Machine Learning-Powered ATR-FTIR Spectroscopic Clinical Evaluation for Rapid Typing of Salmonella enterica O-Serogroups and Salmonella Typhi
Abstract
1. Introduction
2. Methods
2.1. Setting and Surveillance System
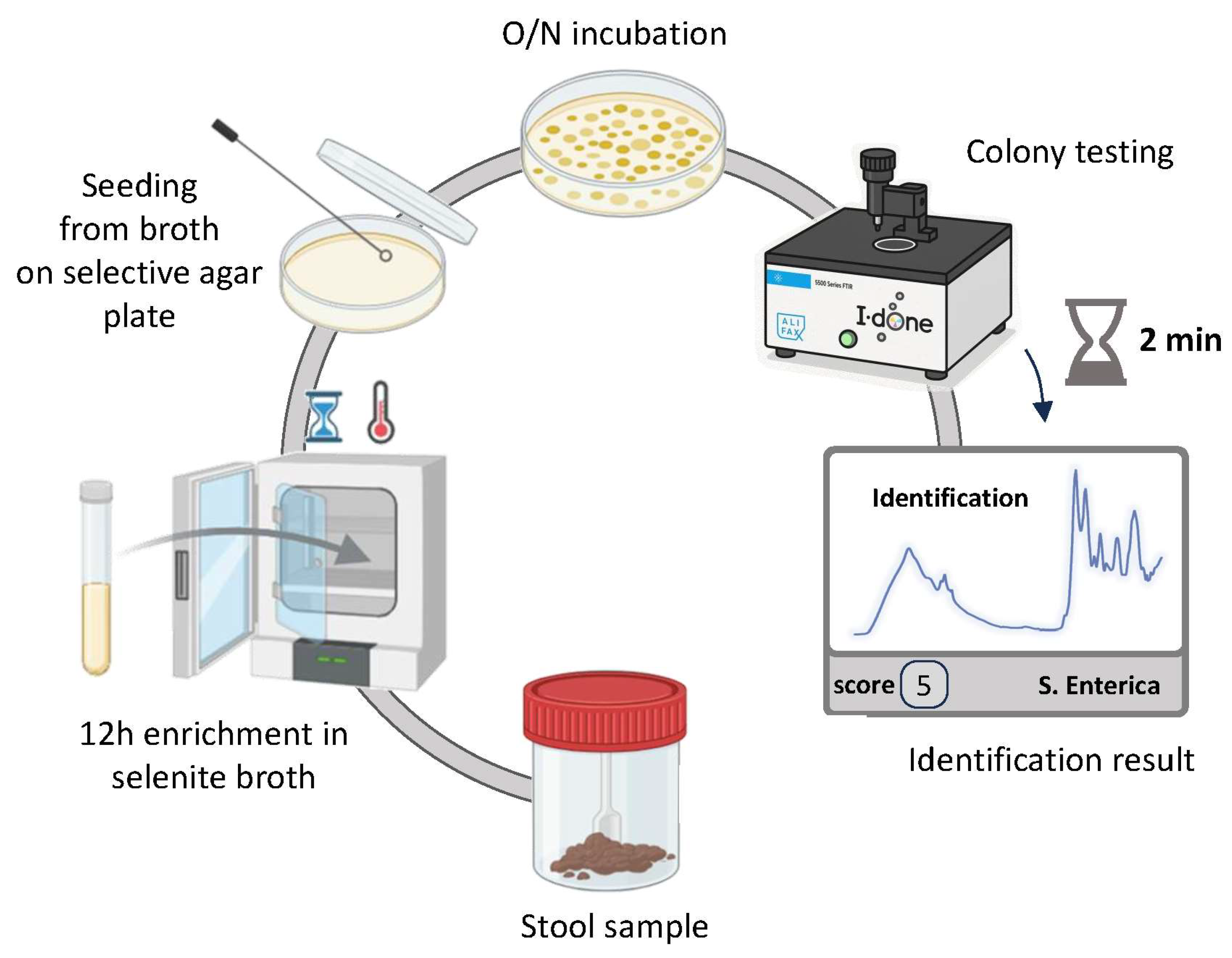
2.2. Bacterial Isolates and Culture Conditions
2.3. ATR-FTIR Spectroscopy
2.4. Ethical Considerations
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tauxe, R.V.; Doyle, M.P.; Kuchenmuller, T.; Schlundt, J.; Stein, C.E. Evolving public health approaches to the global challenge of foodborne infections. Int. J. Food Microbiol. 2010, 139 (Suppl. S1), S16–S28. [Google Scholar] [CrossRef]
- Newell, D.G.; Koopmans, M.; Verhoef, L.; Duizer, E.; Aidara-Kane, A.; Sprong, H.; Opsteegh, M.; Langelaar, M.; Threfall, J.; Scheutz, F.; et al. Food-borne diseases-the challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 2010, 139 (Suppl. S1), S3–S15. [Google Scholar] [CrossRef] [PubMed]
- Quested, T.E.; Cook, P.E.; Gorris, L.G.; Cole, M.B. Trends in technology, trade and consumption likely to impact on microbial food safety. Int. J. Food Microbiol. 2010, 139 (Suppl. S1), S29–S42. [Google Scholar] [CrossRef] [PubMed]
- Bula-Rudas, F.J.; Rathore, M.H.; Maraqa, N.F. Salmonella infections in childhood. Adv. Pediatr. 2015, 62, 29–58. [Google Scholar] [CrossRef]
- Ford, L.; Glass, K.; Veitch, M.; Wardell, R.; Polkinghorne, B.; Dobbins, T.; Lal, A.; Kirk, M.D. Increasing incidence of Salmonella in Australia, 2000–2013. PLoS ONE 2016, 11, e0163989. [Google Scholar] [CrossRef] [PubMed]
- Andino, A.; Hanning, I. Salmonella enterica: Survival, colonization, and virulence differences among serovars. Sci. World J. 2015, 2015, 520179. [Google Scholar] [CrossRef]
