Abstract
The winery sector represents one of the most important agricultural industries in Mediterranean country. Wine production processes generate a large amount of wastes and wastewaters that must be treated before their release in the environment. Among these wastes, wine lees, defined as the viscous material that settles on the bottom of fermenters, represent about 13% of the total wine production. The wine lees do not have applications within a circular economy approach, due to their low value; ethanol and tartaric acid are the only compounds recovered, while the rest is usually not valorized. The aim of this study is to explore the possible cultivation of microalgae on a liquid fraction of wine lees in a batch test at different substrate/inoculum dilutions. The results highlighted that Chlorella vulgaris can grow on wine lees at 1:10 and 1:5 dilutions, but a spontaneous yeast-microalgae consortium is observed (biomass production of 2 g l−1). A high lipid and protein storage was detected in the yeast-microalgae consortium (34.56 ± 13.70% and 39.73 ± 4.49%, respectively), associated with a high sCOD and polyphenols removal (99.95 ± 0.01% and 92.31 ± 0.02%, respectively), encouraging biological wine lees treatment.
1. Introduction
Worldwide wine production reached 300 millions of hectoliter in 2018 [1], and in Italy it is one of the most important agro-industrial sectors, with 17% of wine produced per year located in the Veneto region [2,3]. The winemaking process, after the grape harvest, takes place in autumn and is characterized by fermentation and maceration steps that produce several wastes, such as bagasse, grape marc and lees [4]. During the fermentative process, wine lees interact with polyphenol compounds, providing the color and organoleptic properties required to obtain high-quality wine [5,6]. The composition of wine lees is usually related to the type of grapes used for wine production, microorganisms (bacteria and yeasts), and organic and inorganic compounds; it has very low pH values and a high chemical and biological oxygen demand [4]. Based on Italian Law 238/2016 [7,8], winemakers must cover winery waste disposal and storage-related costs, with a negative effect on their economy. Although there are a few very significant examples of industrial applications of lees (as well as other byproducts) following the principles of circular economy (e.g., Caviro sca, Italy [9]), the valorization of lees is by no means a generalized process and a highly covered research topic [10]. Ioannidou et al. [11] studied wine lees application in a circular economy approach with the application of several chemical solvents and enzymatic extraction to recover polyphenols, ethanol, tartrate salt and hydrolysate rich in nutrients for polymer production. The limit of this treatment is associated with the high cost required to recover the products. In addition, publications dealing with the recovery of matter from wine lees by means of biotechnological treatments are limited [12]. There is great interest in biorefinery development focused on a complete by-product treatment, and an innovative approach could be the application of wine lees as a substrate for microalgal cultivation. No further research has been focused on wine lees as a direct substrate for microalgae growth, because the high concentration of polyphenols, ethanol and other compounds could limit microalgae survival. Bibliographic research identified only two studies that used wine lees as a substrate; however, these studies were generic and used a single type of wine lees (white wine lees, WL), with the lowest polyphenol concentration, as a substrate [13,14].
The aim of this study is to exploit different types of wine lees for microalgae growth, adopting Chlorella vulgaris as the microalgal strain, in order to identify a new wine lees-based process that could be deployed in loco by winemakers (either small or cooperative wineries) to reduce their waste transportation and disposal. The culturing of microalgae comes with a phytoremediation effect on wine lees and allows one to produce a microalgal biomass that could be exploited in a circular economy approach (biorefineries).
2. Materials and Methods
2.1. Winery Lees Characterization
White (WL) and red (RL) wine lees were obtained from a winemaking company located in the Prosecco wine production area (North-East of Italy) and were chemically and physically characterized according to the APHAT and APAT-IRSA/CNR methodology (Table 1) [15,16]. Before the microalgae growth test, only centrifugation (at 10,161× g for 5 min) was applied on WL and RL as pretreatment to remove the solid fraction. No sterilization was applied on either the WL or RL liquid fraction.

Table 1.
White wine lees (WL) and red wine lees (RL) characterization.
