Abstract
The presence of nontuberculous mycobacteria in biofilms on the surface of medical devices may affect the opportunistic pathogens that are common inhabitants of such biofilms. This study assessed the effect of Mycolicibacterium iranicum cell-free supernatants on biofilm formation and antibiotic susceptibility of Escherichia coli and Staphylococcus epidermidis differing in the anti-hemoglobin activity level. The cell-free supernatants have been shown to stimulate biofilm formation and also help reduce susceptibility of opportunistic pathogens to a number of antibiotics.
1. Introduction
Nontuberculous mycobacteria (NTM) are a group of gram-positive microorganisms whose natural habitats are soil and water, as well as aerosols generated above the earth’s surface [1]. Among the NTM, there are saprophytic species, opportunistic and pathogenic species of animals and humans [2,3]. In recent years, the researchers’ interest in this group of microorganisms has been increasing, which is associated with the spread of diseases caused by NTB worldwide [4]. The recorded increase in nontuberculous mycobacterial infections is associated firstly with the improvement of molecular diagnostic methods and their wider use in clinical practice, and secondly with an increase in patients with compromised immune system and hematological or oncological diseases [5,6,7]. Most diseases caused by NTM are associated with the ingress of these bacteria into the human body from the environment [8,9,10], but cases of iatrogenic infection of patients due to the presence of NTM in biofilms on the surface of implants, catheters, probes, and other medical devices have also been described [11,12,13]. It is known that one of the main mechanisms of virulence of opportunistic pathogens (E. coli, S. epidermidis, and other bacterial species) and the spread of nosocomial infections is the biofilm formation on the surface of medical devices and implants [14]. The process of bacterial biofilm formation is a complex phenomenon mediated by quorum-sensing molecules. Implant-related infections are among the most dangerous complications after surgery [15]. Therefore, the NTM presence in biofilms and, in particular, their effect on biofilm formation and antibiotic susceptibility in opportunistic microflora is of great practical interest. As it was shown earlier, adhesion and intraerythrocytic penetration of E. coli and S. epidermidis increase in the NTB presence [16]. Therefore, the objective of this study was to evaluate the effect of cell-free NTB supernatants on the antibiotic resistance and biofilm-forming ability of opportunistic pathogens—E. coli and S. epidermidis.
2. Results
2.1. The Effect of Cell-Free M. iranicum Supernatants on Biofilm Formation of E. coli and S. epidermidis
In this study, used S. epidermidis and E. coli cells were different in antihemoglobin activity (AntiHbA). The anti-hemoglobin activity of the strains characterizes their ability to inactivate hemoglobin in human blood by capturing heme iron. Briefly, AntiHbA was studied by a method based on the residual hemoglobin determination in a substrate after its interaction with a bacterial culture. Strains S. epidermidis IKVS 7 and E. coli IKVS 1 were characterized by high AntiHbA values (15.7 and 8.9 g/L, respectively), while S. epidermidis IKVS 13 and E. coli IKVS 18 showed low levels of this activity (2.0 and 0.7 g/L, respectively). AntiHbA levels had previously been determined [16] and were hence used for further study. Under the influence of NTMB supernatants in the studied cells of S. epidermidis and E. coli, the ability to form biofilms and the sensitivity to antibiotics was studied.
Under the effect of the cell-free supernatant of M. iranicum IKVS S1, an enhanced biofilm formation was observed in the strains of E. coli IKVS 1 and E. coli IKVS 18 relative to the control values by 31.0 ± 2.9% and 20.4 ± 2.6%, respectively (Figure 1). Table S1 presents the average OD values of the studied samples used in calculations of biofilm formation. Moreover, it should be noted that an evident 1.5-fold enhancement in biofilm formation was observed in the E. coli IKVS 1 strain exposed to the M. iranicum IKVS S1 supernatant vs. the strain of E. coli IKVS 18 (at p ≤ 0.05). The M. iranicum IKVS S1 supernatant had a similar effect on the strains of S. epidermidis IKVS 7 and 13—the biofilm formation increased by 25.6 ± 2.6% and 28.7 ± 2.7%, respectively. However, no significant difference was observed between strains.
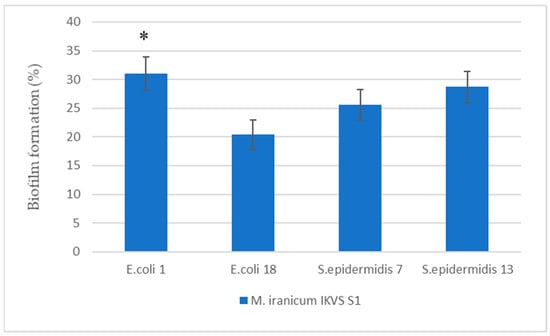
Figure 1.