- Gerner-Smidt, P.; Hise, K.; Kincaid, J.; Hunter, S.; Rolando, S.; Hyytiä-Trees, E.; Ribot, E.M.; Swaminathan, B.; Taskforce, P. PulseNet USA: A five-year update. Foodborne Pathog. Dis. 2006, 3, 9–19. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA). European Centre for Disease Prevention and Control (ECDC) The European Union One Health 2023 Zoonoses report. EFSA J. 2024, 22, e9106. [Google Scholar] [CrossRef]
- Tindall, B.J.; Grimont, P.A.; Garrity, G.M.; Euz_eby, J.P. Nomenclature and taxonomy of the genus Salmonella. Int. J. Syst. Evol. Microbiol 2005, 55, 521–524. [Google Scholar] [CrossRef]
- Mezal, E.H.; Sabol, A.; Khan, M.A.; Ali, N.; Stefanova, R.; Khan, A.A. Isolation and molecular characterization of Salmonella enterica serovar Enteritidis from poultry house and clinical samples during 2010. Food Microbiol. 2014, 38, 67–74. [Google Scholar] [CrossRef]
- Xiong, D.; Song, L.; Pan, Z.; Jiao, X. Identification and discrimination of Salmonella enterica serovar Gallinarum biovars Pullorum and Gallinarum based on a one-step multiplex PCR assay. Front. Microbiol. 2018, 9, 1718. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, H.; Söderlund, R.; Ernholm, L.; Melin, L.; Jansson, D.S. Diagnostics, epidemiological obser vations and genomic subtyping in an outbreak of pullorum disease in non-commercial chickens. Vet. Microbiol. 2018, 217, 47–52. [Google Scholar] [CrossRef] [PubMed]
- ISO 6579; Microbiology of food and animal feeding stuffs—Horizontal method for the detection of Salmonella spp. ISO: Geneva, Switzerland, 2017.
- Classification and Identification of Bacteria by Fourier-Transform Infrared Spectroscopy|Microbiology Society. Available online: https://www.microbiologyresearch.org/content/journal/micro/10.1099/00221287-137-1-69 (accessed on 23 April 2024).
