A Past Genetic Bottleneck from Argentine Beans and a Selective Sweep Led to the Race Chile of the Common Bean (Phaseolus vulgaris L.)
Abstract
1. Introduction
2. Results
2.1. SNP Genotyping
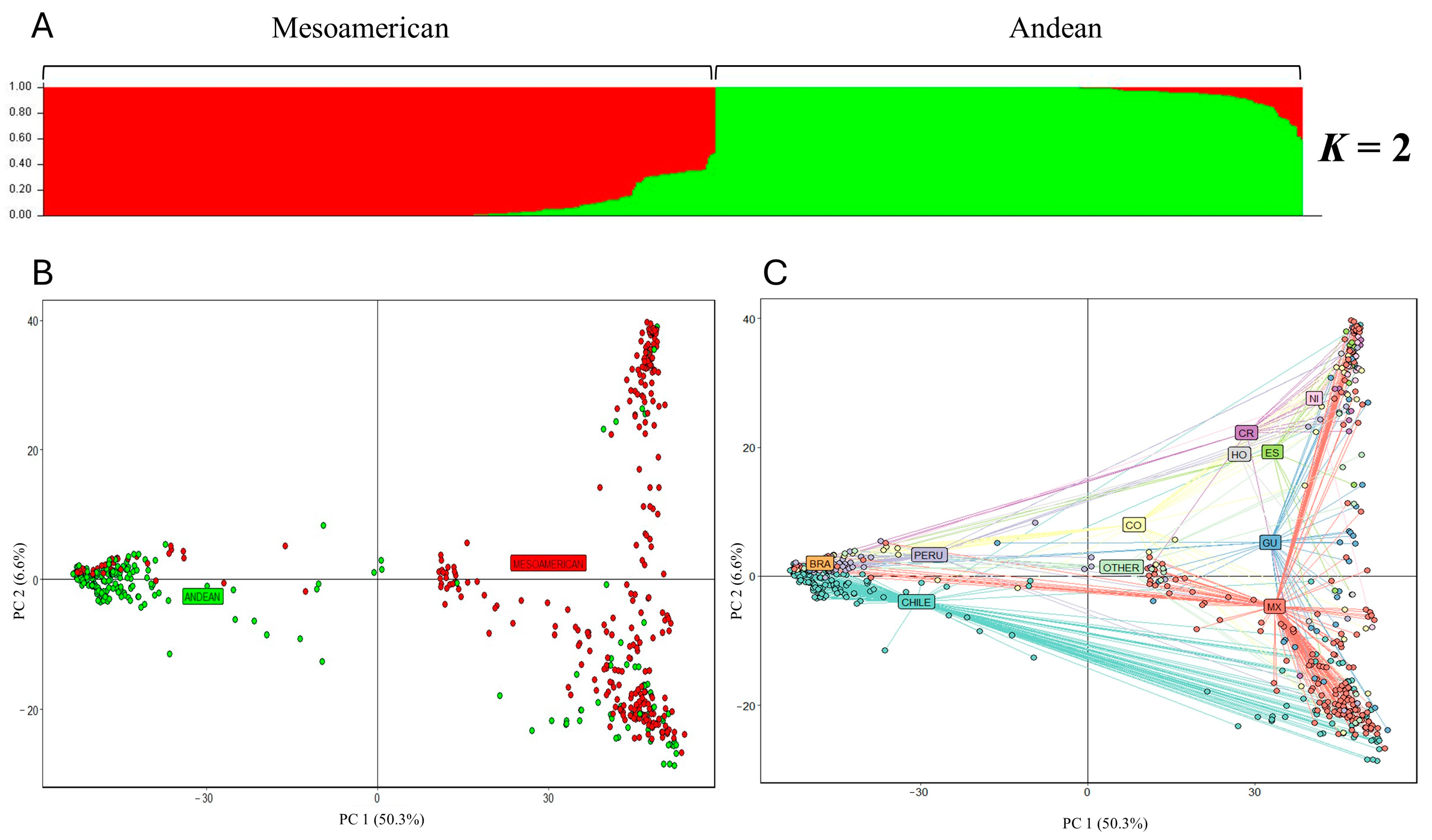
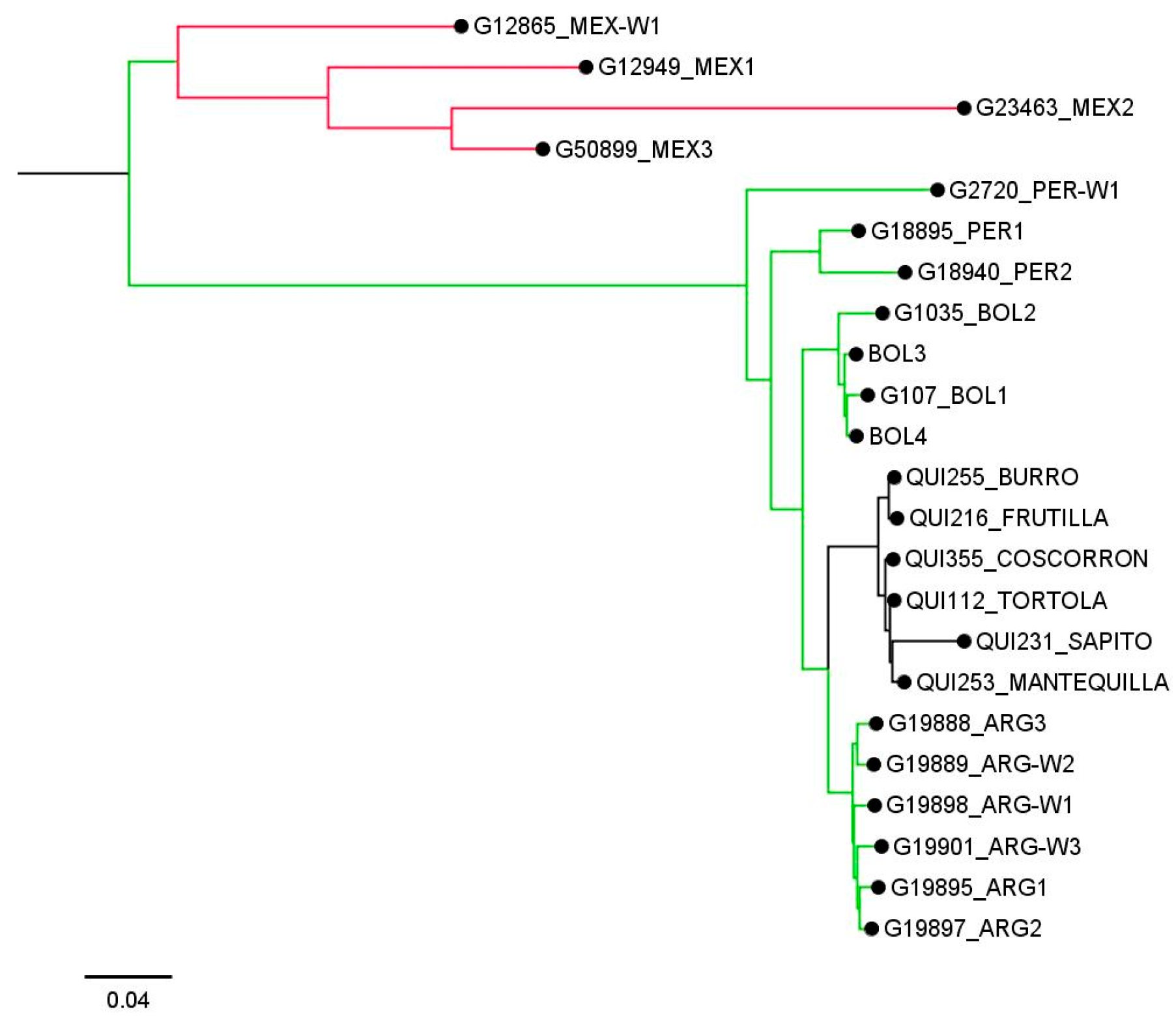
2.2. Population Structure and Relationships among Individuals
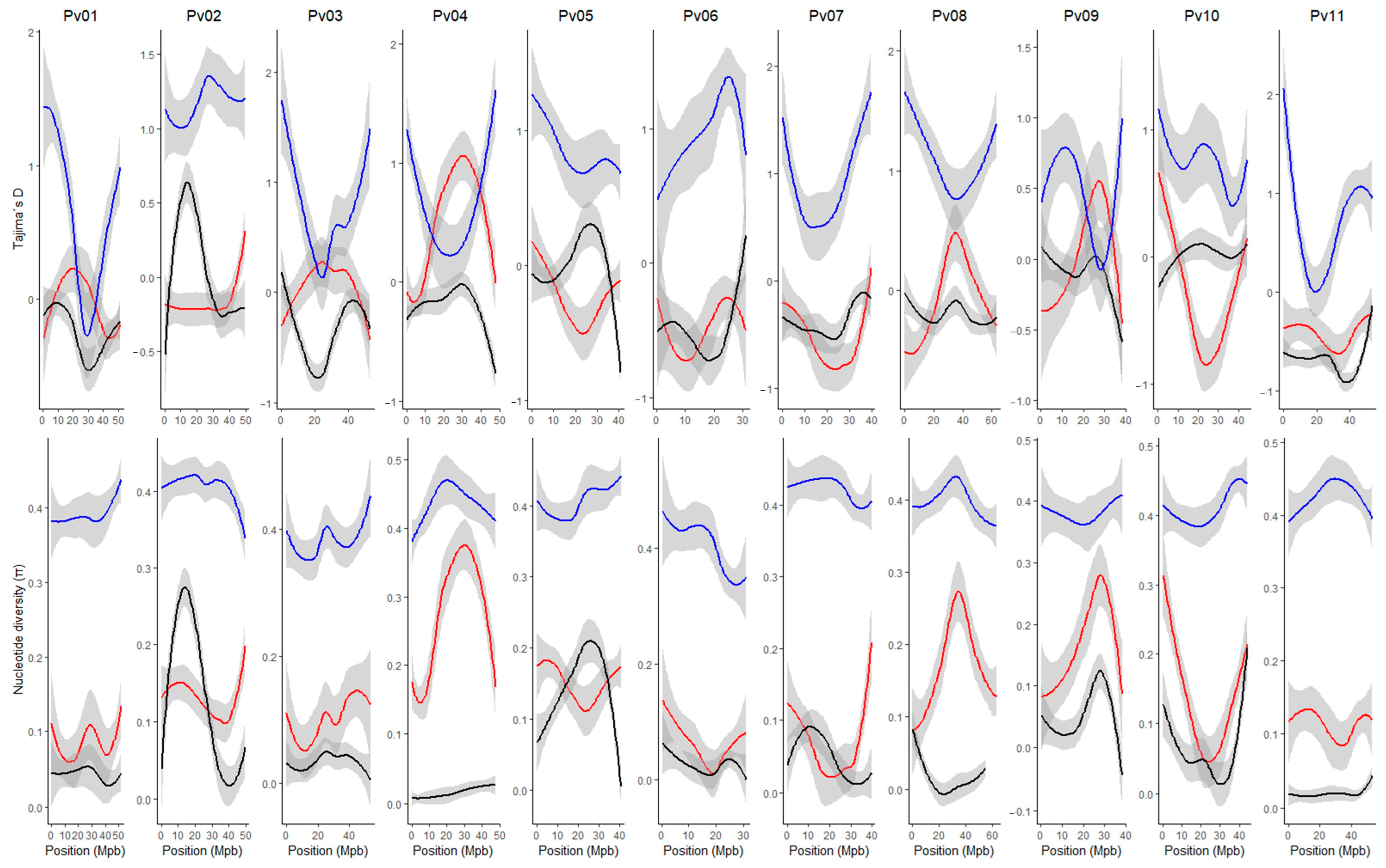
2.3. Genetic Diversity According to Population Structure
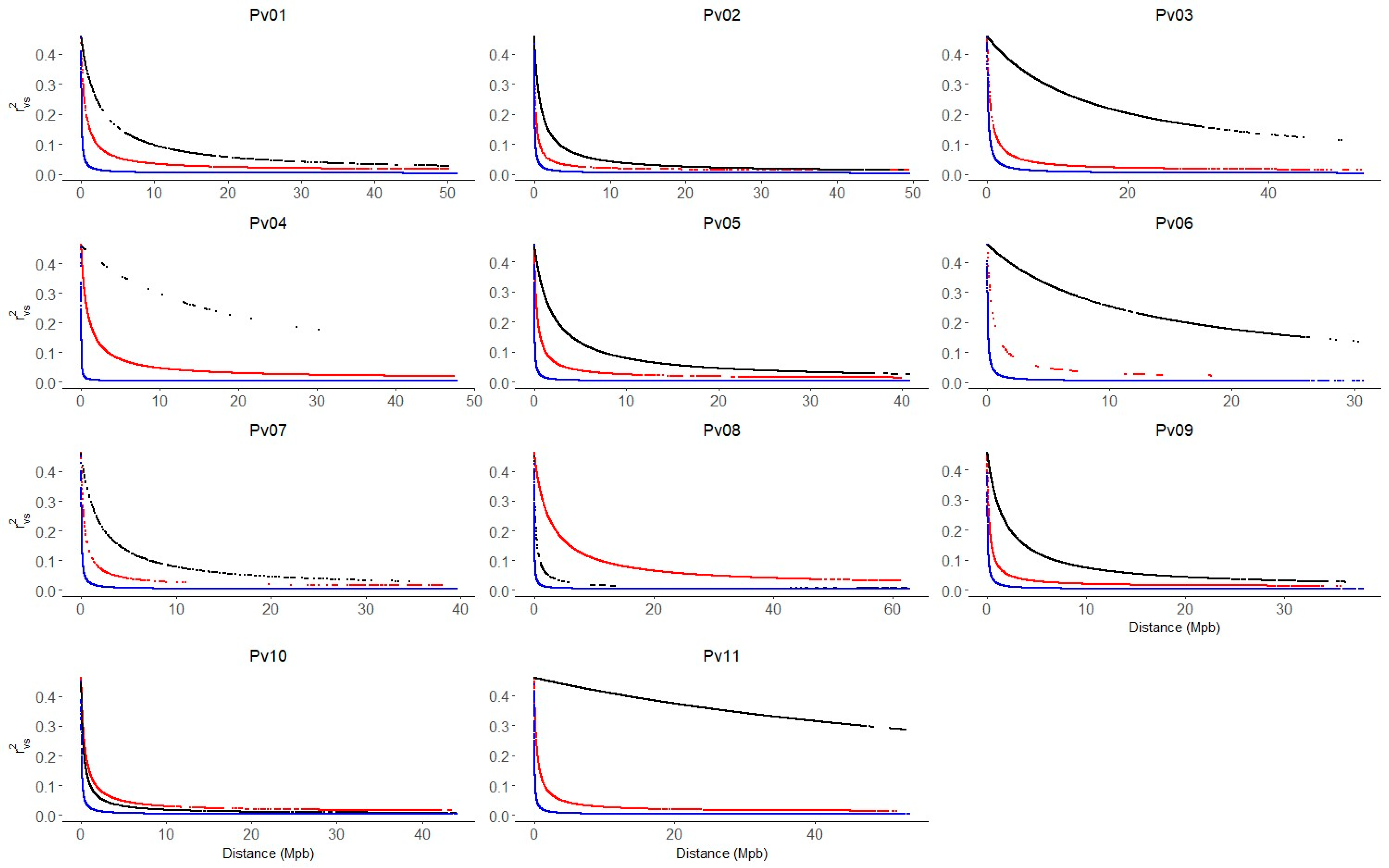
2.4. Linkage Disequilibrium and Haplotypes Blocks
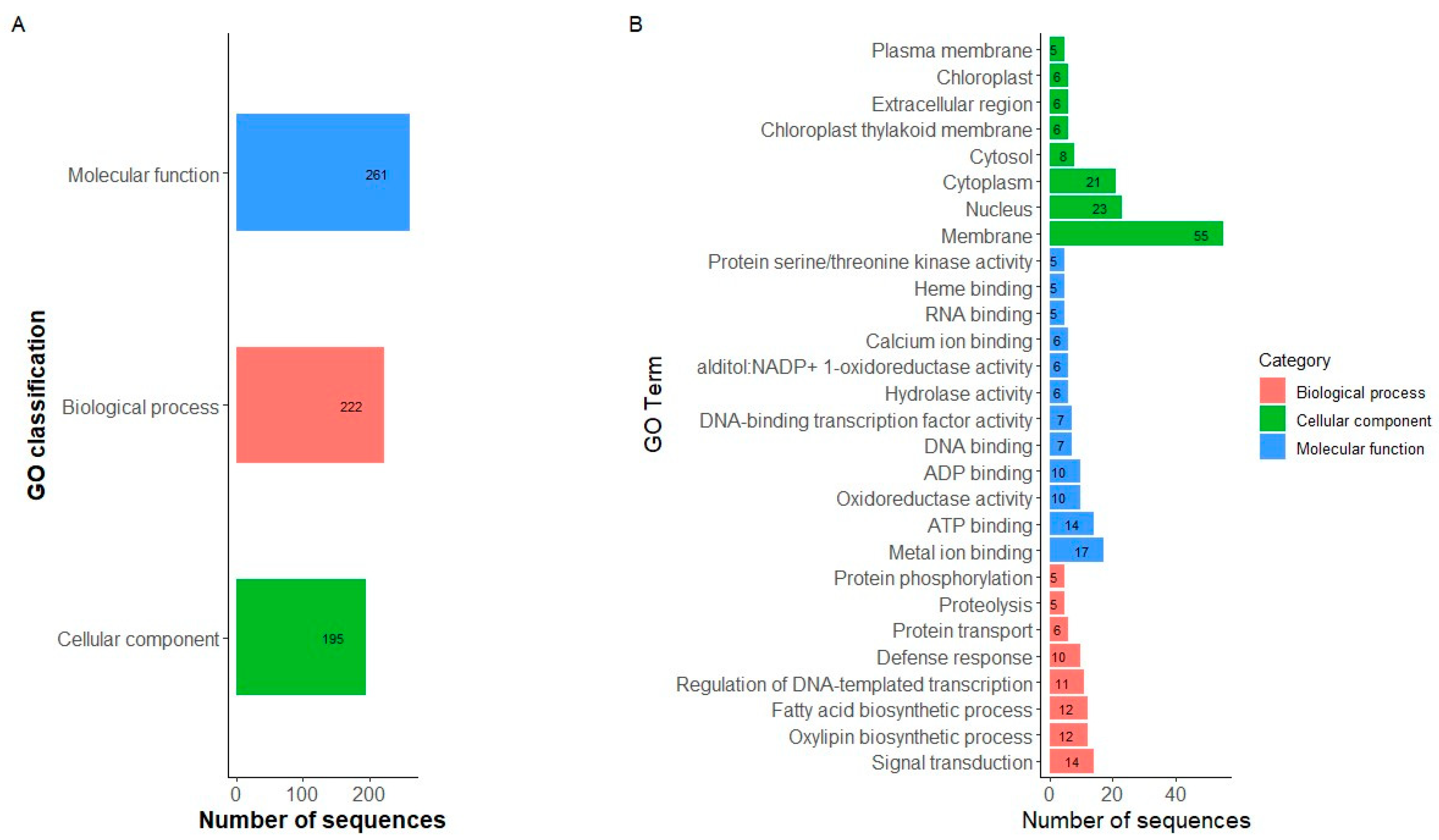
2.5. Candidate Gene Identification and GO Gene Enrichment
3. Discussion
3.1. Archaeological Records of Common Bean in Chile
3.2. Population Structure and Genetic Diversity
3.3. Local Adaptation
3.4. Identification of Candidate Genes and GO Analyses
4. Materials and Methods
4.1. Plant Material
4.2. SNP Genotyping and Filtering
4.3. Population Structure Analysis
4.4. Genetic Diversity by Population and Along the Genome
4.5. Linkage Disequilibrium and Haplotype Blocks
4.6. Candidate Genes Identification and Gene Enrichment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Trucchi, E.; Benazzo, A.; Lari, M.; Iob, A.; Vai, S.; Nanni, L.; Bellucci, E.; Bitocchi, E.; Raffini, F.; Xu, C.; et al. Ancient genomes reveal early Andean farmers selected common beans while pre-serving diversity. Nat. Plants 2021, 7, 123–128. [Google Scholar] [CrossRef]
- Bellucci, E.; Benazzo, A.; Xu, C.; Bitocchi, E.; Rodriguez, M.; Alseekh, S.; Di Vittori, V.; Gioia, T.; Neumann, K.; Cortinovis, G.; et al. Selection and adaptive in-trogression guided the complex evolutionary history of the European common bean. Nat. Commun. 2023, 14, 1908. [Google Scholar] [CrossRef]
- Rivera, M. Recent dated C-14 by AMS of bean samples from the San Pedro Viejo de Pichasca. Boletín Soc. Chil. Arqueol. 1995, 21, 27. [Google Scholar]