Enhancement of biofilm formation by E. coli and S. epidermidis under the influence of cell-free supernatants of M. iranicum IKVS S1. *—significance of differences between strains according to Student’s t-test, p < 0.05.
The cell-free M. iranicum IKVS S2 supernatant affected the biofilm formation in opportunistic strains to a significantly lesser extent compared to M. iranicum IKVS S1 (Figure 2).
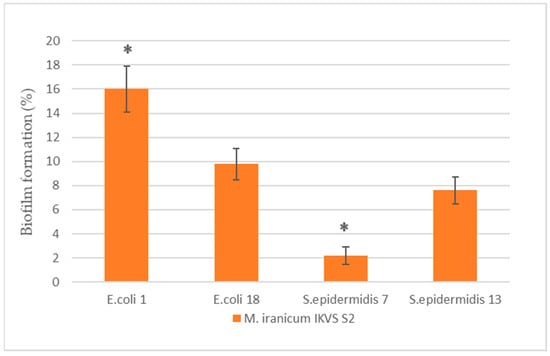
Figure 2.
Enhancement of biofilm formation by E. coli and S. epidermidis under the influence of cell-free supernatants of M. iranicum IKVS S2. *—significance of differences between strains according to Student’s t-test, p < 0.05.
An increase in biofilm formation was observed in the strains of E. coli IKVS 1 and E. coli IKVS 18 relative to the control values by 16 ± 1.9% and 9.8 ± 1.3%, respectively. Moreover, an evident 1.6-fold enhancement in biofilm formation was observed in the E. coli IKVS 1 strain exposed to the M. iranicum IKVS S2 supernatant vs. the strain of E. coli IKVS 18 (at p ≤ 0.05). The M. iranicum IKVS S2 supernatant had the least effect on the strains of S. epidermidis IKVS 7 and 13. An increase in biofilm formation relative to the control was 2.2 ± 0.73% and 7.6 ± 1.1%, respectively. If we compare the ability of the studied strains to form biofilms with each other, the S. epidermidis IKVS 13 cells showed a 3.4-fold increase in biofilm formation compared to S. epidermidis IKVS 7 (at p ≤ 0.05) exposed to the M. iranicum IKVS S2 supernatant.
In strains with high levels of AntiHbA (except for S. epidermidis IKVS 7), increased biofilm formation was observed under the influence of supernatants. We consider antihemoglobin activity as one of the pathogenicity factors. Strains characterized by a high level of this property contribute to the development of an infectious–inflammatory process, as shown in the works of [17]. It was previously shown that strains with high levels of AntiHbA are characterized by the presence of other pathogenicity factors (hemolytic, gelatinase, adhesive activity, etc.) [18]. This study revealed increased biofilm formation in strains with high levels of AntiHbA (with the exception of S. epidermidis IKVS 7), and the authors suggest that opportunistic microorganisms in the body can also lead to the development of an infectious process when influenced by NTM.
2.2. The Effect of the Cell-Free M. iranicum Supernatants on Antibiotic Susceptibility of the Strains of E. coli IKVS 1 and 18
Under the effect of the cell-free supernatants of M. iranicum IKVS S1 and M. iranicum IKVS S2, a decrease in antibiotic susceptibility was observed in the E. coli IKVS 1 strain (Table 1). According to the study results, a significant decrease (at p ≤ 0.05) in the antibiotic susceptibility of E. coli IKVS 1 was observed under the effect of the M. iranicum IKVS S1 supernatant relative to the control to vancomycin (11.3 ± 1.7 vs. 7 ± 1.4) and cefotaxime (26 ± 2.1 vs. 21 ± 1.4), respectively. On the other hand, a 2.4-fold increase in antibiotic susceptibility to clindamycin was observed relative to the control; the strain growth restriction was 20.3 ± 1.7 vs. 8.6 ± 1.6 (at p ≤ 0.05).

Table 1.
Effect of cell-free supernatants of NTM M. iranicum IKVS S1 and IKVS S2 on the antibiotic susceptibility of the strains E. coli IKVS 1.
The most evident significant decrease in the antibiotic susceptibility of the E. coli IKVS 1 strain exposed to M. iranicum IKVS S2 was detected to cefepime, gentamicin, and cefotaxime. A significant increase in antibiotic susceptibility was observed to clindamycin relative to the control—growth restriction was 13 ± 1.4 vs. 8.6 ± 1.6 (at p ≤ 0.05).