- Horbach, I.; Naumann, D.; Fehrenbach, F.J. Simultaneous Infections with Different Serogroups of Legionella Pneumophila Investigated by Routine Methods and Fourier Transform Infrared Spectroscopy. J. Clin. Microbiol. 1988, 26, 1106–1110. [Google Scholar] [CrossRef] [PubMed]
- Mariey, L.; Signolle, J.P.; Amiel, C.; Travert, J. Discrimination, Classification, Identification of Microorganisms Using FTIR Spectroscopy and Chemometrics. Vib. Spectrosc. 2001, 26, 151–159. [Google Scholar] [CrossRef]
- Naumann, D.; Helm, D.; Labischinski, H. Microbiological Characterizations by FT-IR Spectroscopy. Nature 1991, 351, 81–82. [Google Scholar] [CrossRef]
- Zarnowiec, P.; Lechowicz, L.; Czerwonka, G.; Kaca, W. Fourier Transform Infrared Spectroscopy (FTIR) as a Tool for the Identification and Differentiation of Pathogenic Bacteria. Curr. Med. Chem. 2015, 22, 1710–1718. [Google Scholar] [CrossRef]
- Van Belkum, A.; Tassios, P.T.; Dijkshoorn, L.; Haeggman, S.; Cookson, B.; Fry, N.K.; Fussing, V.; Green, J.; Feil, E.; Gerner-Smidt, P.; et al. Guidelines for the Validation and Application of Typing Methods for Use in Bacterial Epidemiology. Clin. Microbiol. Infect. 2007, 13, 1–46. [Google Scholar] [CrossRef]
- Quintelas, C.; Ferreira, E.C.; Lopes, J.A.; Sousa, C. An Overview of the Evolution of Infrared Spectroscopy Applied to Bacterial Typing. Biotechnol. J. 2018, 13, 1700449. [Google Scholar] [CrossRef]
- Vallieres, E.; Quach, C.; Lam, L.; Rallu, F.; Langella, M.; Sedman, J.; Raymond, M.; Lebel, P.; Ismail, A. Attenuated Total Reflectance Fourier Transform Infrared Spectroscopy for Rapid Identification of Non-Fermenting Gram-Negative Bacillated from Patients with Cystic Fibrosis. Open Forum Infect. Dis. 2017, 4, S592. [Google Scholar] [CrossRef][Green Version]
- Napoleoni, M.; Ceschia, S.; Mitri, E.; Beneitez, E.E.; Silenzi, V.; Staffolani, M.; Rocchegiani, E.; Blasi, G.; Gurian, E. Identification of Salmonella Serogroups and Distinction Between Typhoidal and Non-Typhoidal Salmonella Based on ATR-FTIR Spectroscopy. Microorganisms 2024, 12, 2318. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.G.; Otto, G.; Colby, L.A. Chapter 28—Selected Zoonoses. In Laboratory Animal Medicine, 3rd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2015. [Google Scholar] [CrossRef]
- Odoch, T.; Wasteson, Y.; L’Abée-Lund, T.; Muwonge, A.; Kankya, C.; Nyakarahuka, L.; Tegule, S.; Skjerve, E. Prevalence, antimicrobial susceptibility and risk factors associated with non-typhoidal Salmonella on Ugandan layer hen farms. BMC Vet. Res. 2017, 13, 365. [Google Scholar] [CrossRef] [PubMed]
- Niki, M.; Shakeel, A.; Zahid, K.; Konstantinos, C.K. Prevalence, Risks and Antibiotic Resistance of Salmonella in Poultry Production Chain. In Current Topics in Salmonella and Salmonellosis; InTechOpen: London, UK, 2017; pp. 216–234. [Google Scholar]
