First Culturing of Potential Bacterial Endophytes from the African Sahelian Crop Fonio Grown Under Abiotic Stress Conditions
Abstract
1. Introduction
2. Materials and Methods
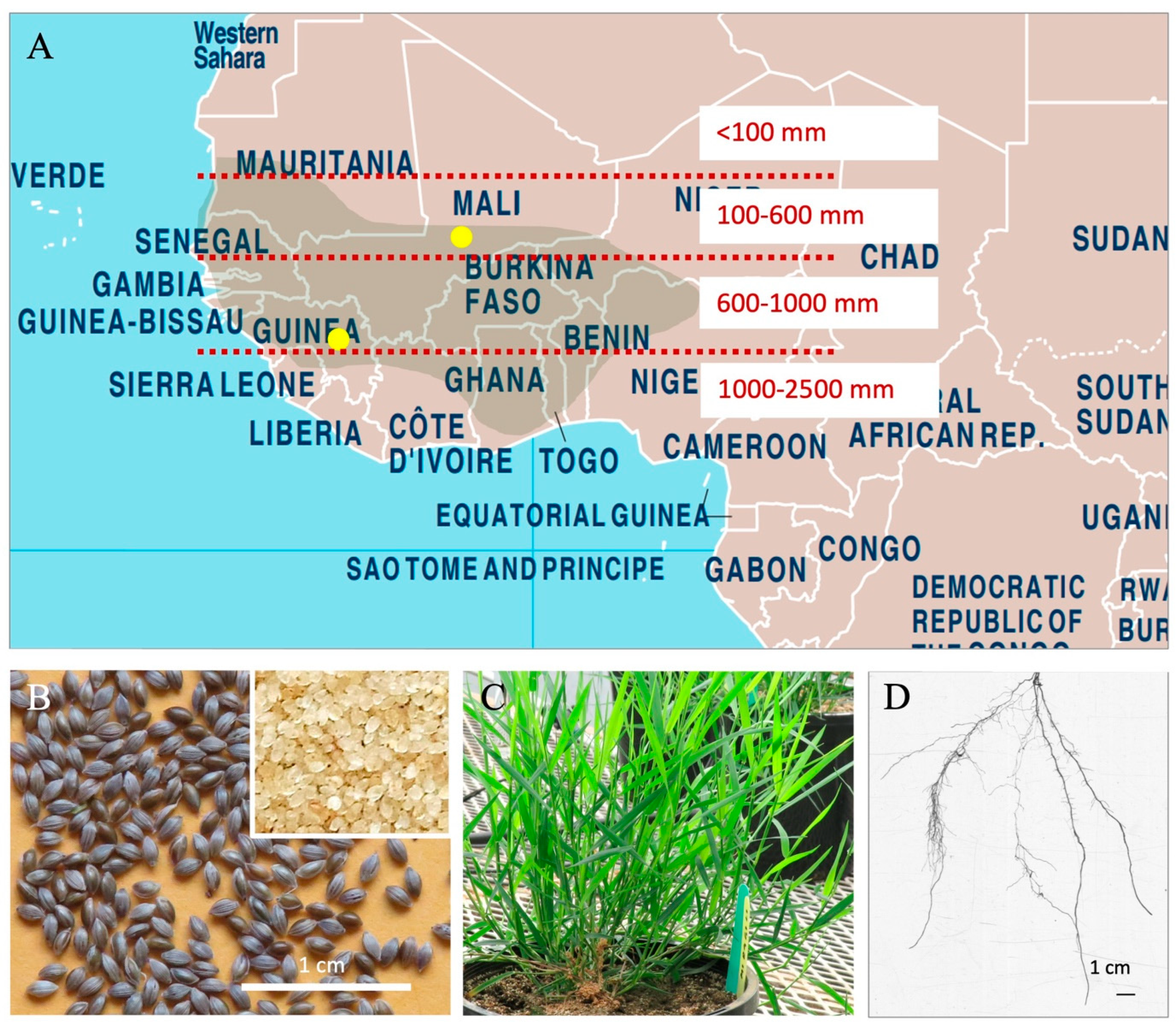
2.1. Plant Genetic Material
2.2. Plant Growth Conditions and Experimental Design
2.3. Tissue Sampling
2.4. Bacterial Isolation Media
2.5. Bacterial Culturing
2.6. Bacterial DNA Isolation and Sequencing
2.7. 16S rRNA Gene Analysis and Phylogenetic Tree Construction
2.8. Bacterial Transmission Bioinformatic Analysis
2.9. GenBank Accession Numbers
2.10. Functional Abiotic Stress Tolerance Experiments
2.10.1. Growth on Nitrogen-Free Media
2.10.2. Drought Tolerance
2.10.3. Acid and Aluminum Tolerance
3. Results
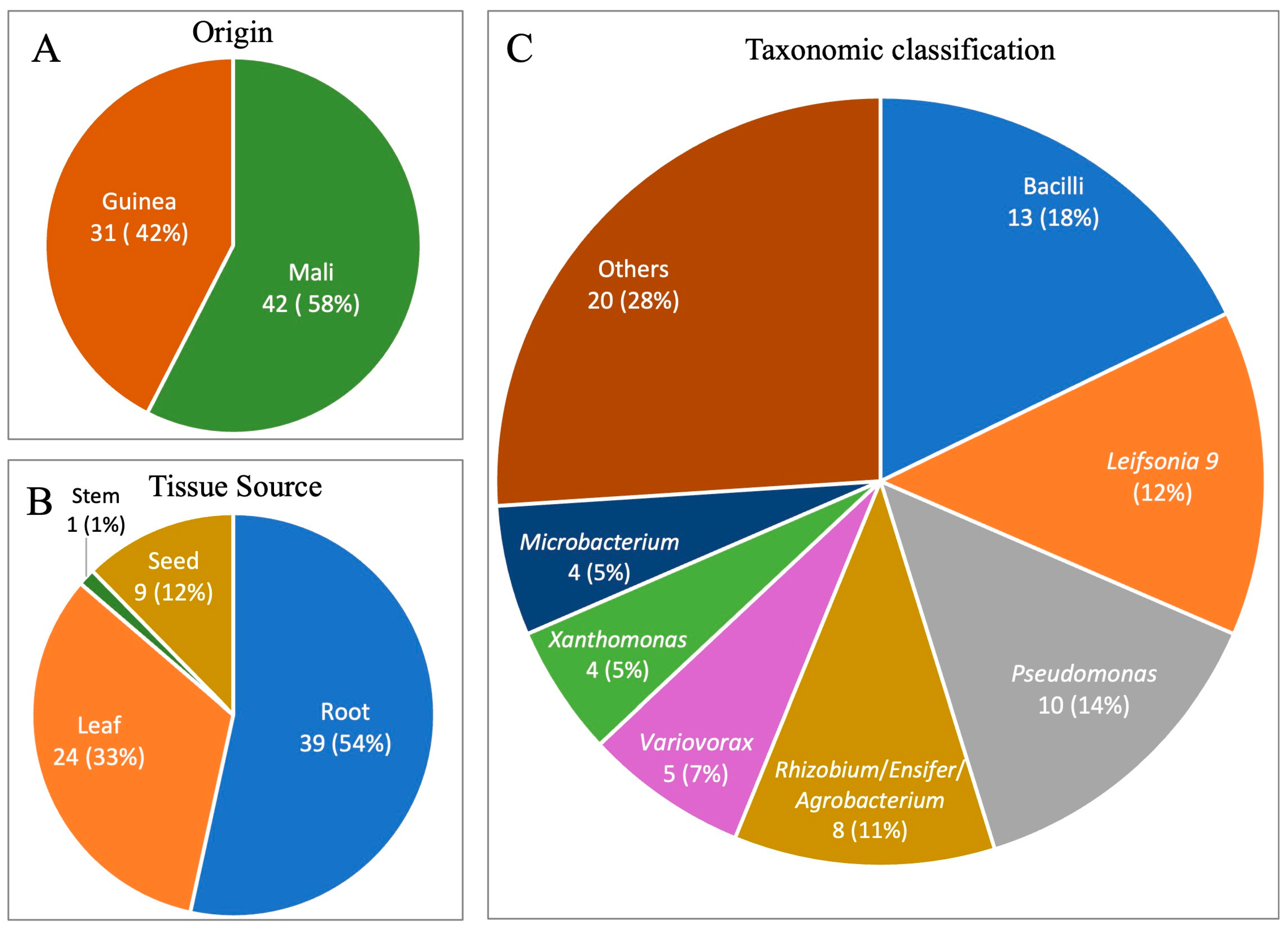
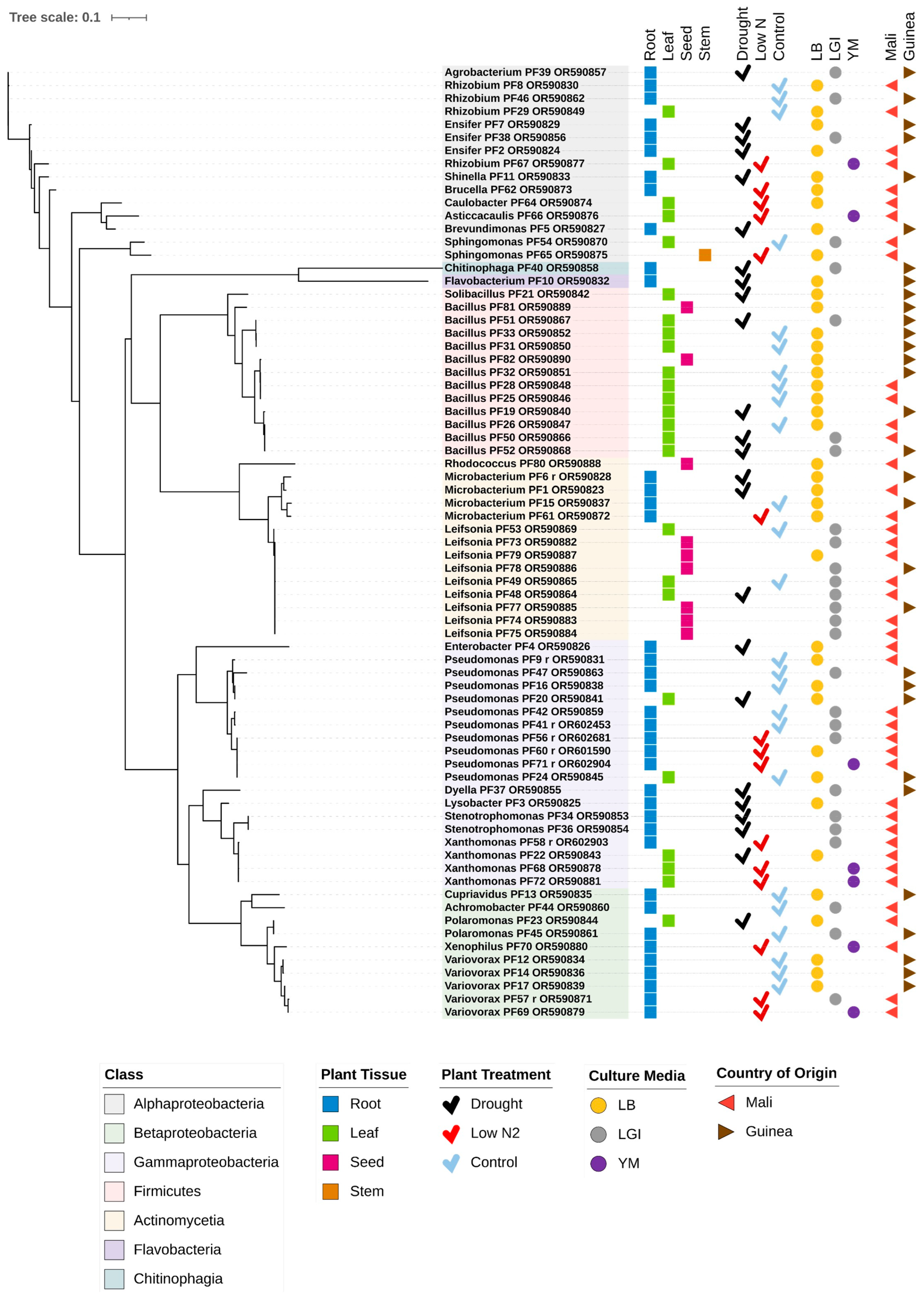
3.1. Overview of Bacterial Isolates from Fonio
3.2. Potential Bacterial Endophytes Isolated from White Fonio Leaves and Roots in the Drought Versus the Optimal Water Experiment
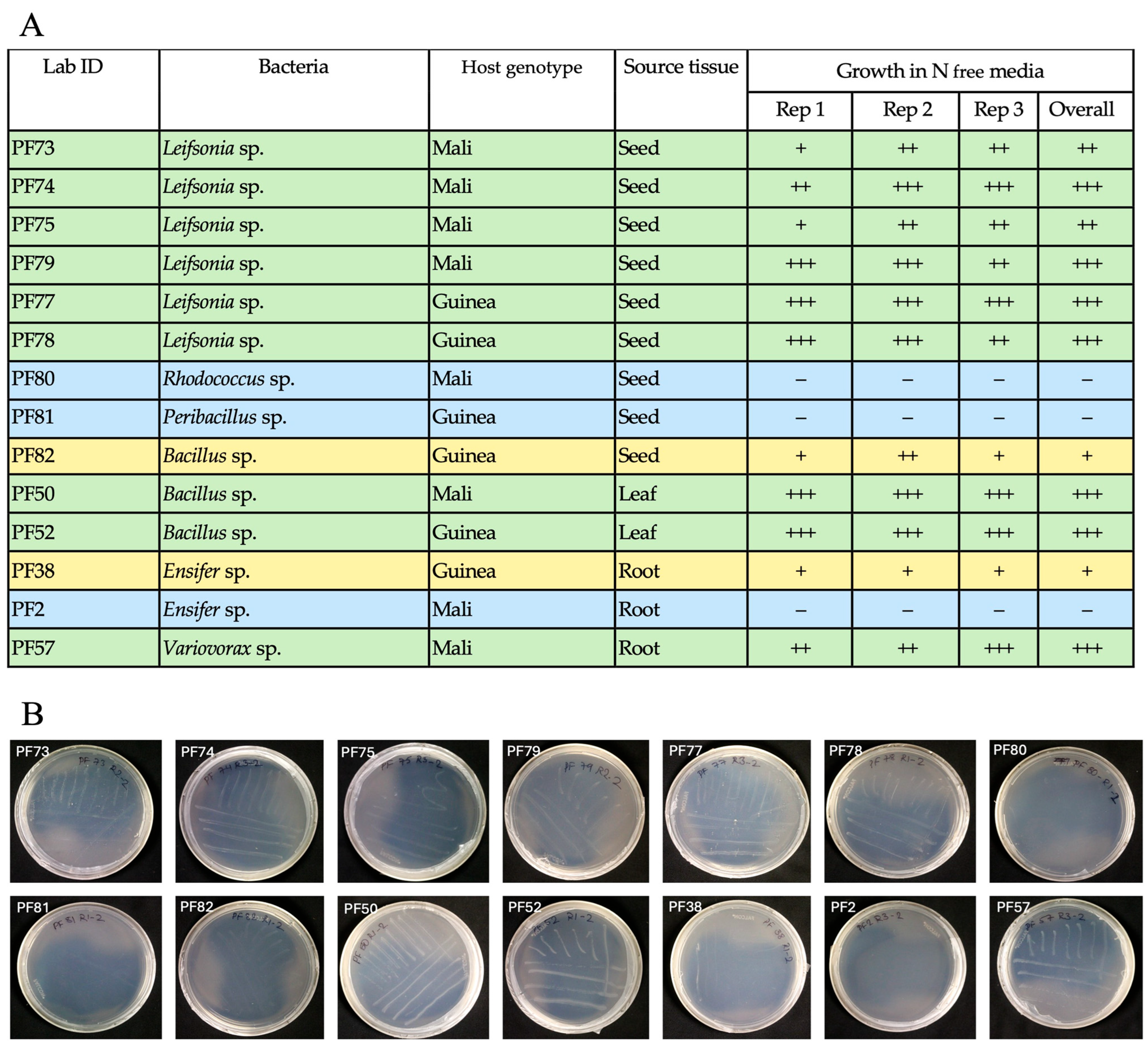
3.3. Potential Bacterial Endophytes from White Fonio Grown Under Low Nitrogen Stress
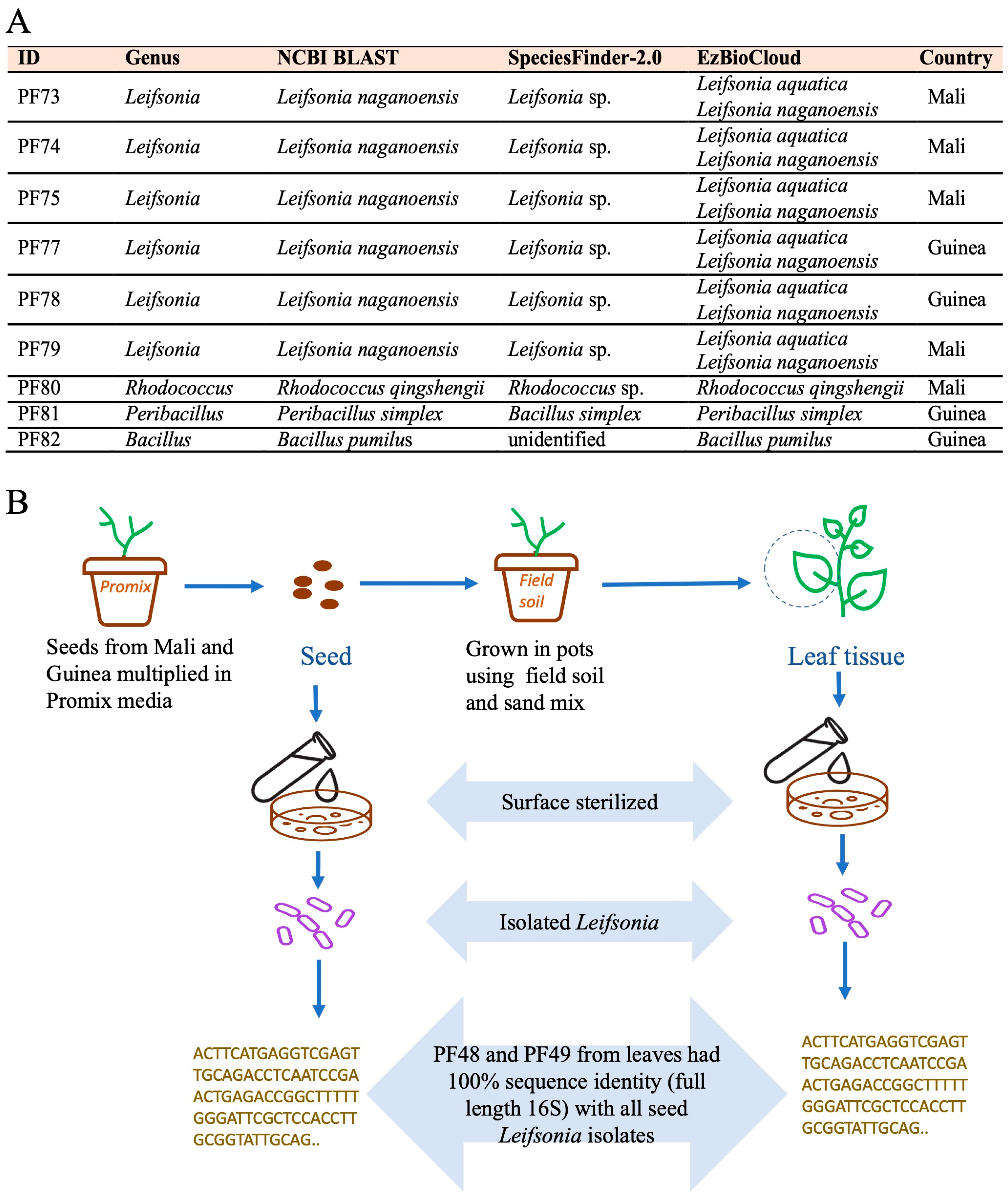
3.4. Potential Bacterial Endophytes from White Fonio Seeds and Possible Transmission Dynamics
3.5. Functional Abiotic Stress Tolerance Experiments for Seed Isolates
4. Discussion
4.1. General Discussion
4.2. The Fonio Endosphere Contains Bacteria Known to Tolerate Abiotic Stress and Promote Plant Growth
4.3. Similarities and Differences Between Fonio Seed and Leaf/Root Endophytes
4.4. Leifsonia Is a Candidate Seed-to-Leaf Transmitted Endophyte in Fonio
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- National Research Council. Lost Crops of Africa, Volume I: Grains; The National Academies Press: Washington, DC, USA, 1996; pp. 59–76. [CrossRef]
- Blench, R.M. Vernacular names for African millets and other minor cereals and their significance for agricultural history. Archaeol. Anthropol. Sci. 2016, 8, 1–8. [Google Scholar] [CrossRef]
- Cruz, J.; Béavogui, F. Fonio, An African Cereal; The French Agricultural Research Centre for International Development CIRAD: Montpellier, France, 2016; pp. 13–41. [Google Scholar]
- Jideani, I.A.; Jideani, V.A. Developments on the cereal grains Digitaria exilis (acha) and Digitaria iburua (iburu). J. Food Sci. Technol. 2011, 48, 251–259. [Google Scholar] [CrossRef]
- Ballogou, V.Y.; Soumanou, M.M.; Toukourou, F.; Hounhouigan, J.D. Indigenous knowledge on landraces and fonio-based food in Benin. Ecol. Food Nutr. 2014, 53, 390–409. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F. Fonio grains: Physicochemical properties, nutritional potential, and food applications. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3365–3389. [Google Scholar] [CrossRef]
- Temple, V.J.; Bassa, J.D. Proximate Chemical Composition of Acha (Digitaria exilis) Grain. J. Sci. Food Agric. 1991, 56, 561–563. [Google Scholar] [CrossRef]
- Vodouhe, S.R.; Achigan-Dako, G.E.; Dansi, A.; Adoukonou-Sagbadja, H. Fonio: A treasure for West Africa. In Proceedings of the Plant Genetic Resources and Food Security in West and Central Africa, Regional Conference, Ibadan, Nigeria, 26–30 April 2004; Vodouhe, R.S., Atta-Krah, K., Achigan-Dako, G.E., Eyog-Matig, O., Avohou, H., Eds.; Bioversity International: Rome, Italy, 2007; pp. 219–223. [Google Scholar]
- Wang, X.; Chen, S.; Ma, X.; Yssel, A.E.J.; Chaluvadi, S.R.; Johnson, M.S.; Gangashetty, P.; Amidou, F.; Sanogo, M.D.; Zwaenepoel, A.; et al. Genome sequence and genetic diversity analysis of an under-domesticated orphan crop, white fonio (Digitaria exilis). GigaScience 2021, 10, giab013. [Google Scholar] [CrossRef]