2.2. Microalgae Strain and Experimental Set-Up
Chlorella vulgaris (provided by the Algal Collection of University of Federico II, Napoli, Italia) was cultivated in batch tests and cultured under mixotrophic growth, under continuous irradiation (3.9 klux), mechanical agitation (330 rpm) and air bubbling (2.3 lh−1 vvm). Erlenmeyer flasks (volume = 200 mL) were used for the experimental growth tests. During the tests, the biomass growth was monitored twice per week by cell count and dry weight analyses.
The experimental set-up was divided into three different experiments:
- RUN 1: batch growth of C. vulgaris in WL and RL liquid fractions at a 1:10 dilution ratio;
- RUN 2: batch growth of C. vulgaris in RL at reduced dilution ratios (1:5 and 1:2).
- RUN 3: batch growth of C. vulgaris in filtered RL at 1:5 and 1:2 dilution ratios.
The RL liquid fraction was filtered by using acetate cellulose filters (Whatman) with 0.2 µm porosity. Control conditions were obtained by growing C. vulgaris on the BG11 synthetic medium at 1:10 (RUN1), 1:5 and 1:2 dilution ratios (RUN 2 and RUN 3, respectively). At the end of each batch growth test, the obtained biomass was recovered and lyophilized for subsequent lipid, starch, and protein quantification. Soluble chemical oxygen demand (sCOD), pH, polyphenols and ionic chromatography analyses were performed on the liquid fraction in accordance with the APHA and APAT-IRSA/CNR methodologies [15,16]. All analyses and tests were performed in duplicate for each experimental condition.
2.3. Biomass Analysis
The monitoring of biomass growth was carried out using the cell count with a Bürker chamber observed under a Leika microscope, at a 40× magnification ratio. Dry weight was measured using acetate cellulose filters (0.45 µm, Whatman, Maidstone, UK) dried at 105 °C. Lipid, starch, and protein quantifications were performed on lyophilized biomass obtained by an EDWARDS Freeze Dryer Modulyo at −40 °C and EDWARDS A653-01-903 vacuum pump. Lipid gravimetrical quantification was carried out on the recovered biomass by the Folch et al. methodology [17] using a 2:1 methanol:chloroform solution. The total lipid percentage was obtained by applying Equation (1):
The total protein percentage was obtained using the Biuret reagent (Merk KGaA, Darmstadt, Germany) [18] and an absorbance measurement at 540 nm. Proteins extraction was carried out following the Safafar et al. methodology [19] with NaOH 0.5 M. The calibration line for protein quantification was obtained using Albumin fraction V from Bovine serum for biochemistry, at 2 mg ml−1 (Merk KGaA, Germany). Total Starch Kit Analysis, Method 76-13.01 (Meganzyme, Wicklow, Ireland) was used for starch quantification in lyophilized microalgae biomass, with GOPOD reagent. Spectrophotometric analysis was carried out at 510 nm.
Pigment quantification was performed using the Lichtenhaler et al. methodology [20] with methanol as solvent. Samples were analyzed on a spectrophotometer UV-VIS at three different λ: 665.2 nm, 652.4 nm and 470 nm.
3. Results and Discussion
3.1. Wine Lees as Substrate for Microalgae Growth
3.1.1. RUN1: 1:10-Diluted WL and RL as Substrate for C. vulgaris Growth
Typically, wine lees as a substrate for microalgae cultivation require pretreatments to remove yeast and fungal contamination caused by wine-native microorganisms (Saccharomyces bayanus, Candida albicans, Hanseniaspora and Merchnikowia) [13,14,21]. In this study, different tests were carried out using only centrifugation as pretreatment to investigate microalgal survival on winery waste and whether spontaneous yeast-microalgae consortia were effective in waste remediation. In RUN 1, WL and RL were diluted at 1:10 and tested as a substrate for C. vulgaris cultivation without sterilization. The results obtained in this test showed that C. vulgaris was able to grow and was predominant. As reported in Figure 1, the dry weight obtained in the presence of WL and RL was 1.71 ± 0.02 g l−1 and 1.61 ± 0.09 g l−1, respectively, values comparable with the control condition (1.77 ± 0.09 g l−1). The increase of dry weight on day 4 under the RL condition was associated with the proliferation of native microorganism, but during the long period, C. vulgaris became predominant in the batch test. The effect of WL and RL on microalgae was detected mainly in the biomass composition, where the application of RL increased the lipid and protein concentration (>40%) (Table 2).