- Elkenany, R.M.; Eladl, A.H.; El-Shafei, R.A. Genetic characterization of class 1 integrons among mul tidrug-resistant Salmonella serotypes in broiler chicken farms. J. Glob. Antimicrob. Resist. 2018, 14, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Liljebjelke, K.A.; Hofacre, C.L.; White, D.G.; Ayers, S.; Lee, M.D.; Maurer, J.J. Diversity of antimicrobial resistance phenotypes in Salmonella isolated from commercial poultry farms. Front. Vet. Sci. 2017, 4, 96. [Google Scholar] [CrossRef]
- Iwamoto, M.; Reynolds, J.; Karp, B.E.; Tate, H.; Fedorka- Cray, P.J.; Plumblee, J.R.; Hoekstra, R.M.; Whichard, J.M.; Mahon, B.E. Ceftriaxone-resistant nontyphoidal Salmonella from humans, retail meats, and food animals in the United States, 1996–2013. Foodborne Pathog. Dis. 2017, 14, 74–83. [Google Scholar] [CrossRef]
- Faruq, A.A.; Hassan, M.M.; Uddin, M.M.; Rahman, M.L.; Rakib, T.M.; Alam, M.; Islam, A. Prevalence and multidrug resistance pattern of Salmonella isolated from resident wild birds of Bangladesh. Int. J. One Health 2016, 2, 35–41. [Google Scholar] [CrossRef]
- Omoshaba, E.O.; Olufemi, F.O.; Ojo, O.E.; Sonibare, A.O.; Agbaje, M. Multidrug-resistant Salmonellae iso lated in Japanese quails reared in Abeokuta, Nigeria. Trop. Anim. Health Prod. 2017, 49, 1455–1460. [Google Scholar] [CrossRef] [PubMed]
- Migura-Garcia, L.; Ramos, R.; Cerdà-Cuéllar, M. Antimicrobial resistance of Salmonella serovars and Campylobacter spp. Isolated from an opportunistic gull species, yellow-legged gull (Larus michahellis). J. Wildl. Dis. 2017, 53, 148–152. [Google Scholar] [CrossRef]
- Anjum, M.F.; Duggett, N.A.; AbuOun, M.; Randall, L.; Nunez-Garcia, J.; Ellis, R.J.; Rogers, J.; Horton, R.; Brena, C.; Williamson, S.; et al. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J. Antimicrob. Chemother. 2016, 71, 2306–2313. [Google Scholar] [CrossRef]
- Phoon, Y.W.; Chan, Y.Y.C.; Koh, T.H. Isolation of multidrug-resistant Salmonella in Singapore. Singap. Med. J. 2015, 56, 142–144. [Google Scholar] [CrossRef]
- Andoh, L.A.; Ahmed, S.; Olsen, J.E.; Obiri-Danso, K.; Newman, M.J.; Opintan, J.A.; Barco, L.; Dalsgaard, A. Prevalence and characterization of Salmonella among humans in Ghana. Trop. Med. Health 2017, 45, 3. [Google Scholar] [CrossRef]
- Afema, J.A.; Mather, A.E.; Sischo, W.M. Antimicrobial resistance profiles diversity in Salmonella from humans cattle, 2004–2011. Zoonoses Public Health 2015, 62, 506–517. [Google Scholar] [CrossRef]
- Brands, D.A.; Inman, A.E.; Gerba, C.P.; Maré, C.J.; Billington, S.J.; Saif, L.A.; Levine, J.F.; Joens, L.A. Prevalence of Salmonella spp. In oysters in the United States. Appl. Environ. Microbiol. 2005, 71, 893–897. [Google Scholar] [CrossRef]
- Chao, G.; Zhou, X.; Jiao, X.; Qian, X.; Xu, L. Prevalence and antimicrobial resistance of foodborne pathogens isolated from food products in China. Foodborne Pathog. Dis. 2007, 4, 277–284. [Google Scholar] [CrossRef]
- Gebreyes, W.A.; Thakur, S. Multidrug-resistant Salmonella enterica serovar Muenchen from pigs and humans and potential interserovar transfer of antimicrobialresistance. Antimicrob. Agents Chemother. 2005, 49, 503–511. [Google Scholar] [CrossRef]
