Abstract
Maize silks (style) receive wind-transmitted pollen. Thereafter, male gametes travel through the silks to reach ovules. Pollinated silks contain a microbiome, members of which are predicted to promote host reproduction and abiotic/biotic stress tolerance during fertilization. It is unknown whether pollinated silk bacteria originate environmentally (air/pollen) or from maternal tissue. Methods are lacking to test microbial colonization of silks in their native habitat [on intact maize inflorescences (cobs) encased by husk leaves]. Current methods focus on naked silks attached to dehusked cob pieces. Here, two novel methods are presented to enable research on silk microbes in their native habitat. Method 1 tests whether silk-associated bacteria with potential environmental origins are attracted toward ovules. Method 2 distinguishes whether a microbe colonizes silks from the environment or maternal parent. Biosafety containment was enabled by housing microbe-treated cobs in large jars. Using these methods, a model bacterial isolate from fertilization-stage silks (DsRed-tagged Pantoea-E04) was shown to colonize husk-covered silks after inoculating exposed silk tips; E04 could not colonize from the cob base, suggesting an environmental origin. In support, E04 colonized silks more frequently when cobs were uncut and oriented vertically. These protocols will enable more biologically relevant investigation of silk microbiomes and pathogens.
Keywords:
microbiome; endophyte; beneficial microbe; cob; pollen; fertilization; reproduction; colonization; fluorescence 1. Introduction
Styles form the female reproductive tract of flowering plants, through which male gametes travel to eventually fertilize an ovule [1]. The male gametes originate from pollen and use elongating pollen tubes that grow through the style to achieve fertilization [2,3]. Maize has uniquely long styles, commonly known as silks, that make it a model system for studying style biology. Maize silk initiates from the ovule, protected by husk leaves, and grows toward the environment to capture pollen, eventually emerging from the tip of the ear of maize; hence, it exhibits clear directionality (polarity) in its growth [2].
There is emerging interest in the microbes that live on or in maize silks, which may promote silk health and host reproduction [4] or may protect developing seeds (derived from ovules) from environmental pathogens that use silks as a passageway [5,6,7,8]. A microbe that inhabits a sub-surface tissue is known as an endophyte [9,10].
Some studies have investigated the vertical transmission of endophytic microbes in plants, where bacteria are transmitted from a parent to progeny via the seeds [11,12]. Endophytes are also transmitted horizontally, for example, from the surrounding environment, sometimes with insect vectors, similar to the horizontal transmission of pathogens [13,14]. Pollen-based microbe transmission has also been proposed [15,16,17]. There are many endophytic microbes in pollinated maize silks, many of which may be beneficial to maize [4,6,7,8]. Bacterial taxa that have the potential for plant growth promotion and nitrogen fixation have also been found in maize pollen [18,19]. It is possible that silk endophytes come from multiple sources, including the environment (horizontal transmission), pollen/pollen tubes (vertical transmission), and endogenously from the maternal plant (possibly originating from seed or soil via the vascular system). However, the origin of maize silk-associated microbes is unknown. The fate of bacteria associated with fertilization-stage silks is also unknown, specifically whether they migrate toward the seed, which is of importance to understanding their vertical transmission to progeny (e.g., if derived from pollen).
We hypothesize that the origin and fate of specific silk-associated microbes can be investigated by tracking the direction of bacterial migration on cobs and silks, specifically to differentiate whether (1) they are competent to migrate from silk tips (environment) to the base (toward the seed), suggestive of transmission from the environment or pollen; or alternatively, (2) they are competent to migrate from the base of the cob through its vascular system into the silk base toward the tip, suggestive of a maternal origin.
The general purpose of this study was to design methods to track bacterial migration on cobs and silks. Previous microscopic protocols have observed bacteria and/or pathogenic fungi on maize silks that remained attached to dissected cob segments [20,21], but methods for observing microbial colonization on silks attached to intact cobs are lacking. Presumably, intact cobs with their protective husk leaves more closely resemble the native habitat of silk endophytes. Furthermore, the use of young cobs may ensure that the host material is metabolically alive to support microbes. To our knowledge, there are no existing techniques for tracking bacterial migration on intact cobs. Briefly, the methods that will be presented here involve the application of fluorescently tagged bacteria to intact maize cobs, which are incubated in large glass jars. The cobs are then dissected, and silk segments are ground and plated onto selective media to determine the colonization patterns of the bacteria.
Maize silks have native bacteria [4,6,7,8], as already mentioned, and hence, to distinguish any bacterium of interest, it must be tagged, such as in this study, with a fluorescent protein. Here, a model bacterial isolate was tagged with DsRed (Addgene, 111257) and used in this case study to demonstrate the new methods. The isolate was referred to as E04 (Pantoea sp.) and was selected for a number of reasons. First, it was isolated from fertilization-stage maize silks and exhibited the potential to provide multiple benefits to the host maize, as well as adaptations to stressors in the silk/pollen environments, and potential for vertical transmission [4,7,8]. Bacterial isolate E04 was recently shown to be tolerant to 10% poly(ethylene glycol) (PEG-6000) as a proxy for desiccation/drought tolerance; to produce indole compounds as a proxy for indoleacetic acid (IAA; an auxin), which may stimulate egg maturation/silk growth; and could grow anaerobically without nitrogen, which may indicate nitrogen fixation ability or capacity to survive in low-nutrient environments [4]. Isolate E04 also contains genes involved in osmoprotectant biosynthesis and transport, indole-3-acetic acid biosynthesis, gamma-aminobutyric acid (GABA, γ-aminobutyrate) import, and aluminum and acid tolerance; these genes may help with bacterial survival and/or promote host reproduction [4]. The E04 genetic sequence matches a 16S V4-MiSeq OTU sequence that was previously predicted to be a member of the core maize silk microbiome and a Fusarium graminearum indicator based on analyses of the silks of 14 North American maize genotypes [6,7,8]. Isolate E04 was observed colonizing maize silks externally and internally, including at wound sites [8,21]. Isolate E04 was also concluded to be safe based on susceptibility to multiple antibiotics, whole-genome mining for virulence genes, and biosafety risk group. Thus, this bacterium has potential as a multi-functional microbial treatment in plants, and its interactions with maize should be studied. Whether strain E04 enters silks from maternal parent tissue or the environment was unknown, and hence, it was tested here using both original methods.