- McRostie, V.; Gallardo, F.; Vidal, A.; Croxatto, S. Early archaeobotanical evidence in the Calama oasis, the Topater 1 Cemetery (Middle Formational, 500 B.C.-100 A.F. Atacama Desert, northern Chile). Darwiniana 2019, 7, 16–38. [Google Scholar] [CrossRef][Green Version]
- Roa, C.; Silva, C.; Campbell, R. The contribution of Isla Mocha to knowledge about the use of plants with nutritional value in Southern Chile (1000–1700 B.P.). In Proceedings of the Actas Del XIX Congreso Nacional de Arqueología Chilena, Arica, Chile, 8–12 October 2012; p. 559. [Google Scholar]
- Campbell, R.; Roa, C.; Santana-Sagredo, F. More southern than beans: First 14C AMS dates for the Cueva de los Catalanes site. Boletín Soc. Chil. Arqueol. 2018, 48, 85–89. [Google Scholar]
- Schmutz, J.; McClean, P.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domes-tications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef]
- Singh, S.P.; Gepts, P.; Debouck, D.G. Races of common bean (Phaseolus vulgaris, Fabaceae). Econ. Bot. 1991, 45, 379–396. [Google Scholar] [CrossRef]
- Becerra, V.V.; Paredes, C.M.; Rojo, M.C.; Díaz, L.M.; Blair, M.W. Microsatellite marker characterization of Chilean common bean (Phaseolus vulgaris L.) germplasm. Crop. Sci. 2010, 50, 1932–1941. [Google Scholar] [CrossRef]
- Paredes, M.; Becerra, V.; Tay, J. Inorganic nutritional composition of common bean (Phaseolus vulgaris L.) genotypes race Chile. Chil. J. Agric. Res. 2009, 69, 486–495. [Google Scholar] [CrossRef]
- Márquez, K.; Arriagada, O.; Pérez-Díaz, R.; Cabeza, R.A.; Plaza, A.; Arévalo, B.; Meisel, L.A.; Ojeda, D.; Silva, H.; Schwember, A.R.; et al. Nutritional Characterization of Chilean Landraces of Common Bean. Plants 2024, 13, 817. [Google Scholar] [CrossRef]
- Paredes, M.; Gepts, P. Extensive introgression of Middle American germplasm into Chilean common bean cultivars. Genet. Resour. Crop. Evol. 1995, 42, 29–41. [Google Scholar] [CrossRef]