- Glenn, L.M.; Lindsey, R.L.; Folster, J.P.; Pecic, G.; Boerlin, P.; Gilmour, M.W.; Harbottle, H.; Zhao, S.; McDermott, P.F.; Fedorka-Cray, P.J. Antimicrobial resistance genes in multidrug-resistant Salmonella enterica isolated from animals, retail meats, and humans in the United States and Canada. Microb. Drug Resist. 2013, 19, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Voss-Rech, D.; Potter, L.; Vaz, C.S.L.; Pereira, D.I.B.; Sangioni, L.A.; Vargas, Á.C.; de Avila, B.S. Antimicrobial resistance in nontyphoidal Salmonella isolated from human and poultry-related samples in Brazil: 20-year meta-analysis. Foodborne Pathog. Dis. 2017, 14, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Michael, G.B.; Schwarz, S. Antimicrobial resis tance in zoonotic nontyphoidal Salmonella: An alarming trend? Clin. Microbiol. Infect. 2016, 22, 968–974. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, K.L.; Pant, N.D.; Bhandari, R.; Khatri, S.; Shrestha, B.; Lekhak, B. Re-emergence of the susceptibility of the Salmonella spp. Isolated from blood samples to conventional first-line antibiotics. Antimicrob. Resist. Infect. Control 2016, 5, 22. [Google Scholar] [CrossRef]
- Angelo, K.M.; Reynolds, J.; Karp, B.E.; Hoekstra, R.M.; Scheel, C.M.; Friedman, C. Antimicrobial resis tance among nontyphoidal Salmonella isolated from blood in the United States, 2003–2013. J. Infect. Dis. 2016, 214, 1565–1570. [Google Scholar] [CrossRef]
- Farmer, J.J. Enterobacteriaceae: Introduction and identification. In Manual of Clinical Microbiology, 6th ed; Murray, P.R., Baron, E.J., Pfaller, M.A., Eds.; American Society for Microbiology: Washington, DC, USA, 1995; pp. 438–449. [Google Scholar]
- Tauxe, R.V.; Pavia, A.T. Salmonellosis: Nontyphoidal. In Bacterial Infections of Humans: Epidemiology and Control, 3rd ed; Evans, A.S., Brachman, P.S., Eds.; Plenum Medical Book Co.: New York, NY, USA, 1998; pp. 613–630. [Google Scholar]
- Field, H.I. Salmonellosis in animals. Vet. Res. 1958, 70, 1050–1052. [Google Scholar]
- Saphra, I.; Wassermann, M. Salmonella choleraesuis. A clinical and epidemiological evaluation of 329 infections identified between 1940 and 1954 in the New York Salmonella Center. Am. J. Med. Sci. 1954, 228, 525–533. [Google Scholar] [CrossRef]
- Wilcock, B.P.; Schwartz, K. Salmonellosis. In Diseases in Swine, 7th ed; Leman, A.D., Straw, B.E., Mengeling, W.E., D’Allaire, S., Taylor, D.J., Eds.; Iowa State University Press: Ames, IA, USA, 1992; pp. 570–583. [Google Scholar]
- Fang, F.C.; Fierer, J. Human infection with Salmonella dublin. Medicine 1991, 70, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, B.D.; Zuckerman, M.J.; Foland, J.A.; Polly, S.M.; Marwah, R.K. Disseminated Salmonella arizonae infection associated with rattlesnake meat ingestions. Am. J. Gastroenterol. 1989, 84, 433–435. [Google Scholar]
- Waterman, S.H.; Juarez, G.; Carr, S.J.; Kilman, L. Salmonella arizonae infections in Latinos associated with rattlesnake folk medicine. Am. J. Public Health 1990, 80, 286–289. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J.; Feldman, R.A. Salmonella bacteremia: Reports to the Centers for Disease Control, 1968–1979. J. Infect. Dis. 1981, 143, 743–746. [Google Scholar] [CrossRef]
