Abstract
We used machine-learning algorithms to evaluate demographic and clinical data in an administrative data set to identify relevant predictors of mortality due to Listeria monocytogenes infection. We used the Spanish Minimum Basic Data Set at Hospitalization (MBDS-H) to estimate the impacts of several predictors on mortality. The MBDS-H is a mandatory registry of clinical discharge reports. Data were coded with International Classification of Diseases, either Ninth or Tenth Revisions, codes. Diagnoses and clinical conditions were defined using recorded data from these codes or a combination of them. We used two different statistical approaches to produce two predictive models. The first was logistic regression, a classic statistical approach that uses data science to preprocess data and measure performance. The second was a random forest algorithm, a strategy based on machine learning and feature selection. We compared the performance of the two models using predictive accuracy and the area under the curve. Between 2001 and 2016, a total of 5603 hospitalized patients were identified as having any clinical form of listeriosis. Most patients were adults (94.9%). Among all hospitalized individuals, there were 2318 women (41.4%). We recorded 301 pregnant women and 287 newborns with listeriosis. The mortality rate was 0.13 patients per 100,000 population. The performance of the model produced by logistic regression after intense preprocessing was similar to that of the model produced by the random forest algorithm. Predictive accuracy was 0.83, and the area under the receiver operating characteristic curve was 0.74 in both models. Sepsis, age, and malignancy were the most relevant features related to mortality. Our combined use of data science, preprocessing, conventional statistics, and machine learning provides insights into mortality due to Listeria-related infection. These methods are not mutually exclusive. The combined use of several methods would allow researchers to better explain results and understand data related to Listeria monocytogenes infection.
1. Introduction
1.1. Listeriosis
Infection by Listeria monocytogenes is also known as listeriosis. This microorganism, a gram-positive rod, can cause a wide spectrum of clinical syndromes in humans, from neonatal to neuroinvasive disease. These clinical syndromes are uncommon in humans, because Listeria infection is more commonly seen in ruminants [1,2]. Among the genus Listeria, L. monocytogenes regularly affects humans, whereas L. ivanovii is more frequently seen in ruminants. Immunosuppression, the presence of solid or hematologic malignancy, extreme age (young or old), and pregnancy are the main risk factors for severe syndromes. Regarding pregnancy, L. monocytogenes often targets placental tissue, can produce chorioamnionitis and bacteremia in pregnant women, and can penetrate the placenta to infect the fetus [3]. Healthy individuals can also be affected, usually in the form of febrile gastroenteritis. Although the prognosis for pregnant women is usually good, listeriosis during pregnancy can cause spontaneous abortion, premature delivery, fetal mortality, and severe neonatal morbidity [4,5]. Meningitis, meningoencephalitis, and brain abscess are the most severe forms of listeriosis and are also risk factors for morbidity and mortality. A diagnosis of listeriosis is usually made upon positive isolation of the pathogen in blood cultures, in some types of tissue (e.g., placental tissue), or in cerebrospinal fluid (CSF) in the case of neuroinvasive infection. If available, polymerase chain reaction (PCR) should be performed in all patients clinically suspected of listeriosis of the central nervous system (CNS). A definitive diagnosis of neurolisteriosis can be made when CSF cultures or PCR are positive for Listeria [3,6,7].
1.2. Statistical Approaches to Predicting Outcomes from Electronic Health Records (EHR)
Early detection of severe listeriosis makes it possible to change the natural course of the disease, as it can help physicians individualize care for a given patient to better manage the condition or to avoid morbidity [8]. When a data set, such as an EHR, has many features, or relationships between these features and a given outcome are nonlinear, selecting a good predictive model is not easy. The most common solution is to rely on classic, well-known methods, such as logistic regression (LR). An alternative is to use other methods, such as machine learning, to produce well-fitting models and to select relevant and interpretable features.
Chi-square and LR are conventional modeling methods in analytical biomedical studies with binary (yes/no) outcomes. They are based on 2 × 2 or contingency tables. LR provides a simple, straightforward interpretation of the effect of independent variables on a proposed outcome by allowing researchers to calculate an odds ratio (OR) to evaluate the final predictive model. However, this approach can have limitations. For example, if one of the cells in the 2 × 2 table is zero, the standard errors will be too large, the model will be biased, mathematical convergence will not be achieved, and the whole model will be unreliable [9]. Also, in health care it is critical to improve predictive accuracy by finding statistical patterns. Although these patterns are often overlooked, or do not always correspond to an identifiable metabolic, physiological, or underlying biological pathway, they can still help physicians gain insight into the mechanisms of disease [10,11].
Machine learning has been suggested to address the limitations of conventional modeling [8,10,12] and can be an alternative approach used in medical investigations. Using machine learning in clinical research allows clinicians to recognize patterns in health-related data sets that could otherwise be overlooked. This enables them to better understand and explain data and to obtain better insights from them [10,12].
One of the benefits of machine learning is that it can produce interpretable, comprehensive, non-complex models whose simple, parsimonious structures allow clinicians to understand their intrinsic workings. Another important advantage of machine learning has to do with feature selection. Given a subset of features or predictors, any clinician can easily address a medical condition. When irrelevant or redundant features are included in a data set, this noise can affect the effectiveness of the predictive model [13]. In clinical research, certain features are often highly correlated with other features. If two features are perfectly correlated, only one of them should be included in the final model, because one alone is sufficient to describe the data. Including extra features does not provide further information but rather introduces noise into the final model. Dropping irrelevant, redundant features improves both the predictive model and computation time. Algorithms such as random forest (RF) can accurately deal with feature selection.
An exhaustive literature review is beyond the scope of this research, but it is worth noting some publications on the use of machine learning in several clinical scenarios. Parikh et al. [14] applied RF and gradient boosting algorithms to a data set based on structured EHR to accurately identify oncology patients at risk for mortality. In that study, machine-learning algorithms outperformed conventional modeling. Recent studies on morbidity and mortality in patients with heart failure [15,16] have shown the high predictive accuracy of algorithms such as RF or support vector machines. In another study, a RF model was the best of several algorithms compared for predicting mortality in patients in intensive care units [17]. Regarding infectious diseases, such as the recognition of temporal patterns in dengue [18] or early diagnosis of HIV [19,20], RF provided the best results in terms of predictive accuracy. These studies demonstrate the usefulness of modeling approaches for predicting early diagnosis and mortality in several clinical scenarios.
Better performance, better understanding, and better insights are the main benefits of applying machine learning to clinical research. Therefore, our aim in this study was not only to describe and analyze demographic and clinical features of L. monocytogenes infection but also to determine the features most associated with mortality due to Listeria-related infections to improve insight into these infections. Our study aims to predict mortality, giving clinicians the opportunity to change the natural course of listeriosis.