- Blair, M.W.; Díaz, J.M.; Hidalgo, R.; Duque, M.C. Microsatellite characterization of Andean races of common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2007, 116, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Jia, G.; Hyten, D.L.; Jenkins, J.; Hwang, E.-Y.; Schroeder, S.G.; Osorno, J.M.; Schmutz, J.; A Jackson, S.; E McClean, P.; et al. SNP Assay Development for Linkage Map Construction, Anchoring Whole-Genome Sequence, and Other Genetic and Genomic Applications in Common Bean. G3 Genes Genomes Genet. 2015, 5, 2285–2290. [Google Scholar] [CrossRef]
- Bitocchi, E.; Nanni, L.; Bellucci, E.; Rossi, M.; Giardini, A.; Zeuli, P.S.; Logozzo, G.; Stougaard, J.; McClean, P.; Attene, G.; et al. Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proc. Natl. Acad. Sci. USA 2012, 109, E788–E796. [Google Scholar] [CrossRef] [PubMed]
- Bitocchi, E.; Bellucci, E.; Giardini, A.; Rau, D.; Rodriguez, M.; Biagetti, E.; Santilocchi, R.; Zeuli, P.S.; Gioia, T.; Logozzo, G.; et al. Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol. 2013, 197, 300–313. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.; Rau, D.; Bitocchi, E.; Bellucci, E.; Biagetti, E.; Carboni, A.; Gepts, P.; Nanni, L.; Papa, R.; Attene, G. Landscape genetics, adaptive diversity and population structure in Phaseolus vulgaris. New Phytol. 2016, 209, 1781–1794. [Google Scholar] [CrossRef] [PubMed]
- Williams, V. Formaciones sociales en el noroeste argentino: Variabilidad prehispánica en el surandino durante el Periodo de Desarrollos Regionales y el estado Inca. Rev. Haucaypata Investig. Arqueol. Tahuantinsuyo 2015, 9, 62–76. [Google Scholar]
- Blair, M.W.; Díaz, L.M.; Buendía, H.F.; Duque, M.C. Genetic diversity, seed size associations and population structure of a core collection of common beans (Phaseolus vulgaris L.). Theor. Appl. Genet. 2009, 119, 955–972. [Google Scholar] [CrossRef] [PubMed]
