
|---|---|---|---|---|
| All Sgcg–/– mice | 28 | 26 | −0.67 | 0.01 |
| Sgcg–/–NucMMito%M mice | 17 | 15 | −0.718 | 0.01 |
| Sgcg–/–NucDMito%M mice | 11 | 9 | −0.643 | 0.05 |
| Primer | Sequence |
|---|---|
| M07 T3900C | 5′ TTT CTT TAC CAA TTT TTA CAG GGG GG 3′ |
| M29 T3900C | 5′ GGG GAT AAT TGC TAG TAG GCT GAA TA 3′ |
| M30 A tract at 9821 | 5′ GCT CTT CTA CTT CCA CTA CCA TGA GC 3′ |
| M31 A tract at 9821 | 5′ GCT ATG GAG CTT ATG GAG TT GAG TT 3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holley-Cuthrell, J.; Iqbal, A.; Heydemann, A. The MRL Mitochondrial Genome Decreases Murine Muscular Dystrophy Severity. Muscles 2023, 2, 37-50. https://doi.org/10.3390/muscles2010005
Holley-Cuthrell J, Iqbal A, Heydemann A. The MRL Mitochondrial Genome Decreases Murine Muscular Dystrophy Severity. Muscles. 2023; 2(1):37-50. https://doi.org/10.3390/muscles2010005
Chicago/Turabian StyleHolley-Cuthrell, Jenan, Aqsa Iqbal, and Ahlke Heydemann. 2023. "The MRL Mitochondrial Genome Decreases Murine Muscular Dystrophy Severity" Muscles 2, no. 1: 37-50. https://doi.org/10.3390/muscles2010005
APA StyleHolley-Cuthrell, J., Iqbal, A., & Heydemann, A. (2023). The MRL Mitochondrial Genome Decreases Murine Muscular Dystrophy Severity. Muscles, 2(1), 37-50. https://doi.org/10.3390/muscles2010005






