Role of Curing Agents in the Adaptive Response of the Bioprotective Latilactobacillus curvatus CRL 705 from a Physiologic and Proteomic Perspective
Abstract
:1. Introduction
2. Results
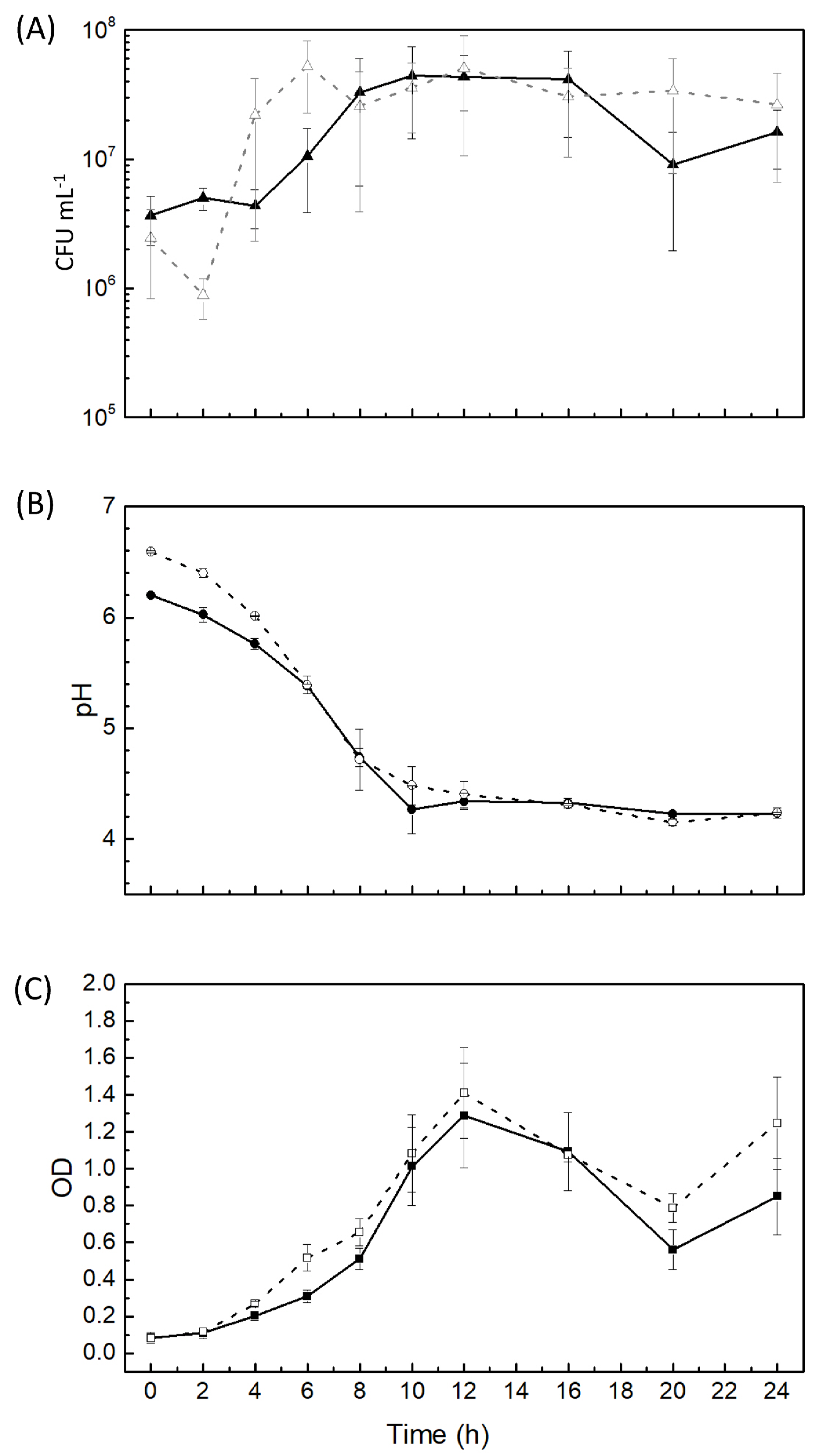
2.1. Growth of L. curvatus CRL 705 in a CDM with and without Curing Additives
2.2. Bacteriocin Activity
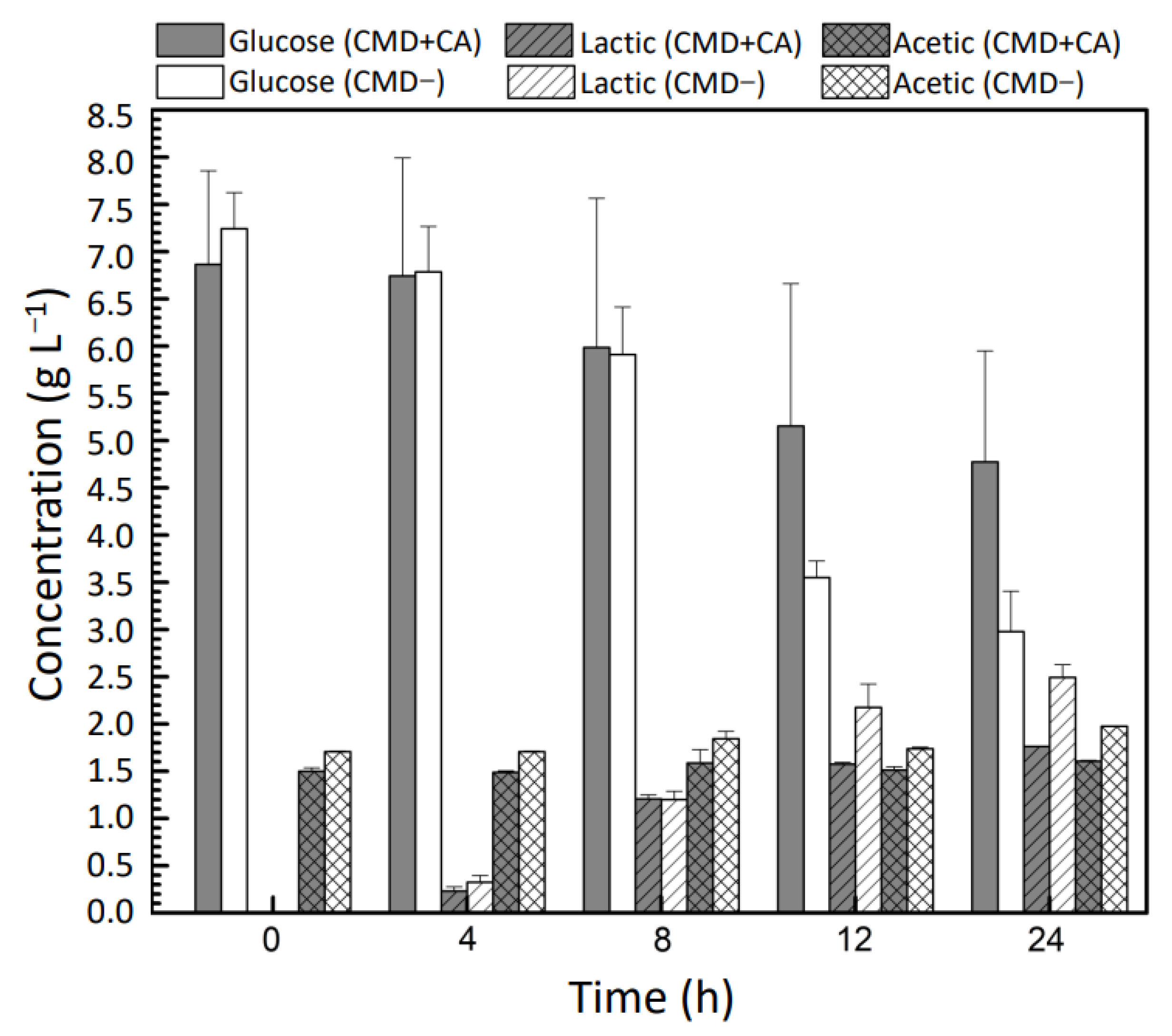
2.3. Consumption of Carbon Sources and Production of Acids
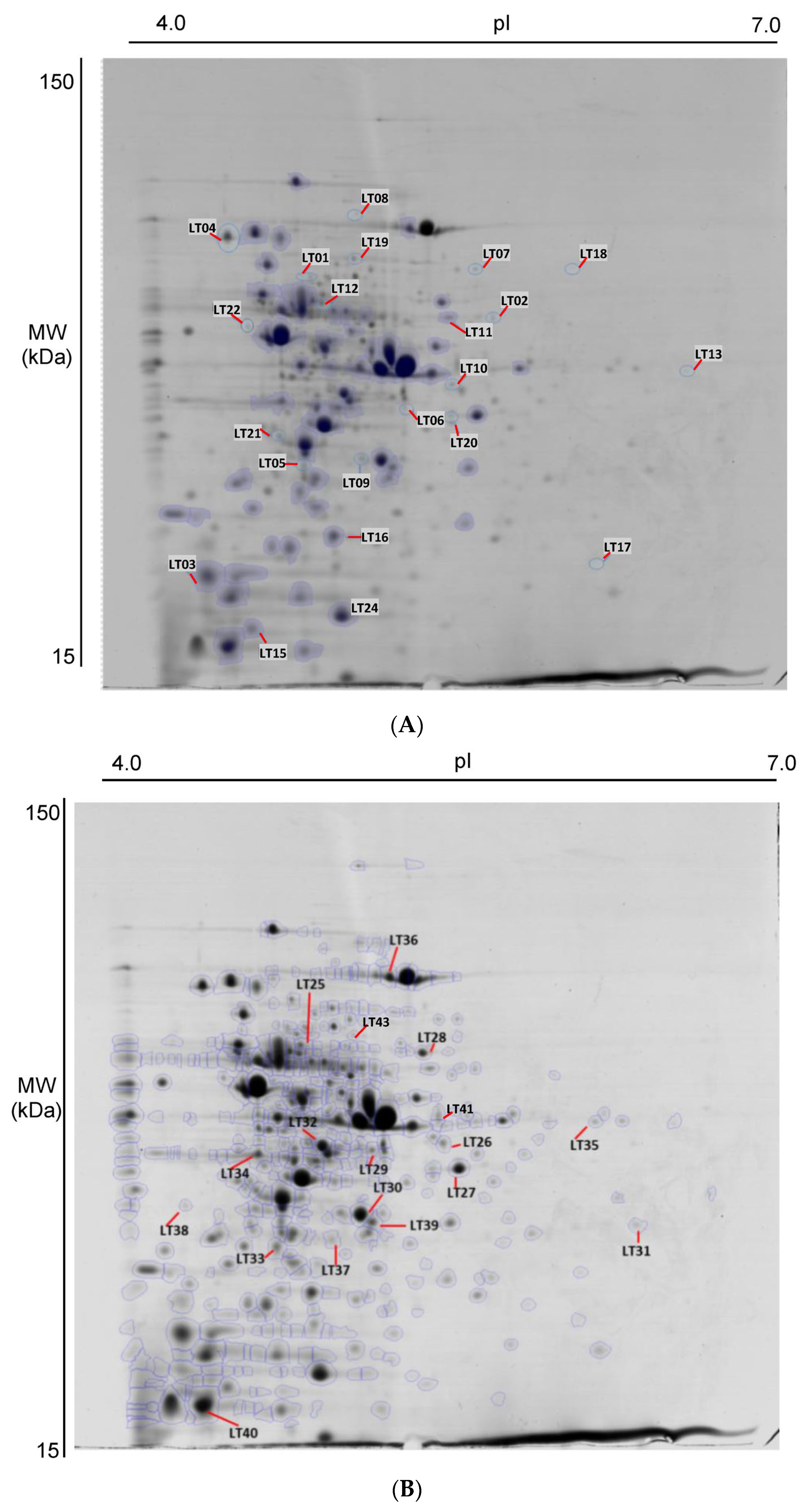
2.4. Differential Protein Expression by L. curvatus CRL 705 in the Presence of Curing Additives
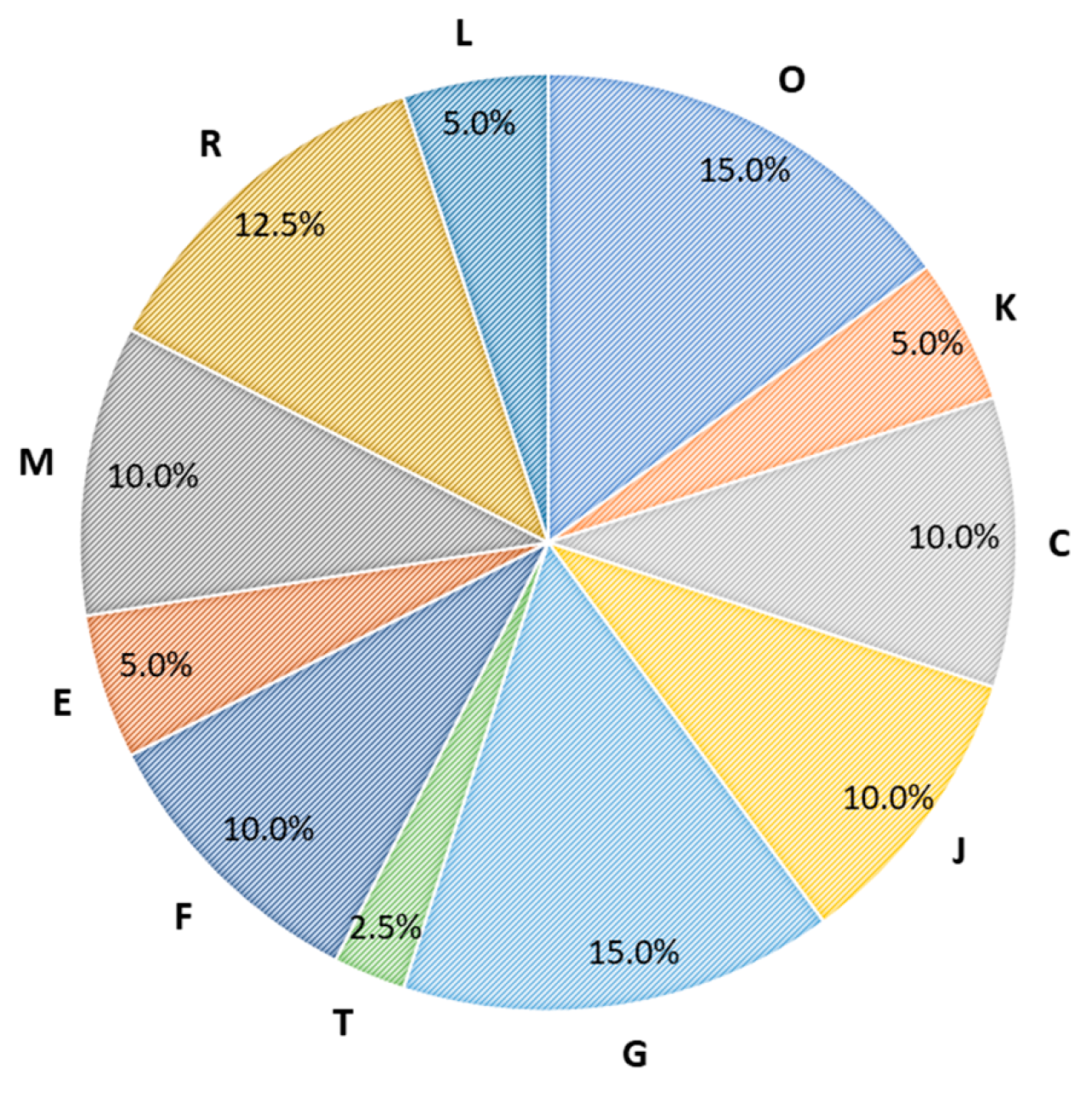
2.5. Functional Analysis and Protein Interaction
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Bacterial Growth
4.3. Determination of the Activity of Lac705 and AL705 Bacteriocins
4.4. Consumption of Carbon Sources and Production of Lactic Acid, Acetic Acid, and Ethanol
4.5. Differential Protein Expression Analysis
4.5.1. L. curvatus CRL705 Cells Recovery
4.5.2. Cell-Free Protein Extraction
4.5.3. Two-Dimensional Gel Electrophoresis (2DE)
4.5.4. Image Acquisition and Data Analysis
4.5.5. Mass Spectrometry Protein Identification
4.6. Functional Analysis and Interaction of the Differentially Expressed Proteins
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.; Wu, J.; Lv, M.; Shao, Z.; Hungwe, M.; Wang, J.; Geng, W. Metabolism characteristics of lactic acid bacteria and the expanding applications in food industry. Front. Bioeng. Biotechnol. 2021, 9, 612285. [Google Scholar] [CrossRef]