2. Materials and Methods
2.1. Data Collection
We conducted a retrospective nationwide cohort study to describe and analyze demographic and clinical features of patients with listeriosis and to describe the burden borne by hospitals due to L. monocytogenes infection between 2001 and 2016. We included data from the Spanish Minimum Basic Data Set at Hospitalization (MBDS-H), a mandatory administrative registry of hospital discharges covering more than 95% of hospitals in the National Spanish Health System and private hospitals. Therefore, nearly 97% of total hospital discharges are covered [21]. The MBDS-H is built from discharge reports. Patient data included, among other data, sex, age, date of admission and date of discharge, type of discharge, primary and secondary diagnoses at discharge, length of stay, and surgical or obstetric procedures. Other administrative data were recorded by default, such as the province where the hospitalization occurred, place of residence, and cost of hospitalization. Data were obtained from the statistical site of the Spanish National Health System [22]. Data were deidentified to ensure patient privacy: no names or personal information were recorded by default.
2.2. Feature Engineering and the Building of the Data Set
Feature engineering is a preprocessing procedure that allows researchers to use data to define features that enable both statistical and machine-learning algorithms to work properly. We were provided with two data sets. The first one was coded using the International Classification of Diseases, Ninth Revision (ICD-9), which covered 2001 to 2015. The second set used the International Classification of Diseases, Tenth Revision, Clinical Modification (ICD-10-CM) and covered 2016 [23]. Because diagnoses and procedures were coded differently, both data sets were preprocessed separately and then merged into one data set.
All diagnoses and medical conditions were coded yes/no (yes = 1, no = 0). In this procedure, called one hot encoding, categorical variables are converted into a form that can be provided to machine-learning algorithms to do a better job at prediction. Although LR and RF can work directly with categorical data depending on their implementation, some functions require input variables to be integers, or numeric in value. This means that any categorical data must be mapped to integers. In both of our data sets, one hot encoding was used to prepare the data for the proposed algorithms and improve prediction. Using one hot encoding, we converted each categorical value (e.g., diabetes) into a new categorical column and assigned a binary value of 1 or 0 (1 = yes, 0 = no) to this column.
We used death as the outcome or response variable, which was a binary variable (yes/no). We also used several predictors: age, sex, pregnancy, newborn, sepsis/septic shock, bacteremia, endocarditis, invasive infection of the central nervous system, peritonitis, febrile gastroenteritis, malignancy, chronic kidney disease (CKD), cirrhosis of the liver, and immunosuppression. These clinical conditions were extracted from main or secondary diagnoses established in medical discharge summaries, coded with either the ICD-9 or ICD-10-CM.
2.3. Definitions of Variables: ICD-9 Codes
Listeriosis was coded as 027.0 in either the main diagnosis or secondary diagnosis field. Age, sex, main and secondary diagnoses, date of admission, and date of discharge were also collected. Other demographic variables, such as insurance/billing data or hospital features, were excluded from our research. Data collected in the MBDS-H are listed in Table 1. Critical data that may be potentially used to identify patients are coded with a hash function, so a hash value was returned instead of the critical value (patient or hospital unique identifiers). Nevertheless, we dropped any columns other than the aforementioned medical conditions.

Table 1.
Data collected in the MBDS-H.
On the basis of 027.0 as the code for listeriosis, we identified patients with meningitis or meningoencephalitis (code 320.7), encephalomyelitis (code 323.0), intracranial abscess (code 324.0), bacterial endocarditis (code 421), febrile gastroenteritis (codes 008, 009, and 558), and sepsis and septic shock (codes 995 and 785) [24]. Other medical conditions analyzed as predictors were also collected: type 1 and type 2 diabetes mellitus (code 255), solid and hematologic malignancy (codes 140–239), immunosuppression (codes 042, 200–208, 279, 288, 289, 795, 140–199, 209, 235–239, 996, V42, 135, 277, 340, 341, 357, 422, 446, 495.9, 516, 555–558, 695, 710–714, 720, and 725), CKD (codes 585 and 403), chronic liver disease (codes 571 and 572), and chronic obstructive pulmonary disease (codes 490–496).
2.4. Definitions of Variables: ICD-10-CM Codes
Regarding the ICD-10-CM, diagnoses were defined by code A32 (listeriosis) in primary diagnosis and then sorted according to the following codes in primary and/or secondary diagnoses: A32.1 (listerial meningitis and meningoencephalitis), A32.11 (listerial meningitis), A32.12 (listerial meningoencephalitis), A32.7 (listerial sepsis), A32.8 (other forms of listeriosis), A32.81 (oculoglandular listeriosis), A32.82 (listerial endocarditis), A32.89 (other forms of listeriosis), and A32.9 (listeriosis, unspecified). We also included patients with code P37.2 (neonatal or congenital disseminated listeriosis), which applies only to newborns and should not appear on the maternal record.
In line with the ICD-10-CM, pregnancy in our data set was coded along a continuum from O03 to O99, although only codes O03, O30, O36, O41, O42, O63, O68, O75, O98, and O99 were present in the data. Newborns were coded as P07, P55, or Q03. Sepsis and septic shock were registered as R65, J96, or A41. Bacteremia was recorded as A32.82 or R78. Endocarditis was coded as I33 or A32.82. Invasive CNS infection was a composite predictor (meningitis, meningoencephalitis, and brain abscess) coded as A32.1, A32.11, A32.12, A87, G00, G01, G03, G04, G05, G06, or G91. Peritonitis was recorded as K35, K61, K65, K80, or K83. Febrile gastroenteritis was coded as K52 or R50. Malignancy was registered as C15, C16, C34, C50, C61, C64, C67, C7A, C77, C78, C85, C90, C91, or C92. CKD was recorded as N18. Cirrhosis of the liver was coded as F10, K70, K71, K72, or K74. Immunosuppression was registered as D61, D70, G35, K51, B20, B55, or M46 [25].
2.5. Statistical Analyses
2.5.1. Descriptive Analyses and Bivariate Analyses
We used descriptive statistics to summarize means and standard deviations, medians and interquartile ranges, or percentages when appropriate. p ≤ 0.05 was considered statistically significant. Statistical analyses were performed with R version 3.5.2 (20 December 2018) [26] and Python 3.7.3 using scikit-learn [27]. The Mann–Whitney–Wilcoxon U test was used to evaluate the correlation between age and mortality (the only continuous variable), whereas the chi-square test was used on the rest of features (categorical variables).
2.5.2. Multivariate Analysis Using LR
Our first approach was a standard statistical approach used in the medical sciences that covers bivariate analyses, chi-square and Mann–Whitney–Wilcoxon U tests, also called filter methods. These methods select features based on the association between explanatory features and the response feature. These classic approaches are often used as screeners: unrelated features are excluded when a multivariate analysis such as LR is performed in a second stage. After an exploratory descriptive analysis, a subset of selected features is used to build a predictive model using LR.
LR is widely used in health care and medical research and can control for the confounding of multiple predictors simultaneously when the outcome is binary (i.e., yes/no). The goal of LR is to create a mathematical model to evaluate the likelihood of an event occurring. LR is a classic method used in medical research to solve classification problems, and medical researchers often use it to obtain an OR to better understand and explain a model. An OR represents the odds that an outcome will occur given the presence of a predictor compared to the odds of that outcome occurring in the absence of that predictor. Clinicians like to use the OR because it can reveal whether a given feature can predict a particular outcome and because it provides a magnitude against which to compare predictors.