- Lyra, M.L.; Kirchhof, S.; Goutte, S.; Kassie, A.; Boissinot, S. Crossing the Rift valley: Using complete mitogenomes to infer the diversification and biogeographic history of ethiopian highlands Ptychadena (anura: Ptychadenidae). Front. Genet. 2023, 14, 1215715. [Google Scholar] [CrossRef]
- Papa, R.; Acosta, J.; Delgado-Salinas, A.; Gepts, P. A genome-wide analysis of differentiation between wild and domesticated Phaseolus vulgaris from Mesoamerica. Theor. Appl. Genet. 2005, 111, 1147–1158. [Google Scholar] [CrossRef]
- Kuzay, S.; Hamilton-Conaty, P.; Palkovic, A.; Gepts, P. Is the USDA core collection of common bean representative of genetic diversity of the species, as assessed by SNP diversity? Crop. Sci. 2019, 60, 1398–1414. [Google Scholar] [CrossRef]
- Bascur, G.; Tay, J. Collection, characterization, and use of genetic variation in Chilean bean germplasm (Phaseolus vulgaris L.). Agric. Técnica 2005, 65, 135–146. [Google Scholar]
- Tuteja, N.; Sopory, S.K. Plant signaling in stress. Plant Signal. Behav. 2008, 3, 79–86. [Google Scholar] [CrossRef]
- Mao, X.; Zhang, H.; Tian, S.; Chang, X.; Jing, R. TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. J. Exp. Bot. 2009, 61, 683–696. [Google Scholar] [CrossRef]
- Tyagi, H.; Jha, S.; Sharma, M.; Giri, J.; Tyagi, A.K. Rice SAPs are responsive to multiple biotic stresses and overexpression of OsSAP1, an A20/AN1 zinc-finger protein, enhances the basal resistance against pathogen infection in tobacco. Plant Sci. 2014, 225, 68–76. [Google Scholar] [CrossRef]
- Zhang, Y.; Lan, H.; Shao, Q.; Wang, R.; Chen, H.; Tang, H.; Zhang, H.; Huang, J. An A20/AN1-tpe zinc finger protein modulates gibberellins and abscisic acid contents and increases sensitivity to abiotic stress in rice (Oryza sativa). J. Exp. Bot. 2016, 67, 315–326. [Google Scholar] [CrossRef]