- Vignolo, G.; Castellano, P.; Fontana, C.; Fadda, S. Lactic acid bacteria in meat fermentations. Role of autochthonous starter cultures on quality, safety and health. In Lactic Acid Bacteria: Microbiological and Functional Aspects, 5th ed.; Vinderola, G., Ouwehand, A., Salminen, S., von Wright, A., Eds.; Taylor and Francis Group, L.L.C.: Boca Raton, FL, USA, 2019; Chapter 14; pp. 215–234. ISBN 9780815366485. [Google Scholar]
- Barcenilla, C.; Ducic, M.; López, M.; Prieto, M.; Álvarez-Ordóñez, A. Application of lactic acid bacteria for the biopreservation of meat products: A systematic review. Meat Sci. 2022, 183, 108661. [Google Scholar] [CrossRef]
- Eisenbach, L.; Janßen, D.; Ehrmann, M.A.; Vogel, R.F. Comparative genomics of Lactobacillus curvatus enables prediction of traits relating to adaptation and strategies of assertiveness in sausage fermentation. Int. J. Food Microbiol. 2018, 286, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Terán, L.C.; Coeuret, G.; Raya, R.; Zagorec, M.; Champomier-Vergès, M.-C.; Chaillou, S. Phylogenomic analysis of Lactobacillus curvatus reveals two lineages distinguished by genes for fermenting plant-derived carbohydrates. Genome Biol. Evol. 2018, 10, 1516–1525. [Google Scholar] [CrossRef]
- Vignolo, G.M.; Suriani, F.; Holgado, A.P.d.R.; Oliver, G. Antibacterial activity of Lactobacillus strains isolated from dry fermented sausages. J. Appl. Microbiol. 1993, 75, 344–349. [Google Scholar] [CrossRef]
- Castellano, P.; Gonzalez, C.; Carduza, F.; Vignolo, G. Protective action of Lactobacillus curvatus CRL705 on vacuum-packaged raw beef. Effect on sensory and structural characteristics. Meat Sci. 2010, 85, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Castellano, P.; Raya, R.; Vignolo, G. Mode of action of lactocin 705, a two-component bacteriocin from Lactobacillus casei CRL705. Int. J. Food Microbiol. 2003, 85, 35–43. [Google Scholar] [CrossRef]