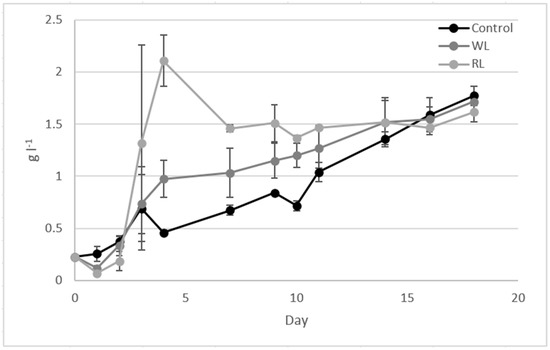
Figure 1.
Dry weight analysis during RUN 1 using 1:10-diluted WL and RL.

Table 2.
Biomass characterization during RUN1 using 1:10-diluted WL and RL.
3.1.2. RUN2: RL 1:5 and 1:2 Diluted as Substrate
Promising results in the biomass composition obtained by RL cultivation (RUN1) encourage the application of centrifugated and diluted (1:5 and 1:2) RL as a substrate (RUN2). The highest concentration of RL in the growth medium caused an increase of microorganism contamination, and C. vulgaris was not the dominant strain at 1:5 dilution. The direct microscope observation highlighted the presence of yeasts (Figure 2b), probably belonging to the Candida genus, which is commonly present in must and wine, and the formation of a yeast-microalgae consortium. Upon application of 1:2-diluted RL, C. vulgaris underwent total inhibition, and yeast became the dominant type of microbial biomass (Figure 2a, biomass had a dark color); with the application of 1:5-diluted RL, microalgae growth was detected, with the establishment of a microalgae-yeast consortium (Figure 2c, biomass with green color).

Figure 2.
(a) Batch test with RL 1:2; (b) view at 40X of yeast (probably Candida spp.); (c) batch test with RL 1:5.
As reported in Figure 3 and Table 3, the yeast-microalgae consortium biomass obtained at the end of the test was lower than the control condition, but the lipid and protein concentration was comparable to the control. However, the dry weight analysis highlighted the faster increase of yeast biomass under all tested conditions with RL compared to the control condition. At the end of yeast’s exponential phase, after 9 days of cultivation, microalgae growth was only detected for the RL 1:5 condition. An interesting lipid percentage was obtained for the yeast biomass under the 1:2-diluted RL condition: the lipid concentration was significantly higher than the control condition with C. vulgaris (p value = 0.05). Lipid storage detected in yeast biomass could be correlated with the experimental conditions insofar as the yeast lipids’ accumulation was strictly associated with cultivation conditions such as the incubation temperature [22,23]. The effect of culture conditions on winery yeasts (S. cerevisiae, S. uvarum and Candida sp.) was detected in other studies, where lipids storage resulted in about 34% [22,23]. As reported by Mishra et al. [23], lipid concentration in yeast biomass can range between 5–60%, depending on the cultivation temperature acting on lipids’ composition and accumulation in yeast strains.
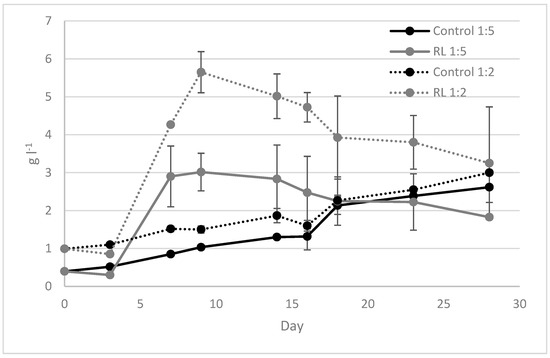
Figure 3.