- Cohen, J.I.; Bartlett, J.A.; Corey, R. Extra-intestinal manifestations of salmonella infections. Medicine 1987, 66, 349–388. [Google Scholar] [CrossRef] [PubMed]
- Tanui, C.K.; Karanth, S.; Niage, P.M.K.; Meng, J.; Pradhan, A.K. Machine learning-based predictive modeling to identify genotypic traits associated with Salmonella enterica disease endpoints in isolates from ground chicken. LWT 2022, 154, 112701. [Google Scholar] [CrossRef]
- Bolinger, H.; Tran, D.; Harary, K.; Paoli, G.C.; Guron, G.K.P.; Namazi, H.; Khaksar, R. Utilizing the microbiota and machine learning algorithms to assess risk of Salmonella contamination in poultry Rinsate. J. Food Prot. 2021, 84, 1648–1657. [Google Scholar] [CrossRef]
- Munck, N.; Njage, P.M.K.; Leekitcharoenphon, P.; Litrup, E.; Hald, T. Application of whole-genome sequences and machine learning in source attribution of Salmonella typhimurium. Risk Anal. 2020, 40, 1693–1705. [Google Scholar] [CrossRef]
- Nguyen, M.; Long, S.W.; McDermott, P.F.; Olsen, R.J.; Olson, R.; Stevens, R.L.; Tyson, G.H.; Zhao, S.; Davis, J.J. Using machine learning to predict antimicrobial MICs and associated genomic features for Nontyphoidal Salmonella. J. Clin. Microbiol. 2019, 57, e01260–e01328. [Google Scholar] [CrossRef]
- Baldauf, N.A.; Rodriguez-Romo, L.A.; Yousef, A.E.; Rodriguez-Saona, L.E. Differentiation of selected Salmonella enterica serovars by Fouriertransform mid-infrared spectroscopy. Appl. Spectrosc. 2006, 60, 592–598. [Google Scholar] [CrossRef]
- Männig, A.; Baldauf, N.A.; Rodriguez-Romo, L.A.; Yousef, A.E.; Rodríguez-Saona, L.E. Differentiationm of Salmonella enterica serovars and strains in cultures and food using infrared spectroscopic and microspectroscopic techniques combined with soft independent modeling of class analogy pattern recognition analysis. J. Food Protec. 2008, 71, 2249–2256. [Google Scholar] [CrossRef] [PubMed]
- Campos, J.; Sousa, C.; Mourão, J.; Lopes, J.; Antunes, P.; Peixe, L. Discrimination of non-typhoid Salmonella serogroups and serotypes by Fourier transform infrared spectroscopy: A comprehensive analysis. Int. J. Food Microbiol. 2018, 285, 34–41. [Google Scholar] [CrossRef]
- Cordovana, M.; Mauder, N.; Join-Lambert, O.; Gravey, F.; LeHello, S.; Auzou, M.; Pitti, M.; Zoppi, S.; Buhl, M.; Steinmann, J.; et al. Machine Learning-Based Typing of Salmonella Enterica O-Serogroups by the Fourier-Transform Infrared (FTIR) Spectroscopy-Based IR Biotyper System. J. Microbiol. Methods 2022, 201, 106564. [Google Scholar] [CrossRef] [PubMed]
- Johler, S.; Stephan, R.; Althaus, D.; Ehling-Schulz, M.; Grunert, T. High-resolution subtyping of Staphylococcus aureus strains by means of Fourier-transform infrared spectroscopy. Syst. Appl. Microbiol. 2016, 39, 189–194. [Google Scholar] [CrossRef] [PubMed]

| Group | Serovar |
|---|---|
| B | S. Abony |
| S. Agona (2) | |
| S. Derby (2) | |
| S. Paratyphi B | |
| S. Typhimurium (6) | |
| Monophasic variant of S. Typhimurium (16) | |