- Castellano, P.; Vignolo, G. Inhibition of Listeria innocua and Brochothrix thermosphacta in vacuum-packaged meat by addition of bacteriocinogenic Lactobacillus curvatus CRL705 and its bacteriocins. Lett. Appl. Microbiol. 2006, 43, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Cuozzo, S.A.; Castellano, P.; Sesma, F.J.; Vignolo, G.M.; Raya, R.R. Differential roles of the two-component peptides of lactocin 705 in antimicrobial activity. Curr. Microbiol. 2003, 46, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Hebert, E.M.; Saavedra, L.; Taranto, M.P.; Mozzi, F.; Magni, C.; Nader, M.E.; Font de Valdez, G.; Sesma, F.; Vignolo, G.; Raya, R. Genome sequence of the bacteriocin-producing Lactobacillus curvatus strain CRL705. J. Bacteriol. 2012, 194, 538–539. [Google Scholar] [CrossRef]
- Terán, L.C.; Raya, R.; Zagorec, M.; Champomier-Vergès, M.-C. Genetics and genomics of Lactobacillus sakei and Lactobacillus curvatus. In Lactobacillus Genomics and Metabolic Engineering; Ruzal, S., Ed.; Caister Academic Press: Poole, UK, 2019; pp. 19–30. [Google Scholar]
- Eisenbach, L.; Geissler, A.J.; Ehrmann, M.A.; Vogel, R.F. Comparative genomics of Lactobacillus sakei supports the development of starter strain combinations. Microbiol. Res. 2019, 221, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Janßen, D.; Dworschak, L.; Ludwig, C.; Ehrmann, M.A.; Vogel, R.F. Interspecies assertiveness of Lactobacillus curvatus and Lactobacillus sakei in sausage fermentations. Int. J. Food Microbiol. 