Potential of Pseudarthrobacter chlorophenolicus BF2P4-5 as a Biofertilizer for the Growth Promotion of Tomato Plants
Abstract
1. Introduction
2. Results
2.1. Morphological and Biochemical Identification of the Bacterium Strain BF2P4-5
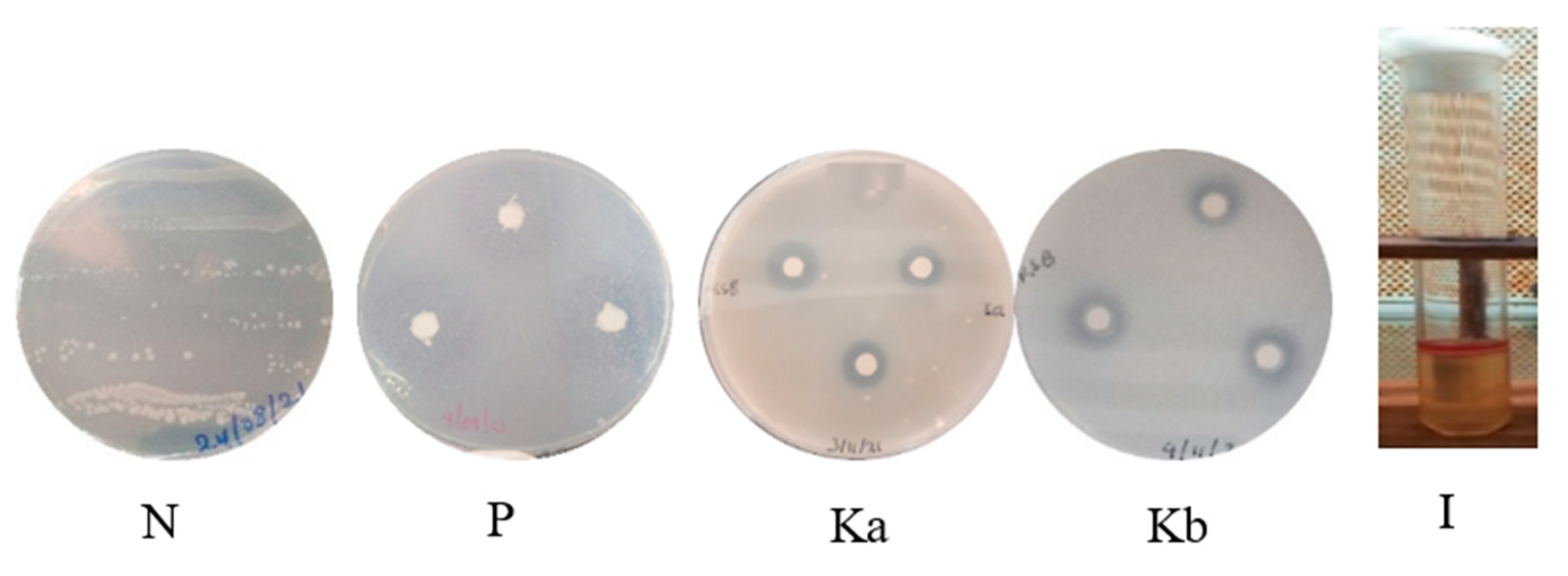
2.2. In Vitro Screening of Strain BF2P4-5 for Potential Plant Beneficial Traits
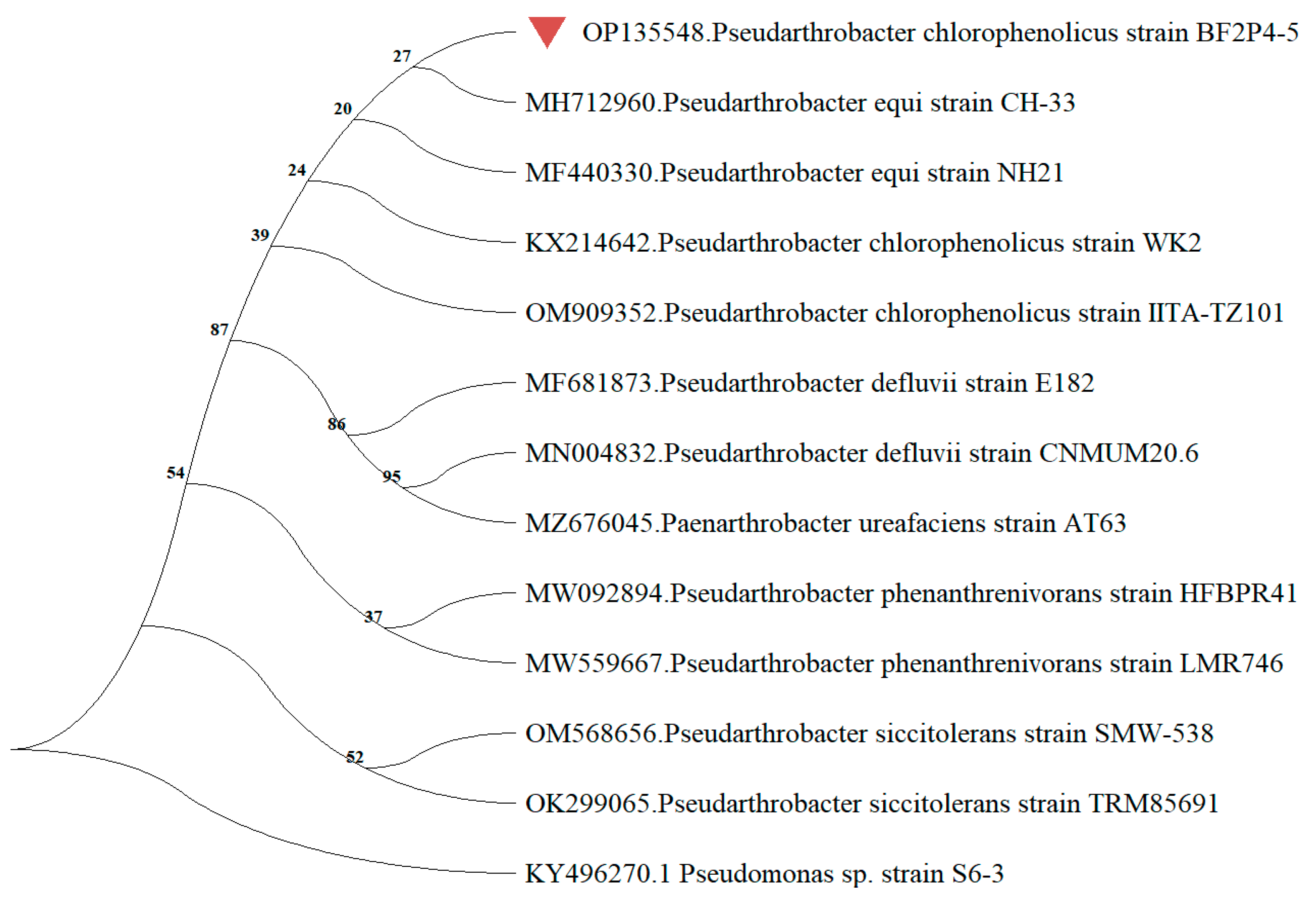
2.3. Sequencing of 16S rRNA and Phylogenetic Analysis of Strain BF2P4-5
2.4. Physiological Analysis of the Bacterial Strain BF2P4-5
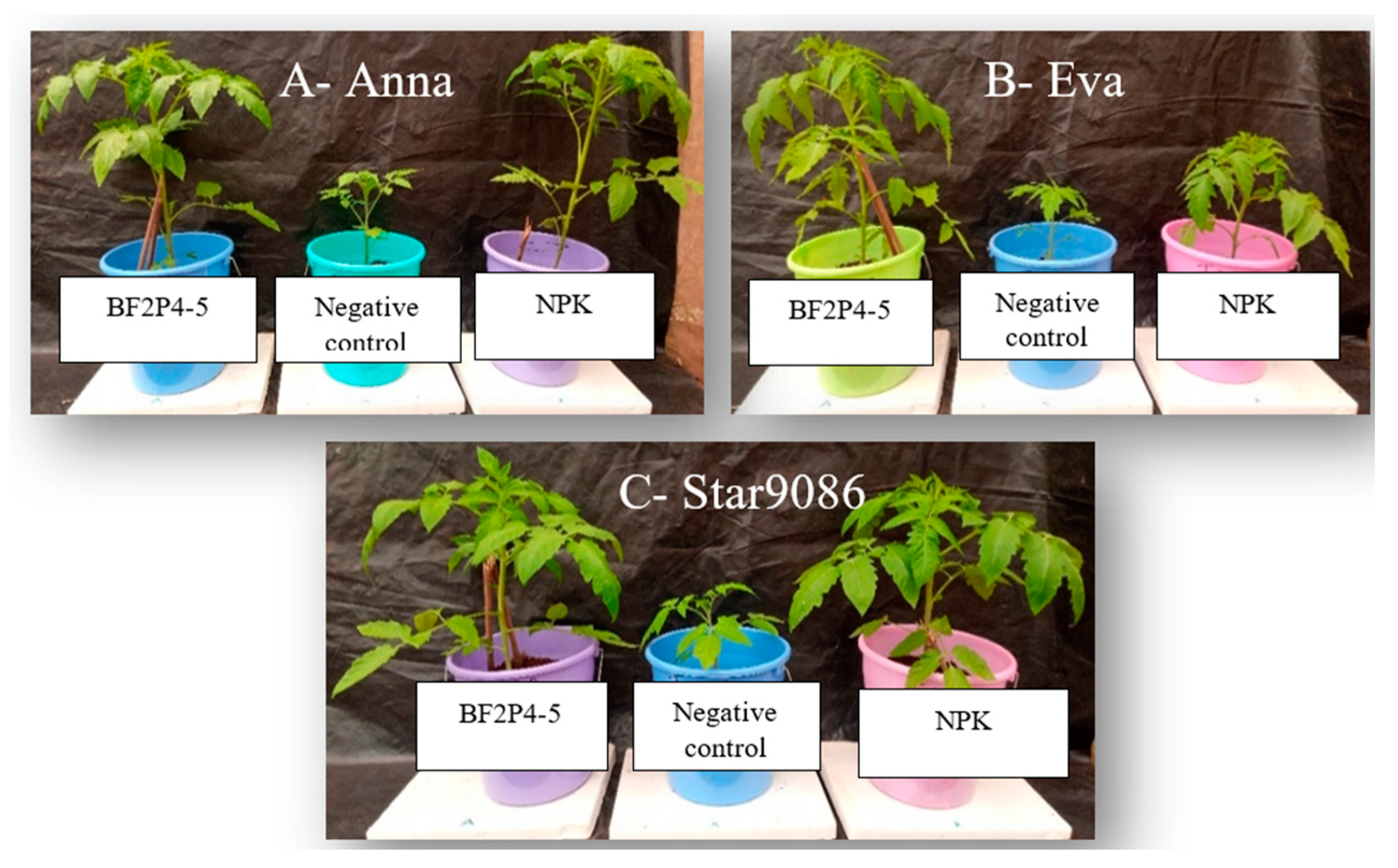
2.5. Validation of BF2P4-5 Strain Inoculation in Tomato Plants under Greenhouse Conditions
2.6. Analysis of Cocopeat Growing Medium
3. Materials and Methods
3.1. Isolation of Bacterial Strain BF2P4-5 from Rhizosphere Soil
3.2. Morphological and Biochemical Observation of Bacterial Strain BF2P4-5
3.3. In Vitro Screening of Strain BF2P4-5 for Potential Plant Beneficial Traits
3.4. DNA Extraction, PCR Amplification, Sequencing, and Phylogenetic Analysis of 16S rRNA
3.5. Physiological Analysis of the Bacterial Strain BF2P4-5
3.6. Analysis of the Cocopeat Growing Medium
3.7. In Plant Tests
3.7.1. Plant Material, Seed Sterilization, and Germination
3.7.2. Inoculation of Solanum lycopersicum with BF2P4-5 Strain
3.7.3. Determination of Plant Phenology and Growth Attributes
3.8. Statistical Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Devi, R.; Kaur, T.; Kour, D.; Yadav, A.N.; Suman, A. Potential Applications of Mineral Solubilizing Rhizospheric and Nitrogen Fixing Endophytic Bacteria as Microbial Consortium for the Growth Promotion of Chilli (Capsicum Annum L.). Biologia 2022, 1–11. [Google Scholar] [CrossRef]
- Amaresan, N.; Kumar, M.S.; Annapurna, K.; Kumar, K.; Sankaranarayanan, A. (Eds.) Beneficial Microbes in Agro-Ecology: Bacteria and Fungi; Academic Press: Amsterdam, The Netherlands, 2020; ISBN 978-0-12-823414-3. [Google Scholar]
- Egamberdieva, D.; Shrivastava, S.; Varma, A. (Eds.) Plant-Growth-Promoting Rhizobacteria (PGPR) and Medicinal Plants; Soil Biology; Springer International Publishing: Cham, Switzerland, 2015; Volume 42, ISBN 978-3-319-13400-0. [Google Scholar]
- Abdeljalil, N.O.-B.; Vallance, J. Characterization of Tomato-Associated Rhizobacteria Recovered from Various Tomato-Growing Sites in Tunisia. J. Plant Pathol. Microbiol. 2016, 7, 12p. [Google Scholar] [CrossRef]