- Soler-Garzón, A.; McClean, P.E.; Miklas, P.N. Coding Mutations in Vacuolar Protein-Sorting 4 AAA+ ATPase Endosomal Sorting Complexes Required for Transport Protein Homologs Underlie bc-2 and New bc-4 Gene Conferring Resistance to Bean Common Mosaic Virus in Common Bean. Front. Plant Sci. 2021, 12, 769247. [Google Scholar] [CrossRef]
- Cortés, A.J.; This, D.; Chavarro, C.; Madriñán, S.; Blair, M.W. Nucleotide diversity patterns at the drought-related DREB2 encoding genes in wild and cultivated common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2012, 125, 1069–1085. [Google Scholar] [CrossRef]
- Konzen, E.R.; Recchia, G.H.; Cassieri, F.; Caldas, D.G.G.; Teran, J.C.B.M.Y.; Gepts, P.; Tsai, S.M. DREBGenes from Common Bean (Phaseolus vulgarisL.) Show Broad to Specific Abiotic Stress Responses and Distinct Levels of Nucleotide Diversity. Int. J. Genom. 2019, 2019, 9520642. [Google Scholar] [CrossRef]
- Cichy, K.A.; Porch, T.G.; Beaver, J.S.; Cregan, P.; Fourie, D.; Glahn, R.P.; Grusak, M.A.; Kamfwa, K.; Katuuramu, D.N.; McClean, P.; et al. A Phaseolus vulgaris Diversity Panel for Andean Bean Improvement. Crop. Sci. 2015, 55, 2149–2160. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Chessel, D.; Dufour, A.B.; Thioulouse, J. The R Journal: The ade4 package—I: One-table methods. R News 2004, 4, 5–10. [Google Scholar]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Wickham, H. A Layered Grammar of Graphics. J. Comput. Graph. Stat. 2010, 19, 3–28. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef]
- Peakall, R.O.D.; Smouse, P.E. genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 1989, 3, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Eckshtain-Levi, N.; Weisberg, A.J.; Vinatzer, B.A. The population genetic test Tajima’s D identifies genes encoding pathogen-associated molecular patterns and other virulence-related genes in Ralstonia solanacearum. Mol. Plant Pathol. 2018, 19, 2187–2192. [Google Scholar] [CrossRef]