Under the effect of supernatants of M. iranicum IKVS S1 and IKVS S2, a significant decrease in antibiotic susceptibility to ceftriaxone, cefepime, gentamicin, cefotaxime, azithromycin, and ciprofloxacin was observed in the E. coli IKVS 18 strain relative to control values (Table 2).

Table 2.
Effect of cell-free supernatants of NTM M. iranicum IKVS S1 and IKVS S2 on the antibiotic susceptibility of the strains E. coli IKVS 18.
Thus, the supernatants of M. iranicum IKVS S1 and IKVS S2 had an evident effect on the antibiotic susceptibility of both E. coli strains. It should be noted that, under the effect of the M. iranicum IKVS S1 supernatant, the strain E. coli with the higher anti-HbA values showed a lesser decrease in the susceptibility to antibiotics of the group of β-lactams, aminoglycosides, fluoroquinolones, and azalides compared to the strain E. coli characterized by the lower anti-HbA values. A similar trend was observed under the effect of the cell-free supernatant of M. iranicum IKVS S2—E. coli IKVS 1 with the higher anti-HbA values did not change the susceptibility to ciprofloxacin (fluoroquinolone) and ceftriaxone (cephalosporin) in contrast to the E. coli IKVS 18 strain with the lower anti-HbA values. It is of particular interest to note that with the E. coli 1 strain, an increase in clindamycin susceptibility was observed under the effect of the cell-free M. iranicum supernatants. Moreover, an increase in susceptibility was higher under the effect of M. iranicum IKVS S1 compared to M. iranicum IKVS S2.
2.3. The Effect of the Cell-Free M. iranicum Supernatants on Antibiotic Susceptibility of the Strains of S. epidermidis IKVS 7 and 13
The S. epidermidis IKVS 7 strain showed a significant decrease in susceptibility to ceftriaxone, gentamicin, vancomycin, clindamycin, cefotaxime, meropenem, rifampicin, azithromycin, and clarithromycin when exposed to the cell-free supernatants of M. iranicum IKVS S1 and IKVS S2 relative to the control values (at p ≤ 0.05) (Table 3).

Table 3.
Effect of cell-free supernatants of NTM M. iranicum IKVS S1 and IKVS S2 on the antibiotic susceptibility of the strains S. epidermidis IKVS 7.
S. epidermidis IKVS 13 showed a significant decrease in susceptibility to ceftriaxone, gentamicin, clindamycin, cefotaxime, meropenem, rifampicin, azithromycin, ciprofloxacin, and clarithromycin when exposed to the cell-free M. iranicum IKVS S1 supernatant (Table 4), and to clindamycin, cefotaxime, meropenem, azithromycin, and clarithromycin when exposed to the M. iranicum IKVS S2 supernatant, relative to the control values (at p ≤ 0.05).

Table 4.
Effect of cell-free supernatants of NTM M. iranicum IKVS S1 and IKVS S2 on the antibiotic susceptibility of the strains S. epidermidis IKVS 13.
Thus, the most pronounced effect of reducing antibiotic sensitivity in S. epidermidis strains, regardless of the level of AntiHbA, was observed under the influence of the cell-free supernatant of M. iranicum IKVS S1: in S. epidermidis strains IKVS 7 and 13, there was a significant decrease in sensitivity to ceftriaxone, cefotaxime, miropinem, gentamicin, clindamycin, azithromycin, and clarithromycin. Differences in reduced antibiotic susceptibility between strains with different anti-HbA levels were insignificant. It was noted that benzylpenicillin susceptibility, under the effect of supernatants of M. iranicum IKVS S2 and IKVS S1 strains, did not change in both strains of S. epidermidis.