Dry weight analysis during RUN2 using 1:5- and 1:2-diluted RL.

Table 3.
Biomass characterization during RUN2 with 1:5- and 1:2-diluted RL.
3.1.3. RUN3: 1:5- and 1:2-Diluted RL (Filtered) as Substrate
Microalgae inhibition growth and yeast proliferation at low RL dilution suggested the need to add another pretreatment step to wine lees to observe the effect on microalgae growth. During RUN 3, two mechanical pretreatments were carried out (centrifugation and filtration at 0.2 µm), and the filtered RL fraction was diluted as in the previous set of runs (1:5 and 1:2). During the test, the presence of yeasts was identified in all test conditions, just as in RUN SET 2. Yeast-microalgae consortium formation and an increase of the C. vulgaris cell size, from 0.05 mm to 0.1 mm, were detected under the RL 1:5 condition (Figure 4). The yeast presence after filtration could be correlated with environmental microorganism contamination, as during winemaking processes. As reported for RUN SET 2, the application of 1:2-diluted RL resulted in the thriving of yeasts and in the total inhibition of the microalgae biomass. Microalgae inhibition and yeast dominance in the culture could be a consequence of the xenic condition and chemical characteristics of wine lees, where a low pH and high polyphenol concentration could limit microalgae survival, while they are favorable for yeast proliferation. The biomass obtained at the end of the 1:5-diluted RL test showed a composition comparable with that of the control biomass (constituted only of C. vulgaris), with a high protein and lipid percentage of 79.67 ± 2.06% and 45.26 ± 11.14%, respectively (Table 4).
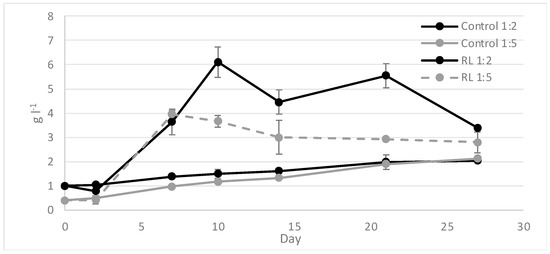
Figure 4.
Dry weight analysis during RUN3 with RL 1:5 and 1:2 after centrifugation and filtration.

Table 4.
Biomass monitoring during RUN 3 with RL 1:5 and 1:2 after centrifugation and filtration.
3.2. Phytoremediation Effect
The direct release of wine lees in the environment shall be avoided, pursuant to Italian decree D. Lgs 152/06 [24], so that the storage and transport cost of wine lees burdens wine producers. Wine lees’ use as a substrate for microorganism cultivation could be a solution to avoiding their economic disposal and transport costs if, in addition to biomass production with an economic value, the biological treatment had a phytodepuration effect.
The sCOD and ammonia concentration at the end of RUN1 and RUN2 (Table 5a,b) were below the limits reported in Italian D.Lgs 152/06 for surface water release; on the contrary, the application of filtration as pretreatment (Table 5c, RUN3) negatively influenced the phytoremediation effect. The filtration probably removes some microorganism that has a synergic effect on organic and inorganic removal. As reported by the Policastro et al. study [25], the degradation of complex compounds from winery waste to produce energy or biomass has some limitations if this process is performed aseptically with monoculture strains. Instead, the use of mixed cultures could enhance the remediation of winery waste due to the syntrophic effect of different metabolisms. Co-culture processes could be applied using native or specific microorganisms capable of removing compounds from waste substrate, and the processes do not require costly sterile conditions.

Table 5.
sCOD, polyphenols and ion removal at the end of (a) RUN 1, (b) RUN 2 and (c) RUN 3.
Although the phytoremediation effect detected during tests was not enough for environmental release, the removal of positive compounds by microalgae-yeast consortium and yeast from wine lees was identified and requires further studies. The different biomass proliferations permitted the removal of different ions. In fact, during RUN 3 1:5, microalgae-yeast consortium proliferation increased Mg2+ removal insofar as this ion represents the chlorophyll core. On the other hand, yeast-native proliferation in RUN 3 1:2 showed strong Ca2+ removal insofar as this ion is the activation regulation factor for the citric acid cycle.