- Leach, J.E.; Triplett, L.R.; Argueso, C.T.; Trivedi, P. Communication in the Phytobiome. Cell 2017, 169, 587–596. [Google Scholar] [CrossRef]
- Busby, P.E.; Soman, C.; Wagner, M.R.; Friesen, M.L.; Kremer, J.; Bennett, A.; Morsy, M.; Eisen, J.A.; Leach, J.E.; Dangl, J.L. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017, 15, e2001793. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- White, J.F.; Kingsley, K.L.; Zhang, Q.; Verma, R.; Obi, N.; Dvinskikh, S.; Elmore, M.T.; Verma, S.K.; Gond, S.K.; Kowalski, K.P. Review: Endophytic microbes and their potential applications in crop management. Pest Manag. Sci. 2019, 75, 2558–2565. [Google Scholar] [CrossRef]
- Babalola, O.O.; Adedayo, A.A. Endosphere microbial communities and plant nutrient acquisition toward sustainable agriculture. Emerg. Top. Life Sci. 2023, 7, 207–217. [Google Scholar] [CrossRef]
- Card, S.; Johnson, L.; Teasdale, S.; Caradus, J. Deciphering endophyte behaviour: The link between endophyte biology and efficacious biological control agents. FEMS Microbiol. Ecol. 2016, 92, 114. [Google Scholar] [CrossRef]
- Mao, W.; Wu, Y.; Li, F.; Tang, W.; Gong, W.; Han, X.; White, J.F.; Ji, X.; Li, H. Seed endophytes and their roles in host plant stress resistance. J. Soil Sci. Plant Nutr. 2023, 23, 2927–2937. [Google Scholar] [CrossRef]
- Nadal, M.C.; dos Reis Ferreira, G.M.; Andrade, G.V.S.; Buttrós, V.H.; Rodrigues, F.A.; da Silva, C.M.; Martins, A.D.; Rufato, L.; Luz, J.M.Q.; Dória, J.; et al. Endophytic bacteria can replace the need for synthetic auxin during in vitro rooting of Pyrus communis. Agronomy 2022, 12, 1226. [Google Scholar] [CrossRef]
- Abideen, Z.; Cardinale, M.; Zulfiqar, F.; Koyro, H.W.; Rasool, S.G.; Hessini, K.; Darbali, W.; Zhao, F.; Siddique, K.H.M. Seed Endophyte bacteria enhance drought stress tolerance in Hordeum vulgare by regulating, physiological characteristics, antioxidants and minerals uptake. Front. Plant Sci. 2022, 13, 980046. [Google Scholar] [CrossRef]
- Orozco-Mosqueda, M.d.C.; Santoyo, G.; Glick, B.R. Recent advances in the bacterial phytohormone modulation of plant growth. Plants 2023, 12, 606. [Google Scholar] [CrossRef]
- Sajjad Mirza, M.; Ahmad, W.; Latif, F.; Haurat, J.; Bally, R.; Normand, P.; Malik, K.A. Isolation, partial characterization, and the effect of plant growth-promoting bacteria (PGPB) on micro-propagated sugarcane in vitro. Plant Soil 2001, 237, 47–54. [Google Scholar] [CrossRef]
- Johnston-Monje, D.; Raizada, M.N. Plant and endophyte relationships: Nutrient management. In Comprehensive Biotechnology, 2nd ed.; Moo-Young, M., Ed.; Elsevier B.V.: Amsterdam, The Netherlands, 2011; pp. 713–727. [Google Scholar]
- Szilagyi-Zecchin, V.J.; Ikeda, A.C.; Hungria, M.; Adamoski, D.; Kava-Cordeiro, V.; Glienke, C.; Galli-Terasawa, L.V. Identification and characterization of endophytic bacteria from corn (Zea mays L.) roots with biotechnological potential in agriculture. AMB Express 2014, 4, 26. [Google Scholar] [CrossRef]
- Siddikee, M.A.; Chauhan, P.S.; Anandham, R.; Han, G.H.; Sa, T. Isolation, characterization, and use for plant growth promotion under salt stress, of ACC deaminase-producing halotolerant bacteria derived from coastal soil. J. Microbiol. Biotechnol. 2010, 20, 1577–1584. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Vardharajula, S.; Ali, S.Z.; Grover, M.; Reddy, G.; Bandi, V. Drought-tolerant plant growth promoting Bacillus spp.: Effect on growth, osmolytes, and antioxidant status of maize under drought stress. J. Plant Interact. 2011, 6, 1–14. [Google Scholar] [CrossRef]