The specific objective of the current study was to test and present two novel methods to track bacteria in young, intact cobs with their husk leaves using model isolate E04. Method 1 is intended to test whether a silk-associated bacterium may have an environmental origin (including pollen) and/or is attracted to the ovule (future seed after fertilization) by applying bacteria to the tip of the cob. Method 1 also tests the impact of cob orientation and silk cutting on bacterial colonization of silks. In this method, bacteria are inoculated from the exposed silk tip, and then colonization is measured along silk segments. Method 2 is broader and is intended to distinguish whether a microbe is able to colonize silks if inoculated from the silk tips (implicating an environmental/pollen origin) versus the base of the cob (implicating a maternal parent origin). These methods are intended to allow researchers to investigate a variety of silk-inhabiting microbes and to improve the collective understanding of the plant reproductive microbiome.
2. Materials and Methods
2.1. Sources of Biological Materials
Source of maize cobs: For the first experiment, PHRE1 inbred maize (Pioneer Hi-Bred, Johnston, IA, USA) was planted at the Elora Research Station (University of Guelph, Elora, ON, Canada) as described in [21], and cobs with young, emerged silks were harvested. All cobs were harvested on the same day and were grouped by similar maturity for each replicate. For the second experiment, LH82 inbred maize was sequentially planted at the University of Guelph Crop Science Greenhouse Facility under the conditions described in [8] and [21]. Cobs were young, intact, and husk-covered to ensure silks were alive and growing and to reflect the native habitat of silk-associated microbes.
Source of bacterial strain: Isolate E04 (Pantoea sp., NCBI GenBank biosample number SAMN37538673, accession number JAWILD000000000) was isolated from pollinated, field-grown maize silks [7,8]. This isolate was selected based on previous microbiome and functional analyses as described above, making E04 a highly relevant strain for future crop treatments and microbiome breeding programs. The isolate was transformed with tetracycline (Tet)-resistant plasmids pSW002-PpsbA-DsRed-Express2 (DsRed) (Addgene, 111257), as previously described [8,21].
2.2. Method 1: Testing the Ability of a Microbe to Be Taken Up from the Environment, Colonize Silks, and Subsequently Migrate Toward Ovules
Method 1 is intended to test whether a silk-associated bacterium is able to colonize from the environment (implicating a possible air or pollen origin) and subsequently migrate toward the ovule (silk base, future seed after fertilization). In this method, fluorescently tagged bacteria were inoculated on the exposed silk tips of intact, husk-covered maize cobs, which were incubated in large jars (Figure 1), and then colonization was measured by dissecting silk segments and culturing them on selective media. Bacteria were inoculated onto cut/uncut silks attached to intact, husk-covered maize silks on cobs that were placed either vertically or horizontally (Figure 1). Uncut silks were tested to mimic their natural field condition. Cut silks were also tested in case this impacted bacterial colonization at the exposed wound site; here, the emerged silk tips were trimmed down to the husk. The cut silks continued to grow during the 5 d incubation and extended beyond the end of the husks, indicating that the silks were still alive after the ear was removed from the stalk (Figure 1C, inset). Hence, the approach used represents an in vitro live tissue assay. It was also tested whether orienting the cobs vertically or horizontally affected the assay. Vertically oriented cobs reflect their positioning when attached to the maize stalk in modern hybrids. If a tip-inoculated bacterium migrates toward the base when the cob is positioned vertically, it would be significant since the transpiration stream and parental metabolites flow in the opposite direction [22]. The horizontal cob orientation was added to understand, for example, the natural assistance of gravity on bacterial migration.

Figure 1.
Method 1: Methodology for testing whether a microbe can colonize maize silks from the environment and whether it is potentially attracted to developing seeds (silk base). Bacteria were inoculated onto cut or uncut, exposed, living maize silks of cobs oriented vertically (typical of modern hybrid maize cobs) or horizontally. (A,B) Shown are young cobs harvested for the experiment. The exposed silks were either left intact or cut with a razor blade ~1 cm inward from the end of the husk, as marked with red lines. (C) DsRed-tagged bacteria were applied to the silk tips (cut or uncut) in either horizontal or vertical positions, and after 5 d of incubation, the cobs were dissected. As seen in the magnified inset (blue rectangle), the silks on the cut cobs grew during the 5 d period, extending beyond the husks. The husks were carefully removed, and approximately 2.5 cm segments were cut near the silk tips (beginning ~1 cm inward from the husk end), middle, and near the base (beginning ~1 cm inward from the base of the cob). (D) Jars in horizontal position. (E) Jar in vertical position, with a Falcon tube taped against the inner wall to hold a young cob. (F) Horizontal and vertical jars containing cobs in the process of being inoculated with DsRed-tagged bacteria.