2.5.3. Feature Selection Using RF
RF is a classifier based on decision trees [28,29,30]. We used the RF implementation in the R package randomForest [31]. Several biomedical publications, such as ones on steatohepatitis, have shown that RF is an excellent, reliable, stable, and robust nonlinear classifier compared to other machine-learning classifiers [32,33,34]. RF works by building many decision trees in which features predict a categorical outcome (e.g., mortality). In the current study, 5000 decision trees were randomly and independently generated. Then these decision trees were combined to generate a single output. We also used the package caret [35] to fine-tune the hyperparameters used by RF by means of resampling. Although several methods are available, such as bootstrapping or leave-one-out, we used 10-fold cross-validation for resampling.
Machine-learning models are often considered black boxes because of their complex inner workings [36]. This phenomenon is typical of accurate nonlinear algorithms such as RF. Greater accuracy often comes at the expense of interpretability, which is key for medical decision making, for instance. Merely building a machine-learning model is not enough, because in certain cases, such as in the biomedical sciences, it is necessary to interpret the output of the model. Moreover, researchers must often explain the predictions of a model to people who do not understand much about machine learning. There are multiple tools to help explain both models and predictions [37,38]. Thus, it is important to understand a given machine-learning model on a global scale (global interpretation) and also to focus predictions on a single observation to derive local explanations (local interpretation).
From among the available tools, we chose feature importance from the RF implementation for global interpretation and local interpretable model-agnostic explanations (LIME) for local interpretation [39]. Although LR can yield a subset of relevant features, and the impact on mortality based on the magnitude of the OR, RF can be used even if nonlinear relationships are present among variables. As mentioned, it can also produce a feature importance analysis, which can be used to identify the most important features when a classification process is computed. With LIME, our aim was to understand the impact of risk factors around a single instance of interest (i.e., a single patient)–that is, to understand the local behavior of a complex model such as RF on a given patient.
As part of the preprocessing stage, we addressed class imbalance [40,41]. Working with health-related data can be challenging. When there are unequal instances for the positive class, there are usually more survivors than death cases. In our data set, the “survivor” class outweighed the “death” class 6 to 1. This phenomenon, called data imbalance, can have a great impact in evaluation metrics, which can be quite poor in the case of imbalanced classes, as most predictions would belong to the majority class. However, there are techniques for dealing with this and improving predictive performance. We used the synthetic minority over-sampling technique (SMOTE) to address imbalanced data by oversampling the minority class [42,43]. SMOTE was applied before we ran either LR or RF.
Correctly interpreting a well-fitting model is just as important as building one. Predictions may be accurate, but most of the time researchers need to know which features are the most predictive. Here we present several approaches for making a model more interpretable. One is to calculate ORs, as mentioned previously. Another is to select features using RF. Feature importance is one of the most useful interpretation tools in RF. Moreover, feature importance will be reliable only if the model has been trained with suitable hyperparameters. We used an RF implementation called permutation importance [44,45]. This approach was introduced by Breiman and Cutler [29,30]. After the baseline accuracy is refolded by passing a testing set through the model, a single column (i.e., one single feature) is permuted, the testing samples are passed back through the model, and accuracy is recomputed. The importance of the permuted feature is the difference between the baseline accuracy and the accuracy computed after the column is permuted. This permutation strategy is computationally expensive, but the results are acceptably reliable.
2.5.4. Data Splitting and Performance Metrics
Once SMOTE was applied, we split the whole merged data set into two samples to test the performance of the model. A training sample with 70% of the observations and a testing sample with 30% of the observations were created. Then we fit LR and RF models on the training set and measured their performance on the testing set. As a metric of the models we used mathematical accuracy, expressed as the number of correctly classified individuals (both true positives and true negatives) among the total number of cases. We also used the area under the receiver operating characteristic curve as a performance metric to evaluate both classification models and then plotted the curve. Figure 1 shows the workflow we used to assess our data. A final dataset with 5603 observations was produced.
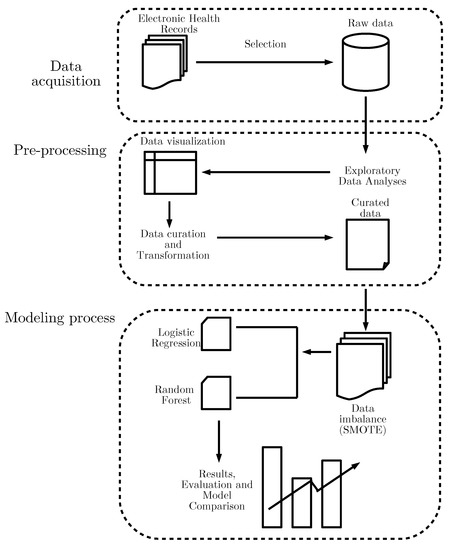
Figure 1.
Workflow of the modeling approach used for our dataset.
3. Results
3.1. Descriptive Analyses
A total of 5603 patients were admitted to Spanish hospitals and registered using the MBDS-H between 2001 and 2016. Table 2 summarizes the descriptive results. Most of the hospitalized patients were adults (94.9%), and 2318 were women (41.4%). Age had a skewed distribution (Figure 2), so medians and interquartile ranges were calculated. The median age of the whole cohort was 65 years, but it was 67 years when we excluded newborns. Most of the patients were discharged alive, either to home or to a convalescent institution. We calculated mortality among both hospitalized patients and the global population. Case mortality among hospitalized patients was 16.8% but varied by year. Mean mortality per 100,000 population over the observation period was 0.13.

Table 2.
Main characteristics of our cohort.

Figure 2.
Age distribution among patients with listeriosis.
We display the trend in hospitalizations in Figure 3, with a rising incidence over the later years of the observation period, and the trend in mortality in Figure 4 and Figure 5. Figure 6 and Figure 7 show incidence and mortality trends per 100,000 population, respectively, compared to the general population. Table 3 summarizes the data on incidence and mortality by year.
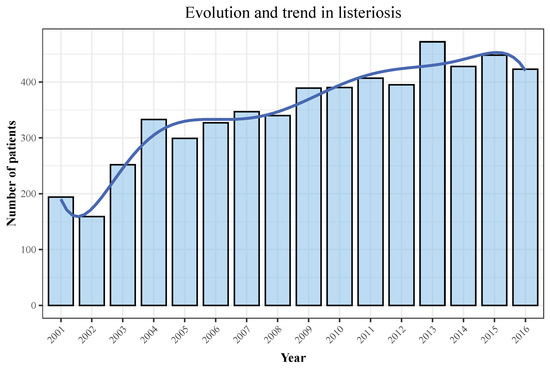
Figure 3.
Distribution of hospitalizations across the observation period. Blue line represents the rising trend of hospitalizations, whereas bars represent the number of patients.