- Ali, S.Z.; Sandhya, V.; Rao, L.V. Isolation and characterization of drought-tolerant ACC deaminase and exopolysaccharide-producing fluorescent Pseudomonas sp. Ann. Microbiol. 2014, 64, 493–502. [Google Scholar] [CrossRef]
- Tabassum, N.; Ahmed, H.I.; Parween, S.; Sheikh, A.H.; Saad, M.M.; Krattinger, S.G.; Hirt, H. Host genotype, soil composition, and geo-climatic factors shape the fonio seed microbiome. Microbiome 2024, 12, 11. [Google Scholar] [CrossRef]
- Xu, L.; Naylor, D.; Dong, Z.; Simmons, T.; Pierroz, G.; Hixson, K.K.; Kim, Y.M.; Zink, E.M.; Engbrecht, K.M.; Wang, Y.; et al. Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, E4284–E4293. [Google Scholar] [CrossRef] [PubMed]
- Walitang, D.I.; Kim, C.G.; Kim, K.; Kang, Y.; Kim, Y.K.; Sa, T. The influence of host genotype and salt stress on the seed endophytic community of salt-sensitive and salt-tolerant rice cultivars. BMC Plant Biol. 2018, 18, 51. [Google Scholar] [CrossRef]
- Dastogeer, K.M.G.; Tumpa, F.H.; Sultana, A.; Akter, M.A.; Chakraborty, A. Plant microbiome–an account of the factors that shape community composition and diversity. Curr. Plant Biol. 2020, 23, 100161. [Google Scholar] [CrossRef]
- Adoukonou-Sagbadja, H.; Wagner, C.; Dansi, A.; Ahlemeyer, J.; Daïnou, O.; Akpagana, K.; Ordon, F.; Friedt, W. Genetic diversity and population differentiation of traditional fonio millet (Digitaria spp.) landraces from different agro-ecological zones of West Africa. Theor. Appl. Genet. 2007, 115, 917–931. [Google Scholar] [CrossRef]
- Olodo, K.F.; Gueye, M.C.; Calatayud, C.; Diop, B.M.; Kane, N.A.; Ngom, A.; Ntui, V.O.; Barreto, M.M.S.; Uyoh, E.A.; Abraham, S.; et al. EST-SSR development for Digitaria exilis and its relatives D. iburua and D. longiflora from transcriptome sequences. Plant Genet. Resour. Characterisation Util. 2019, 17, 280–284. [Google Scholar] [CrossRef]
- Abrouk, M.; Ahmed, H.I.; Cubry, P.; Šimoníková, D.; Cauet, S.; Pailles, Y.; Bettgenhaeuser, J.; Gapa, L.; Scarcelli, N.; Couderc, M.; et al. Fonio millet genome unlocks African orphan crop diversity for agriculture in a changing climate. Nat. Commun. 2020, 11, 4488. [Google Scholar] [CrossRef]
- Sidibé, D. Mali Agricultural Pilot Soil Baseline and Background Research. Mali Agricultural Pilot, Research Report. OXFAM America. 2013. Available online: http://www.oxfamamerica.org/static/media/files/Mali_Agricultural_Study_Report_-_Soil_Baseline_and_Background_Research.pdf (accessed on 17 December 2023).
- Touré, I.; Larjavaara, M.; Savadogo, P.; Bayala, J.; Yirdaw, E.; Diakite, A. Land degradation along a climatic gradient in Mali: Farmers’ perceptions of causes and impacts. Land Degrad. Dev. 2020, 31, 2804–2818. [Google Scholar] [CrossRef]
- FAOSTAT. Crop and Livestock Products. Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/faostat/en/#data (accessed on 29 November 2023).
- Shehata, H.R.; Lyons, E.M.; Jordan, K.S.; Raizada, M.N. Bacterial endophytes from wild and ancient maize are able to suppress the fungal pathogen Sclerotinia homoeocarpa. J. Appl. Microbiol. 2016, 120, 756–769. [Google Scholar] [CrossRef] [PubMed]
- Chojnacki, S.; Cowley, A.; Lee, J.; Foix, A.; Lopez, R. Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res. 2017, 45, 550–553. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; McBeigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Ha, S.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Lukjancenko, O.; Saputra, D.; Rasmussen, S.; Hasman, H.; Sicheritz-Pontén, T.; Aarestrup, F.M.; Ussery, D.W.; Lund, O. Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 2012, 52, 1529–1539. [Google Scholar] [CrossRef]