3. Discussion
Infections associated with medical devices are most often caused by biofilms formed by S. epidermidis and E. coli [19]. Such infections are more difficult to treat because microorganisms within biofilms actively exchange metabolites that enhance microbial survival, regulate biofilm formation and maturation, modulate the host immune response, and cause genetic changes [20,21,22]. The ability of bacterial metabolites to regulate biofilm formation has been described for a number of clinically or environmentally significant strains [23,24]. For example, Yersinia enterocolitica metabolites were shown to stimulate biofilm formation in E. coli strains with limited biofilm-forming ability [24]. The presence of M. mucogenicum in multispecies biofilms stimulated biofilm formation, but its cell-free supernatant reduced the ability to form biofilms in a number of bacteria, including Staphylococcus sp. [23]. In recent years, information has appeared in the literature about the presence of M. iranicum in biofilms on the surfaces of medical instruments [11]. M. iranicum in biofilms can interact with S. epidermidis and E. coli and affect the biofilm formation and/or the antibiotic susceptibility of opportunistic human microflora. Our study showed that E. coli and S. epidermidis exhibited a significant increase in biofilm formation when exposed to the cell-free M. iranicum supernatant. The most evident effect was observed under exposure to the M. iranicum IKVS S1 supernatant. Noteworthy is the fact that an increase in biofilm formation in E. coli IKVS 1 strain characterized by the higher anti-HbA levels was twice higher under the effect of the cell-free supernatants of both NTM strains compared to a similar strain with the lower values of anti-hemoglobin activity. With S. epidermidis strains differing in anti-HbA, significant differences in biofilm formation between strains were only observed under the effect of the M. iranicum IKVS S2 supernatant, while biofilm formation increased most significantly in the S. epidermidis IKVS 13 strain characterized by the lower anti-HbA values. The data obtained suggest the presence of a synergistic relationship between M. iranicum and E. coli and S. epidermidis; this type of relationship between microorganisms is species-specific. Therefore, the simulation of biofilm formation in some species requires intercellular contact, while for others, signaling molecules are sufficient for interspecies communication [25,26].
The ability to form biofilms is one of the leading virulence factors in bacteria and a factor contributing to the infection transition to a chronic form [14,27]. The formation of bacterial biofilms mostly leads to increased bacterial resistance to antibiotics [28,29,30]. Our study showed a significant decrease in susceptibility of E. coli and S. epidermidis to a number of antibiotics, when exposed to the cell-free M. iranicum supernatants. The most evident effect of reducing antibiotic susceptibility was observed in S. epidermidis strains, regardless of the anti-HbA level, and in the case of E. coli strains, in a strain with the lower AntiHbA values. It is known that antibiotic resistance in biofilms and at the cellular level is ensured by different mechanisms. However, despite the differences, these processes are largely synergistic: traditional resistance mechanisms of planktonic cells can also act in biofilms, and biofilms enhance resistance at the cellular level [31,32,33]. The study results were also confirmed in the works of Zakirov I. I. et al. and Carcione D. et al. [33,34]. They show the mechanisms of antibiotic resistance of microorganisms and mark the important role of biofilms in the formation of resistance to antimicrobial drugs, as well as difficulties in the treatment of such diseases contributing to the infection-associated mortality.
The intensification of the process of biofilm formation of opportunistic bacteria and the increase in their resistance to a number of antibiotics under the exposure to M. iranicum discovered in this study could potentially affect the virulence of these strains, as well as their antibiotic resistance, which may be beneficial during the surface colonization and further contribute to a chronic course of the infectious process.
4. Materials and Methods
4.1. Biophysiochemical and Cultural–Morphological Properties of NTM Strains
Collection strains of nontuberculous mycobacteria IKVS S1 and IKVS S2 previously identified as Mycobacterium iranicum (IKVS S1) and Mycobacterium rutilum (IKVS S2) of the Institute of Cellular and Intracellular Symbiosis, Ural Branch of the Russian Academy of Sciences (ICIS, UrB RAS) were used in the work. Both isolates were isolated from the salty Lantsug River, which flows into the hyperhaline Lake Elton (Volgograd region, Russian Federation). We clarified the species identity of archival strains based on the 16S rRNA gene sequence. Genomic DNA from bacterial cultures was isolated by enzymatic lysis [35]. The fragments of the nucleotide sequences of the DNA samples were submitted to SINTOL (Russia) for further analysis. The obtained sequences of the 16S rRNA gene fragments of the studied strains were 407 nucleotides in length. They were compared with homologous sequences of the 16S rRNA gene in the BLAST U.S. database. National Library of Medicine NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 30 May 2023). Based on the alignment results in the BLAST program, the maximum similarity of the sequences of the studied isolates with the type strains—Mycobacterium iranicum strain M05 HQ009482.1 and Mycobacterium iranicum strain H39 KX390634.1, was established and their 100% coincidence confirmed (Table 5). As a result of recent revision of the genus Mycobacterium and based on phylogenetic and genomic analysis, Mycobacterium iranicum species were attributed to the new genus Mycolicibacterium under the new name of Mycolicibacterium iranicum [36], which allows the IKVS S1 and IKVS S2 isolates to be attributed to Mycolicibacterium iranicum.

Table 5.
The results of sequence alignment of the studied IKVS S1 and IKVS S2 strains using the 16S rRNA gene fragment in the BLAST program.
The sequences of the studied strains of Mycolicibacterium iranicum IKVS S1 (accession number OR058793) and IKVS S2 (accession number OR058794) are found in the BLAST U.S. database. National Library of Medicine NCBI (https://www.ncbi.nlm.nih.gov/nuccore/?term=OR058793:OR058794[accn] (accessed on 30 May 2023)).