2020, 331, 108689. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Guo, L.; Yang, Q. Partial replacement of nitrite with a novel probiotic Lactobacillus plantarum on nitrate, color, biogenic amines and gel properties of Chinese fermented sausages. Food Res. Int. 2020, 137, 109351. [Google Scholar] [CrossRef] [PubMed]
- Orihuel, A.; Bonacina, J.; Vildoza, M.J.; Bru, E.; Vignolo, G.; Saavedra, L.; Fadda, S. Biocontrol of Listeria monocytogenes in a meat model using a combination of a bacteriocinogenic strain with curing additives. Food Res. Int. 2018, 107, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Leroy, F.; De Vuyst, L. Temperature and pH conditions that prevail during the fermentation of sausages are optimal for the production of the antilisterial bacteriocin sakacin K. Appl. Environ. Microbiol. 1999, 65, 974–981. [Google Scholar] [CrossRef] [PubMed]
- Fraqueza, M.J.; Laranjo, M.; Elias, M.; Patarata, L. Microbiological hazards associated with salt and nitrite reduction in cured meat products: Control strategies based on antimicrobial effect of natural ingredients and protective microbiota. Curr. Opin. Food Sci. 2021, 38, 32–39. [Google Scholar] [CrossRef]
- Belfiore, C.; Raya, R.R.; Vignolo, G.M. Identification, technological and safety characterization of Lactobacillus sakei and Lactobacillus curvatus isolated from Argentinean anchovies (Engraulis anchoita). SpringerPlus 2013, 2, 257. [Google Scholar] [CrossRef]
- Mbye, M.; Baig, M.A.; AbuQamar, S.F.; El-Tarabily, K.A.; Obaid, R.S.; Osaili, T.M.; Al-Nabulsi, A.A.; Turner, M.S.; Shah, N.P.; Ayyash, M.M. Updates on understanding of probiotic lactic acid bacteria responses to environmental stresses and highlights on proteomic analyses. Com. Rev. Food Sci. Food Saf. 2020, 19, 1110–1124. [Google Scholar] [CrossRef]