- Edgar, R.C.; Drive, R.M.; Valley, M. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acid. Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.E.H.; Raizada, M.N. The microbiome of fertilization-stage maize silks (style) encodes genes and expresses traits that potentially promote survival in pollen/style niches and host reproduction. Microorganisms 2024, 12, 1473. [Google Scholar] [CrossRef]
- Hernández-Fernández, G.; Galán, B.; Carmona, M.; Castro, L.; García, J. Transcriptional response of the xerotolerant Arthrobacter sp. Helios strain to PEG-induced drought stress. Front. Microbiol. 2022, 13, 1009068. [Google Scholar] [CrossRef]
- Latif, M.; Bukhari, S.A.H.; Alrajhi, A.A.; Alotaibi, F.S.; Ahmad, M.; Shahzad, A.N.; Dewidar, A.Z.; Mattar, M.A. Inducing drought tolerance in wheat through exopolysaccharide-producing rhizobacteria. Agronomy 2022, 12, 1140. [Google Scholar] [CrossRef]
- Huang, S.; Wang, X.; Liu, X.; He, G.; Wu, J. Isolation, identification, and characterization of an aluminum-tolerant bacterium Burkholderia sp. SB1 from an acidic red soil. Pedosphere 2018, 28, 905–912. [Google Scholar] [CrossRef]
- Lim, J.C.; Goh, K.M.; Shamsir, M.S.; Ibrahim, Z.; Chong, C.S. Characterization of aluminum resistant Anoxybacillus sp. SK 3–4 isolated from a hot spring. J. Basic Microbiol. 2015, 55, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Small, E. Teff & Fonio—Africa’s sustainable cereals. Biodiversity 2015, 16, 27–41. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Tack, A.J.M.; Lobato, C.; Wassermann, B.; Berg, G. From seed to seed: The role of microbial inheritance in the assembly of the plant microbiome. Trends Microbiol. 2023, 31, 346–355. [Google Scholar] [CrossRef]
- Etesami, H.; Jeong, B.R.; Glick, B.R. Potential use of Bacillus spp. as an effective biostimulant against abiotic stresses in crops—A review. Curr. Res. Biotech. 2023, 5, 100128. [Google Scholar] [CrossRef]
- Hilhorst, T.; Muchena, E.M. (Eds.) Nutrients on the Move: Soil Fertility Dynamics in African Farming Systems; International Institute for Environment and Development: London, UK, 2000; pp. 83–117. [Google Scholar]
- Sissoko, K.; van Keulen, H.; Verhagen, J.; Tekken, V.; Battaglini, A. Agriculture, livelihoods and climate change in the West African Sahel. Reg. Environ. Change 2011, 11, 119–125. [Google Scholar] [CrossRef]
- Lopes, R.; Tsui, S.; Gonçalves, P.J.R.O.; de Queiroz, M.V. A look into a multifunctional toolbox: Endophytic Bacillus species provide broad and underexploited benefits for plants. World J. Microbiol. Biotechnol. 2018, 34, 94. [Google Scholar] [CrossRef]
- Abdelaal, K.; Alkahtani, M.; Attia, K.; Hafez, Y.; Király, L.; Künstler, A. The role of plant growth-promoting bacteria in alleviating the adverse effects of drought on plants. Biology 2021, 10, 520. [Google Scholar] [CrossRef] [PubMed]
- Poveda, J.; González-Andrés, F. Bacillus as a source of phytohormones for use in agriculture. Appl. Microbiol. Biotechnol. 2021, 105, 8629–8645. [Google Scholar] [CrossRef] [PubMed]
- Tsotetsi, T.; Nephali, L.; Malebe, M.; Tugizimana, F. Bacillus for plant growth promotion and stress resilience: What have we learned? Plants 2022, 11, 2482. [Google Scholar] [CrossRef] [PubMed]
- Mahapatra, S.; Yadav, R.; Ramakrishna, W. Bacillus subtilis impact on plant growth, soil health and environment: Dr. Jekyll and Mr. Hyde. J. Appl. Microbiol. 2022, 132, 3543–3562. [Google Scholar] [CrossRef]
- Doso Jnr, S. Land degradation and agriculture in the Sahel of Africa: Causes, impacts and recommendations. J. Agric. Sci. Appl. 2014, 3, 67–73. [Google Scholar] [CrossRef]
- Xing, Y.X.; Wei, C.Y.; Mo, Y.; Yang, L.T.; Huang, S.L.; Li, Y.R. Nitrogen-fixing and plant growth-promoting ability of two endophytic bacterial strains isolated from sugarcane stalks. Sugar Tech. 2016, 18, 373–379. [Google Scholar] [CrossRef]