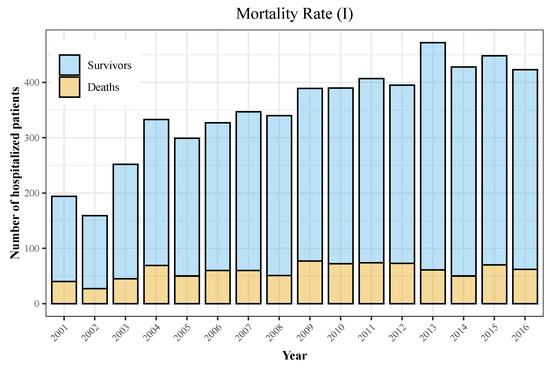
Figure 4.
Mortality rate among hospitalized patients (I). Yellow portion of the bars represents deaths among individuals during the observation period, whereas blue portion of the bar shows patients that were discharged alive.
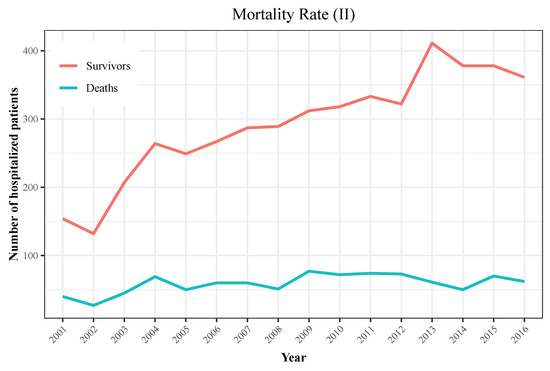
Figure 5.
Mortality rate among hospitalized patients (II). Both lines show the trend of survival patients (red line) and deaths (blue line). Both lines are decoupling along time, which suggests mortality rate decreased in recent years.

Figure 6.
Incidence per 100,000 population.

Figure 7.
Mortality per 100,000 population.

Table 3.
Incidence and mortality per 100,000 population.
Our cohort included 301 pregnant women (5.4%). Regarding clinical presentation, 296 newborns presented with listeriosis. Meningitis was the most common clinical presentation, with 2,388 cases (42.6%). One quarter of all patients presented with malignancy, either solid tumor or hematologic disease, as a comorbidity (1401 cases), but immunosuppression was the most common (39.8% of all patients).
3.2. Bivariate Analyses and LR
Table 4 summarizes the variables included in bivariate analyses of mortality. Only sex, endocarditis, and peritonitis were not associated with mortality. Given that age had a skewed distribution, we repeated the bivariate analyses of age categorizing this continuous variable into ranges, as shown in Table 5.

Table 4.
Bivariate analyses of clinical variables associated with mortality.

Table 5.
Bivariate analyses for mortality with age as a categorical variable.
According to Table 5, the age ranges 51–60 and greater than 70 were associated with increased mortality. However, certain ranges were associated with greater survival, such as the range from <10 years old to <50 years old.
Before performing LR and plotting the receiver operating characteristic curve, we first applied the SMOTE strategy and then split the whole data set, as described in the Methods section. We fit the model using the training set and tested its performance on the testing set. Regarding multivariate analysis, we decided to perform LR on the subset of features associated with mortality in Table 4 and all age ranges in Table 5. We performed both forward and backward stepwise selection to produce a subset of variables significantly associated with mortality, shown in Table 6. Finally, we plotted a receiver operating characteristic curve, which gave us an area under the curve of 0.740 (see Figure 8). The fitted model had a diagnostic accuracy of 0.832.

Table 6.
Multivariate analyses for mortality with age as a categorical variable.
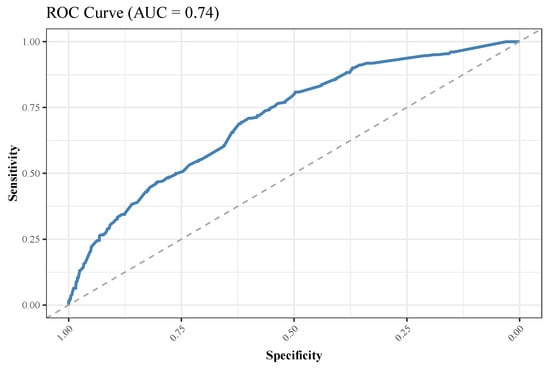
Figure 8.
Receiver operating characteristic (ROC) curve for mortality for the logistic regression model. AUC, area under the curve.
3.3. RF-Based Analyses and Interpretation
As mentioned in the Methods section, we addressed the problem of imbalanced data using SMOTE prior to running the RF algorithm. Then we used 10-fold cross-validation as a resampling strategy to fine-tune hyperparameters for RF. Although all variables were included in the RF algorithm, only the most important features are shown and plotted in Figure 9, which shows the importance of variables. The area under the curve of the model was 0.741 (Figure 10), and the diagnostic accuracy was 0.831. We used feature importance to understand the model from a global perspective and LIME to produce a local interpretation to explain why an individual prediction was made for a given patient or observation. Instead of providing a numerical value such as an OR, RF ranks features in a plot (Figure 9) that shows the impact of a given variable in the model. In line with the results of LR, sepsis/septic shock was the clinical feature most linked to the occurrence of death. Plots such as the one in Figure 9 give clinicians an easy way to evaluate visually the importance of clinical conditions. Features are ranked in terms of importance on the y-axis (features of highest importance at the top). X-axis means the impact in mean accuracy of dropping a certain variable from the random forest model. Mean decrease accuracy shows how much the accuracy decreases when in the model without that feature. Features of high importance are key to the outcome, and their values have a significant impact on mortality. By contrast, features of low importance are omitted from the final model, which makes it simpler and faster to fit and predict. For example, the age range 61–70, which was considered a risk factor in LR, had little impact when the results from RF were plotted. The remaining variables were not plotted, as they had no impact on the final model.

Figure 9.
The importance of features based on permutation in a parsimonious model with nine features. CKD, chronic kidney disease; CNS, central nervous system.

Figure 10.
Receiver operating characteristic curve for mortality for the random forest model. AUC, area under the curve.
To illustrate local interpretation and LIME, we plotted results for two observations (patients) to find the justifications of such interpretations. In both cases, “1” means that the patient has a condition, whereas “0” means that the patient has not that condition. The length of the bars in Figure 11 indicates the magnitude of the impact (y-axis), whereas the color indicates the sign of the estimated coefficient (red for negative, blue for positive). We plotted the 10 most influential variables and whether the variable increased the probability (“supports”) or decreased it (“contradicts”). This plot also revealed the model fit (i.e., how well the model explained mortality or survival in the patient). For the first case (patient 5), our proposed model predicted the probability of death with 88% accuracy and an explanation fit of 93%. Sepsis, age 71–80, and CKD were the three variables that most influenced the high probability. Lack of malignancy, lack of immunosuppression, and age <80 had little impact on the final probability. In contrast, patient 14 had a 96% probability of surviving because that individual had no sepsis, no malignancy, no immunosuppression, and no chronic disease.