- Hammami, I.; Hsouna, A.B.; Hamdi, N.; Gdoura, R.; Triki, M.A. Isolation and Characterization of Rhizosphere Bacteria for the Biocontrol of the Damping-off Disease of Tomatoes in Tunisia. Comptes Rendus Biol. 2013, 336, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-M.; Shahzad, R.; Bilal, S.; Khan, A.L.; Park, Y.-G.; Lee, K.-E.; Asaf, S.; Khan, M.A.; Lee, I.-J. Indole-3-Acetic-Acid and ACC Deaminase Producing Leclercia Adecarboxylata MO1 Improves Solanum lycopersicum L. Growth and Salinity Stress Tolerance by Endogenous Secondary Metabolites Regulation. BMC Microbiol. 2019, 19, 80. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, Y.; Ramassamy, V. Influence of Pseudomonas Fluorescens as Biofertilizer in Secondary Hardening of Tissue Cultured Banana Var. Poovan. Int. J. Appl. Sci. Biotechnol. 2015, 3, 38–41. [Google Scholar] [CrossRef]
- Roy, P.; Kumar, A. Arthrobacter. In Beneficial Microbes in Agro-Ecology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 3–11. ISBN 978-0-12-823414-3. [Google Scholar]
- Jiang, Y.; Song, Y.; Jiang, C.; Li, X.; Liu, T.; Wang, J.; Chen, C.; Gao, J. Identification and Characterization of Arthrobacter Nicotinovorans JI39, a Novel Plant Growth-Promoting Rhizobacteria Strain from Panax Ginseng. Front. Plant Sci. 2022, 13, 873621. [Google Scholar] [CrossRef] [PubMed]
- Bjerketorp, J.; Röling, W.F.M.; Feng, X.-M.; Garcia, A.H.; Heipieper, H.J.; Håkansson, S. Formulation and Stabilization of an Arthrobacter Strain with Good Storage Stability and 4-Chlorophenol-Degradation Activity for Bioremediation. Appl. Microbiol. Biotechnol. 2018, 102, 2031–2040. [Google Scholar] [CrossRef]
- Krishnan, R.; Menon, R.R.; Tanaka, N.; Busse, H.-J.; Krishnamurthi, S.; Rameshkumar, N. Arthrobacter pokkalii sp. Nov, a Novel Plant Associated Actinobacterium with Plant Beneficial Properties, Isolated from Saline Tolerant Pokkali Rice, Kerala, India. PLoS ONE 2016, 11, e0150322. [Google Scholar] [CrossRef]
- Scheublin, T.R.; Leveau, J.H.J. Isolation of Arthrobacter Species from the Phyllosphere and Demonstration of Their Epiphytic Fitness. Microbiologyopen 2013, 2, 205–213. [Google Scholar] [CrossRef]
- Dsouza, M.; Taylor, M.W.; Turner, S.J.; Aislabie, J. Genomic and Phenotypic Insights into the Ecology of Arthrobacter from Antarctic Soils. BMC Genom. 2015, 16, 36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ma, Y.; Yu, H. Arthrobacter Cupressi Sp. Nov., an Actinomycete Isolated from the Rhizosphere Soil of Cupressus sempervirens. Int. J. Syst. Evol. Microbiol. 2012, 62, 2731–2736. [Google Scholar] [CrossRef]