- Fadda, S.; Anglade, P.; Baraige, F.; Zagorec, M.; Talon, R.; Vignolo, G.; Champomier-Vergès, M.C. Adaptive response of Lactobacillus sakei 23K during growth in the presence of meat extracts: A proteomic approach. Int. J. Food Microbiol. 2010, 142, 36–43. [Google Scholar] [CrossRef]
- Belfiore, C.; Fadda, S.; Raya, R.; Vignolo, G. Molecular basis of the adaption of the anchovy isolate Lactobacillus sakei CRL1756 to salted environments through a proteomic approach. Food Res. Int. 2013, 54, 1334–1341. [Google Scholar] [CrossRef]
- Orihuel, A.; Terán, L.; Renaut, J.; Planchon, S.; Valacco, M.P.; Masias, E.; Minahk, C.; Vignolo, G.; Moreno, S.; De Almeida, A.; et al. Physiological and proteomic response of Escherichia coli O157: H7 to a bioprotective lactic acid bacterium in a meat environment. Food Res. Int. 2019, 125, 108622. [Google Scholar] [CrossRef] [PubMed]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; De Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.; Ross, P.; Stanton, C.; et al. Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef] [PubMed]
- Lauret, R.; Morel-Deville, F.; Berthier, F.; Champomier-Verges, M.; Postma, P.; Ehrlich, S.D.; Zagorec, M. Carbohydrate utilization in Lactobacillus sake. Appl. Environ. Microbiol. 1996, 62, 1922–1927. [Google Scholar] [CrossRef] [PubMed]
- Salvucci, E.; Saavedra, L.; Sesma, F. Short peptides derived from the NH2-terminus of subclass IIa bacteriocinenterocin CRL35 show antimicrobial activity. J. Antimicrob. Chemother. 2007, 59, 1102–1108. [Google Scholar] [CrossRef]