- Patel, J.K.; Archana, G. Diverse culturable diazotrophic endophytic bacteria from Poaceae plants show cross-colonization and plant growth promotion in wheat. Plant Soil 2017, 417, 99–116. [Google Scholar] [CrossRef]
- Rosenblueth, M.; Ormeño-Orrillo, E.; López-López, A.; Rogel, M.A.; Reyes-Hernández, B.J.; Martínez-Romero, J.C.; Reddy, P.M.; Martínez-Romero, E. Nitrogen fixation in cereals. Front. Microbiol. 2018, 9, 1794. [Google Scholar] [CrossRef]
- Fall, F.; le Roux, C.; Bâ, A.M.; Fall, D.; Bakhoum, N.; Faye, M.N.; Kane, A.; Ndoye, I.; Diouf, D. The rhizosphere of the halophytic grass Sporobolus robustus Kunth hosts rhizobium genospecies that are efficient on Prosopis juliflora (Sw.) DC and Vachellia seyal (Del.) P.J.H. Hurter seedlings. Syst. Appl. Microbiol. 2019, 42, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Wu, X.; Yao, L.; Chen, Z. Heavy metal-immobilizing bacteria combined with calcium polypeptides reduced the uptake of Cd in wheat and shifted the rhizosphere bacterial communities. Environ. Pollut. 2020, 267, 115432. [Google Scholar] [CrossRef]
- Xi, J.; Qian, K.; Shan, L.; Huang, J.; Yan, Y. The potential of mineral weathering of halophilic-endophytic bacteria isolated from Suaeda salsa and Spartina anglica. Arch. Microbiol. 2022, 204, 561. [Google Scholar] [CrossRef] [PubMed]
- Bahulikar, R.A. Prevalence of Deltaproteobacterial sequences in nifH gene pools associated with the rhizosphere of native switchgrass from Tall Grass Prairie (Oklahoma, USA). Biotech 2023, 13, 210. [Google Scholar] [CrossRef]
- Andrews, M.; Andrews, M.E. Specificity in legume-rhizobia symbioses. Int. J. Mol. Sci. 2017, 18, 705. [Google Scholar] [CrossRef]
- Finkel, O.M.; Salas-González, I.; Castrillo, G.; Conway, J.M.; Law, T.F.; Teixeira, P.J.P.L.; Wilson, E.D.; Fitzpatrick, C.R.; Jones, C.D.; Dangl, J.L. A single bacterial genus maintains root growth in a complex microbiome. Nature 2020, 587, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Duong, B.; Xuan Nguyen, H.; Viet Phan, H.; Colella, S.; Quang Trinh, P.; Thi Hoang, G.; Thi Nguyen, T.; Marraccini, P.; Lebrun, M.; Duponnois, R. Identification and characterization of Vietnamese coffee bacterial endophytes displaying in vitro antifungal and nematicidal activities. Microbiol. Res. 2021, 242, 126613. [Google Scholar] [CrossRef]
- Liaqat, F.; Eltem, R. Identification and characterization of endophytic bacteria isolated from in vitro cultures of peach and pear rootstocks. 3 Biotech 2016, 6, 120. [Google Scholar] [CrossRef]
- Battu, L.; Ulaganathan, K. Whole genome sequencing and identification of host-interactive genes in the rice endophytic Leifsonia sp. ku-ls. Funct. Integr. Genom. 2020, 20, 237–243. [Google Scholar] [CrossRef]
- Jiang, X.; Li, W.W.; Han, M.; Chen, G.; Wu, J.; Lai, S.; Fu, Z.; Zhang, S.; Deng, W.W.; Gao, L.; et al. Aluminum-tolerant, growth-promoting endophytic bacteria as contributors in promoting tea plant growth and alleviating aluminum stress. Tree Physiol. 2022, 42, 1043–1058. [Google Scholar] [CrossRef]
- Verma, A.; Shameem, N.; Jatav, H.S.; Sathyanarayana, E.; Parray, J.A.; Poczai, P.; Sayyed, R.Z. Diversity of bacterial endophytes of maize (Zea mays) and their functional potential for micronutrient biofortification. Curr. Microbiol. 2022, 79, 6. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.M.; Khan, A.L.; You, Y.H.; Kim, J.G.; Kamran, M.; Lee, I.J. Gibberellin production by newly isolated strain Leifsonia soli SE134 and its potential to promote plant growth. J. Microbiol. Biotechnol. 2014, 24, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Nordstedt, N.P.; Jones, M.L. Isolation of rhizosphere bacteria that improve quality and water stress tolerance in greenhouse ornamentals. Front. Plant Sci. 2020, 11, 826. [Google Scholar] [CrossRef] [PubMed]