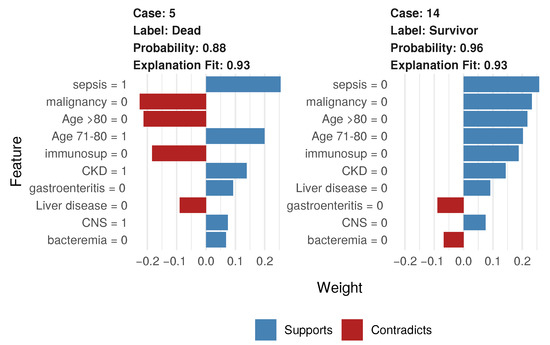
Figure 11.
Local interpretation for two individual patients.
4. Discussion
The first aim of this study was to describe and analyze demographic and clinical features of listeriosis over a 16-year observation period. The second objective was to identify relevant predictors of mortality due to Listeria-related infection. The main finding is that a combination of LR and RF models produced a high predictive value and selected the most relevant subset of features related to mortality due to infection by L. monocytogenes. Sepsis, malignancy, and age > 70 were the main risk factors. Other medical conditions, such as immunosuppression, CKD, and CNS-related infections, also had a great impact on mortality. This feature selection provides better insight into the prognosis for Listeria-related infection.
4.1. Descriptive Analyses
Our work extends the Spanish study by Herrador et al. [46], who performed an epidemiological investigation of incidence and mortality of listeriosis based on MBDS-H data from 1997 to 2015. Those authors demonstrated an increasing trend in hospitalization in the observation period, highlighting the need for awareness of this emergent public health problem. They used nationwide electronic records based on the ICD-9 and explored several medical conditions. Our contribution is to identify risk factors by using a combination of data science, conventional statistics, and machine learning beyond a merely exploratory analysis.
From 2001 to 2016, the annual incidence of listeriosis increased. In 2016, the last year recorded in our data set, the incidence was 0.9 patients per 100,000 inhabitants in Spain, greater than the reported 0.47 patients per 100,000 population in the European Union [47]. It is worth noting that there is actually a certain degree of underreporting because in Spain and Belgium it is not mandatory to report cases of listeriosis. The mean death rate in our study was 0.13 patients per 100,000 population but 16.8% among hospitalized patients, similar to the rest of the European Union but far below the 25% rate reported elsewhere [48].
In line with other studies [49], we found that in Spain listeriosis is rare, but it may be responsible for severe clinical syndromes and death in certain populations. We replicated the findings by Herrador et al. [46], whose epidemiological study demonstrated population demographics identified three main clinical groups: newborns, pregnant women, and remaining adults. Also, we demonstrated that sepsis/septic shock and invasive CNS listeriosis (either meningitis or brain abscess) were the most frequent and severe forms of infection, but malignancy, old age, and immunosuppression were the comorbidities most associated with mortality [50].
4.2. Statistics and Machine Learning
Regarding correlation analyses, bivariate tests such as the chi-square test rely on linear relationships between two variables [51]. However, a strong relationship can be overlooked if the correlation is nonlinear. These approaches perform an evaluation function, in which general characteristics are analyzed without the involvement of a learning algorithm. The benefit of this simple structure is that the insights are comprehensive and easy for researchers to understand. The main disadvantage is that they cannot handle nonlinearity [52]. Some results derived from bivariate analyses can be misleading, because although they show statistically significant differences in certain variables (bacteremia, pregnancy, gastroenteritis), these features cannot be considered to be associated with an increased risk for mortality. Likewise, a data analyst might have overlooked the role of age. In our study, age required deeper analyses, as its real impact on mortality was only revealed when the variable was categorized.
In contrast, combining preprocessing data, classic LR, and machine-learning algorithms such as RF can yield reliable models and select relevant features. We consider RF an excellent, reliable, stable, and robust nonlinear classifier compared to conventional approaches. RF can address nonlinearity, as has been demonstrated previously [34,53]. In our study, RF not only performed fairly well but also selected some interesting features as most relevant to Listeria-related mortality. RF uses built-in methods to select and validate important features as part of the fitting process. It then optimizes this subset of features by assigning them weights in the final model. In our study, sepsis and age were key features predicting mortality in the RF model. The LR and RF models performed similarly, and we cannot claim that one is better than the other. However, the subsets of selected features differed slightly by method.
As mentioned previously, in our study we used RF not only as a classifier but also as a method of evaluating the importance of individual features, which is essentially a measure of how well the model would perform if these features were not included. RF was used not only to accurately predict mortality but also to optimize the identification of features most important for predicting death among our patients.
It is worth mentioning that LR can be considered both a conventional statistical method and a machine-learning algorithm. When used as a conventional method, it provides an OR, a metric commonly used by medical researchers. In our study we used LR to calculate ORs, but by splitting the whole data set into two samples, fitting the LR model on a training set, and then measuring its performance on a testing set, we also used it as a machine-learning algorithm. Generally speaking, LR is the first classifier a data analyst should run if machine learning is used on a new data set.
4.3. Relevant Features of Listeria-Related Mortality
Healthy middle-age patients are less prone to die, whereas the elderly and patients with any kind of malignancy or immunosuppression are more prone to have a bad outcome. Recent publications [46,49] have found that malignancy, either solid organ neoplasm or hematologic disease, and immunosuppression are the most frequently recorded comorbidities in non-pregnant patients and have a great impact on mortality and clinical sequelae. The MONALISA study found that patients with CNS infection, along with immunosuppression and malignancy, had threefold higher mortality [49]. Other studies have found that patients with no underlying conditions usually have a good prognosis [7,54,55]. However, these studies could not establish a relationship between mortality and age. One explanation for this could be the nonlinear relationship between age and mortality. Previous studies may have overlooked this relationship because the correlation between these variables is nonlinear, as revealed by our research. Therefore, clinicians should not consider age a linear function, as doing so provides only partial support for a predictive model for mortality. However, nonlinearity can be properly handled with machine-learning algorithms. Although far from perfect, there was a reasonably coherent relationship between age and mortality in this study.
A concern may arise regarding age as a confounding factor, and this deserves to be discussed. Several factors may affect the trend in mortality at critical ages, such as among newborns or the elderly. These include underlying vulnerabilities and medical factors associated with critical ages, such as hypertension, cholesterol, diabetes, or the vulnerability of newborn infants. However, our research found that newborns are not at high risk despite their critical age. Future studies should further explore the impact of and interactions between underlying conditions, age, and other medical factors when developing models to predict outcomes of listeriosis. More knowledge of these processes will increase our understanding of severity in listeriosis.