Mycolicibacterium iranicum IKVS S1 strain is an aerobe, immobile, and does not form spores. The cells are rod-shaped, sometimes curved, and eventually shortened to coccoid ones. According to the Runyon classification, the strain belongs to group IV of fast-growing NTM—visible growth was observed on days 3–4. The strain is scotochromogenic, the pigment is bright orange, gram-positive, catalase-positive, urease-positive, has an oxidative metabolic pattern, and produces lipase. It grows on standard culture media—1.5% meat infusion agar, Czapek medium. The strain is a moderate halophile capable of growing on media containing up to 10% sodium chloride. Growth temperature: 23–40 °C.
When cultivating on Czapek medium at a temperature of 25 °C for 3 days, IKVS S1 strain forms the convex, flat-topped, isolated, paste-like colonies colored orange with an uneven edge on the medium surface. It does not form aerial mycelium.
Mycolicibacterium iranicum IKVS S2 strain is an aerobe, immobile, and does not form spores. The cells are rod-shaped, sometimes curved, and eventually shortened to coccoid ones. According to the Runyon classification, it belongs to group IV of fast-growing NTM—visible growth is observed on days 3–4. Scotochromogen, orange pigment. The strain is gram-positive, catalase-positive, urease-positive, has an oxidative metabolic pattern, and produces lipase. It grows on standard culture media—1.5% meat infusion agar, Czapek medium. The strain is a moderate halophile capable of growing on media containing up to 7% sodium chloride. Growth temperature: 23–40 °C; optimal temperature: 37 °C.
When cultivating on Czapek medium at a temperature of 25 °C for 3 days, it forms the convex, flat, isolated, mucoid colonies colored orange with an uneven edge on the medium surface. With further cultivation, mucous “crowding” growth is observed.
4.2. Characteristics of Opportunistic Pathogens
To study the effect of NTM supernatants on opportunistic microflora, archival strains of opportunistic bacteria such Staphylococcus epidermidis (IKVS 7, IKVS 13) and Escherichia coli (IKVS 1, IKVS 18), differing in the level of anti-hemoglobin activity (anti-HbA), were selected. The strains of S. epidermidis IKVS 7 and E. coli IKVS 1 were characterized by higher anti-HbA (15.7 and 8.9 g/L, respectively), while S. epidermidis IKVS 13 and E. coli IKVS 18 strains were characterized by a lower activity (2.0 and 0.7 g/L, respectively). Anti-hemoglobin activity of strains characterizes their ability to inactivate hemoglobin in human blood by capturing heme iron. Anti-HbA was studied using a method based on the determination of residual hemoglobin in the substrate after interaction with a bacterial culture. The method for determining this property is detailed in previous works [16,37]
4.3. Receiving the Cell-Free M. iranicum Supernatant
To obtain M. iranicum cell-free supernatant, IKVS S1 and IKVS S2 were cultivated in meat-peptone broth (MPB) for 48 h at 21 °C, and then obtained the supernatant by centrifugation at 9000 g/rev for 20 min and were passed through sterile filters (30 mm, PVDF membrane, 0.22 µm, Jet Biofil (Guangzhou, China)).
4.4. Determination of Bacteria Antibiotic Susceptibility
Antibiotic susceptibility testing was performed using the Kirby–Bauer disk diffusion method on Mueller–Hinton agar according to EUCAST 2021 guidelines [38]. For this purpose, 24 h agar cultures of S. epidermidis and E. coli were suspended in 2 mL of sterile saline to prepare a density of 0.5 according to the McFarland turbidity standard. To assess the effect of nontuberculous Mycobacterium supernatants on the antibiotic susceptibility of opportunistic pathogens, test and control samples were prepared. In test samples, sterile cell-free supernatant of M. iranicum IKVS S1 or IKVS S2 in a volume of 2 mL was added to 1 mL of a suspension of the studied opportunistic pathogens; in control samples, 2 mL of MPB was added to 1 mL of a suspension of the studied opportunistic pathogens. Test and control samples were incubated for 24 h at 37 °C. Then, 0.1 mL of bacterial suspension was applied to the surface of the Mueller–Hinton medium (HiMedia, Mumbai, India) in a Petri dish and distributed evenly with a spatula. The discs containing antibiotics were placed on different areas of the inoculated agar using forceps. The inoculated dishes were placed in a thermostat for 24 h at 37 °C, and then the results were assessed by measuring the inhibition zone (in millimeters) of the studied microorganisms. The following discs were used for this study: ceftriaxone (30 μg), cefepime (30 μg), gentamicin (10 μg), vancomycin (30 μg), clindamycin (2 μg), cefotaxime (30 μg), meropenem (10 μg), rifampicin (5 μg), benzylpenicillin (10 units), azithromycin (15 μg), ciprofloxacin (5 μg), clarithromycin (15 μg). All experiments were performed in triplicate.