- Nordstedt, N.P.; Roman-Reyna, V.; Jacobs, J.M.; Jones, M.L. Comparative genomic understanding of gram-positive plant growth-promoting Leifsonia. Phytobiomes J. 2021, 5, 263–274. [Google Scholar] [CrossRef]
- Cox, C.M.; Koo, J. Soil fertility. In Atlas of African Agriculture Research and Development: Revealing Agriculture’s Place in Africa; Sebastian, K., Ed.; International Food Policy Research Institute (IFPRI): Washington, DC, USA, 2014; pp. 40–41. [Google Scholar] [CrossRef]
| ID | Genus | NCBI BLAST | SpeciesFinder-2.0 | EzBioCloud | Origin | Tissue Source | Treatment |
|---|---|---|---|---|---|---|---|
| PF44 | Achromobacter | Achromobacter spanius | Achromobacter xylosoxidans | Achromobacter mucicolens | M | Root | Optimum water |
| PF39 | Agrobacterium | Agrobacterium tumefaciens | Agrobacterium sp. | Agrobacterium tumefaciens | G | Root | Drought |
| PF5 | Brevundimonas | Uncultured bacterium | Brevundimonas vesicularis | Brevundimonas vesicularis | G | Root | Drought |
| PF40 | Chitinophaga | Chitinophaga sp. | Chitinophaga sp. | Chitinophaga ginsengisoli | G | Root | Drought |
| PF13 | Cupriavidus | Cupriavidus sp. | Cupriavidus sp. | Cupriavidu campinensis | G | Root | Optimum water |
| PF37 | Dyella | Dyella jiangningensis | Dyella jiangningensis | Dyella jiangningensis | G | Root | Drought |
| PF2 | Ensifer | Ensifer adhaerens | Ensifer adhaerens | Ensifer morelensis | M | Root | Drought |
| PF38 | Ensifer | Ensifer sp. | Unknown | Ensifer adhaerens | G | Root | Drought |
| PF7 | Ensifer | Ensifer adhaerens | Sinorhizobium sp. | Ensifer adhaerens | G | Root | Drought |
| PF4 | Enterobacter | Enterobacter ludwigii | Enterobacter sp. | Enterobacter ludwigii | M | Root | Drought |
| PF10 | Flavobacterium | Flavobacterium hauense | Unknown | Flavobacterium sp. | G | Root | Drought |
| PF3 | Lysobacter | Lysobacter sp. | Lysobacter soli | Lysobacter panacisoli | M | Root | Drought |
| PF1 | Microbacterium | Microbacterium sp. | Microbacterium sp. | Microbacterium saccharophilum | M | Root | Drought |
| PF62 | Microbacterium | Microbacterium chocolatum | Microbacterium sp. | Microbacterium atlanticum | G | Root | Drought |
| PF15 | Microbacterium | Microbacterium foliorum | Microbacterium sp. | Microbacterium aerolatum | G | Root | Optimum water |
| PF45 | Polaromonas | Polaromonas sp. | Polaromonas sp. | Polaromonas ginsengisoli | G | Root | Optimum water |
| PF16 | Pseudomonas | Pseudomonas sp. | Pseudomonas sp. | Pseudomonas solani | G | Root | Optimum water |
| PF41 | Pseudomonas | Pseudomonas mosselii | Pseudomonas mosselii | Pseudomonas mosselii | M | Root | Optimum water |
| PF42 | Pseudomonas | Pseudomonas mosselii | Pseudomonas sp. | Pseudomonas mosselii | M | Root | Optimum water |
| PF47 | Pseudomonas | Pseudomonas nitroreducens | Pseudomonas sp. | Pseudomonas nitritireducens | G | Root | Optimum water |
| PF9 | Pseudomonas | Pseudomonas paralcaligenes | Pseudomonas alcaligenes | Pseudomonas alcaligenes | M | Root | Optimum water |
| PF46 | Rhizobium | Agrobacterium tumefaciens, Rhizobium sp. | Rhizobium sp. | Agrobacterium radiobacter | G | Root | Optimum water |
| PF8 | Rhizobium | Agrobacterium tumefaciens | Rhizobium sp. | Agrobacterium radiobacter | M | Root | Optimum water |
| PF11 | Shinella | Unknown | Unknown | Shinella curvata | G | Root | Drought |
| PF34 | Stenotrophomonas | Stenotrophomonas maltophilia | Stenotrophomonas maltophilia | Stenotrophomonas sp. | M | Root | Drought |