A single, fluorescently tagged isolate of particular interest (DsRed-tagged E04, Pantoea) was evaluated for its ability to colonize silks when inoculated on exposed silk tips attached to young, freshly harvested, intact maize cobs in autoclaved glass jars with lids (1 gallon, ~3.785 L, Catalog #S-19317B-M, Uline, Canada). The bacterium was evaluated for its ability to colonize the husk-enclosed silks after being applied onto uncut or cut silks of cobs, which were placed horizontally or upright.
For upright cobs, 50 mL Falcon tubes were taped to the inside of the glass jar. E04 bacterial liquid cultures were centrifuged at 4000× g for 4 min, washed in 10 mM Tris-HCl thrice, and resuspended in 10 mM Tris-HCl to an OD600 reading of 0.2–0.5 (typically ~0.4). Subsequently, 2 mL of resuspended E04 liquid culture was applied via pipette to the exposed silks (cut or uncut) of cobs (placed horizontally or vertically) from which only the first layer of outer husk leaves had been removed, leaving the cob covered with many layers of husk leaves.
Jars were sealed for 5 d, and then the cobs were dissected. The external silks were discarded, the husk leaves were carefully removed, and silks that had been contained within the husk were cut into base, middle, and tip segments using sterile razor blades (see Figure 1). Silks were transferred into sterile mortar and pestles, ground with 2 mL Tris-HCl, and 100 µL was immediately plated onto lysogeny broth (LB) plates containing Tet (150 µL at 0.3 µg/µL pipetted on top and dried) and incubated at 30 °C for 5 d. The plates were then examined for fluorescent colonies under UV light, photographed, and quantified.
2.3. Method 2: Method to Test the Competency of a Microbe to Colonize Silks from the Environment (Silk Tips) Versus from the Maternal Parent (Cob Base)
Method 2 is intended to distinguish whether a microbe is able to colonize silks when inoculated from the silk tips (inferring a possible environmental origin including pollen) versus the base of the cob (inferring a maternal parent origin). In this secondary experiment, 2 mL of 10 mM Tris-HCl-washed bacteria with an OD600 reading of ~0.45 (0.41–0.49) was dispensed into the bottom of the Falcon tubes in some of the jars, and the base of the cob was placed into the bacterial liquid culture (no treatment onto the exposed silks). In parallel, the silk tips were inoculated with additional precautions to prevent contamination at the cob base. The cobs for these silk tip inoculations were temporarily placed with the silk-end submerged in separate Falcon tubes containing E04 bacterial liquid culture for 6 min, followed by the cobs sitting horizontally for 5 min to allow any excess bacteria to drip off, and then the cobs were placed vertically upright in a clean, dry, empty Falcon tube inside the jar. This experiment was also used to control the artifact of excess bacterial liquid culture dripping down the outside of husks in Method 1 so that one could be confident that the bacteria had entered via the exposed silk tips and were not taken up through the vascular system at the base of the cobs. All cobs in this test were uncut and vertical, as these conditions were found to promote silk colonization in Method 1, and these conditions also represent the natural state of cobs in the field. Jars were sealed for 5 d, and then the cobs were dissected. Subsequent steps to recover E04 were the same as in Method 1.
3. Results
3.1. Results from Method 1: Testing the Ability of a Microbe to Be Taken up from the Environment, Colonize Silks, and Subsequently Migrate Toward Ovules
Pantoea strain E04 was inoculated onto the cut/uncut emerged silks of intact, husk-covered maize ears that had been placed either vertically or horizontally in jars (Figure 1). After inoculation and incubation, red fluorescent colonies were cultured from all three cobs that had been vertical and had silks that were left uncut; in 2/3 replicates, E04 was successfully cultured from silks near the base of the cobs (Figure 2). One cob from each of the remaining treatment combinations (vertical cut, horizontal cut, and horizontal uncut) had one or more red fluorescent colonies cultured from a single segment of silks (Figure 2). Additional non-fluorescent colonies were cultured (Figure 2), likely members of the diverse silk microbiome [6,8]. These results suggest that E04 can colonize silks when introduced from the environment and can migrate toward the ovule, but this is most consistent when cobs are uncut and in a vertical orientation.
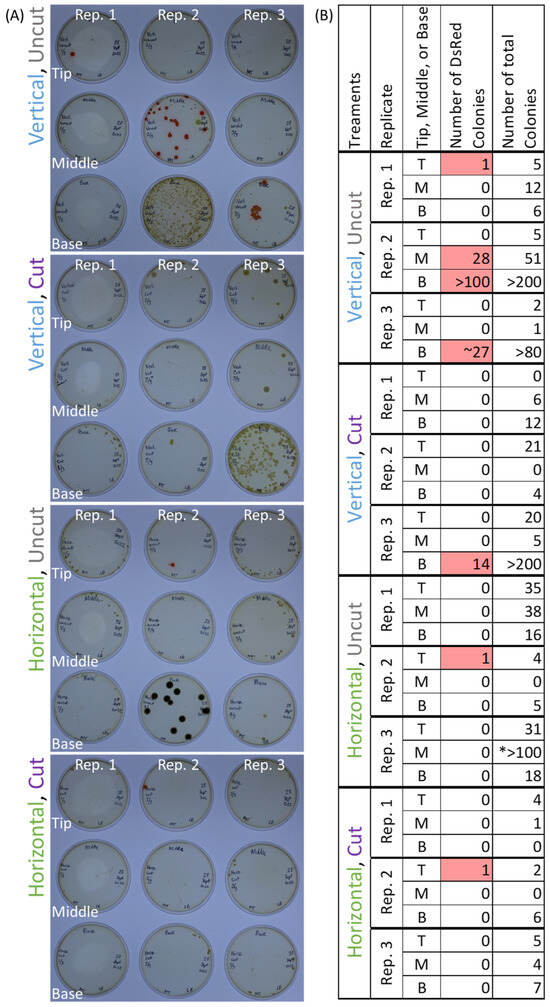
Figure 2.