- Mangin, B.; Siberchicot, A.; Nicolas, S.; Doligez, A.; This, P.; Cierco-Ayrolles, C. Novel measures of linkage disequilibrium that correct the bias due to population structure and relatedness. Heredity 2011, 108, 285–291. [Google Scholar] [CrossRef]
- Endelman, J.B. Ridge Regression and Other Kernels for Genomic Selection with R Package rrBLUP. Plant Genome 2011, 4, 255–258. [Google Scholar] [CrossRef]
- Hill, W.; Weir, B. Variances and covariances of squared linkage disequilibria in finite populations. Theor. Popul. Biol. 1988, 33, 54–78. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Gabriel, S.B.; Schaffner, S.F.; Nguyen, H.; Moore, J.M.; Roy, J.; Blumenstiel, B.; Higgins, J.; DeFelice, M.; Lochner, A.; Faggart, M.; et al. The Structure of Haplotype Blocks in the Human Genome. Science 2002, 296, 2225–2229. [Google Scholar] [CrossRef]
- Smedley, D.; Haider, S.; Ballester, B.; Holland, R.; London, D.; Thorisson, G.; Kasprzyk, A. BioMart–biological queries made easy. BMC Genom. 2009, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S. Blast2GO: A Comprehensive Suite for Functional Analysis in Plant Genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef] [PubMed]
| Gene Pool | N | Na | Ne | I | uHe | Npa | D | π | PPL(%) |
|---|---|---|---|---|---|---|---|---|---|
| Andean | 310 | 1.886 | 1.138 | 0.177 | 0.099 | - | −0.962 | 0.097 | 88.64 |
| A1 | 10 | 1.708 | 1.401 | 0.366 | 0.269 | 342 | 0.370 | 0.268 | 18.52 |
| A2—Peru | 86 | 1.549 | 1.158 | 1.165 | 1.102 | 61 | −0.220 | 0.102 | 59.14 |
| A3—Chile | 187 | 1.538 | 1.073 | 0.096 | 0.053 | 13 | −1.371 | 0.053 | 87.06 |
| Mesoamerican | 350 | 1.949 | 1.520 | 0.446 | 0.300 | - | 3.203 * | 0.300 | 94.93 |
| M1 | 235 | 1.939 | 1.485 | 0.431 | 0.287 | 348 | 2.687 * | 0.286 | 93.87 |