4.5. Determination of the Biofilm Formation Intensity
The intensity of biofilm formation by opportunistic bacteria exposed to NTM supernatants was determined by the crystal violet binding degree in sterile 96-well plates. For this purpose, a suspension of the studied microorganisms—S. epidermidis (IKVS 7, IKVS 13) and E. coli (IKVS 1, IKVS 18)—at a concentration of 109 CFU/mL was prepared from 24 h agar cultures. The resulting suspension was added in a volume of 100 μL into each well in three replicates. Then, 100 μL of sterile cell-free supernatant of M. iranicum IKVS S1 or IKVS S2 was transferred to the test wells. As a control, 100 μL of a suspension of the studied microorganisms—S. epidermidis (IKVS 7, IKVS 13) and E. coli (IKVS 1, IKVS 18) —and 100 μL of MPB were also transferred to the control wells. The plate was incubated for 48 h at 37 °C. After 48 h, MPB was removed from the test and control wells and the wells were washed several times with sterile distilled water. The wells were stained with 0.1% crystal violet solution for 10 min, washed three times with distilled water, and air dried. Then, 95% ethanol was added to the dry wells and incubated for 15 min, 125 μL of alcohol was transferred into the wells of a clean plate, and the optical density was measured at 540 nm wavelength [39].
Biofilm formation calculation formula:
where
Biofilm formation enhancement rate (%) = ((OD540test/OD540control) − 1) × 100%,
OD540test = optical density in the test;
OD540control = optical density in the control.
All experiments were performed in triplicate, and the results are expressed as the mean ± SD. Student’s t-test was performed to determine the significance of the results obtained. Statistical analysis was performed using XLSTAT Statistical Analysis Software 2016 (2020.4.1.1018).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/bacteria2040013/s1, Table S1: Average optical densities of test and control samples.
Author Contributions
E.A.S. experimental work to study biofilm formation and antibiotic resistance of strains, statistical analysis, writing—original draft preparation. O.A.G. genetic analysis of 16S rRNA gene fragment sequences, literature review, preparation of the manuscript for publication. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zimina, V.N.; Degtyareva, S.Y.; Beloborodova, E.N.; Kulabukhova, E.I.; Rusakova, L.I.; Fesenko, O.V. Mycobacterioses: The current state of the problem. CMAC 2017, 19, 276–282. [Google Scholar]
- Delghandi, M.R.; El-Matbouli, M.; Menanteau-Ledouble, S. Mycobacteriosis and infections with non-tuberculous mycobacteria in aquatic organisms: A Review. Microorganisms 2020, 8, 1368. [Google Scholar] [CrossRef] [PubMed]
- Morgado, S.; Ramos, N.V.; Pereira, B.B.D.N.; Freitas, F.; da Fonseca, E.L.; Vicente, A.C. Multidrug-resistant Mycolicibacterium fortuitum infection in a companion cat (Felis silvestris catus) in Brazil. Access Microbiol. 2022, 4, 000317. [Google Scholar] [CrossRef] [PubMed]
- Lopeman, R.C.; Harrison, J.; Desai, M.; Cox, J.A.G. Mycobacterium abscessus: Environmental bacterium turned clinical nightmare. Microorganisms 2019, 7, 90. [Google Scholar] [CrossRef] [PubMed]
- Dolezalova, K.; Strachan, T.; Matej, R.; Ricna, D.; Bloomfield, M. Manifestations of cutaneous mycobacterial infections in patients with inborn errors of IL-12/IL-23-IFNγ immunity. Eur. J. Dermatol. 2022, 32, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Seidel, A.; Nunes, D.H.; Fernandes, C.; Funchal, G.D.G. Skin infection by Mycobacterium marinum—Diagnostic and therapeutic challenge. An. Bras. Dermatol. 2022, 97, 366–368. [Google Scholar] [CrossRef]
- Pampaloni, A.; Tosto, S.; Locatelli, M.E.; Gentile, A.; Scuderi, D.; Marino, A.; Cosentino, F.; Moscatt, V.; Nunnari, G.; Cacopardo, B. Skin and soft tissue infection by Mycobacterium intracellulare in an immunocompetent patient. IDCases 2020, 19, e00720. [Google Scholar] [CrossRef]