Results from Method 1: (A) After dissection, the silks were ground with buffer and plated onto LB agar plates that had been treated with tetracycline and then incubated for 2 d. The images show the results of 3 replicates from each treatment combination. (B) Summary of numbers of microbial colonies present from panel A. The DsRed-tagged colonies (highlighted in pink) fluoresced under UV light and were E04 that had entered into/colonized the husk-covered silks, whereas the untagged colonies were members of the naturally occurring silk microbiome. The asterisk (*) indicates that the colonies were all small.
Microbe studies have a need for containment, and there are restrictions on the use of fluorescently tagged transgenic microbes. An advance of this study was the use of large (~3.8 L) glass jars to contain the large maize cobs treated with transgenic microbes (e.g., Figure 1E).
3.2. Results from Method 2: Testing the Competency of a Microbe to Colonize Silks from the Environment (Silk Tips) Versus Maternal Parent (Cob Base)
Pantoea strain E04 was inoculated onto vertical cobs, on the uncut silk tips, and in parallel onto the cob base (Figure 3). Fluorescent red colonies were cultured from all three replicate cobs that had been inoculated at silk tips (Figure 3D, left side), and E04 colonized all of the way down the silks to the silk tissue at the base in replicate 2. No red fluorescent colonies were isolated from samples that had been inoculated at the base of the cobs (Figure 3D, right side). Combined with the results of Figure 2, these results suggest that strain E04 colonizes silks from the environment via exposed silk tips.
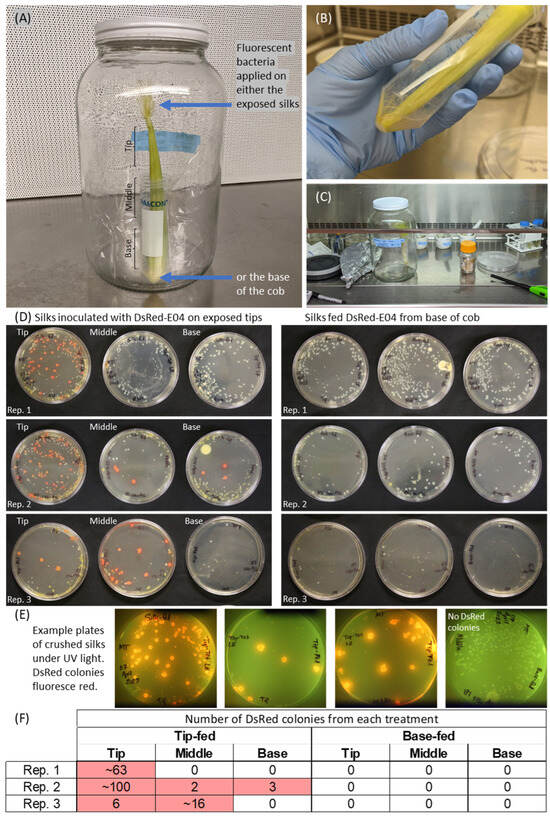
Figure 3.
Method 2: Methodology and results to distinguish whether a microbe is able to colonize silks if inoculated from the silk tips (and hence may have an environmental origin) versus the base of the cob (and hence may have a maternal parent origin). (A) Shown is the jar system containing a young, intact, and husk-covered maize cob to mimic the native habitat of silk-associated microbes. (B) For applications to the silk tips, the silks were temporarily submersed in the buffer-washed DsRed bacteria in a separate Falcon tube, then laid horizontally on a large Petri dish to allow excess liquid to drip off, and the cobs were then transferred, base-down, into a clean, dry Falcon tube taped to the inside of a jar. (C) After 5 d, the cobs were dissected in a biosafety hood. The external silks were discarded, and silks that had been contained within the husk were dissected into tip, middle, and base segments and ground with buffer in a mortar and pestle, as shown here. (D) Results of incubating the plated, crushed silk solutions on tetracycline-coated LB plates for 5 d. The bright pink colonies are DsRed-E04, which fluoresce under UV light. The non-DsRed bacteria are expected to be native members of the silk microbiome because maize silks are known to contain a rich diversity of bacteria. (E) Example plates showing the verification screening method used to distinguish true DsRed colonies from background colonies. Plates show colonies that grew from crushed silks, pictured under UV light to reveal bright DsRed fluorescence of E04 colonies. (F) Summary of numbers of DsRed colonies present from panel D. In some cases, the colonies were small and/or grew together, which is the reason that approximate numbers were sometimes reported. Pink cells indicate non-zero values, i.e., the presence of DsRed.