Several studies have highlighted the importance of neurological syndromes for the severity of listeriosis. Arslan et al. [6] found that CNS infections were an independent risk factor for mortality, and parenchymal involvement in neuroimaging was an independent risk factor for severe infection. Brouwer et al. [56] found a mortality rate of 17%, with immunocompromised status as the main risk factor for mortality. Goulet et al. [57] reported that more than 50% of their patients developed severe sepsis, with 20–25% having neurological infections. Although they have a great impact, we could not demonstrate that immunosuppression or neuroinvasive listeriosis are the risk factors most associated with an increased risk of mortality. We believe that the low mortality rate among our patients made it difficult to establish prognostic factors. Another reason for the lack of findings is discussed below as a limitation of our study.
4.4. Limitations: The Reliability of Administrative Data
The main limitation of this study is that it was based on administrative data. Although EHR have improved the collection and availability of data, a major limitation of clinical research is that clinical data are not readily available. Furthermore, EHR usually contain administrative data, which are focused on economic management rather than clinical relevance [58,59,60]. In the current study, we used administrative data from the MBDS-H. One of its objectives is to facilitate clinical research. Its main advantage is that it covers more than 95% of Spanish hospitals. However, the validity of the data set relies on accurate medical discharge reports and on correct recording of variables. Primary and secondary diagnoses can be provisional, incorrect, or incomplete; some clinical conditions can be either excluded from diagnoses or redundant. Moreover, the data may have been collected by administrative staff with no medical training. The reliability of the MBDS-H may not be guaranteed [61].
We identified some issues in our data set regarding the coding of certain clinical conditions, as many conditions were recorded unevenly or not included as diagnoses. We could not assess some comorbidities, types of solid tumors or hematologic malignancy, immunosuppressive drugs, types of pathogen isolation, antibiotic therapies, neuroimaging techniques, or surgical treatments, as they were not recorded. This undercoding may explain the lack of significance of certain features. This misclassification is inherent to the MBDS-H because of the nature of the coding process.
A key factor in the diagnosis of listeriosis is the positive isolation of L. monocytogenes in blood, CSF, or tissue. We could not report positive isolations of L. monocytogenes in CSF or tissue, simply because they were not recorded. Isolation of the microorganism in blood cultures was indirect data, collected through, for instance, ICD-10-CM codes A32.82 (listerial endocarditis) and R78 (findings of drugs and other substances, not normally found in blood). Thus, bacteremia was inferred based on unreliable data. In our study, bacteremia was not a relevant predictor and did not provide any new knowledge of the disease, either because it was incorrectly recorded or because finding this microorganism in blood has no clinical value.
This discrepancy between clinically correct diagnoses established by clinicians and incorrect diagnoses recorded in the MBDS-H is a major limitation of our study. Demographic and administrative data, such as age, sex, hospital stay, and type of discharge, can be easily collected, but data on clinical conditions rely on the accuracy of diagnoses (both primary and secondary). However, although a certain amount of misclassification is expected, reliability could be improved with strategies such as examining secondary diagnoses [62,63]. Some have argued that the MBDS-H provides sufficiently valid information and can be a useful tool in epidemiological and clinical studies [64,65]; nevertheless, researchers should take these limitations into account.
The main strength of our study is that we obtained useful insights into Listeria-related infection using administrative data and machine-learning approaches. Both administrative data sets and machine-learning analyses have proven to be efficient tools in clinical and epidemiological studies of infectious diseases.
5. Conclusions
Mortality due to listeriosis is lower in Spain than in the European Union, but it remains at a high level. Aging, sepsis, malignancy, chronic diseases such as liver or kidney disease, and immunosuppression are the most important predictors of mortality in patients with listeriosis. The increasing trend in listeriosis observed in the past decade should prompt health providers and state governments to combine the use of preventive actions and sanitary food safety procedures, especially among populations at risk.
Regarding data analysis, combining LR and RF can help clinicians make a reliable and accurate prognosis for mortality, as it makes it possible to identify critical ages and other conditions associated with an increase in mortality. Both conventional statistics and machine learning can be complementary methods of analyzing high-dimensional EHR, and data analysts should not consider these methods mutually exclusive. In our study we highlighted the use of complex machine-learning algorithms such as RF; several strategies for preprocessing data prior to analyzing them; and several tools for interpreting, understanding, and explaining results. We developed a reliable, comprehensive predictive model, demonstrating the usefulness of some optimal techniques for analyzing the real data of patients at high risk for mortality due to listeriosis. Performance (in terms of predictive accuracy) and the area under the receiver operating characteristic curve can be considered acceptable. Features considered most relevant were consistent with their physiological roles. From a technical point of view, we consider our main contribution to have provided a machine-learning strategy to make algorithms such as RF more interpretable and to produce simple, well-fitting predictive models to identify populations at risk. Clinical researchers can benefit from a framework in which data science, conventional statistics, and machine learning are combined.
Author Contributions
R.G.-C. conceived and designed the study, wrote the first draft of the manuscript, and preprocessed and analyzed the data. Experts on the Spanish Minimum Basic Data Set at Hospitalization, J.R.-G. and P.R.-M. collected demographic and clinical data. O.V.-G. made substantial contributions to the interpretation of the results, critically reviewed the first draft of the manuscript, and made valuable suggestions. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
All procedures involving human participants were performed in accordance with the ethical standards of the responsible institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Ethical review and approval were not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
A contract signed with Mostoles University Hospital and the Spanish Ministry of Health, which provided the data set, prohibits the authors from providing their data to any other researcher. Furthermore, they must destroy the data upon the conclusion of their investigation. The data cannot be uploaded to any public repository. However, the data can be obtained from the Spanish Ministry of Health at Portal Estadístico at https://pestadistico.inteligenciadegestion.mscbs.es/publicoSNS/Comun/DefaultPublico.aspx (last accessed on 6 July 2019).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| CKD | chronic kidney disease |
| CNS | central nervous system |
| CSF | cerebrospinal fluid |
| EHR | electronic health records |