- Gerez, C.; Carbajo, M.; Rollán, G.; Torres Leal, G.; Font de Valdez, G. Inhibition of citrus fungal pathogens by using lactic acid bacteria. J. Food Sci. 2010, 75, M354–M359. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, M.E.; Fornaguera, M.J.; Raya, R.R.; Mozzi, F. Lactobacillus reuteri CRL 1101 highly produces mannitol from sugarcane molasses as carbon source. Appl. Microbiol. Biotechnol. 2012, 95, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Orihuel, A.; Terán, L.; Renaut, J.; Vignolo, G.M.; De Almeida, A.M.; Saavedra, M.L.; Fadda, S. Differential proteomic analysis of lactic acid bacteria—Escherichia coli O157: H7 interaction and its contribution to bioprotection strategies in meat. Front. Microbiol. 2018, 9, 1083. [Google Scholar] [CrossRef] [PubMed]
- Candiano, G.; Bruschi, M.; Musante, L.; Santucci, L.; Ghiggeri, G.M.; Carnemolla, B.; Orecchia, P.; Righetti, P.G. Blue silver: A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 2004, 25, 1327–1333. [Google Scholar] [CrossRef]
- Nally, J.E.; Grassmann, A.A.; Planchon, S.; Sergeant, K.; Renaut, J.; Seshu, J.; McBride, A.J.; Caimano, M.J. Pathogenic leptospires modulate protein expression and post-translational modifications in response to mammalian host signals. Front. Cell. Infect. Microbiol. 2017, 7, 362. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015, 43, D261–D269. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafu, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Taboada, B.; Estrada, K.; Ciria, R.; Merino, E. Operon-mapper: A web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics 2018, 34, 4118–4120. [Google Scholar] [CrossRef]