4. Discussion
Here, two novel methods were successfully developed and tested with the objective of creating biologically relevant assays that could track the bacterial migration on cobs and silks, which may help determine the origin of the large numbers of silk-associated bacteria in maize [4,6,7] and whether they are attracted to ovules and hence vertically transmitted to progeny. Intact, growing cobs with attached husk leaves were successfully used to mimic the native habitat of silk-associated microbes. As maize cobs are large and microbial studies have a need for containment, particularly for transgenic microbes such as those that are fluorescently tagged, an advance of this study was the successful use of large glass jars to incubate cobs and their silks. A diverse and abundant microbiome is known to exist in maize silks [6,8], including many strains with resistance to multiple antimicrobials [8], so it was expected that some colonies without the Tet-resistant fluorescent plasmid would appear on the Tet-LB plates. The use of the fluorescent tag with UV light made it possible to distinguish colonies of E04. On a cautionary note, a limitation of this study was that, although replicated, each method was only validated in a single trial. However, the general biological findings from Method 1 were similar to those from Method 2; these experiments were conducted weeks apart and completely independent, and support the robustness of the findings as well as both approaches.
4.1. Directional Colonization of Maize Silks by Pantoea Strain E04
The methods presented for testing the directional colonization of silks in intact maize cobs were successful in determining that Pantoea strain E04 can enter silks when applied to the exposed silk tips of a vertical cob in a jar but not when applied at the cob base (Figure 3). This result suggests that this microbe, which was cultured from open-pollinated maize silks, may have possibly originated from maize pollen or the environment, although further experiments would be required to confirm this hypothesis, including whether E04 inhabits maize pollen. Many Pantoea strains are known to be beneficial to plants [23,24,25,26,27,28,29], and some are likely vertically transmitted [12,30]. The results here, combined with the previous gene-mining and in vitro study [4], suggest that E04 may hold potential as a multi-functional microbial treatment that could be sprayed onto exposed silks in season.
As to the reason that E04 was unable to colonize silks via the cob base, the base was essentially a wound site where the cob had been snapped off from the stalk. Though this should have created an entry point for E04 into the cob, on a cautionary note, the wounded end may have acted differently than an intact connection point between a cob and the main stalk of maize. Perhaps the plant defenses in response to the wounding [31,32,33] prevented bacterial colonization from the base. In this regard, the cob base inoculation assay should be viewed cautiously until bacteria have been found that can successfully colonize silks from a severed cob base. Cob base-entering microbes would also be challenged by the obstacles on the path to reach the silks, including the cob itself and the ovule, which originates silks [2] and which, presumably, by the fertilization stage would have defenses against microbes [34].
4.2. The Validity of Bacterial Inoculation of Silk Tips Attached to Intact Maize Cobs In Vitro
The results from Method 2 showing directional colonization from the exposed silk tips (Figure 3) validated Method 1 (Figure 1 and Figure 2) and, more specifically, the reliability of inoculating silk tips of intact cobs in vitro. This is because the application of DsRed-tagged E04 to silk tips in Method 1 allowed some E04 liquid culture to drip down to the base of the cobs in the vertical orientation and pool at the bottom of the Falcon tubes. This left the results unclear as to whether the E04 colonies that were cultured from these silks had entered the enclosed silks via the exposed silk tips or traveled upward through the vasculature of the cob from the base. Since in Method 2, all the deliberately base-fed cobs (Figure 3) were free from E04 in the silks, and all of the silk tip fed cobs contained E04 in the silks, it can be assumed that all of the E04 cultured from silks in the first experiment (Method 1) entered the enclosed silks via the exposed silk tips.
4.3. The Effect of Cob Orientation on Bacterial Colonization
The assay was successful in determining the conditions in which bacterial isolate E04 would colonize maize silks on intact, detached cobs. The combination of uncut silks and a vertical cob orientation resulted in the greatest silk colonization (Figure 1 and Figure 2). As to the underlying reason for greater colonization on these silks within vertical cobs, the vertical orientation mimics the position of cobs on a stalk in modern maize hybrids that are adapted to a high field density [35], and it is possible that gravity contributes to the rapid downward colonization of the silks [36]. In pathogenic P. ananatis, flagella aid the bacteria in attaching to and spreading across the plant surfaces [37]; perhaps gravity aids the efficacy of flagella-based motility.
4.4. The Effect of Cutting Silks on Bacterial Colonization
The cut silks were observed to have grown out beyond the ends of the husk during the incubation period, indicating that this in planta assay employing young cobs utilizes living tissues, making it a contained, resource-efficient model for testing colonization of metabolically-active silks (Figure 1C). The uncut silks were more readily colonized than cut silks, perhaps due to a host wound response preventing entry of E04 and possibly related to the migration of silk cell nuclei to wound sites [21,38]. This is especially relevant in conjunction with maize breeding in general, and specifically microbiome-associated breeding, because controlled crosses are often performed by cutting the exposed silks before deliberate pollination [39]. These cuts could potentially reduce the transmission of microbes inherited via pollen, altering the seed microbiome of the progeny, and this warrants further research. It was interesting that here, E04 typically did not colonize far into silks that had been cut, whereas previous work observed E04 colonizing the inside of short segments of silks via wound sites [8,21]. Perhaps internal colonization via wound sites occurs across short distances, and further transit along the length of a silk is more challenging.