| M2 | 103 | 1.687 | 1.288 | 0.273 | 0.177 | 4 | 1.157 | 0.176 | 83.60 |
| Overall | 667 | 2.000 | 1.497 | 0.473 | 0.308 | - | 5.504 * | 0.403 | - |
| Chr | Total # of Blocks | Average SNP/Block | Mean Block Size (kb) | Unique Blocks * | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MESO | AND | Chile | MESO | AND | Chile | MESO | AND | Chile | MESO | AND | Chile | |
| Pv01 | 20 | 2 | 3 | 3.35 | 2 | 4.3 | 122.2 | 9.5 | 228.32 | 18 | 1 | 0 |
| Pv02 | 29 | 24 | 16 | 2.51 | 4.7 | 4.56 | 58.68 | 269.37 | 319.18 | 4 | 5 | 3 |
| Pv03 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 10 | 0 | 0 | 2 | 0 |
| Pv04 | 22 | 19 | 0 | 2.13 | 3.84 | 0 | 22 | 190.42 | 0 | 13 | 12 | 0 |
| Pv05 | 20 | 25 | 14 | 3.05 | 5.04 | 5.07 | 73.75 | 258.48 | 402.07 | 5 | 7 | 2 |
| Pv06 | 12 | 0 | 0 | 3.33 | 0 | 0 | 119.75 | 0 | 0 | 12 | 0 | 0 |
| Pv07 | 15 | 3 | 2 | 2.53 | 2.66 | 2 | 38.33 | 58 | 6.5 | 15 | 1 | 0 |
| Pv08 | 20 | 8 | 8 | 2.3 | 2.5 | 2.5 | 53.8 | 128.5 | 128.5 | 19 | 0 | 0 |
| Pv09 | 8 | 8 | 1 | 2.25 | 4 | 4 | 20.12 | 209.75 | 495 | 4 | 5 | 1 |
| Pv10 | 6 | 11 | 6 | 2.33 | 4.36 | 7.5 | 88.16 | 214.36 | 361.83 | 3 | 6 | 5 |
| Pv11 | 20 | 8 | 1 | 2.75 | 6.12 | 4 | 67.25 | 210 | 290 | 12 | 3 | 1 |
| Total | 172 | 110 | 51 | 2.41 | 3.38 | 3.08 | 60.37 | 141.67 | 202.85 | 105 | 42 | 12 |
| Gene Name | Description |
|---|---|
| Phvul.005G124800 | SERINE/THREONINE-PROTEIN KINASE RIO |
| Phvul.005G125000 | AN1-TYPE ZINC FINGER PROTEIN |
| Phvul.005G125100 | vacuolar protein-sorting-associated protein 4 (VPS4) |
| Phvul.005G125200 | PPR repeat (PPR)//PPR repeat family (PPR_2)//DYW family of nucleic acid deaminases (DYW_deaminase) |
| Phvul.005G125300 | protein HIRA/HIR1 (HIRA, HIR1) |
| Phvul.005G125400 | FATTY ACYL-COA REDUCTASE 3-RELATED |
| Phvul.005G125500 | Long-chain-fatty-acyl-CoA reductase/Acyl-CoA reductase |
| Phvul.005G125700 | histidine-containing phosphotransfer protein (AHP) |