- Franco-Paredes, C.; Marcos, L.A.; Henao-Martínez, A.F.; Rodríguez-Morales, A.J.; Villamil-Gómez, W.E.; Gotuzzo, E.; Bonifaz, A. Cutaneous mycobacterial infections. Clin. Microbiol. Rev. 2018, 32, e00069-18. [Google Scholar] [CrossRef]
- Máiz Carro, L.; Barbero Herranz, E.; Nieto Royo, R. Respiratory infections due to nontuberculous mycobacterias. Med. Clin. 2018, 150, 191–197. [Google Scholar] [CrossRef]
- Acharya, S.; Anwar, S.; Medina, Y.; Thapa, S.; Glaser, A. Mycobacterium abscessus pneumonia in severe alcoholism. Cureus 2022, 14, e26251. [Google Scholar] [CrossRef]
- Ranson, E.L.; Tsevat, R.K.; von Bredow, B.; Kamau, E.; Yang, S.; Prabaker, K.K. Catheter-Related bloodstream infection caused by Mycolicibacterium iranicum, California, USA. Emerg. Infect Dis. 2023, 29, 217–219. [Google Scholar] [CrossRef] [PubMed]
- Senozan, E.A.; Adams, D.J.; Giamanco, N.M.; Warwick, A.B.; Eberly, M.D. A catheter-related bloodstream infection with Mycobacterium frederiksbergense in an immunocompromised child. Pediatr. Infect Dis. J. 2015, 34, 445–447. [Google Scholar] [CrossRef] [PubMed]
- Tagashira, Y.; Kozai, Y.; Yamasa, H.; Sakurada, M.; Kashiyama, T.; Honda, H. A cluster of central line-associated bloodstream infections due to rapidly growing nontuberculous mycobacteria in patients with hematologic disorders at a Japanese tertiary care center: An outbreak investigation and review of the literature. Infect. Control Hosp. Epidemiol. 2015, 36, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiang, J.; Gao, Y.; Sun, Y.; Dai, J.; Wu, Y.; Qu, D.; Ma, G.; Fang, X. Staphylococcus epidermidis small basic protein (Sbp) forms amyloid fibrils, consistent with its function as a scaffolding protein in biofilms. J. Biol. Chem. 2018, 293, 14296–14311. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, R.; Dhawan, B.; Garg, B.; Shankar, V.; Chandra Nag, T. A Comparison of bacterial adhesion and biofilm formation on commonly used orthopaedic metal implant materials: An in vitro study. Indian J. Orthop. 2019, 53, 148–153. [Google Scholar] [CrossRef]
- Gogoleva, O.A.; Shchuplova, E.A. The influence of non-tuberculous mycobacteria on the interaction of opportunistic microorganisms with erythrocytes. Folia Microbiol. 2020, 65, 417–421. [Google Scholar] [CrossRef]
- Pashkova, T.M.; Morozova, N.V.; Kuzmin, M.D.; Kartashova, O.L.; Popova, L.P. Characteristics of the pathogenic potential of Escherichia coli isolated from patients with calculous pyelonephritis. Urology 2021, 4, 19–24. [Google Scholar] [CrossRef]
- Shchuplova, E.A.; Herzen, N.V.; Fadeev, S.B.; Valyshev, A.V. Antihemoglobin activity of enterococci. Bull. OSU 2014, 13, 139–142. [Google Scholar]
- Mack, D.; Davies, A.P.; Harris, L.G.; Knobloch, J.K.; Rohde, H. Staphylococcus epidermidis biofilms: Functional molecules, relation to virulence, and vaccine potential. Top. Curr. Chem. 2009, 288, 157–182. [Google Scholar] [CrossRef]
- Sharma, G.; Sharma, S.; Sharma, P.; Chandola, D.; Dang, S.; Gupta, S.; Gabrani, R. Escherichia coli biofilm: Development and therapeutic strategies. J. Appl. Microbiol. 2016, 121, 309–319. [Google Scholar] [CrossRef]
- Dheilly, A.; Soum-Soutera, E.; Klein, G.L.; Bazire, A.; Compere, C.; Haras, D.; Dufour, A. Antibiofilm activity of the marine bacterium Pseudoalteromonas sp. strain 3J6. Appl. Environ. Microbiol. 2010, 76, 3452–3461. [Google Scholar] [CrossRef] [PubMed]
- Blango, M.G.; Mulvey, M.A. Bacterial landlines: Contact-dependent signaling in bacterial populations. Curr. Opin. Microbiol. 2009, 12, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Simões, L.C.; Simões, M.; Vieira, M.J. The effects of metabolite molecules produced by drinking water-isolated bacteria on their single and multispecies biofilms. Biofouling 2011, 27, 685–699. [Google Scholar] [CrossRef]