| C1 | S. Choleraesuis (2) |
| S. Infantis (6) | |
| S. Virchow | |
| S. Thomson | |
| C2 | S. Goldcoast |
| S. Manhattan | |
| D | S. Enteritidis (6) |
| S. Typhi | |
| S. Napoli (2) | |
| E | S. Senftenberg |
| S. Goelzau | |
| S. Give | |
| F | S. Veneziana |
| G | S. Poona |
| Y | S. enterica subsp. diarizonae |
| Wards | Isolates (%) |
|---|---|
| Pediatric | 22% |
| External patients | 20% |
| General Medicine | 11% |
| Hematology | 8% |
| Surgery | 7% |
| Emergency Department | 6% |
| Infectious Disease | 4% |
| N° | Serovar | Group (Agglutination Test) | Group (I-dONE) | Score (I-dONE) |
|---|---|---|---|---|
| 1 | S. Abony | B | B | 5 |
| 2 | S. Agona | B | B | 5 |
| 3 | S. Agona | B | B | 5 |
| 4 | S. Choleraesuis | C1 | C1 | 5 |
| 5 | S. Choleraesuis | C1 | C1 | 5 |
| 6 | S. Derby | B | B | 5 |
| 7 | S. Derby | B | B | 5 |
| 8 | S. Enteritidis | D1 | D1 | 5 |
| 9 | S. Enteritidis | D1 | D1 | 5 |
| 10 | S. Enteritidis | D1 | D1 | 5 |
| 11 | S. Enteritidis | D1 | D1 | 5 |
| 12 | S. Enteritidis | D1 | D1 | 5 |
| 13 | S. Enteritidis | D1 | D1 | 5 |
| 14 | S. Goelzau | E | E | 5 |
| 15 | S. Goldcoast | C2 | C2 | 5 |
| 16 | S. Infantis | C1 | C1 | 5 |
| 17 | S. Infantis | C1 | C1 | 5 |
| 18 | S. Infantis | C1 | C1 | 5 |
| 19 | S. Infantis | C1 | C1 | 5 |
| 20 | S. Infantis | C1 | C1 | 5 |
| 21 | S. Infantis | C1 | C1 | 5 |
| 22 | S. Napoli | D1 | D1 | 5 |
| 23 | S. Napoli | D1 | D1 | 5 |
| 24 | S. Poona | G | G | 5 |
| 25 | S. Senftenberg | E | E | 5 |
| 26 | S. Thomson | C1 | C1 | 5 |
| 27 | S. Typhi | D1 | D1-S. Typhi | 5 |
| 28 | S. Typhimurium | B | B | 5 |
| 29 | S. Typhimurium | B | B | 5 |
| 30 | S. Typhimurium | B | B | 5 |
| 31 | S. Typhimurium | B | B | 5 |
| 32 | S. Typhimurium | B | B | 5 |
| 33 | S. Typhimurium | B | B | 5 |
| 34 | Monophasic variant of S. Typhimurium | B | B | 5 |
| 35 | Monophasic variant of S. Typhimurium | B | B | 5 |
| 36 | Monophasic variant of S. Typhimurium | B | B | 5 |
| 37 | Monophasic variant of S. Typhimurium | B | B | 5 |
| 38 | Monophasic variant of S. Typhimurium | B | B | 5 |
| 39 | Monophasic variant of S. Typhimurium | B | B | 5 |
| 40 | Monophasic variant of S. Typhimurium | B | B | 5 |
| 41 | Monophasic variant of S. Typhimurium | B | B | 5 |
| 42 | Monophasic variant of S. Typhimurium | B | B | 5 |
| 43 | Monophasic variant of S. Typhimurium | B | B | 5 |
| 44 | Monophasic variant of S. Typhimurium | B | B | 5 |
| 45 | Monophasic variant of S. Typhimurium | B | B | 5 |
| 46 | Monophasic variant of S. Typhimurium | B | B | 5 |
| 47 | Monophasic variant of S. Typhimurium | B | B | 5 |
| 48 | Monophasic variant of S. Typhimurium | B | B | 5 |
| 49 | Monophasic variant of S. Typhimurium | B | B | 5 |
| 50 | S. Virchow | C1 | C1 | 5 |
| 51 | Not known | C1/C2 | C1 | 5 |
| 52 | Not known | D1 | D | 5 |
| 53 | Not known | C1 | C | 4.5 |
| 54 | Not known | E G | E1 | 5 |
| 55 | Not known | B | B | 5 |
| 56 | Not known | D1 | D | 5 |
| 57 | S. Typhi | D1 | D1-S. Typhi | 4.75 |
| 58 | Not known | D1 | D | 5 |
| 59 | Not known | D1 | D | 5 |