4. Conclusions
The use of wine lees as a substrate for microbial cultivation could open a new circular-economy opportunity for waste treatment in the winery domain. This research has improved the application potential of microalgae cultivation in wine lees and its phytoremediation effect. Microalgae growth was detected when a high dilution of white and red wine lees was applied, using centrifugation as the pretreatment. A low dilution of wine lees allowed for the proliferation of the endogenous yeast population, while at a 1:5 dilution of wine lees, a microalgae-yeast consortium was detected. The consortium showed a promising composition from a biorefinery perspective (41.63 ± 10.11% and 44.58 ± 6.30% of lipids and protein, respectively). A phytoremediation effect of microalgae and yeast on wine lees was detected in all the test conditions, with promising sCOD, polyphenols and ions removal. The data obtained in this study showed a promising application of microalgae and native yeast from wine lees for a new circular-economy approach to the treatment of winery waste such as dedicated biorefineries.
Author Contributions
Conceptualization, methodology, investigation, data curation and writing—original draft preparation: P.S.; writing—review and editing: M.B. and C.C.; Conceptualization, validation, supervision, funding acquisition: C.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Ethical review and approval were waived for this study due to not being applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data will be available upon request.
Acknowledgments
Serena Wine Spa’s support is gratefully acknowledged.
Conflicts of Interest
The authors declare no conflict of interest.
References
- OIV. 2019 Statistical Report on World Vitiviniculture. 2019. Available online: https://www.oiv.int/sites/default/files/documents/oiv-2019-statistical-report-on-world-vitiviniculture.pdf (accessed on 15 January 2023).
- Bonari, E.; Ercoli, L.; Silvestri, N.; Carcea, G.; Barresi, F. Linee Guida per L’utilizzazione Agronomica Delle Acque di Vegetazione e Delle Acque Reflue da Aziende Agroalimentari; APAT: Rome, Italy, 2007; ISBN 978-88-448-0301-8. [Google Scholar]
- ISTAT. Statistiche on Line: Settore Agricoltura. Vari anni. Available online: https://www.istat.it (accessed on 10 August 2022).
- Devesa-Rey, R.; Vecino, X.; Varela-Alende, J.L.; Barral, M.T.; Cruz, J.M.; Moldes, A.B. Valorization of winery waste vs. the costs of not recycling. Waste Manag. 2011, 31, 2327–2335. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Serradilla, J.A.; de Castro, M.D.L. Role of lees in wine production: A review. Food Chem. 2008, 111, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.J.; Maicas, S. Valorization of winery and oil mill wastes by microbial technologies. Food Res. Int. 2015, 73, 13–25. [Google Scholar] [CrossRef]
- D. Lgs 479/2008. Disposizioni di Attuazione dei Regolamenti (CE) n. 479/2008 del Consiglio e (CE) n. 555/2008 Della Commissione per Quanto Riguarda L’applicazione Della Misura Della Distillazione dei Sottoprodotti Della Vinificazione. Available online: https://www.tuttocamere.it/files/camcom/2008_11_27.pdf (accessed on 22 November 2022).
- Ministero Delle Polite Agricole Alimentari e Forestali. Testo Unico Della Vite e del Vino. Legge 238 del 12/12/2016. 2016. Masaf—Testo Unico Vite e Vino L. 238/16 (politicheagricole.it). Available online: https://www.politicheagricole.it/flex/cm/pages/ServeBLOB.php/L/IT/IDPagina/12012 (accessed on 22 November 2022).
- Available online: https://www.caviro.com/en/ (accessed on 13 May 2023).