| Bacteriocin | ||||||||
|---|---|---|---|---|---|---|---|---|
| Lactocin 705 * | AL 705 ** | |||||||
| Growth Medium/Time (h) | 2 | 8 | 16 | 24 | 2 | 8 | 16 | 24 |
| MRS | - | + | +++ | ++ | + | ++ | +++ | +++ |
| CDM− | - | + | +++ | ++ | + | ++ | +++ | +++ |
| CDM + CA | - | ++ | +++ | ++ | + | +++ | +++ | +++ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terán, L.C.; Orihuel, A.; Bentencourt, E.; Raya, R.; Fadda, S. Role of Curing Agents in the Adaptive Response of the Bioprotective Latilactobacillus curvatus CRL 705 from a Physiologic and Proteomic Perspective. Bacteria 2023, 2, 142-154. https://doi.org/10.3390/bacteria2040011
Terán LC, Orihuel A, Bentencourt E, Raya R, Fadda S. Role of Curing Agents in the Adaptive Response of the Bioprotective Latilactobacillus curvatus CRL 705 from a Physiologic and Proteomic Perspective. Bacteria. 2023; 2(4):142-154. https://doi.org/10.3390/bacteria2040011
Chicago/Turabian StyleTerán, Lucrecia C., Alejandra Orihuel, Emilse Bentencourt, Raúl Raya, and Silvina Fadda. 2023. "Role of Curing Agents in the Adaptive Response of the Bioprotective Latilactobacillus curvatus CRL 705 from a Physiologic and Proteomic Perspective" Bacteria 2, no. 4: 142-154. https://doi.org/10.3390/bacteria2040011
APA StyleTerán, L. C., Orihuel, A., Bentencourt, E., Raya, R., & Fadda, S. (2023). Role of Curing Agents in the Adaptive Response of the Bioprotective Latilactobacillus curvatus CRL 705 from a Physiologic and Proteomic Perspective. Bacteria, 2(4), 142-154. https://doi.org/10.3390/bacteria2040011