| 60 | Not known | B | B | 5 |
| 61 | Not known | D1 | D | 4.5 |
| 62 | Not known | D1 | D | 5 |
| 63 | Not known | D1 | D | 5 |
| 64 | Not known | B | B | 5 |
| 65 | Not known | B | B | 5 |
| 66 | Not known | B | B | 5 |
| 67 | Not known | C1 | C1 | 5 |
| 68 | Not known | B | B | 4.5 |
| 69 | Not known | B | B | 5 |
| 70 | Not known | C1 | C1 | 5 |
| 71 | S. Anatum | E1 | E1 | 5 |
| 72 | S. Anatum | E1 | E1 | 5 |
| 73 | Not known | C1 | C1 | 5 |
| 74 | S. Anatum | E1 | E1 | 5 |
| 75 | S. Anatum | E1 | E1 | 5 |
| 76 | Not known | D1 | D1 | 5 |
| 77 | Not known | D1 | D1 | 5 |
| 78 | Not known | D1 | D1 | 5 |
| 79 | Not known | D1 | D1 | 4.75 |
| 80 | Not known | B | B | 5 |
| 81 | Not known | B | B | 5 |
| 82 | S. Typhi | D1 | D1-S. Typhi | 5 |
| 83 | Not known | C1 | C1 | 5 |
| 84 | Not known | D1 | D1 | 5 |
| 85 | Not known | B | B | 5 |
| 86 | Not known | C1 | C1 | 4, 5 |
| 87 | Not known | D1 | D1 | 5 |
| 88 | S. Anatum | E1 | E1 | 5 |
| 89 | S. Anatum | E1 | E1 | 5 |
| 90 | S. Enteritidis | D1 | D1 | 4.75 |
| 91 | S. London | E1 | E1 | 4.75 |
| 92 | S. Anatum | E1 | E1 | 5 |
| 93 | S. Anatum | E1 | E1 | 5 |
| 94 | S. Typhi | D1 | D1-S. Typhi | 5 |
| 95 | Monophasic variant of S. Typhimurium | B | B | 5 |
| Serogroup | Sensitivity | Specificity |
|---|---|---|
| B | 100.0% | 100.0% |
| C1 | 100.0% | 98.7% |
| C2 | 0.0% | 100.0% |
| D1 | 100.0% | 100.0% |
| D1-S. Typhi | 100.0% | 100.0% |
| E1 | 100.0% | 98.8% |
| E4 | 0.0% | 100.0% |
| G | 100.0% | 100.0% |
| Predicted Data | |||||||
|---|---|---|---|---|---|---|---|
| B | C1 | D1 | D1-S. Typhi | E1 | G | ||
| Reference data | B | 38 | 0 | 0 | 0 | 0 | 0 |
| C1 | 0 | 17 | 0 | 0 | 0 | 0 | |
| C2 | 0 | 1 | 0 | 0 | 0 | 0 | |
| D1 | 0 | 0 | 22 | 0 | 0 | 0 | |
| D1-S. Typhi | 0 | 0 | 0 | 4 | 0 | 0 | |
| E G | 0 | 0 | 0 | 0 | 1 | 0 | |
| E1 | 0 | 0 | 0 | 0 | 10 | 0 | |
| E4 | 0 | 0 | 0 | 0 | 1 | 0 | |
| G | 0 | 0 | 0 | 0 | 0 | 1 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giordano, C.; Del Conte, F.; Napoleoni, M.; Barnini, S. Machine Learning-Powered ATR-FTIR Spectroscopic Clinical Evaluation for Rapid Typing of Salmonella enterica O-Serogroups and Salmonella Typhi. Bacteria 2025, 4, 45. https://doi.org/10.3390/bacteria4030045
Giordano C, Del Conte F, Napoleoni M, Barnini S. Machine Learning-Powered ATR-FTIR Spectroscopic Clinical Evaluation for Rapid Typing of Salmonella enterica O-Serogroups and Salmonella Typhi. Bacteria. 2025; 4(3):45. https://doi.org/10.3390/bacteria4030045
Chicago/Turabian StyleGiordano, Cesira, Francesca Del Conte, Maira Napoleoni, and Simona Barnini. 2025. "Machine Learning-Powered ATR-FTIR Spectroscopic Clinical Evaluation for Rapid Typing of Salmonella enterica O-Serogroups and Salmonella Typhi" Bacteria 4, no. 3: 45. https://doi.org/10.3390/bacteria4030045
APA StyleGiordano, C., Del Conte, F., Napoleoni, M., & Barnini, S. (2025). Machine Learning-Powered ATR-FTIR Spectroscopic Clinical Evaluation for Rapid Typing of Salmonella enterica O-Serogroups and Salmonella Typhi. Bacteria, 4(3), 45. https://doi.org/10.3390/bacteria4030045