- Westerberg, K.; Elväng, A.M.; Stackebrandt, E.; Jansson, J.K. Arthrobacter chlorophenolicus sp. Nov., a New Species Capable of Degrading High Concentrations of 4-Chlorophenol. Int. J. Syst. Evol. Microbiol. 2000, 50, 2083–2092. [Google Scholar] [CrossRef]
- Li, Y.; Yu, L.; Li, H.; Xu, L.; Jiao, J.; Hu, F. Isolation, Identification and Characteristics of a Peanut Growth-Promoting Strain of Rhizobacteria. J. Ecol. Rural Environ. 2012, 28, 416–421. [Google Scholar]
- Kageyama, A.; Morisaki, K.; Omura, S.; Takahashi, Y. Arthrobacter oryzae sp. Nov. and Arthrobacter humicola sp. Nov. Int. J. Syst. Evol. Microbiol. 2008, 58, 53–56. [Google Scholar] [CrossRef]
- Kotoučková, L.; Schumann, P.; Durnová, E.; Spröer, C.; Sedláček, I.; Neča, J.; Zdráhal, Z.; Němec, M. Arthrobacter nitroguajacolicus sp. Nov., a Novel 4-Nitroguaiacol-Degrading Actinobacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 773–777. [Google Scholar] [CrossRef]
- Cheng, J.; Zhang, M.-Y.; Zhao, J.-C.; Xu, H.; Zhang, Y.; Zhang, T.-Y.; Wu, Y.-Y.; Zhang, Y.-X. Arthrobacter ginkgonis sp. Nov., an Actinomycete Isolated from Rhizosphere of Ginkgo biloba L. Int. J. Syst. Evol. Microbiol. 2017, 67, 319–324. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Fayaz, U.; Banday, S.; Shahnaz, E.; Khan, N.A.; Zargar, S.M.; Bhat, A.H. Morpho-Cultural, Physiological and Molecular Characterisation of Sphaceloma ampelinum Causing Anthracnose of Grapes in Temperate Region of India and Its Management. Indian Phytopathol. 2021, 74, 949–957. [Google Scholar] [CrossRef]
- Tripathi, N.; Sapra, A. Gram Staining. In StatPearls; StatPearls Publishing: Tampa, FL, USA, 2022. [Google Scholar]
- Abiola, C.; Oyetayo, V.O. Isolation and Biochemical Characterization of Microorganisms Associated with the Fermentation of Kersting’s Groundnut (Macrotyloma geocarpum). Res. J. Microbiol. 2016, 11, 47–55. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, S. Identification and Characterization of the Phosphate-Solubilizing Bacterium Pantoea sp. S32 in Reclamation Soil in Shanxi, China. Front. Microbiol. 2019, 10, 2171. [Google Scholar] [CrossRef]
- Bechtaoui, N.; Raklami, A.; Tahiri, A.-I.; Benidire, L.; El Alaoui, A.; Meddich, A.; Göttfert, M.; Oufdou, K. Characterization of Plant Growth Promoting Rhizobacteria and Their Benefits on Growth and Phosphate Nutrition of Faba Bean and Wheat. Biol. Open 2019, 8, bio043968. [Google Scholar] [CrossRef]