| PF36 | Stenotrophomonas | Stenotrophomonas maltophilia | Stenotrophomonas maltophilia | Stenotrophomonas sp. | M | Root | Drought |
| PF12 | Variovorax | Variovorax beijingensis | Variovorax sp. | Variovorax paradoxus | G | Root | Optimum water |
| PF14 | Variovorax | Variovorax beijingensis | Variovorax sp. | Variovorax paradoxus | G | Root | Optimum water |
| PF17 | Variovorax | Variovorax beijingensis | Variovorax sp. | Variovorax paradoxus | G | Root | Optimum water |
| PF19 | Bacillus | Bacillus subtilis | Bacillus subtilis | Bacillus tequilensis | G | Leaf | Drought |
| PF50 | Bacillus | Bacillus sp. | Bacillus subtilis | Bacillus siamensis | M | Leaf | Drought |
| PF51 | Bacillus | Bacillus thuringiensis | Bacillus sp. | Bacillus proteolyticus | G | Leaf | Drought |
| PF52 | Bacillus | Bacillus siamensis | Bacillus subtilis | Bacillus siamensis | G | Leaf | Drought |
| PF25 | Bacillus | Bacillus safensis | Bacillus safensis | Bacillus safensis | M | Leaf | Optimum water |
| PF26 | Bacillus | Bacillus sp. | Bacillus sp. | Bacillus siamensis | M | Leaf | Optimum water |
| PF28 | Bacillus | Bacillus sp. | unidentified | Bacillus safensis | M | Leaf | Optimum water |
| PF31 | Bacillus | Bacillus toyonensis | Bacillus cereus | Bacillus toyonensis | G | Leaf | Optimum water |
| PF32 | Bacillus | Bacillus safensis | unidentified | Bacillus safensis | G | Leaf | Optimum water |
| PF33 | Bacillus | Bacillus sp. | Bacillus cereus | Bacillus toyonensis | G | Leaf | Optimum water |
| PF48 | Leifsonia | Leifsonia naganoensis | Leifsonia sp. | Leifsonia aquatica Leifsonia naganoensis | M | Leaf | Drought |
| PF49 | Leifsonia | Leifsonia naganoensis | Leifsonia sp. | Leifsonia aquatica Leifsonia naganoensis | M | Leaf | Optimum water |
| PF53 | Leifsonia | Leifsonia naganoensis | Leifsonia sp. | Leifsonia aquatica Leifsonia naganoensis | M | Leaf | Optimum water |
| PF23 | Polaromonas | Polaromonas sp. | Polaromonas sp. | Polaromonas ginsengisoli | M | Leaf | Drought |
| PF20 | Pseudomonas | Pseudomonas sp. | Pseudomonas sp. | Pseudomonas solani | G | Leaf | Drought |
| PF24 | Pseudomonas | Pseudomonas jessenii | Pseudomonas sp. | Pseudomonas moorei | G | Leaf | Optimum water |
| PF29 | Rhizobium | Agrobacterium tumefaciens | Rhizobium sp. | Agrobacterium radiobacter | M | Leaf | Optimum water |
| PF21 | Solibacillus | Solibacillus sp. | Solibacillus sp. | Solibacillus isronensis | G | Leaf | Drought |
| PF5 | Sphingomonas | Sphingomonas echinoides | Sphingomonas sp. | Sphingomonas echinoides | M | Leaf | Optimum water |
| PF22 | Xanthomonas | Xanthomonas sp. | Xanthomonas oryzae pv. Oryzae | Xanthomonas maliensis | M | Leaf | Drought |
| ID | Genus | NCBI BLAST | SpeciesFinder-2.0 | EzBioCloud | Tissue Source |
|---|---|---|---|---|---|
| PF62 | Brucella | Ochrobactrum sp. | Ochrobactrum sp. | Brucella tritici | Root |
| PF61 | Microbacterium | Microbacterium oxydans | Microbacterium oxydans | Microbacterium oxydans | Root |
| PF56 | Pseudomonas | Pseudomonas chlororaphis | Pseudomonas fluorescens | Pseudomonas moorei | Root |
| PF60 | Pseudomonas | Pseudomonas moorei | Pseudomonas sp. | Pseudomonas vancouverensis | Root |
| PF71 | Pseudomonas | Pseudomonas moorei | Pseudomonas sp. | Pseudomonas vancouverensis | Root |
| PF57 | Variovorax | Variovorax paradoxus | Variovorax sp. | Variovorax sp. | Root |
| PF69 | Variovorax | Variovorax sp. | Variovorax sp. | Variovorax sp. | Root |
| PF58 | Xanthomonas | Xanthomonas maliensis | Xanthomonas citri pv. Punicae | Xanthomonas euroxanthea | Root |
| PF72 | Xanthomonas | Xanthomonas sp. | Xanthomonas sp. | Xanthomonas maliensis | Root |