5. Limitations
The methods presented here represent an in planta assay that makes testing of novel bacteria possible in a way that keeps the bacteria with unknown human and environmental health impacts and the fluorescent plasmids contained to protect researchers and the environment. A trade-off, in terms of comparing these methods to whole-plant experiments, is that they do not exactly replicate field conditions; the cobs are detached from the maize stalk, and they are expected to have fewer long-term endogenous plant defenses with these methods [40], potentially allowing for more colonization. In terms of the cob base assay, the wounded tissue and some plant defenses at this location [31,32,33] may have also worked against upward colonization of the silks from the base. Furthermore, in this study, the microbes did not face the variable weather conditions as they would in the field. However, these limitations were essential to maintain sealed, contained conditions at a reasonable scale of size in a laboratory.
6. Conclusions and Future Applications
This study presented two novel, biologically relevant methods to track bacterial migration on maize silks from intact cobs to help determine the origin and fate of specific bacteria. A case study was presented for the model bacterial isolate E04 (Pantoea), which colonized silks when applied to the exposed, uncut tips on vertical cobs.
Microbial treatments are increasingly important tools in sustainable agriculture. The methods developed here hold potential for assessing the ability of novel bacterial isolates to colonize silk tissues, which is critical for future studies, including vertical transmission of the floral microbiome, biocontrol against silk-invading pathogens [5], and biostimulants to promote plant reproduction [4]. These methods may be used as a screen prior to investing in greenhouse or field trials. If a strain of bacteria cannot colonize the silks well in a jar experiment, it may be precluded from further experiments. The methods demonstrated the ability of E04 to colonize silks when applied externally, which indicates that this isolate has the potential to colonize silks successfully if it were to be applied as an in-field microbial treatment.
These methods may also be used in the study of microbial consortia [41,42], and different microbial strains could be tagged with different fluorescent tags (e.g., a bacterium with DsRed, another microbe with GFP, and another microbe with Turquoise). Interactions between members of the microbiome could be observed via confocal microscopy.
These methods should also be utilized in conjunction with plant disease studies; there is much potential for research in this area, and a small, biologically relevant, contained system lends itself well to the study of silk–microbe–pathogen interactions since several fungal pathogens are known to invade seeds through the silk channel [5].
Author Contributions
Conceptualization, M.E.H.T.; methodology, M.E.H.T.; formal analysis, M.E.H.T.; investigation, M.E.H.T.; data curation, M.E.H.T.; writing—original draft preparation, M.E.H.T.; writing—review and editing, M.E.H.T. and M.N.R.; visualization, M.E.H.T.; supervision, M.N.R.; project administration, M.N.R.; funding acquisition, M.N.R.; M.E.H.T. conceived of the study, performed all experiments, analyzed all data, and wrote the manuscript; M.N.R. supervised the study and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
Funding was provided by grants to M.N.R. from Grain Farmers of Ontario (054810), the Ontario Ministry of Agriculture, Food and Rural Affairs (OMAFRA 030356 and 030564), and the Natural Sciences and Engineering Research Council of Canada (NSERC 401424, 401663, and 400924).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The whole-genome sequence corresponding to the strain in this study was previously deposited in the NCBI GenBank under biosample number SAMN37538673, accession number JAWILD000000000. Previous whole-genome mining and in vitro assay results for the strain can be found in the previous publication [4].
Acknowledgments
Scholarships were provided to M.E.H.T. by NSERC CGS-D, NSERC CGS-M, and a Food from Thought Research Assistantship. We thank Kamal Khadka for growing maize at the Elora Research Station, Dylan Brettingham for growing maize in the greenhouse and helping to prepare lab supplies, and Julianna Tindall and Sue Couling for their help in the greenhouse. We thank Elaine Corbett (SPARK, Ontario Genomics) for her enthusiasm and support of this project. This research was made possible because of a previously reported silk culture collection and NGS study, and we thank the prior collaborators involved in that research: Anuja Shrestha, Jeffrey Rinne, and Eman Khalaf (all University of Guelph), as well as Victor Limay-Rios, Todd Phibbs, Stacie Dobson, Katiani Eli, and Darrell Galbraith (all Ridgetown Campus, University of Guelph), and finally, Lana Reid (Agriculture and Agri-Food Canada, Ottawa).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sauter, M. A Guided Tour: Pollen Tube Orientation in Flowering Plants. Chin. Sci. Bull. 2009, 54, 2376–2382. [Google Scholar] [CrossRef]