- Siddiqui, A.R.; Shahzad, S.M.; Ashraf, M.; Yasmeen, T.; Kausar, R.; Albasher, G.; Alkahtani, S.; Shakoor, A. Development and Characterization of Efficient K-Solubilizing Rhizobacteria and Mesorhizobial Inoculants for Chickpea. Sustainability 2021, 13, 10240. [Google Scholar] [CrossRef]
- Ghadamgahi, F.; Tarighi, S.; Taheri, P.; Saripella, G.V.; Anzalone, A.; Kalyandurg, P.B.; Catara, V.; Ortiz, R.; Vetukuri, R.R. Plant Growth-Promoting Activity of Pseudomonas Aeruginosa FG106 and Its Ability to Act as a Biocontrol Agent against Potato, Tomato and Taro Pathogens. Biology 2022, 11, 140. [Google Scholar] [CrossRef]
- Khalifa, A.Y.Z.; Alsyeeh, A.-M.; Almalki, M.A.; Saleh, F.A. Characterization of the Plant Growth Promoting Bacterium, Enterobacter Cloacae MSR1, Isolated from Roots of Non-Nodulating Medicago Sativa. Saudi J. Biol. Sci. 2016, 23, 79–86. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012; ISBN 978-1-936113-41-5. [Google Scholar]
- Treves, D.S. Review of Three DNA Analysis Applications for Use in the Microbiology or Genetics Classroom. J. Microbiol. Biol. Educ. 2010, 11, 186–187. [Google Scholar] [CrossRef][Green Version]
- Altschul, S. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, A.; Maurya, B.R.; Raghuwanshi, R. Isolation and Characterization of PGPR and Their Effect on Growth, Yield and Nutrient Content in Wheat (Triticum aestivum L.). Biocatal. Agric. Biotechnol. 2014, 3, 121–128. [Google Scholar] [CrossRef]
- Chhetri, G.; Kim, I.; Kang, M.; So, Y.; Kim, J.; Seo, T. An Isolated Arthrobacter Sp. Enhances Rice (Oryza sativa L.) Plant Growth. Microorganisms 2022, 10, 1187. [Google Scholar] [CrossRef] [PubMed]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [PubMed]
- Unell, M.; Abraham, P.E.; Shah, M.; Zhang, B.; Rückert, C.; VerBerkmoes, N.C.; Jansson, J.K. Impact of Phenolic Substrate and Growth Temperature on the Arthrobacter chlorophenolicus Proteome. J. Proteome Res. 2009, 8, 1953–1964. [Google Scholar] [CrossRef]
- Cochard, B.; Giroud, B.; Crovadore, J.; Chablais, R.; Arminjon, L.; Lefort, F. Endophytic PGPR from Tomato Roots: Isolation, In Vitro Characterization and In Vivo Evaluation of Treated Tomatoes (Solanum lycopersicum L.). Microorganisms 2022, 10, 765. [Google Scholar] [CrossRef]
- Sahu, S.G.; Rawat, A.K.; Dash, A.K.; Panda, N. Effect of Arthrobacter Isolates on Germination, Chlorophyll Content, Nodulation, Yield and Nutrient Uptake by Soybean (Glycine Max) in a Vertisol. Int. J. Curr. Microbiol. Appl. Sci. 2020, 9, 1858–1867. [Google Scholar] [CrossRef]