| PF70 | Xenophilus | Variovorax sp. | Xenophilus aerolatus | Xenophilus aerolatus | Root |
| PF66 | Asticcacaulis | Asticcacaulis sp. | Asticcacaulis sp. | Asticcacaulis benevestitus | Leaf |
| PF64 | Caulobacter | Caulobacter rhizosphaerae | Uncultured bacterium | Caulobacter sp. | Leaf |
| PF67 | Rhizobium | Rhizobium sp. | Rhizobium mongolense | Rhizobium sp. | Leaf |
| PF68 | Xanthomonas | Xanthomonas sp. | Xanthomonas sp. | Xanthomonas maliensis | Leaf |
| PF65 | Sphingomonas | Sphingomonas sp. | Sphingomonas mucosissima | Sphingomonas olei | Stem |
| Lab ID | Bacteria | Host Genotype | Source Tissue | 1 Tolerance [Percent OD600 Versus Negative Control Without Stress] | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10% PEG | 30% PEG | 40% PEG | 0.1 mM AlCl3 at pH 4.5 | 0.4 mM AlCl3 at pH 4.5 | ||||||||||||||
| 2 MAX | MIN | AVG | MAX | MIN | AVG | MAX | MIN | AVG | MAX | MIN | AVG | MAX | MIN | AVG | ||||
| PF73 | Leifsonia sp. | Mali | Seed | 97 | 101 | 99 | 25 | 22 | 23 | 6 | 4 | 5 | 115 | 130 | 114 | 28 | 25 | 27 |
| PF74 | Leifsonia sp. | Mali | Seed | 111 | 108 | 111 | 11 | 6 | 10 | 9 | 10 | 10 | 72 | 57 | 61 | 36 | 8 | 23 |
| PF75 | Leifsonia sp. | Mali | Seed | 67 | 57 | 61 | 5 | 4 | 5 | 4 | 3 | 3 | 121 | 140 | 131 | 44 | 69 | 58 |
| PF79 | Leifsonia sp. | Mali | Seed | 107 | 96 | 104 | 10 | 4 | 7 | 11 | 7 | 9 | 15 | 85 | 31 | 36 | 69 | 44 |
| PF77 | Leifsonia sp. | Guinea | Seed | 94 | 91 | 95 | 8 | 5 | 6 | 6 | 6 | 6 | 96 | 127 | 108 | 88 | 182 | 108 |
| PF78 | Leifsonia sp. | Guinea | Seed | 104 | 117 | 111 | 26 | 9 | 15 | 18 | 9 | 13 | 73 | 60 | 68 | 11 | 9 | 9 |
| PF80 | Rhodococcus sp. | Mali | Seed | 90 | 96 | 94 | 9 | 8 | 9 | 12 | 7 | 9 | 73 | 136 | 122 | 9 | 33 | 17 |
| PF81 | Peribacillus sp. | Guinea | Seed | 67 | 67 | 67 | 14 | 11 | 14 | 5 | 4 | 4 | 29 | 100 | 41 | 12 | 50 | 22 |
| PF82 | Bacillus sp. | Guinea | Seed | 64 | 50 | 59 | 24 | 13 | 17 | 7 | 4 | 5 | 23 | 62 | 37 | 15 | 23 | 22 |
| PF50 | Bacillus sp. | Mali | Leaf | 63 | 60 | 61 | 11 | 6 | 8 | 7 | 5 | 6 | 46 | 28 | 39 | 11 | 5 | 8 |
| PF52 | Bacillus sp. | Guinea | Leaf | 8 | 9 | 9 | 2 | 2 | 2 | 2 | 3 | 3 | 47 | 47 | 46 | 1 | 1 | 1 |
| PF38 | Ensifer sp. | Guinea | Root | 49 | 49 | 49 | 3 | 3 | 3 | 3 | 3 | 3 | 98 | 157 | 114 | 65 | 109 | 76 |
| PF2 | Ensifer sp. | Mali | Root | 47 | 38 | 41 | 9 | 4 | 7 | 8 | 7 | 8 | 72 | 65 | 67 | 67 | 74 | 73 |
| PF57 | Variovorax sp. | Mali | Root | 55 | 46 | 50 | 10 | 7 | 8 | 15 | 2 | 6 | 63 | 50 | 65 | 42 | 175 | 74 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pudasaini, R.; Khalaf, E.M.; Brettingham, D.J.L.; Raizada, M.N. First Culturing of Potential Bacterial Endophytes from the African Sahelian Crop Fonio Grown Under Abiotic Stress Conditions. Bacteria 2025, 4, 31. https://doi.org/10.3390/bacteria4030031
Pudasaini R, Khalaf EM, Brettingham DJL, Raizada MN. First Culturing of Potential Bacterial Endophytes from the African Sahelian Crop Fonio Grown Under Abiotic Stress Conditions. Bacteria. 2025; 4(3):31. https://doi.org/10.3390/bacteria4030031
Chicago/Turabian StylePudasaini, Roshan, Eman M. Khalaf, Dylan J. L. Brettingham, and Manish N. Raizada. 2025. "First Culturing of Potential Bacterial Endophytes from the African Sahelian Crop Fonio Grown Under Abiotic Stress Conditions" Bacteria 4, no. 3: 31. https://doi.org/10.3390/bacteria4030031
APA StylePudasaini, R., Khalaf, E. M., Brettingham, D. J. L., & Raizada, M. N. (2025). First Culturing of Potential Bacterial Endophytes from the African Sahelian Crop Fonio Grown Under Abiotic Stress Conditions. Bacteria, 4(3), 31. https://doi.org/10.3390/bacteria4030031