- Vacheva, A.; Ivanova, R.; Paunova-Krasteva, T.; Stoitsova, S. Released products of pathogenic bacteria stimulate biofilm formation by Escherichia coli K-12 strains. Antonie Van Leeuwenhoek 2012, 102, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Xavier, K.B.; Bassler, B.L. LuxS quorum sensing: More than just a numbers game. Curr. Opin. Microbiol. 2003, 6, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Castonguay, M.H.; Schaaf, S.; Koester, W.; Krooneman, J.; van der Meer, W.; Harmsen, H.; Landini, P. Biofilm formation by Escherichia coli is stimulated by synergistic interactions and co-adhesion mechanisms with adherence-proficient bacteria. Res. Microbiol. 2006, 157, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, C.J., Jr.; Mende, K.; Beckius, M.L.; Akers, K.S.; Romano, D.R.; Wenke, J.C.; Murray, C.K. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect Dis. 2013, 29, 13. [Google Scholar] [CrossRef]
- Cardiliya, A.P.; Chandrasekar, M.J.N.; Nanjan, M.J. Incidence of biofilms among the multidrug resistant E. coli, isolated from urinary tract infections in the Nilgiris district, South India. Braz. J. Microbiol. 2023, 54, 1809–1818. [Google Scholar] [CrossRef]
- Shaikhrazieva, N.D.; Bulycheva, I.A.; Lopushov, D.V.; Sabayeva, F.N. Etiological structure and antibiotic resistance of hospital strains of microorganisms in the department of anesthesiology and intensive care. Medical Almanac 2019, 1, 32–34. [Google Scholar] [CrossRef]
- Penesyan, A.; Gillings, M.; Paulsen, I.T. Antibiotic discovery: Combatting bacterial resistance in cells and in biofilm communities. Molecules 2015, 20, 5286–5298. [Google Scholar] [CrossRef]
- Lynch, S.V.; Dixon, L.; Benoit, M.R.; Brodie, E.L.; Keyhan, M.; Hu, P.; Ackerley, D.F.; Andersen, G.L.; Matin, A. Role of the rapA gene in controlling antibiotic resistance of Escherichia coli biofilms. Antimicrob. Agents Chemother. 2007, 51, 3650–3658. [Google Scholar] [CrossRef] [PubMed]
- Bernard, C.S.; Giraud, C.; Spagnolo, J.; de Bentzmann, S. Biofilms: The secret story of microbial communities. In Bacterial Pathogenesis: Molecular and Cellular Mechanisms; Locht, C., Simonet, M., Eds.; Caister Academic Press: Norfolk, UK, 2012; pp. 129–168. [Google Scholar]
- Zakirov, I.I.; Kadyrova, E.R.; Safina, A.I.; Kayumov, A.R. Antibiotic resistance of Staphylococcus aureus and Pseudomonas aeruginosa on the model of cystic fibrosis as a chronic disease of the bronchopulmonary system. Pediatrics 2018, 97, 176–186. [Google Scholar] [CrossRef]
- Carcione, D.; Leccese, G.; Conte, G.; Rossi, E.; Intra, J.; Bonomi, A.; Sabella, S.; Moreo, M.; Landini, P.; Brilli, M.; et al. Lack of direct correlation between biofilm formation and antimicrobial resistance in clinical Staphylococcus epidermidis isolates from an italian hospital. Microorganisms 2022, 10, 1163. [Google Scholar] [CrossRef] [PubMed]
- Belkova, N.L.; Andreeva, A.M. Introduction to the Molecular Ecology of Microorganisms; Printhouse: Yaroslavl, Russia, 2009; p. 91. [Google Scholar]
- Gupta, R.S.; Lo, B.; Son, J. Phylogenomics and comparative genomic studies robustly support division of the genus mycobacterium into an emended genus mycobacterium and four novel genera. Front. Microbiol. 2018, 13, 9–67. [Google Scholar] [CrossRef]
- Bukharin, O.V.; Usvyatsov, B.Y.; Khanina, E.A. Determination of the Antihemoglobin Activity of Microorganisms. Patent of Russia No. 2262705.2005, 4 August 2005. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing, EUCAST. Available online: http://www.eucast.org (accessed on 18 October 2023).
- Gladysheva, I.V.; Cherkasov, S.V. Antibiofilm activity of cell-free supernatants of vaginal isolates of Corynebacterium amycolatum against Pseudomonas aeruginosa and Klebsiella pneumoniae. Arch. Microbiol. 2023, 205, 158. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).