- Alzate Zuluaga, M.Y.; Martinez de Oliveira, A.L.; Valentinuzzi, F.; Tiziani, R.; Pii, Y.; Mimmo, T.; Cesco, S. Can Inoculation with the Bacterial Biostimulant Enterobacter Sp. Strain 15S Be an Approach for the Smarter P Fertilization of Maize and Cucumber Plants? Front. Plant Sci. 2021, 12, 719873. [Google Scholar] [CrossRef]
- Awang, Y.; Shaharom, A.S.; Mohamad, R.B.; Selamat, A. Chemical and Physical Characteristics of Cocopeat-Based Media Mixtures and Their Effects on the Growth and Development of Celosia Cristata. Am. J. Agric. Biol. Sci. 2009, 4, 63–71. [Google Scholar] [CrossRef]
- Abad, M.; Noguera, P.; Puchades, R.; Maquieira, A.; Noguera, V. Physico-Chemical and Chemical Properties of Some Coconut Coir Dusts for Use as a Peat Substitute for Containerised Ornamental Plants. Bioresour. Technol. 2002, 82, 241–245. [Google Scholar] [CrossRef]
- Espiritu, B.M. Composting and Microbial Inoculation of Coconut Coir Dust-Chicken Manure Mixture for Organic Fertilizer Use. Philipp. J. Crop Sci. P JCS 2011, 36, 47–56. [Google Scholar]
- Wang, J.; Li, R.; Zhang, H.; Wei, G.; Li, Z. Beneficial Bacteria Activate Nutrients and Promote Wheat Growth under Conditions of Reduced Fertilizer Application. BMC Microbiol. 2020, 20, 38. [Google Scholar] [CrossRef]
- Aliyat, F.Z.; Maldani, M.; El Guilli, M.; Nassiri, L.; Ibijbijen, J. Phosphate-Solubilizing Bacteria Isolated from Phosphate Solid Sludge and Their Ability to Solubilize Three Inorganic Phosphate Forms: Calcium, Iron, and Aluminum Phosphates. Microorganisms 2022, 10, 980. [Google Scholar] [CrossRef]
- Kalayu, G. Phosphate Solubilizing Microorganisms: Promising Approach as Biofertilizers. Int. J. Agron. 2019, 2019, 4917256. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial Phosphorus Solubilization and Its Potential for Use in Sustainable Agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef]
- Jimtha John, C.; Jishma, P.; Karthika, N.R.; Nidheesh, K.S.; Ray, J.G.; Mathew, J.; Radhakrishnan, E.K. Pseudomonas Fluorescens R68 Assisted Enhancement in Growth and Fertilizer Utilization of Amaranthus tricolor (L.). 3 Biotech 2017, 7, 256. [Google Scholar] [CrossRef]
| Characteristics | Results | Characteristics | Results |
|---|---|---|---|
| Colony and Cellular Morphology | Biochemical characteristics | ||
| Form | Circular | Oxidase | Positive |
| Elevation | Raised | Catalase | Positive |
| Margin | Entire | Citrate Utilization | Positive |
| Size | Punctiform | Starch Hydrolysis | Positive |
| Pigmentation | Cream | Hydrogen Sulphide (H2S) | Negative |
| Texture | Smooth | Motility | Positive |