- Kiesselbach, T. The Structure and Reproduction of Corn, 50th Anniv; Brown, D., Schaefer, S., Eds.; Cold Spring Harbour Laboratory Press: New York, NY, USA, 1999. [Google Scholar]
- Sankaranarayanan, S.; Higashiyama, T. Capacitation in Plant and Animal Fertilization. Trends Plant Sci. 2018, 23, 129–139. [Google Scholar] [CrossRef]
- Thompson, M.E.H.; Raizada, M.N. The Microbiome of Fertilization-Stage Maize Silks (Style) Encodes Genes and Expresses Traits That Potentially Promote Survival in Pollen/Style Niches and Host Reproduction. Microorganisms 2024, 12, 1473. [Google Scholar] [CrossRef]
- Thompson, M.E.H.; Raizada, M.N. Fungal Pathogens of Maize Gaining Free Passage along the Silk Road. Pathogens 2018, 7, 81. [Google Scholar] [CrossRef] [PubMed]
- Khalaf, E.M.; Shrestha, A.; Rinne, J.; Lynch, M.D.J.; Shearer, C.R.; Limay-Rios, V.; Reid, L.M.; Raizada, M.N. Transmitting Silks of Maize Have a Complex and Dynamic Microbiome. Sci. Rep. 2021, 11, 1–17. [Google Scholar] [CrossRef]
- Thompson, M.E.H.; Shrestha, A.; Rinne, J.; Limay-Rios, V.; Reid, L.; Raizada, M.N. The Cultured Microbiome of Pollinated Maize Silks Shifts after Infection with Fusarium graminearum and Varies by Distance from the Site of Pathogen Inoculation. Pathogens 2023, 12, 1322. [Google Scholar] [CrossRef]
- Thompson, M.E.H. Discovery and Testing of Pollinated Maize Silk-Associated Microbes Including Microbiome Assisted Selection of Biocontrol Agents against Fusarium graminearum. Ph.D. Thesis, University of Guelph, Guelph, ON, Canada, 2023. [Google Scholar]
- Mitter, B.; Pfaffenbichler, N.; Flavell, R.; Compant, S.; Antonielli, L.; Petric, A.; Berninger, T.; Naveed, M.; Sheibani-Tezerji, R.; von Maltzahn, G.; et al. A New Approach to Modify Plant Microbiomes and Traits by Introducing Beneficial Bacteria at Flowering into Progeny Seeds. Front. Microbiol. 2017, 8, 11. [Google Scholar] [CrossRef]
- Turner, T.R.; James, E.K.; Poole, P.S. The Plant Microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef]
- Liu, Y.; Zuo, S.; Xu, L.; Zou, Y.; Song, W. Study on Diversity of Endophytic Bacterial Communities in Seeds of Hybrid Maize and Their Parental Lines. Arch. Microbiol. 2012, 194, 1001–1012. [Google Scholar] [CrossRef]
- Kim, H.; Jeon, J.; Lee, K.K.; Lee, Y.H. Longitudinal Transmission of Bacterial and Fungal Communities from Seed to Seed in Rice. Commun. Biol. 2022, 5, 772. [Google Scholar] [CrossRef]
- Frank, A.C.; Guzmán, J.P.S.; Shay, J.E. Transmission of Bacterial Endophytes. Microorganisms 2017, 5, 70. [Google Scholar] [CrossRef] [PubMed]
- Bacon, C.W.; Yates, I.E.; Hinton, D.M.; Meredith, F. Biological Control of Fusarium moniliforme in Maize. Environ. Health Perspect. 2001, 109, 325–332. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Z.; Zhang, R.; Xu, T.; Zhao, J.; Liu, Y. Diversity of Endophytic Bacteria in Hybrid Maize Seeds and Bacillus mojavensis J2416-7 May Be Capable of Vertical Transmission. Arch. Microbiol. 2022, 204, 213. [Google Scholar] [CrossRef] [PubMed]
- Truyens, S.; Weyens, N.; Cuypers, A.; Vangronsveld, J. Bacterial Seed Endophytes: Genera, Vertical Transmission and Interaction with Plants. Environ. Microbiol. Rep. 2015, 7, 40–50. [Google Scholar] [CrossRef]
- Ambika Manirajan, B.; Ratering, S.; Rusch, V.; Schwiertz, A.; Geissler-Plaum, R.; Cardinale, M.; Schnell, S. Bacterial Microbiota Associated with Flower Pollen Is Influenced by Pollination Type, and Shows a High Degree of Diversity and Species-Specificity. Environ. Microbiol. 2016, 18, 5161–5174. [Google Scholar] [CrossRef] [PubMed]
- Khalaf, E.M.; Shrestha, A.; Reid, M.; McFadyen, B.J.; Raizada, M.N. Conservation and Diversity of the Pollen Microbiome of Pan-American Maize Using PacBio and MiSeq. Front. Microbiol. 2023, 14, 1276241. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, A.; Rios, V.L.; Brettingham, D.J.L.; Raizada, M. Maize Pollen Carry Bacteria That Suppress a Fungal Pathogen That Enters through the Male Gamete Fertilization Route. Front. Plant. Sci. 2024, 14, 1286199. [Google Scholar] [CrossRef]
- Miller, S.S.; Reid, L.M.; Harris, L.J. Colonization of Maize Silks by Fusarium graminearum, the Causative Organism of Gibberella Ear Rot. Can. J. Botany 2007, 85, 369–376. [Google Scholar] [CrossRef]
- Thompson, M.E.H.; Raizada, M.N. Protocols to Enable Fluorescence Microscopy of Microbial Interactions on Living Maize Silks (Style Tissue). J. Microbiol. Methods 2024, 225, 107027. [Google Scholar] [CrossRef] [PubMed]
- Westgate, M.E.; Boyer, J.S. Osmotic Adjustment and the Inhibition of Leaf, Root, Stem. Planta 1985, 164, 540–549. [Google Scholar] [CrossRef]