- De Iseppi, A.; Lomolino, G.; Marangon, M.; Curioni, A. Current and future strategies for wine yeast lees valorization. Food Res. Int. 2020, 137, 109352. [Google Scholar] [CrossRef] [PubMed]
- Ioannidou, S.M.; Pateraki, C.; Ladakis, D.; Papapostolou, H.; Tsakona, M.; Vlysidis, A.; Kookos, I.K.; Koutinas, A. Sustainable production of bio-based chemicals and polymers via integrated biomass refining and bioprocessing in a circular bioeconomy context. Bioresour. Technol. 2020, 307, 123093. [Google Scholar] [CrossRef] [PubMed]
- Calicchio Berardi, P.; Dias, J.M. How Has the Wine Sector Incorporated the Premises of Circular Economy? J. Environ. Sci. Eng. B 2019, 8, 108–117. [Google Scholar] [CrossRef]
- Salati, S.; D’Imporzano, G.; Menin, B.; Veronesi, D.; Scaglia, B.; Abbruscato, P.; Mariani, P.; Adani, F. Mixotrophic cultivation of Chlorella for local protein production using agro-food by-products. Bioresour. Technol. 2017, 230, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Veronesi, D.; D’Imporzano, G.; Menin, B.; Salati, S.; Adani, F. Organic wastes/by-products as alternative to CO2 for producing mixotrophic microalgae enhancing lipid production. Bioprocess Biosyst. Eng. 2020, 43, 1911–1919. [Google Scholar] [CrossRef] [PubMed]
- APAT; IRSA/CNR. Metodologie Analitiche per il Controllo della Qualità Delle Acque [Analytical Methodologies for Water Quality Management]; APAT: Roma, Italy, 2003; ISBN 88-448-0083-7. [Google Scholar]
- APHA/AWWA/WEF. Standard Methods for the Examination of Water and Wastewater, 22nd ed.; American Public Health Association: Washington, DC, USA, 2012; ISBN 978-087553-013-0. [Google Scholar]
- Folch, J.; Lees, M.; Stanley, G.H.S. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Janairo, G.; Linley, M.S.; Yap, L.; Llanos-Lazaro, N.; Robles, J. Determination of the Sensitivity Range of Biuret Test for Undergraduate biochemistry experiments. E-J. Sci. Technol. 2011, 6, 77–83. [Google Scholar]
- Safafar, H.; Hass, M.Z.; Møller, P.; Holdt, S.L.; Jacobsen, C. High-EPA biomass from Nannochloropsis salina cultivated in a flat-panel photo-bioreactor on a process water-enriched growth medium. Mar. Drugs 2016, 14, 144. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K.; Buschmann, C. Chlorophylls and Carotenoids Measurement and UV-VIS characterization Lichtenthaler 2001. Curr. Protoc. Food Anal. Chem. 2001, 1, F4. 3.1–F4. 3.8. [Google Scholar] [CrossRef]
- León-Vaz, A.; León, R.; Díaz-Santos, E.; Vigara, J.; Raposo, S. Using agro-industrial wastes for mixotrophic growth and lipids production by the green microalga Chlorella sorokiniana. New Biotechnol. 2019, 51, 31–38. [Google Scholar] [CrossRef]
- Froissard, M.; Canonge, M.; Pouteaux, M.; Cintrat, B.; Mohand-Oumoussa, S.; Guillouet, S.E.; Chardot, T.; Jacques, N.; Casaregola, S. Lipids containing medium-chain fatty acids are specific to post-whole genome duplication Saccharomycotina yeasts Genome evolution and evolutionary systems biology. BMC Evol. Biol. 2015, 15, 97. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Prasad, R. An overview of lipids of Candida albicans. Prog. Lipid Res. 1990, 29, 65–85. [Google Scholar] [CrossRef] [PubMed]
- INDAM. ACQUE DI SCARICO: Acque Reflue Urbane e Acque Reflue Industriali. All 5, P Terza, DLgs N152 n.d.:152. Available online: https://www.indam.it/fileadmin/user_upload/tabelle/INDAM_acque_scarico.pdf (accessed on 12 January 2023).
- Pilocastro, G.; Carraturo, F.; Compagnone, M.; Giuda MFabbricino, M. Enhancing hydrogen production from winery wastewater through fermentative microbial culture selection. Bioresour. Technol. Rep. 2022, 19, 101196. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).