| Surface | Glistening | Methyl red | Positive |
| Opacity | Translucent | Voges–Proskauer (VP.) | Positive |
| Gram reaction | Positive | Glucose | Positive |
| Cell shape | Staphylococci | Lactose | Negative |
| PGPR attributes | Sucrose | Negative | |
| Nitrogen-fixing | +++ | Urease | Negative |
| Phosphate solubilization | +++ | Nitrate reduction | Positive |
| Potassium solubilization | KCL (−), K2SO4 (+), Mica (+), K2PO4 (−) | Indole Acetic Acid (IAA) | Positive |
| Bacterial Strain | Accession Number | % Coverage | % Identity |
|---|---|---|---|
| Pseudarthrobacter equi strain CH-33 | MH712960.1 | 100 | 98.43 |
| Pseudarthrobacter equi strain NH21 | MF440330.1 | 100 | 98.43 |
| Pseudarthrobacter chlorophenolicus strain WK2 | KX214642.1 | 100 | 98.43 |
| Pseudarthrobacter chlorophenolicus strain IITA-TZ101 | OM909352.1 | 100 | 98.43 |
| Pseudarthrobacter defluvii strain CNMUM20.6 | MN004832.1 | 100 | 98.03 |
| Pseudarthrobacter defluvii strain E182 | MF681873.1 | 100 | 98.04 |
| Pseudarthrobacter phenanthrenivorans strain HFBPR41 | MW092894.1 | 100 | 98.17 |
| Pseudarthrobacter phenanthrenivorans strain LMR746 | MW559667.1 | 100 | 98.03 |
| Pseudarthrobacter siccitolerans strain SMW-538 | OM568656.1 | 100 | 98.03 |
| Pseudarthrobacter siccitolerans strain TRM85691 | OK299065.1 | 100 | 98.03 |
| Paenarthrobacter ureafaciens strain AT63 | MZ676045.1 | 100 | 98.03 |
| Characteristics | Range of Growth | Optimum Growth |
|---|---|---|
| pH for growth | 4 to 9 | 7 |
| Growth at different NaCl concentrations (%) | 0 to 20 | 15 |
| Growth in different temperature ranges (°C) | 20 to 45 | 35 |
| Treatments | Number of Leaves/Plants | Leave Length | Leave Diameter | Chlorophyll Content (SPAD Lower Canopy) | Chlorophyll Content (SPAD Mid Canopy) | Chlorophyll Content (SPAD Upper Canopy) | Plant Height at 55 DAS | Stem Length | Stem Girth (5 cm above) | Stem Girth (15 cm above) | LSD | F Value | CV% |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NPK + Anna F1 | 8.6 ± 0.5 abcf | 22.1 ± 1.3 bd | 14.8 ± 0.8 dee | 31.5 ± 1.1 abcdc | 36.9 ± 1.4 ab | 37.2 ± 1.9 ab | 44.3 ± 2.8 aa | 24.4 ± 1.9 ad | 0.5 ± 0.03 cfg | 0.4 ± 0.04 cg | 4.11 | 118.6 | 14.6 |
| NPK + Eva F1 | 8.0 ± 0.5 cd | 15.5 ± 1.2 cc | 12.8 ± 1.0 ec | 30.0 ± 0.7 bcdb | 34.3 ± 0.3 aba | 34.7 ± 0.9 aa | 32.4 ± 2.3 cab | 14.2 ± 1.1 cc | 0.5 ± 0.03 ce | 0.4 ± 0.05 ce | 2.95 | 173.7 | 12.60 |
| NPK + Star9086 | 8.2 ± 0.4 bce | 22.1 ± 1.3 bc | 17.8 ± 0.8 bcd | 33.3 ± 2.1 abb | 35.3 ± 1.3 abab | 37.4 ± 1.1 aa | 37.8 ± 1.5 bca | 19.6 ± 0.7 bcd | 0.6 ± 0.03 bf | 0.5 ± 0.02 bcf | 3.16 | 175.1 | 11.61 |