| ICD-9 | International Classification of Diseases, Ninth Revision |
| ICD-10-CM | International Classification of Diseases, Tenth Revision, Clinical Modification |
| IQR | interquartile range |
| LIME | Local Interpretable Model-Agnostic Explanations |
| LR | logistic regression |
| MBDS-H | Spanish Minimum Basic Data Set at Hospitalization |
| OR | odds ratio |
| RF | random forest |
| SMOTE | synthetic minority over-sampling technique |
References
- Farber, J.M.; Peterkin, P.I. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 1991, 55, 476–511. [Google Scholar] [CrossRef]
- Swaminathan, B.; Gerner-Smidt, P. The epidemiology of human listeriosis. Microbes Infect. 2007, 9, 1236–1243. [Google Scholar] [CrossRef]
- Lorber, B. Listeria Monocytogenes. In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases, 8th ed.; Bennett, J., Dolin, R., Blaser, M., Eds.; Elsevier/Saunders: Philadelphia, PA, USA, 2015; pp. 2383–2390.e2. [Google Scholar] [CrossRef]
- Elinav, H.; Hershko-Klement, A.; Valinsky, L.; Jaffe, J.; Wiseman, A.; Shimon, H.; Braun, E.; Paitan, Y.; Block, C.; Sorek, R.; et al. Pregnancy-associated listeriosis: Clinical characteristics and geospatial analysis of a 10-year period in Israel. Clin. Infect. Dis. 2014, 59, 953–961. [Google Scholar] [CrossRef]
- Wadhwa Desai, R.; Smith, M.A. Pregnancy-related listeriosis. Birth Defects Res. 2017, 109, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Arslan, F.; Meynet, E.; Sunbul, M.; Sipahi, O.R.; Kurtaran, B.; Kaya, S.; Inkaya, A.C.; Pagliano, P.; Sengoz, G.; Batirel, A.; et al. The clinical features, diagnosis, treatment, and prognosis of neuroinvasive listeriosis: A multinational study. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1213–1221. [Google Scholar] [CrossRef] [PubMed]
- Pagliano, P.; Ascione, T.; Boccia, G.; De Caro, F.; Esposito, S. Listeria monocytogenes meningitis in the elderly: Epidemiological, clinical and therapeutic findings. Le Infez. Med. 2016, 24, 105–111. [Google Scholar]
- Rajkomar, A.; Oren, E.; Chen, K.; Dai, A.M.; Hajaj, N.; Hardt, M.; Liu, P.J.; Liu, X.; Marcus, J.; Sun, M.; et al. Scalable and accurate deep learning with electronic health records. NPJ Digit. Med. 2018, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Eyduran, E. Usage of penalized maximum likelihood estimation method in medical research: An alternative to maximum likelihood estimation method. J. Res. Med. Sci. 2008, 13, 325–330. [Google Scholar]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine Learning in Medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef]
- Beam, A.L.; Kohane, I.S. Big data and machine learning in health care. JAMA 2018, 319, 1317–1318. [Google Scholar] [CrossRef]
- Obermeyer, Z.; Emanuel, E.J. Predicting the Future—Big Data, Machine Learning, and Clinical Medicine. N. Engl. J. Med. 2016, 375, 1216–1219. [Google Scholar] [CrossRef] [PubMed]
- Hameed, S.; Petinrin, O.; Hashi, A.O.; Saeed, F. Filter-Wrapper Combination and Embedded Feature Selection for Gene Expression Data. Int. J. Adv. Soft Comput. Appl. 2018, 10, 90–105. [Google Scholar]
- Parikh, R.B.; Manz, C.; Chivers, C.; Regli, S.H.; Braun, J.; Draugelis, M.E.; Schuchter, L.M.; Shulman, L.N.; Navathe, A.S.; Patel, M.S.; et al. Machine Learning Approaches to Predict 6-Month Mortality Among Patients With Cancer. JAMA Netw. Open 2019, 2, e1915997. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.; Steinhubl, S.R.; DeFilippi, C.; Dey, S.; Stewart, W.F. Early Detection of Heart Failure Using Electronic Health Records: Practical Implications for Time before Diagnosis, Data Diversity, Data Quantity, and Data Density. Circulation. Cardiovasc. Qual. Outcomes 2016, 9, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Angraal, S.; Mortazavi, B.J.; Gupta, A.; Khera, R.; Ahmad, T.; Desai, N.R.; Jacoby, D.L.; Masoudi, F.A.; Spertus, J.A.; Krumholz, H.M. Machine Learning Prediction of Mortality and Hospitalization in Heart Failure with Preserved Ejection Fraction. JACC Heart Fail. 2020, 8, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.H.; Hsieh, M.J.; Chen, C.M.; Hsieh, C.C.; Chao, C.M.; Lai, C.C. Comparison of machine learning models for the prediction of mortality of patients with unplanned extubation in intensive care units. Sci. Rep. 2018, 8, 17116. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, T.M.; Viacrusis, K.M.; Hernandez, L.F.T.; Ho, H.T.; Amalin, D.M.; Watanabe, K. Machine learning methods reveal the temporal pattern of dengue incidence using meteorological factors in metropolitan Manila, Philippines. BMC Infect. Dis. 2018, 18, 183. [Google Scholar] [CrossRef]
- Ahlström, M.G.; Ronit, A.; Omland, L.H.; Vedel, S.; Obel, N. Algorithmic prediction of HIV status using nation-wide electronic registry data. EClinicalMedicine 2019, 17, 100203. [Google Scholar] [CrossRef]
- Marcus, J.L.; Hurley, L.B.; Krakower, D.S.; Alexeeff, S.; Silverberg, M.J.; Volk, J.E. Use of electronic health record data and machine learning to identify candidates for HIV pre-exposure prophylaxis: A modelling study. Lancet HIV 2019, 6, e688–e695. [Google Scholar] [CrossRef]
- España. Real Decreto 69/2015, de 6 de Febrero, por el que se Regula el Registro de Actividad de Atención Sanitaria Especializada. Available online: https://www.boe.es/buscar/pdf/2015/BOE-A-2015-1235-consolidado.pdf (accessed on 6 July 2019).
- Ministerio de Sanidad Consumo y Bienestar Social. Portal Estadístico. Area de Inteligencia de Gestión. Available online: https://pestadistico.inteligenciadegestion.mscbs.es/publicoSNS/comun/ArbolNodos.aspx?idNodo=23525 (accessed on 6 July 2019).
- Ministerio de Sanidad Consumo y Bienestar Social. eCIEMaps-CIE-10-ES Diagnosticos. Available online: https://eciemaps.mscbs.gob.es/ecieMaps/browser/index_10_mc.html (accessed on 6 July 2019).
- De Noordhout, C.M.; Devleesschauwer, B.; De Noordhout, A.M.; Blocher, J.; Haagsma, J.A.; Havelaar, A.H.; Speybroeck, N. Comorbidities and factors associated with central nervous system infections and death in non-perinatal listeriosis: A clinical case series. BMC Infect. Dis. 2016, 16, 256. [Google Scholar] [CrossRef]
- World-Health-Organization. International Statistical Classification of Diseases and Related Health Problems, 10th Revision, 5th ed.; World Health Organization: Geneva, Switzerland, 2015; Volume 1. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Cutler, D.R.; Edwards, T.C., Jr.; Beard, K.H.; Cutler, A.; Hess, K.T.; Gibson, J.; Lawler, J.J. Random forests for classification in ecology. Ecology 2007, 88, 2783–2792. [Google Scholar] [CrossRef] [PubMed]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Sowa, J.P.; Heider, D.; Bechmann, L.P.; Gerken, G.; Hoffmann, D.; Canbay, A. Novel algorithm for non-invasive assessment of fibrosis in NAFLD. PLoS ONE 2013, 8, e62439. [Google Scholar] [CrossRef]
- Sowa, J.P.; Atmaca, Ö.; Kahraman, A.; Schlattjan, M.; Lindner, M.; Sydor, S.; Scherbaum, N.; Lackner, K.; Gerken, G.; Heider, D.; et al. Non-invasive separation of alcoholic and non-alcoholic liver disease with predictive modeling. PLoS ONE 2014, 9, e101444. [Google Scholar] [CrossRef]
- García-Carretero, R.; Holgado-Cuadrado, R.; Barquero-Pérez, Ó. Assessment of Classification Models and Relevant Features on Nonalcoholic Steatohepatitis Using Random Forest. Entropy 2021, 23, 763. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M. Caret: Classification and regression training. Astrophys. Source Code Libr. 2015, 28, 1–26. [Google Scholar]
- Palczewska, A.; Palczewski, J.; Robinson, R.M.; Neagu, D. Interpreting random forest classification models using a feature contribution method. In Integration of Reusable Systems; Springer: Berlin/Heidelberg, Germany, 2014; pp. 193–218. [Google Scholar]
- Saabas, A. Interpreting random forests. Diving Data 2014. Available online: https://blog.datadive.net/interpreting-random-forests/ (accessed on 1 May 2021).