- Mishra, A.; Chauhan, P.S.; Chaudhry, V.; Tripathi, M.; Nautiyal, C.S. Rhizosphere Competent Pantoea agglomerans Enhances Maize (Zea mays) and Chickpea (Cicer arietinum L.) Growth, without Altering the Rhizosphere Functional Diversity. Antonie Van Leeuwenhoek 2011, 100, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Lee, J.H.; Kang, B.R.; Rong, X.; Gardener, B.B.M.; Ji, H.J.; Park, C.S.; Kim, Y.C. Draft Genome Sequence of Pantoea ananatis B1-9, a Nonpathogenic Plant Growth-Promoting Bacterium. J Bacteriol 2012, 194, 729. [Google Scholar] [CrossRef] [PubMed]
- Montañez, A.; Abreu, C.; Gill, P.R.; Hardarson, G.; Sicardi, M. Biological Nitrogen Fixation in Maize (Zea mays, L.) by 15N Isotope-Dilution and Identification of Associated Culturable Diazotrophs. Biol. Fertil. Soils. 2009, 45, 253–263. [Google Scholar] [CrossRef]
- Sammer, U.F.; Reiher, K.; Spiteller, D.; Wensing, A.; Völksch, B. Assessment of the Relevance of the Antibiotic 2-Amino-3-(Oxirane-2,3-Dicarboxamido)-Propanoyl-Valine from Pantoea agglomerans Biological Control Strains against Bacterial Plant Pathogens. Microbiologyopen 2012, 1, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Pusey, P.L.; Stockwell, V.O.; Reardon, C.L.; Smits, T.H.M.; Duffy, B. Antibiosis Activity of Pantoea agglomerans Biocontrol Strain E325 against Erwinia amylovora on Apple Flower Stigmas. Phytopathology 2011, 101, 1234–1241. [Google Scholar] [CrossRef] [PubMed]
- Cañamás, T.P.; Viñas, I.; Abadias, M.; Usall, J.; Torres, R.; Teixidó, N. Acid Tolerance Response Induced in the Biocontrol Agent Pantoea agglomerans CPA-2 and Effect on Its Survival Ability in Acidic Environments. Microbiol. Res. 2009, 164, 438–450. [Google Scholar] [CrossRef]
- Deroo, W.; De Troyer, L.; Dumoulin, F.; De Saeger, S.; De Boevre, M.; Vandenabeele, S.; De Gelder, L.; Audenaert, K. A Novel in Planta Enrichment Method Employing Fusarium graminearum-Infected Wheat Spikes to Select for Competitive Biocontrol Bacteria. Toxins 2022, 14, 222. [Google Scholar] [CrossRef]
- Sheibani-Tezerji, R.; Naveed, M.; Jehl, M.A.; Sessitsch, A.; Rattei, T.; Mitter, B. The Genomes of Closely Related Pantoea ananatis Maize Seed Endophytes Having Different Effects on the Host Plant Differ in Secretion System Genes and Mobile Genetic Elements. Front. Microbiol. 2015, 6, 440. [Google Scholar] [CrossRef]
- Brady, C.; Cleenwerck, I.; Venter, S.; Vancanneyt, M.; Swings, J.; Coutinho, T. Phylogeny and Identification of Pantoea Species Associated with Plants, Humans and the Natural Environment Based on Multilocus Sequence Analysis (MLSA). Syst. Appl. Microbiol. 2008, 31, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Snoeck, S.; Guayazán-Palacios, N.; Steinbrenner, A.D. Molecular Tug-of-War: Plant Immune Recognition of Herbivory. Plant Cell. 2022, 34, 1497–1513. [Google Scholar] [CrossRef]
- Toyota, M.; Betsuyaku, S. In Vivo Imaging Enables Understanding of Seamless Plant Defense Responses to Wounding and Pathogen Attack. Plant Cell Physiol. 2022, 63, 1391–1404. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Mittler, R. Reactive Oxygen Species-Dependent Wound Responses in Animals and Plants. Free. Radic. Biol. Med. 2012, 53, 2269–2276. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Samota, M.K.; Choudhary, M.; Choudhary, M.; Pandey, A.K.; Sharma, A.; Thakur, J. How Do Plants Defend Themselves against Pathogens-Biochemical Mechanisms and Genetic Interventions. Physiol. Mol. Biol. Plants 2022, 28, 485–504. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.N.; Tanveer, M.; Abbas, A.; Yildirim, M.; Shah, A.A.; Ahmad, M.I.; Wang, Z.; Sun, W.; Song, Y. Combating Dual Challenges in Maize under High Planting Density: Stem Lodging and Kernel Abortion. Front. Plant Sci. 2021, 12, 699085. [Google Scholar] [CrossRef] [PubMed]
- Nasir, A.; Le Bail, A.; Daiker, V.; Klima, J.; Richter, P.; Lebert, M. Identification of a Flagellar Protein Implicated in the Gravitaxis in the Flagellate Euglena gracilis. Sci. Rep. 2018, 8, 7605. [Google Scholar] [CrossRef] [PubMed]
- Weller-Stuart, T.; Toth, I.; De Maayer, P.; Coutinho, T. Swimming and Twitching Motility Are Essential for Attachment and Virulence of Pantoea ananatis in Onion Seedlings. Mol. Plant Pathol. 2017, 18, 734–745. [Google Scholar] [CrossRef]
- Griffis, A.H.N.; Groves, N.R.; Zhou, X.; Meier, I. Nuclei in Motion: Movement and Positioning of Plant Nuclei in Development, Signaling, Symbiosis, and Disease. Front. Plant Sci. 2014, 5, 129. [Google Scholar] [CrossRef] [PubMed]
- Debruin, J.L.; Hemphill, B.; Schussler, J.R. Silk Development and Kernel Set in Maize as Related to Nitrogen Stress. Crop. Sci. 2018, 58, 2581–2592. [Google Scholar] [CrossRef]
- Prusky, D.; Romanazzi, G. Induced Resistance in Fruit and Vegetables: A Host Physiological Response Limiting Postharvest Disease Development. Annu. Rev. Phytopathol. 2023, 61, 279–300. [Google Scholar] [CrossRef]
- Woo, S.L.; Pepe, O. Microbial Consortia: Promising Probiotics as Plant Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1801. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).