| BF2P4-5 + AnnaF1 | 8.6 ± 0.4 abcf | 23.3 ± 1.2 bd | 16.8 ± 1.1 cde | 32.1 ± 0.9 abcc | 36.9 ± 0.8 ab | 36.7 ± 1.1 ab | 44.6 ± 2.3 aa | 23.2 ± 1.2 ad | 0.7 ± 0.04 ag | 0.6 ± 0.05 abg | 3.13 | 210.6 | 10.94 |
| BF2P4-5 + Eva F1 | 9.4 ± 0.3 abf | 27.2 ± 0.8 acd | 19.2 ± 1.1 abe | 29.8 ± 0.9 cdc | 33.7 ± 0.7 bb | 34.4 ± 0.8 abb | 46.1 ± 2.3 aa | 24.6 ± 1.2 ad | 0.6 ± 0.03 abg | 0.5 ± 0.03 abg | 2.93 | 215.9 | 10.18 |
| BF2P4-5 + Star9086 | 9.8 ± 0.4 af | 27.8 ± 0.7 ac | 20.3 ± 0.7 ae | 33.8 ± 1.2 ab | 36.2 ± 0.9 abb | 34.3 ± 0.7 abb | 43.4 ± 1.8 aba | 24.1 ± 0.8 ad | 0.6 ± 0.03 abg | 0.6 ± 0.03 ag | 2.49 | 299.1 | 8.43 |
| NC + Anna F1 | 5.2 ± 0.5 dd | 8.58 ± 1.2 dc | 7.06 ± 0.5 fcd | 25.9 ± 1.8 dea | 28.6 ± 1.2 ca | 28.5 ± 1.8 ca | 19.0 ± 1.9 db | 9.20 ± 0.7 dc | 0.3 ± 0.03 de | 0.2 ± 0.03 de | 3.26 | 96.99 | 19.27 |
| NC + Eva F1 | 6.0 ± 0.3 de | 9.60 ± 0.7 dd | 7.70 ± 0.3 fde | 25.6 ± 0.6 eb | 29.6 ± 1.6 ca | 31.1 ± 0.9 bca | 20.9 ± 1.2 dc | 8.6 ± 0.4 dd | 0.3 ± 0.02 df | 0.2 ± 0.02 df | 2.22 | 230.9 | 12.41 |
| NC + Star9086 | 5.0 ± 0.6 dd | 10.6 ± 1.1 dc | 8.60 ± 0.3 fc | 28.1 ± 0.8 ea | 28.6 ± 1.2 ca | 29.1 ± 1.1 ca | 23.0 ± 2.4 db | 8.4 ± 0.6 dc | 0.3 ± 0.03 de | 0.2 ± 0.03 de | 3.04 | 122.5 | 16.77 |
| L.S.D (0.05) | 1.209 | 3.086 | 2.238 | 3.504 | 2.977 | 3.509 | 6.039 | 3.016 | 0.093 | 0.103 | |||
| F value | 18.04 | 49.67 | 42.64 | 5.94 | 11.49 | 7.60 | 27.69 | 47.48 | 22.26 | 19.7 | |||
| p-value | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | |||
| CV% | 12.333 | 12.979 | 12.559 | 9.099 | 6.964 | 8.113 | 13.596 | 13.541 | 14.114 | 20.36 |
| Treatment | pH | Electrical Conductivity (EC) (mS/cm) | % Available Nitrogen | Available Phosphorus (mg/kg) | Exchangeable Potassium (meq/100 g) | % Moisture | L.S.D (0.05) | F Value | p-Value | CV% |
|---|---|---|---|---|---|---|---|---|---|---|
| Pre-treatment | ||||||||||
| Fresh Cocopeat media | 7.19 | 0.40 | 0.12 | 3.00 | 0.77 | 27 | ||||
| Post-treatment | ||||||||||
| Cocopeat + NPK + Anna F1 | 6.16 ± 1.73 acd | 1.33 ± 0.08 bd | 1.21 ± 0.28 ad | 3007.29 ± 8.66 ca | 21.62 ± 1.73 ab | 14.84 ± 5.77 dcb | 13.456 | 78,587.0 | <0.0001 | 1.487 |
| Cocopeat + NPK + Eva F1 | 5.96 ± 1.66 ac | 1.83 ± 0.09 ac | 1.39 ± 1.33 ac | 3148.80 ± 6.65 ba | 21.98 ± 1.66 ab | 20.09 ± 5.78 db | 13.456 | 86,091.6 | <0.0001 | 1.418 |
| Cocopeat + NPK + Star9086 | 6.28 ± 1.73 ac | 1.89 ± 0.07 ac | 1.38 ± 0.82 ac | 3279.82 ± 7.66 aa | 23.13 ± 1.73 ab | 29.72 ± 4.99 dcb | 13.456 | 93,304.1 | <0.0001 | 1.357 |