- Li, X.; Wang, Y.; Basu, S.; Kumbier, K.; Yu, B. A debiased MDI feature importance measure for random forests. arXiv 2019, arXiv:1906.10845. [Google Scholar]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. Model-agnostic interpretability of machine learning. arXiv 2016, arXiv:1606.05386. [Google Scholar]
- Luque, A.; Carrasco, A.; Martín, A.; de las Heras, A. The impact of class imbalance in classification performance metrics based on the binary confusion matrix. Pattern Recognit. 2019, 91, 216–231. [Google Scholar] [CrossRef]
- Krawczyk, B. Learning from imbalanced data: Open challenges and future directions. Prog. Artif. Intell. 2016, 5, 221–232. [Google Scholar] [CrossRef]
- Lusa, L. Improved shrunken centroid classifiers for high-dimensional class-imbalanced data. BMC Bioinform. 2013, 14, 1–13. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Altmann, A.; Toloşi, L.; Sander, O.; Lengauer, T. Permutation importance: A corrected feature importance measure. Bioinformatics 2010, 26, 1340–1347. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.; Rudin, C.; Dominici, F. All Models are Wrong, but Many are Useful: Learning a Variable’s Importance by Studying an Entire Class of Prediction Models Simultaneously. J. Mach. Learn. Res. 2019, 20, 1–81. [Google Scholar]
- Herrador, Z.; Gherasim, A.; López-Vélez, R.; Benito, A. Listeriosis in Spain based on hospitalisation records, 1997 to 2015: Need for greater awareness. Eurosurveillance 2019, 24, 1800271. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, e05500. [Google Scholar] [CrossRef]
- Scallan, E.; Griffin, P.M.; Angulo, F.J.; Tauxe, R.V.; Hoekstra, R.M. Foodborne illness acquired in the United States–unspecified agents. Emerg. Infect. Dis. 2011, 17, 16–22. [Google Scholar] [CrossRef]
- Charlier, C.; Perrodeau, É.; Leclercq, A.; Cazenave, B.; Pilmis, B.; Henry, B.; Lopes, A.; Maury, M.M.; Moura, A.; Goffinet, F.; et al. Clinical features and prognostic factors of listeriosis: The MONALISA national prospective cohort study. Lancet Infect. Dis. 2017, 17, 510–519. [Google Scholar] [CrossRef]
- Garcia-Carretero, R. Clinical Features and Predictors for Mortality in Neurolisteriosis: An Administrative Data-Based Study. Bacteria 2022, 1, 3–11. [Google Scholar] [CrossRef]
- Chandrashekar, G.; Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Garcia-Carretero, R.; Vigil-Medina, L.; Barquero-Perez, O.; Ramos-Lopez, J. Pulse wave velocity and machine learning to predict cardiovascular outcomes in prediabetic and diabetic populations. J. Med Syst. 2020, 44, 16. [Google Scholar] [CrossRef] [PubMed]
- Scobie, A.; Kanagarajah, S.; Harris, R.J.; Byrne, L.; Amar, C.; Grant, K.; Godbole, G. Mortality risk factors for listeriosis–A 10 year review of non-pregnancy associated cases in England 2006–2015. J. Infect. 2019, 78, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Mook, P.; Patel, B.; Gillespie, I. Risk factors for mortality in non-pregnancy-related listeriosis. Epidemiol. Infect. 2012, 140, 706–715. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, M.C.; van de Beek, D.; Heckenberg, S.G.B.; Spanjaard, L.; de Gans, J. Community-acquired Listeria monocytogenes meningitis in adults. Clin. Infect. Dis. 2006, 43, 1233–1238. [Google Scholar] [CrossRef] [PubMed]
- Goulet, V.; Hebert, M.; Hedberg, C.; Laurent, E.; Vaillant, V.; De Valk, H.; Desenclos, J.C. Incidence of Listeriosis and Related Mortality Among Groups at Risk of Acquiring Listeriosis. Clin. Infect. Dis. 2011, 54, 652–660. [Google Scholar] [CrossRef]
- Howe, J.L.; Adams, K.T.; Hettinger, A.Z.; Ratwani, R.M. Electronic Health Record Usability Issues and Potential Contribution to Patient Harm. JAMA 2018, 319, 1276–1278. [Google Scholar] [CrossRef]
- Erickson, S.M.; Rockwern, B.; Koltov, M.; McLean, R.M. Putting Patients First by Reducing Administrative Tasks in Health Care: A Position Paper of the American College of Physicians. Ann. Intern. Med. 2017, 166, 659–661. [Google Scholar] [CrossRef]
- Sinsky, C.; Tutty, M.; Colligan, L. Allocation of Physician Time in Ambulatory Practice. Ann. Intern. Med. 2017, 166, 683–684. [Google Scholar] [CrossRef] [PubMed]
- Calle, J.E.; Saturno, P.J.; Parra, P.; Rodenas, J.; Perez, M.J.; Eustaquio, F.S.; Aguinaga, E. Quality of the information contained in the minimum basic data set: Results from an evaluation in eight hospitals. Eur. J. Epidemiol. 2000, 16, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Redondo-Gonzalez, O.; Tenias-Burillo, J.M. A multifactorial regression analysis of the features of community-acquired rotavirus requiring hospitalization in Spain as represented in the Minimum Basic Data Set. Epidemiol. Infect. 2016, 144, 2509–2516. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Greenberg, J.A.; Hohmann, S.F.; Hall, J.B.; Kress, J.P.; David, M.Z. Validation of a Method to Identify Immunocompromised Patients with Severe Sepsis in Administrative Databases. Ann. Am. Thorac. Soc. 2016, 13, 253–258. [Google Scholar] [CrossRef]
- Fernandez-Navarro, P.; Lopez-Abente, G.; Salido-Campos, C.; Sanz-Anquela, J.M. The Minimum Basic Data Set (MBDS) as a tool for cancer epidemiological surveillance. Eur. J. Intern. Med. 2016, 34, 94–97. [Google Scholar] [CrossRef]
- Hernandez Medrano, I.; Guillan, M.; Masjuan, J.; Alonso Canovas, A.; Gogorcena, M.A. Reliability of the minimum basic dataset for diagnoses of cerebrovascular disease. Neurologia 2017, 32, 74–80. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).