| Phvul.005G126000 | 60S ACIDIC RIBOSOMAL PROTEIN P2-4 |
| Phvul.005G126100 | PEPTIDYLPROLYL ISOMERASE DOMAIN AND WD REPEAT-CONTAINING PROTEIN 1 |
| Phvul.005G126300 | AP2 domain (AP2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arriagada, O.; Arévalo, B.; Pacheco, I.; Schwember, A.R.; Meisel, L.A.; Silva, H.; Márquez, K.; Plaza, A.; Pérez-Diáz, R.; Pico-Mendoza, J.; et al. A Past Genetic Bottleneck from Argentine Beans and a Selective Sweep Led to the Race Chile of the Common Bean (Phaseolus vulgaris L.). Int. J. Mol. Sci. 2024, 25, 4081. https://doi.org/10.3390/ijms25074081
Arriagada O, Arévalo B, Pacheco I, Schwember AR, Meisel LA, Silva H, Márquez K, Plaza A, Pérez-Diáz R, Pico-Mendoza J, et al. A Past Genetic Bottleneck from Argentine Beans and a Selective Sweep Led to the Race Chile of the Common Bean (Phaseolus vulgaris L.). International Journal of Molecular Sciences. 2024; 25(7):4081. https://doi.org/10.3390/ijms25074081
Chicago/Turabian StyleArriagada, Osvin, Bárbara Arévalo, Igor Pacheco, Andrés R. Schwember, Lee A. Meisel, Herman Silva, Katherine Márquez, Andrea Plaza, Ricardo Pérez-Diáz, José Pico-Mendoza, and et al. 2024. "A Past Genetic Bottleneck from Argentine Beans and a Selective Sweep Led to the Race Chile of the Common Bean (Phaseolus vulgaris L.)" International Journal of Molecular Sciences 25, no. 7: 4081. https://doi.org/10.3390/ijms25074081
APA StyleArriagada, O., Arévalo, B., Pacheco, I., Schwember, A. R., Meisel, L. A., Silva, H., Márquez, K., Plaza, A., Pérez-Diáz, R., Pico-Mendoza, J., Cabeza, R. A., Tapia, G., Fuentes, C., Rodríguez-Alvarez, Y., & Carrasco, B. (2024). A Past Genetic Bottleneck from Argentine Beans and a Selective Sweep Led to the Race Chile of the Common Bean (Phaseolus vulgaris L.). International Journal of Molecular Sciences, 25(7), 4081. https://doi.org/10.3390/ijms25074081