| Cocopeat + BF2P4-5 + Anna F1 | 6.21 ± 1.73 acd | 0.73 ± 0.09 cd | 1.28 ± 0.41 abd | 1763.32 ± 8.68 da | 15.38 ± 1.73 bc | 51.64 ± 5.77 abb | 13.461 | 26,713.9 | <0.0001 | 2.469 |
| Cocopeat + BF2P4-5 + Eva F1 | 6.07 ± 1.75 adc | 0.53 ± 0.07 dcd | 1.29 ± 0.40 abdc | 1220.59 ± 7.68 fa | 14.31 ± 1.75 bc | 62.58 ± 5.87 ab | 13.461 | 12,682.1 | <0.0001 | 3.479 |
| Cocopeat + BF2P4-5 + Star9086 | 6.30 ± 1.72 abc | 0.79 ± 0.08 cc | 1.28 ± 0.35 abc | 1371.47 ± 8.68 ea | 16.16 ± 1.72 bb | 15.19 ± 5.67 db | 13.461 | 16,240.9 | <0.0001 | 3.217 |
| Negative control + Anna F1 | 6.5 ± 1.74 ac | 0.21 ± 0.09 ec | 0.02 ± 0.01 cc | 711.21 ± 6.98 ia | 0.22 ± 1.74 cc | 20.24 ± 4.77 db | 13.451 | 5490.97 | <0.0001 | 6.094 |
| Negative control + Eva F1 | 6.31 ± 1.67 ab | 0.31 ± 0.06 deb | 0.07 ± 0.00 bcb | 797.11 ± 8.66 ha | 0.35 ± 1.76 cb | 13.07 ± 5.78 db | 13.451 | 5490.97 | <0.0001 | 5.524 |
| Negative control + Star9086 | 6.06 ± 1.80 ac | 0.28 ± 0.07 dec | 0.02 ± 0.35 bcc | 944.11 ± 7.88 ga | 0.23 ± 1.88 cc | 40.00 ± 5.79 bcb | 13.451 | 7637.63 | <0.0001 | 4.557 |
| L.S.D (0.05) | 5.146 | 0.262 | 0.8545 | 25.73 | 5.146 | 17.154 | ||||
| F value | 0.01 | 53.50 | 3.91 | 14851.7 | 18.40 | 9.59 | ||||
| p-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||||
| CV% | 48.34 | 16.89 | 66.63 | 0.83 | 21.03 | 33.66 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Issifu, M.; Songoro, E.K.; Onguso, J.; Ateka, E.M.; Ngumi, V.W. Potential of Pseudarthrobacter chlorophenolicus BF2P4-5 as a Biofertilizer for the Growth Promotion of Tomato Plants. Bacteria 2022, 1, 191-206. https://doi.org/10.3390/bacteria1040015
Issifu M, Songoro EK, Onguso J, Ateka EM, Ngumi VW. Potential of Pseudarthrobacter chlorophenolicus BF2P4-5 as a Biofertilizer for the Growth Promotion of Tomato Plants. Bacteria. 2022; 1(4):191-206. https://doi.org/10.3390/bacteria1040015
Chicago/Turabian StyleIssifu, Muazu, Edinah K. Songoro, Justus Onguso, Elijah Miinda Ateka, and Victoria Wambui Ngumi. 2022. "Potential of Pseudarthrobacter chlorophenolicus BF2P4-5 as a Biofertilizer for the Growth Promotion of Tomato Plants" Bacteria 1, no. 4: 191-206. https://doi.org/10.3390/bacteria1040015
APA StyleIssifu, M., Songoro, E. K., Onguso, J., Ateka, E. M., & Ngumi, V. W. (2022). Potential of Pseudarthrobacter chlorophenolicus BF2P4-5 as a Biofertilizer for the Growth Promotion of Tomato Plants. Bacteria, 1(4), 191-206. https://doi.org/10.3390/bacteria1040015






